Abstract
The phytochrome-mediated regulation of photomorphogenesis under red and far-red light conditions involves both positively and negatively acting factors. The positively acting factors (e.g. HY5/HFR1/LAF1 and others) are degraded in the dark to prevent photomorphogenesis. By contrast, the negatively acting factors (e.g. phytochrome-interacting factors or PIFs) are degraded in response to light to promote photomorphogenesis. Here, we show that the negatively acting factor PIF1 is also degraded in the dark by direct heterodimerization with the positively acting factor HFR1. Conversely, PIF1 also promotes the degradation of HFR1 in darkness. PIF1 enhances the poly-ubiquitylation of HFR1 by COP1 in vivo and in vitro. In addition, the reciprocal co-degradation of PIF1 and HFR1 is dependent on the 26S proteasome pathway in vivo. Genetic evidence shows that the hfr1 mutant partially suppresses the constitutive photomorphogenic phenotypes of cop1-6 pif1 and of the quadruple mutant pifq both in the dark and in far-red light conditions. Taken together, these data uncover a co-degradation mechanism between PIFs and HFR1 that underlies photomorphogenic development in Arabidopsis thaliana.
KEY WORDS: bHLH transcription factor, E3 ubiquitin ligase, Photomorphogenesis, Reciprocal degradation, 26S proteasome
Summary: Reciprocal degradation of PIFs and HFR1 highlights a novel mechanism by which HLH factors regulate the abundance of bHLH factors to optimize photomorphogenesis in Arabidopsis.
INTRODUCTION
Plants undergo skotomorphogenic development in the dark, which is characterized by elongated hypocotyl and small appressed cotyledons. By contrast, they undergo photomorphogenic development under light, which is characterized by short hypocotyl and expanded open cotyledons. Under red/far-red light conditions, photomorphogenesis is regulated by the phytochrome (phy) family photoreceptors (Bae and Choi, 2008; de Wit, et al., 2016). Encoded by a small five-member family (phyA-phyE) in Arabidopsis, phys can form homo- and heterodimers in vivo (Clack, et al., 2009). They are synthesized as the inactive Pr form in the dark. Upon sensing red light using the billin chromophore, phys undergo a conformational change to the biologically active Pfr form, which can be converted back to the Pr form by exposure to far-red light in a process called low fluence response (LFR). An exception among phys is phyA, for which a response can be triggered on exposure to very low amounts of any light (very low fluence response, VLFR) and for which continuous irradiation with high fluence rate far-red light will also trigger a response (high irradiance response, HIR) (Casal et al., 2003). The Pfr form of all phys migrates into the nucleus with differential kinetics (Klose et al., 2015), and regulates expression of a large number of genes to promote photomorphogenesis (Jiao et al., 2007; Quail 2007).
The phy-mediated light signaling pathways involve both positively and negatively acting factors (Huq and Quail, 2005). For example, HFR1, HY5, LAF1 and others are the major positive regulators (Lau and Deng, 2012; Xu et al., 2015, 2016), whereas phytochrome-interacting factors (PIFs) act as major negative regulators of photomorphogenesis (Castillon et al., 2007; Leivar and Monte, 2014; Leivar and Quail, 2011). There are seven PIF genes (PIF1, PIF3-8), encoding basic helix-loop-helix (bHLH) transcription factors (Toledo-Ortiz et al., 2003). They preferentially bind to the G-box (CACGTG) DNA sequence elements present in gene promoters and repress light-inducible genes while activating light-repressed genes in the dark (Kim et al., 2016a; Leivar and Monte, 2014). The quadruple mutant pifq (pif1 pif3 pif4 pif5) displays constitutively photomorphogenic phenotypes, suggesting that PIFs promote skotomorphogenesis (Leivar et al., 2008; Shin et al., 2009).
The 26S proteasome-mediated degradation of both positively and negatively acting factors plays a central role in phy signaling pathways. In darkness, the positively acting factors (e.g. HY5, LAF1, HFR1 and others) are degraded (Lau and Deng, 2012). In this process, CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase, directly interacts with HY5, HFR1 and LAF1 and induces their degradation via the 26S proteasome pathway (Lau and Deng, 2012; Saijo et al., 2003; Seo et al., 2003; Xu et al., 2015). COP1 also associates with SUPPRESSOR OF PHYA-105 (SPA1-4) family members and CUL4, and the CUL4COP1-SPA complex degrades the positively acting factors to repress photomorphogenesis in the dark (Chen et al., 2010; Hoecker, 2005; Saijo et al., 2003; Zhu et al., 2008). Strikingly, PIFs and the COP1-SPA complex function synergistically to degrade HY5 to repress photomorphogenesis in the dark (Xu et al., 2014). Conversely, PIFs are phosphorylated, poly-ubiquitylated and subsequently degraded under light (Leivar and Quail, 2011; Xu et al., 2015). However, a recent study has shown that phyB can induce degradation of PIF1 non-cell-autonomously (Kim et al., 2016b). Among the candidate kinases, oat phyA has been shown to directly phosphorylate PIFs and regulate the light-induced degradation of PIF3 under far-red light (Shin et al., 2016). CK2 (casein kinase II) and BIN2 have been shown to phosphorylate PIF1 and PIF4, respectively, in a light-independent manner (Bernardo-García et al., 2014; Bu et al., 2011b). In addition, both CUL3 and CUL4-based E3 ligase complexes mediate the light-induced ubiquitylation of PIF3 and PIF1, respectively, during dark-to-light transition (Ni et al., 2014; Zhu et al., 2015). HEMERA has also been shown to induce degradation of PIFs under de-etiolated conditions possibly in a transcription-coupled manner (Qiu et al., 2015). However, PIFs are still degraded in these E3 ligase and kinase mutants, suggesting that additional factors are functioning in these processes.
Although it is known that positively acting factors are degraded in the dark, the dark-induced degradation of negative factors has not yet been shown. Here, we show that PIFs are degraded in the dark via the 26S proteasome pathway. In this process, the positively acting factor HFR1 promotes the degradation of PIF1 in the dark by direct heterodimerization. We further provide biochemical and genetic evidence to support the hypothesis that PIF1 and HFR1 undergo reciprocal co-degradation via the 26S proteasome pathway in the dark to optimize photomorphogenesis.
RESULTS
PIFs are degraded in the dark via the 26S proteasome pathway
In general, PIFs are stable in the dark, and light exposure induces their rapid degradation via the 26S proteasome pathway (Castillon et al., 2007; Leivar and Quail, 2011; Xu et al., 2015). To test whether PIFs are also degraded in the dark, we examined TAP-PIF1, PIF3-myc, PIF4-myc, PIF5-myc and native PIF1 and PIF5 levels in the dark treated without or with the proteasome inhibitor Bortezomib (Bortz) for 3 h. The results show that proteasome inhibitor treatment stabilized all four PIFs (Fig. 1A,B; Fig. S1A-C). We also examined PIF1 protein level from wild-type dark-grown seedlings treated with the protein synthesis inhibitor. Strikingly, the data show that PIF1 is rapidly degraded in the dark in the absence of new protein synthesis (Fig. S1A). Although cellular proteins have a finite half-life, inhibition of PIF degradation by the proteasome inhibitor suggests that PIFs are degraded either directly or indirectly by the 26S proteasome pathway in the dark. These data sharply contrast the prevailing view that PIFs are only degraded in response to light.
Fig. 1.
