Abstract
Targeted cancer therapies, which act on specific cancer-associated molecular targets, are predominantly inhibitors of oncogenic kinases. While these drugs have achieved some clinical success, the inactivation of kinase signaling via stimulation of endogenous phosphatases has received minimal attention as an alternative targeted approach. Here, we have demonstrated that activation of the tumor suppressor protein phosphatase 2A (PP2A), a negative regulator of multiple oncogenic signaling proteins, is a promising therapeutic approach for the treatment of cancers. Our group previously developed a series of orally bioavailable small molecule activators of PP2A, termed SMAPs. We now report that SMAP treatment inhibited the growth of KRAS-mutant lung cancers in mouse xenografts and transgenic models. Mechanistically, we found that SMAPs act by binding to the PP2A Aα scaffold subunit to drive conformational changes in PP2A. These results show that PP2A can be activated in cancer cells to inhibit proliferation. Our strategy of reactivating endogenous PP2A may be applicable to the treatment of other diseases and represents an advancement toward the development of small molecule activators of tumor suppressor proteins.
Introduction
Cancer development and progression involve coordinate changes in both oncogene and tumor suppressor function (1, 2). Specific protein phosphatases, such as protein phosphatase 2A (PP2A), act as critical negative regulators of oncogenic signaling and are modulated by physiological and pathological mechanisms (3–5). PP2A is a serine/threonine phosphatase that controls many cellular functions, such as cell cycle, growth, metabolism, and apoptosis (6). The core enzyme of PP2A is composed of a complex between the scaffolding subunit (A) and the catalytic subunit (C). Both the A and C subunits have 2 possible isoforms, α and β, with Aα and Cα accounting for the majority of each subunit in most cells. The core enzyme associates with a variety of regulatory subunits (B) to form the heterotrimeric holoenzyme complex (7). There are multiple B subunit families (B, B′, B′′, B′′′), each consisting of several isoforms and splice variants, bringing about the diversity of the PP2A holoenzyme. Whereas the B subunits are specific to tissue and cell type/developmental stage, the A and C subunits are ubiquitously expressed (8–10). Posttranslational modifications modulate PP2A activity and B subunit interactions. Signaling via MAPK and PI3K/AKT pathways is regulated, in part, by PP2A, which dephosphorylates and inactivates multiple protein kinases. As a tumor suppressor gene, PP2A is genetically altered or functionally inactivated in many cancers (11–15), and activation of PP2A represents a promising strategy for therapeutic intervention (5).
Results
We investigated whether activation of PP2A in KRAS mutant lung cancer cells could be a viable treatment approach. We developed small-molecule activators of PP2A (SMAPs) by reengineering tricyclic neuroleptics (16) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI89548DS1). Tricyclic neuroleptics have been reported to activate PP2A, (17, 18) and this was proposed to be through direct binding of the PP2A Aα subunit. However, clinical trials evaluating these molecules in the early 1990s were restricted by dose-limiting CNS-related toxicity (19). Developing tricyclic neuroleptics as anticancer agents required neutralization of their potent CNS pharmacology and improvement of their antiproliferative properties. By replacing the basic amine with a neutral polar functional group, the CNS effects were abrogated, and further chemical derivatization improved the anticancer potency of these SMAPs (17).
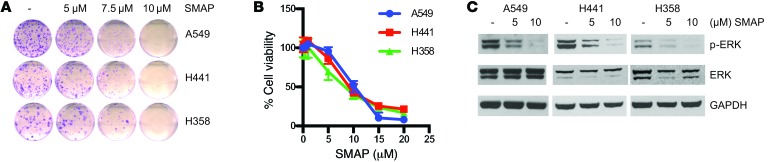
To determine the efficacy of our compounds, KRAS mutant lung cancer cell lines (A549, H441, H358, H23, and CALU-1) were treated with SMAPs. These SMAPs decreased cell survival and induced cell death in each of these cell lines (Figure 1, A–C, Supplemental Figure 2, A–C and Supplemental Figure 3, A–C). SMAP treatment induced a significant increase in annexin V staining, which was prevented by cotreatment with the caspase inhibitor Z-VAD, indicating that cell death was through a caspase-dependent mechanism (Supplemental Figure 2B and Supplemental Figure 3B). Additionally, treatment of these cell lines with SMAPs resulted in increased poly (ADP-ribose) polymerase (PARP) cleavage and decreased phosphorylation of ERK (Figure 1C, Supplemental Figure 2D, and Supplemental Figure 3D).
Figure 1. SMAPs decrease cell viability and inhibit MAPK signaling.
(A) Clonogenic assay of KRAS mutant cell lines (A549, H441, H358) treated with SMAPs for 3 weeks. (B) MTT assay in A549, H441, and H358 cells treated with increasing doses of SMAP at 24 hours. (C) Western blots for p-ERK and ERK normalized to GAPDH in KRAS mutant cell lines treated with SMAP. Data represent mean ± SEM from 3 experiments.