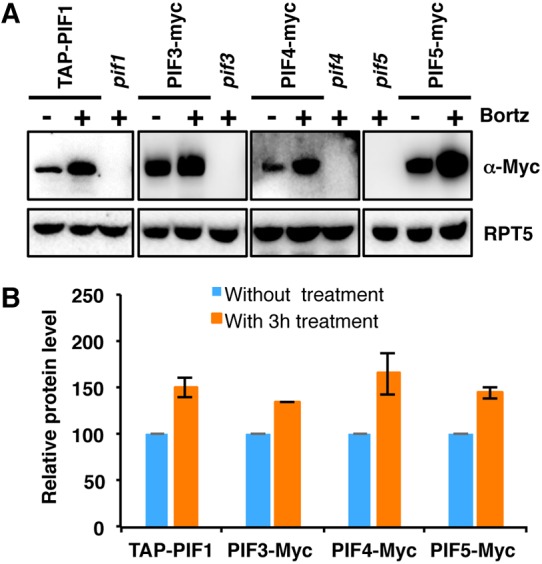
PIF1, PIF3, PIF4 and PIF5 are degraded in the dark via the 26S proteasome pathway. (A) Immunoblots showing the level of PIFs in 4-day-old pPIF1:TAP-PIF1, 35S:PIF3-Myc, 35S:PIF4-Myc and 35S:PIF5-Myc dark-grown seedlings. One batch of seedlings was pretreated with 40 µM Bortezomib (Bortz) for 3 h before protein extraction. pif1, pif3, pif4 and pif5 mutants were used as controls. The blot was probed with anti-Myc or anti-RPT5 antibodies. (B) Quantification of TAP-PIF1, PIF3-Myc, PIF4-Myc and PIF5-Myc protein levels using ImageJ. RPT5 was used as a control. The TAP-PIF1, PIF3-Myc, PIF4-Myc and PIF5-Myc protein levels without proteasome inhibitor treatment were set as 100, respectively. Error bars indicate s.d. (n=3).
Previously, both PIF1 and PIF3 have been shown to be unstable in cop1-4 plants in the dark (Bauer et al., 2004; Zhu et al., 2015). We have examined the PIF5 level in cop1-4 seedlings grown in the dark. Similar to PIF1 and PIF3, the PIF5 level is also lower in cop1-4 compared with wild-type seedlings (Fig. S1C). To examine whether the degradation of PIF1 and PIF5 in cop1-4 depends on the 26S proteasome, we treated cop1-4 dark-grown seedlings with the proteasome inhibitor and measured PIF1 and PIF5 levels by immunoblotting. The results show that both PIF1 and PIF5 are strongly stabilized in cop1-4 by the proteasome inhibitor (Fig. S1B,C), suggesting that PIF1 and PIF5 might be actively degraded in the cop1-4 background.
HFR1 promotes the degradation of PIF1 and PIF5
A recent study showed that overexpression of HECATE2 (HEC2), a HLH transcription factor, stabilizes PIF1 in the dark and reduces the light-induced degradation of PIF1 (Zhu et al., 2016). HFR1, another HLH factor, was originally identified as an important positive regulator of phyA-mediated far-red light signaling and shade avoidance pathways (Fairchild et al., 2000; Fankhauser and Chory, 2000; Hersch et al., 2014; Lorrain et al., 2009). To examine whether HFR1 regulates PIF1 levels, we performed immunoblotting to examine the PIF1 level in the hfr1 background under dark conditions. The results show that PIF1 is stabilized in the hfr1 background under darkness (Fig. 2A,B). PIF5 is also slightly stabilized in the hfr1 single mutant in the dark, similar to PIF1 (Fig. S1D). The HFR1-mediated PIF1 degradation is post-translational as the PIF1 mRNA level is slightly lower in the hfr1 mutant compared with wild-type seedlings (Fig. S1E). These data suggest that HFR1 promotes the degradation of PIF1 and PIF5 under dark conditions.
Fig. 2.
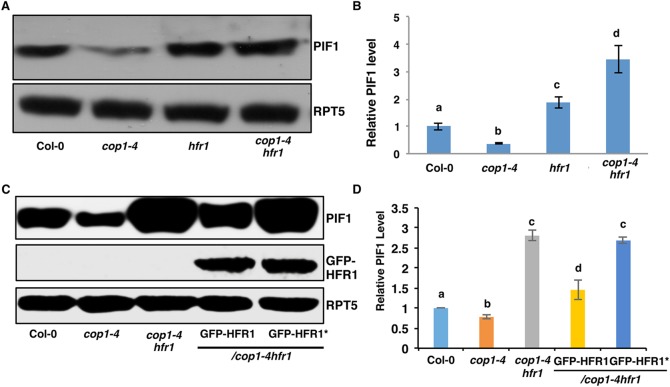
HFR1 promotes PIF1 degradation in the dark. (A) Immunoblot shows higher abundance of PIF1 in the hfr1 and cop1-4 hfr1 backgrounds compared with wild-type seedlings. Four-day-old dark-grown seedlings were used for protein extraction. The blot was probed with anti-PIF1 and anti-RPT5 antibodies. (B) Quantification of PIF1 protein level using RPT5 as a control. The letters a-d indicate statistically significant differences between means of protein levels (P<0.05) based on two-way ANOVA analyses. Error bars indicate s.d. (n=7). (C) Immunoblots show the PIF1 (top panel) and GFP-HFR1 (middle panel) and loading control RPT5 (bottom panel) levels in wild-type Col-0, cop1-4, cop1-4 hfr1, cop1-4 hfr1/GFP-HFR1 and cop1-4 hfr1/GFP-HFR1*. Immunoblot was performed as described in A. (D) Quantification of PIF1 protein level using RPT5 as a control. The letters a-d indicate statistically significant differences between means of protein levels (P<0.05) based on two-way ANOVA analyses. Error bars indicate s.d. (n=3).
Previous studies showed that COP1 promotes the degradation of HFR1 via the 26S proteasome pathway in the dark (Jang et al., 2005; Yang et al., 2005b). Because PIF1 can interact with HFR1 and COP1 directly (Bu et al., 2011a; Shi et al., 2013; Xu et al., 2014; Yang et al., 2005b), it is possible that COP1, PIF1 and HFR1 form a trimolecular complex that promotes the degradation of PIF1 in the dark. To test this hypothesis, we examined the PIF1 level in cop1-4 and cop1-4 hfr1 backgrounds under dark conditions (Fig. 2B). As expected, the PIF1 level is lower in cop1-4 in darkness compared with wild type. Strikingly, in the dark, PIF1 is strongly stabilized in cop1-4 hfr1 compared with cop1-4 and wild-type backgrounds (Fig. 2B). In addition, the PIF5 level is also higher in cop1-4 hfr1 compared with cop1-4 and wild-type seedlings grown in darkness (Fig. S1D). These data strongly suggest that HFR1 promotes PIF1 and PIF5 degradation in the wild-type as well as in cop1-4 backgrounds in the dark. Because cop1-4 expresses a truncated COP1 protein that has been shown to retain residual function (McNellis et al., 1994), the much higher abundance of PIF1 and PIF5 in the cop1-4 hfr1 seedlings compared with cop1-4 and hfr1 suggest that COP1 is required for HFR1-mediated PIF1 and PIF5 turnover in the dark. Consistent with this conclusion, the PIF1 level is slightly higher in cop1-5, a mutant with a null allele of cop1 (Fig. S1F).
PIF1-HFR1 heterodimerization is necessary for HFR1-mediated PIF1 degradation
HFR1 heterodimerizes with PIFs to inhibit their activity. The substitution mutations of Val172Leu173 to Asp172Glu173 in the HLH domain of HFR1 have been shown to eliminate the dimerization between HFR1 and PIF1/PIF4/PIF5 (Fig. S2A) (Hornitschek et al., 2009; Shi et al., 2013). To test whether heterodimerization is necessary for HFR1-mediated degradation of PIF1, we created a mutant version of HFR1 (HFR1*) that interferes with the dimerization between HFR1 and PIF1 as shown previously. Using yeast two-hybrid assays, we confirmed that the mutant HFR1* does indeed lack interaction with PIF1 (Fig. S2B). We made transgenic plants expressing GFP-HFR1* in the hfr1 background and selected homozygous lines expressing similar amounts of the mutant and wild-type GFP-HFR1. We also performed immunoblots to examine whether HFR1* is degraded in the dark similar to wild-type HFR1 as previously reported (Jang et al., 2005; Yang et al., 2005a). Interestingly, the data show that the GFP-HFR1* level is similar under both dark conditions and dark-to-light transition (Fig. S3A-C). In contrast, GFP-HFR1 is degraded in the dark but stabilized under light as previously reported. These data suggest that dimerization is necessary for degradation of HFR1 in the dark.