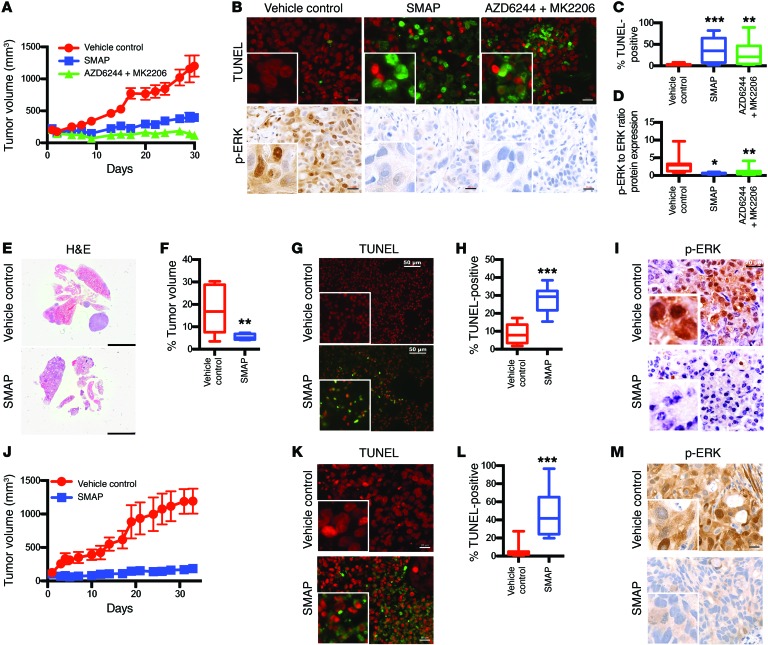
To evaluate the efficacy of SMAPs in vivo, we used xenograft and transgenic mouse models of KRAS-activated lung adenocarcinoma. In the H358 xenograft model, SMAP treatment resulted in significant inhibition of tumor growth (Figure 2A) and the induction of apoptosis (Figure 2, B and C). Western blotting and IHC analyses showed substantial reductions of phosphorylated ERK (p-ERK) in the tumors (Figure 2, B–D, and Supplemental Figure 4A). Single-agent SMAP treatment displayed efficacy similar to that of combination treatment with the kinase inhibitors MK2206 (AKT inhibitor) plus AZD6244 (MEK inhibitor). This suggested that SMAP treatment simultaneously reduced signaling via multiple kinases. Importantly, there were no deaths, behavioral abnormalities, or changes in body weight during the course of treatments, indicating a favorable toxicity profile in these model systems (Supplemental Figure 5A). Additional evaluation of SMAPs administered at relatively high doses for 7 days showed no apparent adverse effects, and SMAP dosing resulted in no changes in clinical chemistry parameters, reflecting no apparent impact on liver function (Supplemental Tables 1–3). Similar studies showed efficacy of SMAPs in xenograft models utilizing A549, H441, and H358 KRAS mutant cells (Supplemental Figure 6, A–L, Supplemental Figure 4B, and Supplemental Figure 5, C–E). Additionally, we examined a transgenic KRASLA2 murine model, which harbors oncogenic KRAS alleles that are activated by spontaneous recombination (20), leading to the development of lung tumors that are phenotypically and histologically similar to those found in human non–small cell lung cancer (NSCLC). SMAP treatment in this model caused a significant decrease in tumor burden and induction of apoptosis (Figure 2, E–H). Treatment with SMAPs decreased p-ERK levels in tumor cells, as revealed by IHC (Figure 2I). We also utilized a KRAS mutant patient-derived xenograft (PDX) model and found that treatment with SMAPs induced growth inhibition of the tumors (Figure 2J). SMAP treatment induced apoptosis (Figure 2, K and L) and decreased phosphorylation of ERK (Figure 2M). These results indicate that SMAPs are well tolerated, orally bioavailable, and active in both transgenic and xenograft mouse models of KRAS mutant lung cancer.
Figure 2. SMAPs promote tumor growth inhibition and inhibit MAPK signaling in KRAS mutant lung cancer.
(A) 1 × 107 H358 cells were subcutaneously injected into nude mice and allowed to grow to an average of 100 mm3. Mice were treated with vehicle control (n = 10), a combination of 6 mg/kg MK2206 and 24 mg/kg AZD6244 (n = 10), or 5 mg/kg SMAP (n = 10) twice a day for 4 weeks. Tumor volume over course of treatment is shown. (B) Tumors were evaluated by sacrificing the mice 2 hours after final treatment. Representative TUNEL staining and p-ERK staining of treated tumors. Scale bars: 20 μM. Original magnification: ×40. (C) Quantification of TUNEL positivity. (D) Quantification of p-ERK levels in xenograft tumors as performed by immunoblotting and densitometry. (E) KRASLA2 mice were randomized into treatment groups after reaching 16 weeks of age. Mice were treated with vehicle control (n = 3) or 15 mg/kg SMAP (n = 3) orally twice a day for 3 weeks. Mice were sacrificed and lungs were extracted 2 hours after final treatment. Representative images of lungs. Scale bar: 5 mM. (F) Percentage of tumor volume was evaluated by ImageJ using 3 sections of H&E for each mouse. (G) Representative TUNEL staining. Scale bars: 50 μM. (H) Quantification of TUNEL positivity in all tumors treated. (I) Immunohistochemistry of p-ERK in treated animals. Scale bars: 20 μM. Original magnification: ×40. (J) KRAS mutant PDX tumor fragments were surgically reimplanted in the right flank of NSG mice. Mice were treated with vehicle control (n = 7) and 5 mg/kg SMAP (n = 6) twice a day for 4 weeks. Tumor volume over course of treatment. (K) Tumors were evaluated by sacrificing mice 2 hours after final treatment. Representative TUNEL staining. Scale bar: 20 μM. Original magnification: ×40. (L) Quantification of TUNEL positivity in all tumors treated. (M) Immunohistochemistry of p-ERK in treated animals. Scale bar: 20 μM. Original magnification: ×40. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
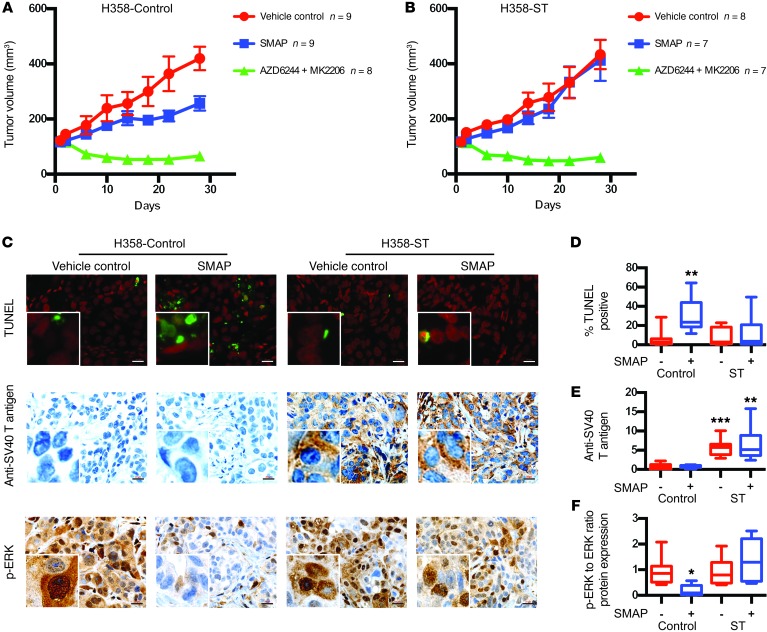
To assess whether the biological effect of SMAPs was through PP2A activation, we utilized SV40 small T antigen as a form of target validation. The small T antigen of SV40 is known to bind the PP2A A subunit at the B subunit binding domain and prevent holoenzyme assembly, altering PP2A activity (21–23). Expression of the small T antigen in lung cancer H358 cells conferred resistance to SMAP treatment in the xenograft model (Figure 3, A–F) while having no impact on response to the PP2A-independent mechanism of MK2206/AZD6244 combination treatment. Western blotting and IHC analyses showed that, while SMAPs reduced p-ERK levels in control tumors, tumors expressing the small T antigen had no significant change (Figure 3, C–F, and Supplemental Figure 4C). This indicated that SMAP-mediated growth inhibition was dependent on functional PP2A holoenzymes.
Figure 3. Expression of the small T antigen confers resistance to SMAPs.