We then crossed both the wild-type GFP-HFR1 and the mutant GFP-HFR1* into the cop1-4 hfr1 background. Phenotypic analyses showed that GFP-HFR1 suppressed the hypocotyl lengths of hfr1 and cop1-4 hfr1 under far-red light, but GFP-HFR1* failed to reduce the hypocotyl lengths of the hfr1 and cop1-4 hfr1 under these conditions (Figs S3C-E and S4A-D), confirming that the mutant HFR1* is non-functional in vivo as previously reported (Hornitschek et al., 2009; Shi et al., 2013). We then examined PIF1 levels in cop1-4 hfr1/GFP-HFR1 and cop1-4 hfr1/GFP-HFR1* by immunoblot. Strikingly, GFP-HFR1 in the cop1-4 hfr1 background reduced the PIF1 level close to that of wild type (Fig. 2C,D). In contrast, GFP-HFR1* failed to reduce PIF1 level, suggesting that HFR1 promotes PIF1 degradation in the dark in a heterodimerization-dependent manner.
Our data, along with that of others, show that PIF1, PIF3 and PIF5 levels are lower in cop1-4 compared with wild type at the seedling stage (Fig. S1) (Bauer et al., 2004; Zhu et al., 2015). However, in imbibed seeds, the PIF1 level is much higher in cop1-4 and the quadruple mutant spaq (spa1 spa2 spa3 spa4) compared with wild type (Zhu et al., 2015). To test whether the expression level of HFR1 contributes to this difference, we measured HFR1 and PIF1 mRNA levels in both wild-type and cop1-4 imbibed seeds and 4-day-old dark-grown seedlings using RT-qPCR. The results show that HFR1 is strongly expressed in dark-grown seedlings with very weak expression in imbibed seeds whereas PIF1 is strongly expressed in imbibed seeds (Fig. S5A,B). HFR1 expression is slightly lower in the cop1-4 seedlings compared with wild-type seedlings, but still much higher than the HFR1 level at the seed stage (Fig. S5B). These data suggest that the lower level of PIFs in the cop1-4 dark-grown seedlings might be due to increased abundance of HFR1, which promotes the degradation of PIFs in the cop1-4 background. Taken together, these data demonstrate that HFR1 regulates the PIF level in the dark in both wild-type and cop1-4 backgrounds.
HFR1 promotes PIF1 degradation via 26S proteasome
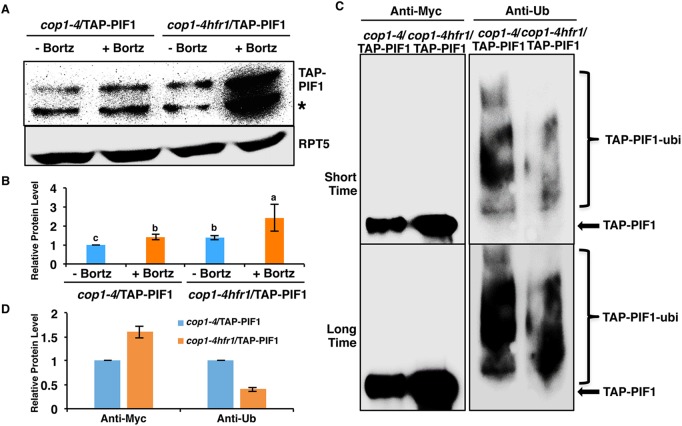
To examine whether HFR1-mediated degradation of PIF1 is proteasome dependent, we created transgenic plants expressing TAP-PIF1 in cop1-4 and cop1-4 hfr1 backgrounds and performed immunoblotting in the presence and absence of the proteasome inhibitor. The results show that TAP-PIF1 degradation is blocked in the presence of the proteasome inhibitor in both the cop1-4 and cop1-4 hfr1 mutant backgrounds under darkness, similar to observations in the wild-type background (Figs 1 and 3A,B). In addition, TAP-PIF1 is higher in the cop1-4 hfr1 background compared with that in the cop1-4 background (Fig. 3A,B), which is consistent with results for the native PIF1 level (Fig. 2B,C). These data suggest that HFR1 promotes the degradation of PIF1 via the 26S proteasome pathway.
Fig. 3.
HFR1-mediated PIF1 degradation is 26S proteasome dependent. (A) Immunoblot shows the TAP-PIF1 level in cop1-4 and cop1-4 hfr1 background. Total protein was extracted from 4-day-old dark-grown seedlings. One batch of seedlings was pretreated with 40 µM Bortezomib (Bortz) for 3 h before protein extraction. The blot was probed with anti-Myc or anti-RPT5 antibodies. Asterisk indicates a cross-reacting band or proteolytically cleaved product. (B) Quantification of TAP-PIF1 protein level using RPT5 as a control. The letters a-c indicate statistically significant differences among four samples and two treatment conditions (P<0.05) based on two-way ANOVA analyses. Error bars indicate s.d. (n=3). (C) TAP-PIF1 level is higher but the ubiquitylation level is lower in the cop1-4 hfr1 compared with cop1-4 background in darkness. Total protein was extracted from 4-day-old dark-grown seedlings with 40 mM Bortezomib pretreatment for 3 h before protein extraction. TAP-PIF1 was immunoprecipitated using anti-Myc antibody from protein extracts. The immunoprecipitated samples were then separated on 6.5% SDS-PAGE gels and probed with anti-Myc (left) or anti-Ub (right) antibodies. The top and bottom panels are low and high exposures, respectively. Arrow indicates TAP-PIF1. (D) Quantification of TAP-PIF1 and TAP-PIF1-ubi protein levels shown in C. The TAP-PIF1 and TAP-PIF1-ubi protein levels in cop1-4 background were set as 1 respectively. Error bars indicate s.d. (n=3).
Recently, it was shown that some proteins are degraded via the 26S proteasome pathway independently of polyubiquitylation due to the presence of an unstructured region or through interaction with another protein containing an unstructured region (Fishbain et al., 2015). To determine whether HFR1-mediated degradation of PIF1 is polyubiquitin dependent or independent, we immunoprecipitated TAP-PIF1 from cop1-4 and cop1-4 hfr1 mutants pretreated with the proteasome inhibitor and then detected with anti-Myc and anti-Ub antibodies. The results show that the immunoprecipitated TAP-PIF1 level is significantly higher in the cop1-4 hfr1 background than that in the cop1-4 background (Fig. 3C, left panel, Fig. 3D). But the ubiquitylation level of the immunoprecipitated TAP-PIF1 is significantly lower in the cop1-4 hfr1 background than that in the cop1-4 background (Fig. 3C, right panel, Fig. 3D). The immunoprecipitated TAP-PIF1 is more abundant but contains less polyubiquitylation in the cop1-4 hfr1 than in the cop1-4 background, which supports the hypothesis that HFR1 promotes the degradation of PIF1 in the dark via the 26S proteasome pathway by increasing the amount of polyubiquitylation of PIF1.
PIFs promote the degradation of HFR1 post-translationally
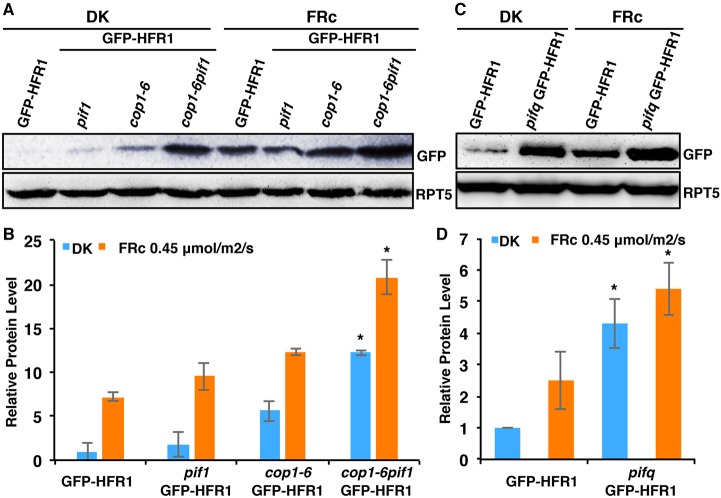
The COP1-SPA complex interacts with HFR1 and induces its degradation via the 26S proteasome pathway in the dark (Jang et al., 2005; Yang et al., 2005b). The COP1-SPA complex and PIFs also synergistically suppress plant photomorphogenesis in the dark by regulating the abundance of HY5 post-translationally (Xu et al., 2014). To determine whether the synergistic promotion of photomorphogenesis observed in the cop1-6 pif1 mutant is also partially due to an increased abundance of HFR1, we generated GFP-HFR1 transgenic plants by crossing GFP-HFR1 into the pif1, cop1-6 and cop1-6 pif1 backgrounds. Immunoblots showed that in both darkness and far-red light conditions, the GFP-HFR1 protein is synergistically stabilized in cop1-6 pif1 compared with GFP-HFR1 in either pif1 or cop1-6 single mutant backgrounds (Fig. 4A,B). This regulation is at the post-translational level as the amount of the GFP-HFR1 mRNA is similar in these backgrounds (Fig. S6A). In addition, as pifq displays constitutive photomorphogenic phenotypes similar to cop1, we also created GFP-HFR1 transgenic plants in the pifq background. Strikingly, the GFP-HFR1 protein level, but not the GFP-HFR1 mRNA level, is increased in the pifq compared with the wild-type background (Fig. 4C,D; Fig. S6A). A recent study also showed that the HFR1 mRNA level is reduced in the pifq compared with the wild type (Fig. S6B) (Zhang et al., 2013), suggesting that PIFs also transcriptionally activate the expression of HFR1. Taken together, these data suggest that HFR1 abundance is also regulated by PIFs and COP1 in a post-translational manner.