(A and B) 1 × 107 H358 control (A) and H358 small T antigen (ST) (B) cells were subcutaneously injected into nude mice and allowed to grow to an average of 100 mm3. Mice were treated with vehicle control (n = 9 for H358 control; n = 8 for H358 ST), a combination of 6 mg/kg MK2206 and 24 mg/kg AZD6244 (n = 8 for H358 control; n = 7 for H358 ST), and 5 mg/kg SMAP (n = 9 for H358 control; n = 7 for H358 ST) twice a day for 4 weeks. Tumor volume over course of treatment. (C) Representative TUNEL staining of treated tumors and immunohistochemistry of SV40 T antigen and p-ERK in treated animals. Scale bars: 20 μM. Original magnification: ×40. (D) Quantification of TUNEL positivity. (E) Quantification of SV40 T antigen levels. (F) Quantification of p-ERK levels. Quantification of SV40 T antigen and p-ERK levels in the xenograft tumors was performed by immunoblotting and densitometry. TUNEL, anti-SV40 T, and ERK signaling in the tumors were evaluated by sacrificing the mice 2 hours after final treatment. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
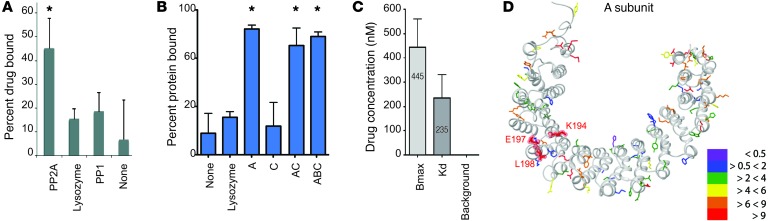
Next, to assess binding of SMAPs to PP2A, a tritiated version of SMAP was prepared. Using equilibrium dialysis, tritiated SMAP bound to the PP2A heterodimer (AC) but not to the related serine/threonine phosphatase PP1 or lysozyme, used as negative control proteins (Figure 4A). Binding assays were performed using purified recombinant A (PP2A A) and C (PP2A C) subunits, the AC dimer (PP2A AC), and the PP2A trimer (PP2A AB56C). The tritiated SMAP compound specifically bound the PP2A A subunit alone as well as the PP2A dimer and trimer that both contained the A subunit, but did not bind to the PP2A C subunit (Figure 4B). We concluded that SMAPs bind the A subunit of PP2A. The KD using the PP2A A subunit was 235 nM (Figure 4C), and the half-maximal binding using the A subunit was at approximately 500 nM (Supplemental Figure 7A). Competition of different SMAPs with tritiated SMAP showed that they all could outcompete the interaction with the PP2A A subunit (Supplemental Figure 7B). Experiments with cell culture or tumors in vivo required substantially higher concentrations of SMAP than those needed to saturate purified PP2A. We attribute this difference to nonspecific binding of SMAPs to albumin and possibly other proteins that reduce the effective free drug concentration available for binding to the target.
Figure 4. SMAPs binding to protein phosphatase PP2A.
(A) Binding studies of radiolabeled SMAPs with PP2A AC dimer, lysozyme, and PP1 using equilibrium dialysis. (B) Binding of radiolabeled SMAPs to lysozyme as negative control and PP2A (A–C) subunits using equilibrium dialysis. (C) Quantification of the KD and binding maximum (Bmax) for SMAP against the PP2A trimer. Data represent mean ± SD of 3 experiments. (D) Projection of changes in solvent exposure based on hydroxylradical modification in the A subunit of PP2A AC upon SMAP ligand addition. The structure of the A subunit (in light gray) is taken from 2A (PP2A) holoenzyme (PDB 2IAE). Modified amino acids are represented by colored side chains. The color codes indicate the changes in rates of modification for each specific site upon SMAP binding to the A subunit of PP2A AC. Purple indicates the residues that showed change in modification of less than 0.5-fold, blue indicates the residues that show minimal to no change (< 2-fold) in modification, green indicates decreases of more than 2-fold and less than 4-fold, yellow indicates decreases of more than 4-fold and less than 6-fold, orange indicates decreases of more than 6-fold and less than 9-fold, and red indicates decreases of more than 9-fold in the modification rate upon SMAP binding. Data represent mean ± SEM. *P < 0.05, Student’s t test.
Photoaffinity labeling (PAL) was used to characterize biochemical binding for the SMAP family of activators to the PP2A core enzyme (PP2A AC) using DT-453, a well-characterized SMAP analog. For the PAL experiments, a photoactivatable nitrene was incorporated in DT-453 such that covalent attachment between drug and protein was generated upon treatment with UV and activation of the nitrene (Supplemental Figure 8 and Supplemental Figure 9, A and B). The samples, which included a PP2A AC experimental sample (+ drug/+UV), PP2A AC (+ drug/–UV) nonactivated control, and PP2A AC (–drug/–UV) negative control, were digested using trypsin and analyzed by liquid chromatography–mass spectrometry (LC-MS). Comparison of the MS and MS/MS spectra of the sets of tryptic peptides identified a common set of unmodified tryptic peptides, with the exception of a single peptide, 184-AAASKLGEFAKVLELDNVK-202 of the A subunit (within the HEAT domain 5–8), which had a mass increase of 439.11 Da only in the +drug/+UV samples for each independently completed experiment. This mass increase matches the predicted mass of the nitrene DT-453 adduct to this peptide within 3 ppm.
Based on the PAL experiments, we tentatively assigned drug binding to the flexible HEAT repeat domains 5–8 in the PP2A A subunit. To more precisely characterize the binding site and to explore drug-induced conformational changes in PP2A, we conducted hydroxyl radical footprinting (HRF) and MS experiments to characterize the SMAPs binding to the scaffolding subunit of PP2A (24, 25). Hydroxyl radicals (▪OH) generated during exposure to x-rays oxidatively modify solvent accessible amino acid side chains and provide high resolution information on solvent accessibility based on the observed extent of modification, which can be quantified at the peptide and side-chain level. Drug-binding sites and ligand-induced conformational changes were visualized using existing 3D protein structure information. We performed HRF on the PP2A AC complex in the absence and presence of various SMAPs, including SMAP, SMAP2, and SMAP3, which have varying efficacy in cancer suppression. Samples of each protein in solution were exposed to x-rays, followed by digestion with both pepsin and trypsin enzymes, and subsequent analysis by LC-MS to determine extents and rates of modification and MS/MS to determine the sites of modification. A protection factor (PF) for each specific side chain was calculated as the ratio of modification rate for PP2A AC divided by the modification rate for the PP2A AC-SMAP complex (Supplemental Table 4). PFs greater than 1 represent drug-dependent reduction in the hydroxyl radical labeling of PP2A AC due to direct binding of the drug to the protein and/or conformational changes induced within the complex upon SMAP binding. In particular, for SMAP binding, the observed PF values ranged from 0.9 to 38.5, with median values of 5.4 for the A subunit. Residues K194, E197, and L198 within the peptic peptide 192-KAVLEL-198 of the A subunit exhibited large PFs of 18, 23, and 18, respectively. These sites are highlighted in Figure 4D and are contained on the outer connecting loop of HEAT repeats 5 and 6. These results, complemented by the equilibrium dialysis, PAL, and molecular modeling, suggest the high PF values for residues K194, E197, and L198 in the A subunit are due to SMAP binding the A subunit within the 194–198 region (Figure 4D). The HRF data suggest a model indicating that SMAP binding to the PP2A A subunit promotes conformational changes, supported by the high PF values outside the putative drug-binding region. Assays of PP2A with a peptide substrate showed a dose-dependent increase in phosphatase activity of AC and AB56C with added SMAP (Supplemental Figure 10, A–D). Thus, the conformational changes due to SMAP binding produce activation of the phosphatase.
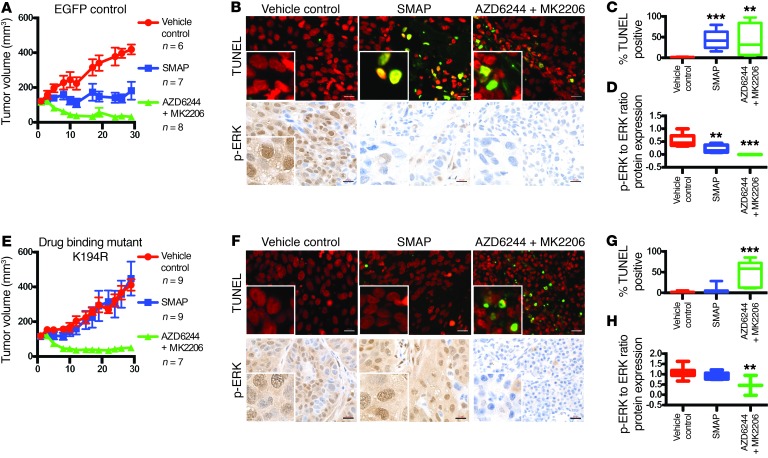
Based on the HRF results, the 194–198 region of the A subunit was identified as a putative SMAP-binding region, and K194, E197, and L198 were identified as potential sites of interaction. Therefore, mutations of residues K194, E197, and L198 were created using site-directed mutagenesis of a pLX304 vector containing a V5-tagged PP2A Aα (Supplemental Figure 11, A–F). H358cells stably overexpressingWT or the mutated PP2A A subunit were compared with cells with pLX304 EGFP as controls. Tumor xenografts in mice using these cell lines, including both EGFP and WT PP2A as controls, were treated with SMAP (Figure 5, A–H, and Supplemental Figure 12, A–L). Tumors with mutations K194R and L198V at the putative drug-binding site in PP2A Aα exhibited resistance to SMAP treatment (Figure 5, E–G, and Supplemental Figure 12, I–K). Treatment response with combination AKT inhibitor and MEK inhibitor (MK2206/AZD6244) was not affected by expression of these mutations, supporting once again a PP2A-dependent mechanism of action for these SMAPs. Western blotting and IHC analyses showed that, while the tumors expressing control EGFP and WT PP2A had reductions of p-ERK, tumors expressing the mutations K194R and L198V did not show significant changes in these markers when treated with SMAPs (Figure 5, B, D, F, and H; Supplemental Figure 12, B, D, F, H, J, L; and Supplemental Figure 13, A–E). Additionally, coimmunoprecipitation analyses and PP2A activity assays showed that the mutant Aα proteins were competent to form holoenzymes that were as active as AC with the WT A subunit (Supplemental Figure 11, A–F). Together, these data indicate that SMAP-mediated growth inhibition was dependent on PP2A activation and that residues K194 and L198 in the A subunit are required for SMAP activity.