Fig. 4.
PIFs promote the degradation of HFR1 post-translationally in the dark and under far-red light. (A) Immunoblot shows HFR1 protein level in GFP-HFR1 transgenic plants and in pif1, cop1-6 and cop1-6 pif1 harboring the GFP-HFR1 transgene. Seedlings are grown either in the dark for 4 days or grown in the dark for 21 h and then transferred to FRc (0.45 μmol/m2/s) for 3 days. The blot was probed with anti-GFP or anti-RPT5 antibodies. (B) Bar graph shows GFP-HFR1 protein level in the mutants indicated. For quantification, GFP-HFR1 band intensities were measured from three independent blots using ImageJ, and then normalized against RPT5 levels. GFP-HFR1 dark level was set as 1 and the relative protein levels were calculated. Error bars indicate s.d. Asterisk indicates significant difference (P<0.05) between double and single mutant background. (C) Immunoblot shows HFR1 protein level in the GFP-HFR1 and pifq/GFP-HFR1. RPT5 was used as loading control. Seedlings were grown in the dark or FRc light as described above. (D) Bar graph shows the quantified GFP-HFR1 levels in GFP-HFR1 and pifq/GFP-HFR1. Error bars indicate s.d. Asterisk indicates significant difference between GFP-HFR1 and pifq/GFP-HFR1 in both conditions, respectively (P<0.05). DK, dark.
PIF1 promotes HFR1 degradation via 26S proteasome
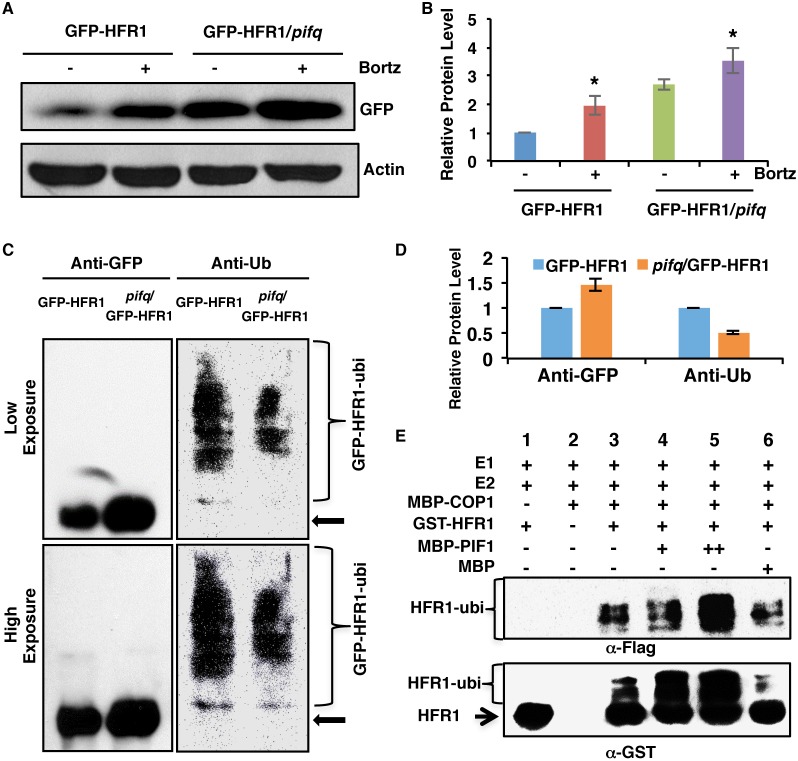
As HFR1 promotes PIF1 degradation by polyubiquitylation via the 26S proteasome pathway (Fig. 3), we hypothesized that PIFs promote HFR1 degradation in a similar manner. To examine whether PIF-mediated degradation of HFR1 is proteasome dependent, we first performed immunoblotting for GFP-HFR1 in the presence and absence of the proteasome inhibitor. The results show that GFP-HFR1 degradation is blocked in the presence of the proteasome inhibitor in the GFP-HFR1 background under dark conditions (Fig. 5A,B). The proteasome inhibitor also blocked GFP-HFR1 degradation in the pifq background but to a lesser degree compared with the GFP-HFR1 background (Fig. 5A,B). Then, we immunoprecipitated GFP-HFR1 fusion protein from GFP-HFR1 and pifq/GFP-HFR1 transgenic seedlings pretreated with proteasome inhibitor and then detected with anti-Ub and anti-GFP antibodies. The results show that the immunoprecipitated GFP-HFR1 level is significantly higher in the pifq/GFP-HFR1 than in the GFP-HFR1 background as observed above (Fig. 5C, left panel; Fig. 5D). However, the polyubiquitylation level of the immunoprecipitated GFP-HFR1 is significantly reduced in the pifq background than in the GFP-HFR1 background (Fig. 5C, right panel; Fig. 5D). These data support the hypothesis that PIFs promote the degradation of HFR1 in the dark via polyubiquitylation followed by 26S proteasome pathway activation in vivo.
Fig. 5.
PIF1 promotes HFR1 degradation in a ubiquitylation-dependent manner. (A) Immunoblot shows the GFP-HFR1 protein level in GFP-HFR1 and GFP-HFR1/pifq backgrounds. Total protein was extracted from 4-day-old seedlings grown in darkness. One batch of seedlings was pretreated with 40 mM Bortezomib (Bortz) for 3 h before protein extraction. The blot was probed with anti-GFP or anti-actin antibodies. (B) Quantification of GFP-HFR1 protein level using actin as a control. Asterisks indicate statistically significant differences compared with non-Bortezomib treatment for GFP-HFR1 and GFP-HFR1/pifq, respectively (P<0.05). Error bars indicate s.d. (n=4). (C) The protein level of GFP-HFR1 is higher but the ubiquitylation level of GFP-HFR1 is lower in the pifq compared with the GFP-HFR1 background in darkness in vivo. Sample preparation is as described in A. GFP-HFR1 was immunoprecipitated using anti-GFP antibody, and then separated on 8% SDS-PAGE gels and probed with anti-GFP (left) or anti-Ub (right) antibodies. The top and bottom panels are low and high exposures, respectively. The arrow indicates the GFP-HFR1 size. (D) Quantification of GFP-HFR1 and GFP-HFR1-ubi levels for the blot shown in C by ImageJ. The GFP-HFR1 and GFP-HFR1-ubi levels were set as 1, respectively. Error bars indicate s.d. (n=3). (E) PIF1 promotes the ubiquitylation of HFR1 by COP1 in vitro. In vitro ubiquitylation assay was performed using MBP-COP1 as E3 ubiquitin ligase, GST-HFR1 as a substrate, Flag-ubiquitin, UBE1 (E1), UbcH5b (E2) and increasing concentrations of MBP-PIF1. MBP was used as a control. Ubiquitylated GST-HFR1 was detected by anti-Flag antibody (top panel) and anti-GST antibody (bottom panel). The arrow indicates non-ubiquitylated GST-HFR1.
PIF1 enhances COP1-mediated ubiquitylation of HFR1
COP1 directly ubiquitylates HFR1 in vitro (Jang et al., 2005; Yang et al., 2005b). The polyubiquitylation level is also reduced in the pifq background in vivo as shown above (Fig. 5C,D), suggesting that PIFs might enhance the ubiquitylation activity of COP1 towards HFR1. To test this hypothesis, we performed an in vitro ubiquitylation assay as described previously (Jang et al., 2005; Xu et al., 2014; Yang et al., 2005b) using MBP-COP1, GST-HFR1 and different amounts of MBP-PIF1 or MBP as a control. The results show that COP1 functions as an E3 ligase to polyubiquitylate HFR1 as previously reported (Fig. 5E, lane 3) (Jang et al., 2005). In addition, PIF1 promotes the polyubiquitylation of HFR1 by COP1 in a concentration-dependent manner (Fig. 5E, lanes 4 and 5). In contrast, addition of the MBP control protein did not affect COP1-mediated ubiquitylation of HFR1 (Fig. 5E, lane 6). Taken together, these results demonstrate that PIF1 promotes COP1-mediated polyubiquitylation of HFR1.
hfr1 partially suppresses the cop1-6 pif1 and pifq phenotypes
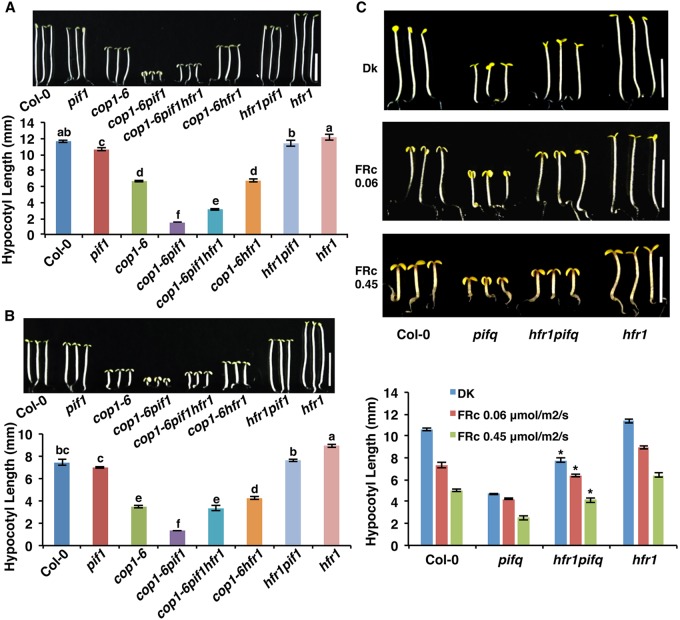
PIFs and HFR1 have a long history of antagonistic functions in regulating seedling de-etiolation, seed germination and shade avoidance (Castillon et al., 2009; Duek and Fankhauser, 2003; Fairchild et al., 2000; Fankhauser and Chory, 2000; Hersch et al., 2014; Lorrain et al., 2009; Oh et al., 2004; Shi et al., 2013). To complement these published data, we examined whether HFR1 can rescue the synergistic phenotype of cop1-6 pif1, we generated a cop1-6 pif1 hfr1 triple mutant. Phenotypic analyses showed that the de-etiolated phenotypes are partially suppressed in the cop1-6 pif1 hfr1 triple mutant compared with those in the cop1-6 pif1 double mutant both in the dark and in far-red light (Fig. 6A,B; Fig. S7A-D). As the partial suppression of the cop1-6 pif1 phenotype by hfr1 might be due to suppression by hfr of the cop1-6 phenotype only, as shown previously (Kim et al., 2002; Yang et al., 2005b), we created a hfr1 pifq quintuple mutant. The constitutive photomorphogenic phenotypes of pifq are also partially suppressed by hfr1 under both dark and far-red light conditions (Fig. 6C). This could be due to suppression of other PIF activity as HFR1-mediated suppression of PIF7 has been shown previously (Hersch et al., 2014). Thus, these phenotypic data are consistent with the high abundance of GFP-HFR1 in cop1-6 pif1 and pifq backgrounds (Figs 4 and 5A,B). Because hfr1 suppresses the pifq phenotype and, conversely, pifq suppresses the hfr1 phenotype under far-red light (Fig. 6C), the hyposensitive phenotype of hfr1 under far-red and blue light might be partly due to the higher amount of PIFs in the hfr1 background suppressing photomorphogenesis (Castillon et al., 2009; Duek and Fankhauser, 2003; Fairchild et al., 2000; Fankhauser and Chory, 2000). Taken together, these genetic and biochemical data suggest that HFR1 acts downstream of COP1 and PIFs in regulating photomorphogenesis.
Fig. 6.
hfr1 partially suppresses the phenotypes of cop1-6 pif1 and pifq. (A,B) Photographs and bar graphs showing hypocotyl lengths of seedlings of wild type, pif1, cop1-6, cop1-6 pif1, cop1-6 pif1 hfr1, cop1-6 hfr1, hfr1 pif1 and hfr1. Seedlings were grown either in the dark for 5 days (A) or grown in the dark for 21 h then transferred to continuous FRc (0.06 μmol/m2/s) for 4 days (B). Error bars indicate s.d. The letters a-f indicate statistically significant differences between means for hypocotyl lengths (P<0.05) based on two-way ANOVA analyses (n>30, three biological replicates). (C) Photographs and bar graph showing hypocotyl lengths of seedlings of wild type, pifq, hfr1 pifq and hfr1. Seedlings were grown either in the dark for 5 days (top panel) or grown in the dark for 21 h then transferred to FRc for four additional days. Error bars indicate s.d. Asterisks indicate significant difference (P<0.05) compared with pifq (n>30, three biological replicates). Scale bars: 5 mm.
Moreover, because HFR1 regulates seed germination under red light by controlling PIF1 activity (Shi et al., 2013), we also performed seed germination assays for hfr1 under an increasing fluence of far-red light conditions. hfr1 displayed reduced seed germination compared wild type, suggesting that HFR1 also functions in phyA-dependent seed germination responses (Fig. S8A). In addition, the hfr1 pif1 double mutant displayed the same phenotype as the pif1 single mutant, suggesting that pif1 is epistatic to hfr1 in the phyA-dependent seed germination response. Consistent with this phenotype, the expression of PIF1 target genes is increased in the hfr1 mutant background compared with wild type under both dark and far-red light conditions (Fig. S8B). These data further support the hypothesis that HFR1 promotes seed germination by regulating the abundance and the DNA-binding activity of PIF1.
DISCUSSION
PIFs are known to be stable in the dark, and have been shown to undergo rapid degradation in response to red, far-red and blue light conditions (Leivar and Quail, 2011; Xu et al., 2015). In this process, phy interaction is necessary for light-induced phosphorylation, polyubiquitylation and subsequent degradation (Leivar and Quail, 2011). Both CUL3-LRB and CUL4-COP1-SPA complexes have been shown to function as E3 ubiquitin ligases for the light-induced degradation of PIF3 and PIF1, respectively (Ni et al., 2014; Xu et al., 2015; Zhu et al., 2015; Zhu and Huq, 2014). In addition, DELLA proteins have been shown to promote degradation of PIFs independently of light (Li et al., 2016). However, the degradation of PIFs in the dark has not yet been shown. Our data showing that PIFs are stabilized in the presence of a proteasome inhibitor suggest that the abundance of PIFs is also regulated in the dark via the 26S proteasome pathway. Thus, PIFs are post-translationally regulated both in the dark and in light.