Figure 5. Effects of mutations in putative drug-binding site.
(A) Male nude mice were subcutaneously injected (1 × 107 cells per injection) in the right flank with the different isogenic cell lines (control EGFP and putative drug-binding mutant K194R). Once the tumors reached a volume of 100 mm3, the mice were randomly enrolled in vehicle control (n = 6 for EGFP; n = 9 for K194R), a combination of 6 mg/kg MK2206 and 24 mg/kg AZD6244 (n = 8 for EGFP; n = 7 for K194R), or 5 mg/kg SMAP (n = 7 for EGFP; n = 9 for K194R) twice a day for 4 weeks. Mouse tumor volume for control EGFP-expressing H358 xenograft over course of treatment. (B) Tumors were evaluated by sacrificing the mice 2 hours after final treatment. Representative TUNEL staining and p-ERK IHC of treated tumors. Scale bars: 20 μM. Original magnification: ×40. (C) Quantification of TUNEL positivity in tumor. (D) Quantification of p-ERK levels in the xenograft tumors as performed by immunoblotting and densitometry. (E) Mouse tumor volume for drug-binding mutant K194R expressing H358 xenograft over course of treatment. Tumor volume over course of treatment. (F) Tumors were evaluated by sacrificing the mice 2 hours after final treatment. Representative TUNEL staining and p-ERK IHC of treated tumors. Scale bar: 20 μM. Original magnification: ×40. (G) Quantification of TUNEL positivity in tumors treated. (H) Quantification of p-ERK levels in xenograft tumors as performed by immunoblotting and densitometry. Data represent mean ± SEM. **P < 0.01; ***P < 0.001, Student’s t test.
Discussion
The studies presented here demonstrate that small molecules derived from the tricyclic neuroleptics phenothiazine and dibenzazepine directly activate PP2A in KRAS mutant lung cancer and thereby inhibit tumor growth and induce apoptosis. KRAS signaling is altered in about 20% of human cancers (26, 27). Small-molecule inhibitors that target protein kinases in the RAS pathway have been developed and are FDA approved (1). However, potential resistance to inhibition of KRAS signaling can involve mutations in targets or activation of compensatory mechanisms (28–30). Inhibiting multiple pathways concurrently with the MEK and AKT inhibitors, AZD6244 and MK2206, respectively, successfully abrogates tumor growth in preclinical models (31). While kinase inhibitor combination therapy is a promising option, the limitation lies in determining the appropriate window with maximal activity and lowest toxicity. PP2A activation by SMAPs simultaneously inhibits multiple oncogenic signaling pathways by inactivation of kinases that are substrates of the phosphatase. Therapeutic activation of a protein phosphatase such as PP2A is a newly emerging tactic, which concurrently indirectly targets several oncogenes with a single agent. We included a combination of the kinase inhibitors arm in our preclinical in vivo studies and demonstrated a comparable activity of SMAPs to the “double-down double-win” effect of combination therapies described by Shimizu et al. (32). These results underscore the advantage of targeting phosphatases such as PP2A with tumor-suppressive potential.
Several lines of evidence support the tumor-suppressor role of PP2A: (a) PP2A is mutated and inactive in several human cancers; (b) overexpression of theendogenous inhibitory proteins of PP2A, namely CIP2A and SET, is observed in tumors; and (c) inhibition of PP2A by small T polyoma virus SV40 and okadaic acid promotes cellular transformation and tumor growth in mice, respectively (13, 33–37). The restoration of PP2A activity has been explored as a therapeutic strategy in the context of pancreatic cancer and acute myeloid leukemia. Fingolimod FTY720 and its chiral deoxy analog drugs were used to indirectly reactivate PP2A by partly blocking the PP2A inhibitor protein SET (38). However, direct activation of the tumor-suppressor PP2A is observed with phenothiazines that directly bind the scaffold PP2A Aα subunit (16). Therefore, we generated a series of SMAPs by decoupling the CNS pharmacology from the antiproliferative properties of phenothiazines. The derivatives with enhanced antiproliferative effect as well as in vivo bioavailability were used in the experiments described in this study. Activation of PP2A via the SMAPs in KRAS-driven lung adenocarcinoma mouse xenografts or PDX models and the transgenic KRASLA2 murine model resulted in substantial tumor-growth inhibition compared with vehicle control. Furthermore, mutation of residues at the putative drug-binding site completely abrogated the antitumorigenic effect of SMAP. These results were predicted by in vitro assays showing that SMAP binds directly to the scaffolding A subunit of PP2A. Aside from the phenothiazines themselves that harbor pharmacological toxicities, our SMAPs are the first generation of anticancer molecules, to our knowledge, that directly bind and activate a tumor-suppressor enzyme. Based on changes seen in the solvent exposure of the PP2A A subunit upon ligand binding, we propose that SMAPs act by inducing conformational changes in the A subunit of PP2A. This is further supported by evidence that the SV40 small T antigen abrogates SMAP activity. The small T antigen acts on PP2A by replacing the B subunits; therefore, the ability of small T to abrogate SMAP activity supports the notion that regulatory subunit function in the ABC holoenzymes is essential for SMAP activity (21–23). These findings may extend to other malignancies driven by oncogenic substrates of PP2A or malignancies characterized by PP2A inactivation due to the overexpression of endogenous inhibitors. Additional development and the combination of SMAPs with other targeted therapies will enable the translation of our findings to patients. Finally, our results and approaches could provide the structural and molecular framework for the pharmacological reactivation of other phosphatases.