Phy-mediated light signaling pathways involve both negatively acting bHLH factors (e.g. PIFs), and positively acting HLH factors (e.g. HFR1, PRE6/KIDARI, PAR1, PAR2, HECs and possibly others) (Fairchild et al., 2000; Fankhauser and Chory, 2000; Hyun and Lee, 2006; Kim et al., 2002; Lorrain et al., 2009; Roig-Villanova et al., 2006; Zhou et al., 2014; Zhu et al., 2016). Similar to the established antagonistic relationship between bHLH and HLH proteins in eukaryotic systems (Littlewood and Evans, 1998; Toledo-Ortiz et al., 2003), HFR1 sequesters PIF1/PIF4/PIF5/PIF7 to regulate red light-induced seed germination and shade avoidance responses (Hersch et al., 2014; Hornitschek et al., 2009; Shi et al., 2013). Here, we show that HFR1 also promotes seed germination under far-red light conditions, consistent with its role under far-red light in seedling de-etiolation (Fairchild et al., 2000; Fankhauser and Chory, 2000; Kim et al., 2002). Thus, HFR1 regulates PIF function not only by sequestration, but also by negatively regulating their abundance post-translationally. These dual mechanisms ensure inhibition of PIF activity to optimize plant development in response to light. This is in contrast with another small family of HLH proteins named HECATE, which stabilizes PIF1 (Zhu et al., 2016). Thus, bHLH-HLH interactions not only result in sequestration, but also post-translational regulation of protein levels.
PIFs have been shown to display nontranscriptional roles in regulating HY5 post-translationally (Xu 2014, 2015). We provide strong biochemical and genetic evidence that PIF1 and COP1 synergistically regulate HFR1 post-translationally. Thus, PIF1 acts as a co-factor for COP1 to regulate multiple COP1 substrates in vivo as predicted (Xu et al., 2015). By contrast, PIFs are not directly ubiquitylated by COP1 in vitro (Jang et al., 2010; Xu et al., 2014; Zhu et al., 2015). However, PIFs directly interact with COP1 and SPA1 in vitro and in vivo in dark and light conditions (Jang et al., 2010; Xu et al., 2014; Zhu et al., 2015). PIF1 is also poly-ubiquitylated by the CUL4-COP1-SPA complex in vivo (Zhu et al., 2015). These data suggest a bifurcation of biochemical function such that COP1 is sufficient to polyubiquitiylate HFR1, HY5 and other positive factors in vitro, whereas COP1 might need to form a CUL4-COP1-SPA complex to polyubiquitylate PIFs in vitro. Further biochemical assays using the CUL4-COP1-SPA complex are necessary to examine this hypothesis.
In summary, PIF1 and HFR1 undergo reciprocal degradation in the dark (Fig. S9, left). Under red and far-red light, PIF1 is degraded by the CUL4-COP1-SPA complex, whereas HFR1 is stabilized by phy-mediated inhibition of COP1-SPA. The increased abundance of HFR1 sequesters residual PIF1 and other PIFs to promote seed germination and seedling de-etiolation under light (Fig. S9, right). Recently, PIF3 and phyB have been shown to undergo co-degradation in response to light via the CUL3LRB complex (Ni et al., 2014; Zhu and Huq, 2014). The co-degradation of PIF3 and phyB appears to attenuate the incoming signals to protect plants by degrading the signal receptor as well as the primary signal acceptor in a mutually destructive manner (Ni et al., 2014; Zhu and Huq, 2014). The co-degradation of PIF1 and HFR1 found in our study also demonstrates a similar mechanism in the dark, by which photomorphogenesis would not be over-repressed by an excessively high abundance of PIF repressors. This mechanism is important because elevated levels of PIF in the dark or during early light exposure distort seedling growth and gene expression during de-etiolation as has been shown previously (Khanna et al., 2004; Krzymuski et al., 2014).
MATERIALS AND METHODS
Plant materials, growth conditions and measurements
Seeds of wild-type Col-0 and various mutant and tagged lines have been described (Castillon et al, 2009; Park et al., 2004; Sakuraba et al., 2014; Xu et al., 2014; Zhu et al., 2015). cop1-6 pif1 hfr1, cop1-6 hfr1, cop1-4 hfr1 and hfr1 pifq were generated by crossing hfr1 with cop1-6 pif1, cop1-6, cop1-4 and pifq. For generation of pif1 GFP-HFR1, pifq GFP-HFR1, cop1-6 GFP-HFR1 and cop1-6 pif1 GFP-HFR1, GFP-HFR1 was crossed into those mutant backgrounds. For generation of cop1-4 hfr1/GFP-HFR1 and cop1-4 hfr1/TAP-PIF1, cop-4 hfr1 was crossed into GFP-HFR1 and TAP-PIF1, respectively. The primers used for genotyping were as previously described (Castillon et al., 2009; Xu et al., 2014). To generate HFR1*GFP, HFR1* was first generated by site-directed mutagenesis with the primers listed in the Table S1 (Shi et al., 2013). The HFR1 open reading frame was cloned into pENTR vector as previously described (Hornitschek et al., 2009). Then it was cloned into the GFP destination vector for transformation into the hfr1 background as described (Bu et al., 2011a). To generate cop1-4 hfr1 HFR1*GFP, cop1-4 hfr1 was crossed to the hfr1/ HFR1*GFP.
Plants were grown in Metro-Mix 200 soil (Sun Gro Horticulture, Bellevue, WA, USA) under 24-h light at 22±0.5°C. Seeds were sterilized with bleach and then plated on the Murashige and Skoog (MS) medium supplemented 0.9% agar without sucrose as described (Shen et al., 2005). After 3-4 days of cold treatment, seeds were exposed to white light for 3 h at room temperature to trigger germination. For GFP-HFR1 immunoblotting, seeds were either placed back into the dark for 4 days or grown in the dark for 21 h then transferred to continuous far-red light (FRc; 0.45 μmol/m2/s) for 3 days. For PIF1 immunoblotting, seeds were placed back into the dark for 4 days for direct protein extraction. For cycloheximide and proteasome inhibitor treatment, 4-day-old dark-grown seedlings were transferred to 5 ml media containing either 20 mM cycloheximide or 40 µM Bortezomib and incubated in the dark for the duration indicated in each figure as previously described (Shen et al., 2005; Zhu et al., 2015). For gene expression and in vivo co-immunoprecipitation assays, seeds were placed back into the dark for 4 days. For de-etiolation phenotypes, seedlings are grown either in the dark for 5 days or grown in the dark for 21 h then transferred to FRc for 4 days before taking pictures. For measurement of hypocotyl lengths, cotyledon areas and cotyledon angles, digital pictures of dark- or FRc-grown seedlings as mentioned above were taken and at least 30 seedlings were measured using ImageJ (http://rsb.info.nih.gov/ij/). The phenotypic assays were replicated as least three times. The phyA-dependent seed germination assays were performed as previously described (Zhu et al., 2015).
RNA isolation and quantitative RT-PCR
Quantitative RT-PCR (RT-qPCR) for seedlings and seeds was performed as previously described (Xu et al., 2014; Zhu et al., 2015, 2016). For seedlings, total RNA of 3- or 4-day-old dark-grown seedlings were extracted with the Spectrum plant total RNA kit (Sigma-Aldrich). For seeds, wild type and hfr1 were plated on MS plates with 100 μM paclobutrazol for 1 h and then treated with far-red light (34 μmol/m2/s) for 5 min and kept in the dark for 2 days before RNA isolation. One microgram of total RNA was used to reverse transcribe into cDNA using SuperScript III (Life Technologies) after DNase I treatment. RT-qPCR was performed using the Power SYBR Green Kit (Applied Biosystems) in a 7900HT Fast Real-Time PCR machine. PP2A (At1g13320) was used as a control. The resulting cycle threshold (Ct) values were used for calculation of the relative expression level for GFP genes relative to PP2A. The value of GFP-HFR1 was set as 1 to calculate the relative values of other genotypes. Primers for RT-qPCR are listed in Table S1.
Protein extraction and immunoblot analyses
For GFP-HFR1 and native PIF1/5 immunoblots, seedlings were grown as described above. For TAP-PIF1, PIF3-Myc, PIF4-Myc and PIF5-myc immunoblots, plates were kept in darkness for 4 days and one batch of seedlings for each genotype was treated with proteasome inhibitor (40 µM Bortezomib) for 3 h before protein extraction. Total protein was extracted in buffer [100 mM MOPS PH 7.6, 5% SDS, 10% glycerol, 40 mM EDTA pH 8, 1×protease inhibitor cocktail (Sigma-Aldrich), 40 mM β-mercaptoethanol, 2 mM PMSF, 25 mM β-GP, 10 mM NaF and 2 mM sodium orthovanadate], followed by boiling in water for 3 mins. The samples were centrifuged at 16,000 g for 10 min and then the supernatant was loaded onto 8% SDS-PAGE gel. After blotting onto polyvinylidene difluoride (PVDF) membranes, the same membrane was first blotted with anti-GFP, anti-PIF1 (Shen et al., 2008), anti-PIF5 (AS12 2112, Agrisera, Vännäs, Sweden) or anti-Myc (1:2000, catalog code OP10, EMD Millipore) antibodies followed by anti-RPT5 or anti-actin antibody after stripping. For the quantification, we used ImageJ software to measure band intensities based on at least three independent blots.
In vivo immunoprecipitation assays
To detect the ubiquitylation of TAP-PIF1 and GFP-HFR1 in the pifq background in vivo, immunoprecipitation from 4-day-old dark-grown seedlings of each genotype were performed as previously described with minor modifications (Shen et al., 2008). Briefly, total protein was extracted from ∼0.4 g 4-day-old dark-grown seedlings pretreated with 40 µM Bortezomib for 3 h before protein extraction. Total protein was extracted from seedling tissues (∼0.4 g) with 1 ml native extraction buffer [100 mM NaH2PO4 phosphate buffer (pH 7.8), 150 mM NaCl, 0.1% NP-40, 1 mM PMSF, 25 mM β-glycerophosphate, 10 mM NaF, 2 mM sodium orthovanadate, 1×protease inhibitor cocktail (catalog code P9599, Sigma-Aldrich), 40 μM bortezomib], and centrifuged in the dark at 16,000 g for 15 min at 4°C. TAP-PIF1 or GFP-HFR1 was immunoprecipitated from the supernatant with Dynabeads Protein A bound to anti-Myc or anti-GFP antibodies, respectively. Then the pellets were washed and heated with SDS buffer for 5 min at 65°C before loading to 6.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels. Same blot was first probed with anti-Ub antibody followed by either anti-Myc (mouse, EMD Millipore) or anti-GFP antibody after stripping for TAP-PIF1 or GFP-HFR1 blot, respectively.
In vitro ubiquitylation assays
The in vitro ubiquitylation assay was performed as previously described with minor modifications (Jang et al., 2005; Xu et al., 2014; Yang et al., 2005b). MBP-PIF1 was purified from Escherichia coli as previously described (Xu et al., 2014). HFR1 was digested from HFR1-GAD (Castillon et al., 2009), and then cloned into pGEX4T-1 to obtain GST-HFR1. Both MBP-COP1 and GST-HFR1 proteins were purified from E. coli as previously described (Hardtke et al., 2000; Xu et al., 2014). Flag-tagged ubiquitin (Flag-Ub), UBE1 (E1) and UbcH5b (E2) were used as previously described (Jang et al., 2005) (Boston Biochem). For the in vitro ubiquitylation reaction, 5 μg of Flag-ubiquitin, ∼25 ng of E1, ∼25 ng of E2, ∼500 ng of MBP-COP1, ∼200 ng of GST-HFR1, and 50 or 100 ng MBP-PIF1 were added to the reaction buffer containing 50 mM Tris, pH7.5, 2 mM ATP, 5 mM MgCl2 and 2 mM DTT. MBP-COP1 was pretreated with 20 μM ZnCl2 for 45 min at 22°C before adding into the reaction system. Reactions were carried out at 30°C for 2 h, and then the samples were heated at 95°C with SDS buffer. Reaction mixtures were then loaded onto 8% SDS-PAGE gel and blotted onto PVDF membrane. Ubiquitylated GST-HFR1 was first detected with α-Flag antibody (F1804; Sigma-Aldrich) and same blot was then probed with anti-GST-HRP conjugate (GE Healthcare Bio-Sciences).
Yeast two hybrid analyses
The full-length HFR1, HFR1* and C-terminal DNA binding domain (bHLH) of PIF1 (C328) open reading frames were amplified by PCR using the primers listed in Table S1. The entry clone containing HFR1* was used as the template to amplify mutant HFR1*. The PIF1-C328 clone has been described (Shen et al., 2008). The full-length HFR1 and mutant HFR1* fragments were cloned into pGAD424 vector. These plasmids were transformed into yeast strain Y187. A β-galactosidase activity assay for quantification of the interaction was performed according to the manufacturer's instructions (Matchmaker Two-Hybrid System; Clontech Laboratories).
Acknowledgements
We thank Drs Xing Wang Deng for sharing the cop1 mutant, Haiyang Wang for GFP-HFR1 seeds and Giltsu Choi for PIF3-myc and PIF5-myc seeds.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.H., X.X., P.K.K., V.N.P.; Methodology: E.H., X.X., P.K.K., V.N.P., A.N., Q.B.; Validation: X.X., P.K.K., V.N.P., A.N., Q.B.; Formal analysis: E.H., X.X., P.K.K., V.N.P., Q.B.; Investigation: E.H., X.X., P.K.K., V.N.P., Q.B.; Resources: E.H., X.X., P.K.K., A.N., Q.B.; Data curation: X.X.; Writing - original draft: E.H., X.X.; Writing - review & editing: E.H., X.X., P.K.K., V.N.P.; Visualization: X.X.; Supervision: E.H.; Project administration: E.H.; Funding acquisition: E.H.
Funding
This work was supported by grants from the National Institutes of Health (NIH) (1R01 GM-114297) and National Science Foundation (MCB-1543813) to E.H. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.146936.supplemental
References
- Bae G. and Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59, 281-311. 10.1146/annurev.arplant.59.032607.092859 [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K. C. S., Adám, E., Fejes E. Schäfer E. et al. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433-1445. 10.1105/tpc.021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.-M. and Prat S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28, 1681-1694. 10.1101/gad.243675.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Castillon A., Chen F., Zhu L. and Huq E. (2011a). Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol. 77, 501-511. 10.1007/s11103-011-9827-4 [DOI] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Dennis M. D., Yu L., Lu S. X., Person M. D., Tobin E. M., Browning K. S. and Huq E. (2011b). Phosphorylation by CK2 enhances the rapid light-induced degradation of PIF1. J. Biol. Chem. 286, 12066-12074. 10.1074/jbc.M110.186882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J. J., Luccioni L. G., Oliverio K. A. and Boccalandro H. E. (2003). Light, phytochrome signalling and photomorphogenesis in Arabidopsis. Photochem. Photobiol. Sci. 2, 625-636. 10.1039/b300094j [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H. and Huq E. (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514-521. 10.1016/j.tplants.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H. and Huq E. (2009). Blue light induces degradation of the negative regulator Phytochrome Interacting Factor 1 to promote photomorphogenic development of Arabidopsis seedlings. Genetics 182, 161-171. 10.1534/genetics.108.099887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O. S., Yanagawa Y., Zhang Y., Li J., Lee J.-H. and Zhu D. et al. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22, 108-123. 10.1105/tpc.109.065490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T., Shokry A., Moffet M., Liu P., Faul M. and Sharrock R. A. (2009). Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21, 786-799. 10.1105/tpc.108.065227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M., Galvão V. C. and Fankhauser C. (2016). Light-mediated hormonal regulation of plant growth and development. Ann. Rev. Plant Biol. 67, 513-537. 10.1146/annurev-arplant-043015-112252 [DOI] [PubMed] [Google Scholar]
- Duek P. D. and Fankhauser C. (2003). HFR1, a putative bHLH-transcription factor, mediates both phytochrome A and cryptochrome signaling. Plant J. 34, 827-836. 10.1046/j.1365-313X.2003.01770.x [DOI] [PubMed] [Google Scholar]
- Fairchild C. D., Schumaker M. A. and Quail P. H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377-2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C. and Chory J. (2000). RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 124, 39-45. 10.1104/pp.124.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbain S., Inobe T., Israeli E., Chavali S., Yu H., Kago G., Babu M. M. and Matouschek A. (2015). Sequence composition of disordered regions fine-tunes protein half-life. Nat. Struct. Molec. Biol. 22, 214-221. 10.1038/nsmb.2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C. S., Gohda K., Osterlund M. T., Oyama T., Okada K. and Deng X. W. (2000). HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. Embo. J. 19, 4997-5006. 10.1093/emboj/19.18.4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M., Lorrain S., de Wit M., Trevisan M., Ljung K., Bergmann S. and Fankhauser C. (2014). Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc. Nat. Acad. Sci. 111, 6515-6520. 10.1073/pnas.1320355111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U. (2005). Regulated proteolysis in light signaling. Curr. Op. Plant. Biol. 8, 469-476. 10.1016/j.pbi.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O. and Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. Embo. J. 28, 3893-3902. 10.1038/emboj.2009.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E. and Quail P. H. (2005). Phytochrome signaling. In Handbook of Photosensory Receptors (ed. Briggs W. R. and Spudich J. L.), pp. 151-170. Weinheim, Germany: Wiley-VCH. [Google Scholar]
- Hyun Y. and Lee I. (2006). KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61, 283-296. 10.1007/s11103-006-0010-2 [DOI] [PubMed] [Google Scholar]