Methods
Cell lines and reagents.
Human lung cancer cell lines were purchased from ATCC. All cells were cultured in RPMI supplemented with 5% FBS and 1% penicillin/streptomycin. The cells were treated with the SMAPs (dissolved in DMSO) and screened for cell viability through the MTT assay using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide kit (Sigma-Aldrich). The CyQUANT Cell Proliferation Kit was purchased from Thermo Fisher Scientific. MK2206 (AKT inhibitor) and AZD6244 (MEK inhibitor) were purchased from Selleck. These compounds were diluted in DMSO and stored at –20°C. All SMAP compounds were diluted with DMSO and stored at room temperature. Cells were treated with 20 μM of SMAPs in all assays unless otherwise stated. PPP2R1A (Homo sapiens) in pLX304 (Gateway V5–tagged lentiviral expression vector) was purchased from the DNASU Plasmid Repository (no. HsCD00444402). Pbabe-small T retroviral plasmid was purchased from Addgene (Addgene plasmid 10673). H358–small T cell line was generated following the Gryphon retroviral expression protocol.
Antibodies.
For immunoblotting, antibodies specific for p-ERK (no. 9272), ERK (no. 4695), PP2A A (no. 2041S), PP2A C (no. 2038), V5-tag (no. 13202S), and vinculin (no. 13901S) were obtained from Cell Signaling Technology. PARP p85 fragment (no. G7341) was purchased from Promega. GAPDH antibody (no. sc-32233) was purchased from Santa Cruz Biotechnology Inc. SV40 small T antigen antibodies (pab280) were purchased from EMD Millipore. For immunohistochemistry, p-ERK antibody (Thr202/Tyr204) XP (no. 4370) was obtained from Cell Signaling Technology.
Western blotting.
Protein was isolated from cells with RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) and from animal tissues with T-PER Tissue Protein Extraction Reagent (no. 78510, Thermo Fisher Scientific) with complete ULTRA Tablets (no. 05892791001, Roche). Isolated protein was quantified and normalized via Bio-Rad assay. Protein was run on 12% SDS–polyacrylamide gel electrophoresis gels (Invitrogen, Life Technologies) and transferred onto nitrocellulose membranes. Membrane was blocked with 5% nonfat milk (LabScientific Inc.) in Tris-buffered saline–Tween buffer. The membranes were probed with the antibodies mentioned above.
Site-directed mutagenesis, RT-PCR, and sequencing.
Site-directed mutagenesis was performed using the QuikChange Lightning Site-Directed Mutagenesis Kit (no. 210519, Agilent Technologies). Primers for mutagenesis, quantitative real-time PCR (qRT-PCR), and sequencing were purchased from Life Technologies (Invitrogen). Mutations were validated using sanger sequencing.
Annexin staining.
Annexin V staining was performed using annexin V–conjugate Alexa Fluor 488 (Invitrogen, Life Technologies) and annexin-binding buffer (no. V13246, Invitrogen, Life Technologies) according to the manufacturer’s protocol. For cell-cycle analysis, cells were stained with propidium iodide (Roche) to ascertain the DNA content and determine cell-cycle distribution within the cell population (39). Each experiment was performed in triplicate.
Clonogenic assay.
For clonogenicity (colony formation) assay, cells were plated at a low density in 6-well plates. After 24 hours, cells were treated with SMAPs and incubated for 3 weeks. Cells were then fixed and stained with 1% crystal violet (Sigma-Aldrich) solution. Quantification was performed through the cell counter function on ImageJ (NIH). Each experiment was plated and repeated in triplicate.