- Jang I.-C., Yang J.-Y., Seo H. S. and Chua N.-H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19, 593-602. 10.1101/gad.1247205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.-C., Henriques R., Seo H. S., Nagatani A. and Chua N.-H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR Proteins Promote Phytochrome B Polyubiquitination by COP1 E3 Ligase in the Nucleus. Plant Cell 22, 2370-2383. 10.1105/tpc.109.072520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O. S. and Deng X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217-230. 10.1038/nrg2049 [DOI] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E. A., Al-Sady B., Lanzatella C. and Quail P. H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16, 3033-3044. 10.1105/tpc.104.025643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-M., Woo J.-C., Song P.-S. and Soh M. S. (2002). HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J. 30, 711-719. 10.1046/j.1365-313X.2002.01326.x [DOI] [PubMed] [Google Scholar]
- Kim J., Kang H., Park J., Kim W., Yoo J., Lee N., Kim J., Yoon T.-Y. and Choi G. (2016a). PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 28, 1388-1405. 10.1105/tpc.16.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Song K., Park E., Kim K., Bae G. and Choi G. (2016b). Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-autonomously. Plant Cell 28, 2770-2785. 10.1105/tpc.16.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C., Viczián A., Kircher S., Schäfer E. and Nagy F. (2015). Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 206, 965-971. 10.1111/nph.13207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzymuski M., Cerdán P. D., Zhu L., Vinh A., Chory J., Huq E. and Casal J. J. (2014). Phytochrome A antagonizes PHYTOCHROME INTERACTING FACTOR 1 to prevent over-activation of photomorphogenesis. Mol. Plant 7, 1415-1428. 10.1093/mp/ssu078 [DOI] [PubMed] [Google Scholar]
- Lau O. S. and Deng X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584-593. 10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Leivar P. and Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26, 56-78. 10.1105/tpc.113.120857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P. and Quail P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19-28. 10.1016/j.tplants.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E. and Quail P. H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815-1823. 10.1016/j.cub.2008.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Yu R., Fan L.-M., Wei N., Chen H. and Deng X. W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7, 11868 10.1038/ncomms11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T. and Evans G. I. (1998). Helix-Loop-Helix Transcription Factors, 3rd edn. New York: Oxford University Press. [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S. and Fankhauser C. (2009). Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 60, 449-461. 10.1111/j.1365-313X.2009.03971.x [DOI] [PubMed] [Google Scholar]
- McNellis T. W., von Arnim A. G., Araki T., Komeda Y., Misera S. and Deng X. W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487-500. 10.1105/tpc.6.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., Tepperman J. M., Stanley D. J., Maltby D. A., Gross J. D., Burlingame A. L., Wang Z.-Y. and Quail P. H. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160-1164. 10.1126/science.1250778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.-I., Kang C. and Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16, 3045-3058. 10.1105/tpc.104.025163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.-I., Liu J. R. and Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968-975. 10.1093/pcp/pch125 [DOI] [PubMed] [Google Scholar]
- Qiu Y., Li M., Pasoreck E. K., Long L., Shi Y., Galvão R. M., Chou C. L., Wang H., Sun A. Y. and Zhang Y. C. et al. (2015). HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis Photomorphogenesis. Plant Cell 27, 1409-1427. 10.1105/tpc.114.136093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P. H. (2007). Phytochrome-regulated gene expression. J. Integr. Plant Biol. 49, 11-20. 10.1111/j.1744-7909.2006.00422.x [DOI] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P. F. and Martinez-Garcia J. F. (2006). Identification of primary target genes of phytochrome signaling. early transcriptional control during shade avoidance responses in arabidopsis. Plant Physiol. 141, 85-96. 10.1104/pp.105.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J. A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U. and Deng X. W. (2003). The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642-2647. 10.1101/gad.1122903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M.-Y., Kim J., Paek N.-C. and Choi G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5, 4636 10.1038/ncomms5636 [DOI] [PubMed] [Google Scholar]
- Seo H. S., Yang J.-Y., Ishikawa M., Bolle C., Ballesteros M. L. and Chua N.-H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995-999. 10.1038/nature01696 [DOI] [PubMed] [Google Scholar]
- Shen H., Moon J. and Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize seedling photomorphogenesis in Arabidopsis. Plant J. 44, 1023-1035. 10.1111/j.1365-313X.2005.02606.x [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L. Castillon A., Majee M., Downie B. and Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME INTERACTING FACTOR 1 depends upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20, 1586-1602. 10.1105/tpc.108.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhong S., Mo X., Liu N., Nezames C. D. and Deng X. W. (2013). HFR1 Sequesters PIF1 to Govern the Transcriptional Network Underlying Light-Initiated Seed Germination in Arabidopsis. Plant Cell 25, 3770-3784. 10.1105/tpc.113.117424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I. S., Bae G., Lee C.-H., Lee D. and Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Nat. Acad. Sci. USA 106, 7660-7665. 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin A.-Y., Han Y.-J., Baek A., Ahn T., Kim S. Y., Nguyen T. S., Son M., Lee K. W., Shen Y., Song P.-S. et al. (2016). Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7, 11545 10.1038/ncomms11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E. and Quail P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749-1770. 10.1105/tpc.013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Bu Q., Huang X., Deng X. W. and Huq E. (2014). PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in arabidopsis. Plant Cell 26, 1992-2006. 10.1105/tpc.114.125591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L. and Huq E. (2015). Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20, 641-650. 10.1016/j.tplants.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Xu D., Jiang Y., Li J., Lin F., Holm M. and Deng X. W. (2016). BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 113, 7655-7660. 10.1073/pnas.1607687113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lin R., Hoecker U., Liu B., Xu L. and Wang H. (2005a). Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J. 43, 131-141. 10.1111/j.1365-313X.2005.02433.x [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X. W. and Wang H. (2005b). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in arabidopsis. Plant Cell 17, 804-821. 10.1105/tpc.104.030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J. M., Speed T. P. and Quail P. H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in arabidopsis. PLoS Genet. 9, e1003244 10.1371/journal.pgen.1003244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Song M., Yang Q., Su L., Hou P., Guo L., Zheng X., Xi Y., Meng F., Xiao Y. et al. (2014). Both PAR1 and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol 164, 841-852. 10.1104/pp.113.227231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. and Huq E. (2014). Suicidal co-degradation of the Phytochrome Interacting Factor 3 and phytochrome B in response to light. Mol. Plant 7, 1709-1711. 10.1093/mp/ssu108 [DOI] [PubMed] [Google Scholar]
- Zhu D. M., Maier A., Lee J.-H., Laubinger S., Saijo Y., Wang H. Y., Qu L.-J., Hoecker U. and Deng X. W. (2008). Biochemical Characterization of Arabidopsis Complexes Containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA Proteins in Light Control of Plant Development. Plant Cell 20, 2307-2323. 10.1105/tpc.107.056580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Bu Q., Xu X., Paik I., Huang X., Hoecker U., Deng X. W. and Huq E. (2015). CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 6, 7245 10.1038/ncomms8245 [DOI] [PubMed] [Google Scholar]
- Zhu L., Xin R., Bu Q., Shen H., Dang J. and Huq E. (2016). A negative feedback loop between PHYTOCHROME INTERACTING FACTORs and HECATE proteins fine tunes photomorphogenesis in Arabidopsis. Plant Cell 28, 855-874. 10.1105/tpc.16.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]