Mouse models and treatment studies.
KrasLA2 mice were purchased from the National Cancer Institute Mouse Repository. For xenograft studies, H358 (1 × 107), A549 (5 × 106), and H441 (5 × 106) cells were injected into the right flanks of 6- to 8-week-old male BALB/c nu/nu mice (Charles River Laboratories). For PDX studies, KRAS mutant PDX tumor models were purchased from the Jackson Laboratory (model TM00231). Tumor fragments were surgically reimplanted in the right flanks of NSG mice. When tumor volumes reached an average of 200 mm3, mice were randomized to treatment groups and tumor volume was assessed by caliper measurement every other day throughout the study. Mice were treated by gavage with vehicle control, MK2206 (6 mg/kg), AZD6244 (24 mg/kg), SMAP (5 mg/kg or 15 mg/kg), and SMAP2 (100 mg/kg). Mouse body weights were recorded weekly, and percentages of mouse body weights during treatment were calculated as follows: weight at each time point/initial weight × 100. Animals were observed for signs of toxicity (mucous diarrhea, abdominal stiffness, and weight loss). Blood and tumor tissue were harvested 2 hours after the final dose of the treatment study. Animals were submitted for toxicology testing at IDEXX Laboratories. Tumors were both formalin fixed, for IHC, and snap-frozen in liquid nitrogen, for immunoblotting.
TUNEL assay and immunohistochemistry.
Tissue was fixed in 10% buffered formalin phosphate (Fisher Scientific, no. SF100-4), transferred to 70% ethanol, and blocked in paraffin. Serial tissue sections (5-μm thick) were cut from the paraffin-embedded blocks and placed on charged glass slides. Tumor sections were stained with H&E and p-ERK (no. 4370, Cell Signaling Technology). Briefly, sections were deparaffinized with xylene and rehydrated through graded alcohol washes, followed by antigen retrieval in a pressure cooker (Dako) in citrate buffer (10 mM, pH 6.0, Vector Laboratories). Slides were then incubated in hydrogen peroxide–methanol, followed by incubation in normal goat serum in PBS. Antibody was applied overnight at 4°C. DAB substrate was applied, followed by counterstaining in hematoxylin. The ApopTag Fluorescein In Situ Apoptosis Detection Kit (Millipore) was used according to the manufacturer’s protocol to perform the TUNEL assay. Prior to the addition of TdT enzyme, sections were deparaffinized with xylene and rehydrated through graded alcohol washes. VECTASHIELD Mounting Medium with Propidium Iodide (Vector Laboratories) was used for counterstaining. Bright-field and fluorescent images were captured using an Olympus MVX10 or Zeiss Axioplan 2 IE microscope. Quantification was completed using the cell counter function of ImageJ (NIH). Imaging was performed at the Microscopy CORE at the Icahn School of Medicine at Mount Sinai.
PP2A-binding assay.
For the equilibrium dialysis binding studies, the RED system (ThermoFisher) was used. Purified protein and radiolabeled compound were incubated using the RED device for 6 to 8 hours or overnight, and drug binding was determined by measuring radioactivity by liquid scintillation counting. The percentage of bound drug was calculated by the following equation: % free = (concentration buffer chamber/concentration protein chamber) × 100 % bound = 100% – % free.
Phosphatase activity assay using DiFMU.
Assay buffer was 50 mM MOPS, pH 7.5, 100 mM NaCl, 0.1% 2-ME, 10 mM MgSO4, and 1 mM MnCl2. 6,8-Difluoro-4-methylumbelliferyl phosphate (DiFMUP, Invitrogen D6567) was added to a final concentration at 100 μM to 1 nM PP2A AC dimer (WT and mutants). Samples were incubated at 25°C, and activity was measured from fluorescence measured at 358/455 nm with plate reader.
Assays of PP2A with ProFluor Ser/Thr peptide substrate.
PP2A AC dimer and recombinant AB56C trimer assembled from purified recombinant subunits were assayed for phosphatase activity using the ProFluor Ser/Thr R110 Substrate System (Promega) following the manufacturer’s instructions. The PP2A were assayed over a range of 0 to 100 ng to demonstrate a linear response range, and 15 ng of AC and 10 ng AB56C were used to test for effects of compounds that were included at the indicated final concentrations (0.1 to 20 μM). Activities are plotted as the percentage relative to control with no added compound, set as 100%. TRC-766 is an inactive compound. Assays were completed in triplicate with calculation of the mean and SEM, shown by the error bars.
Modeling of interactions between SMAP and PP2A.
Crystal structure for PP2A was retrieved from PDB (PDB ID: 2IAE), and the coordinates for SMAP were sketched using PubChem (https://pubchem.ncbi.nlm.nih.gov/). Structures of PP2A and SMAP were prepared for docking using the Protein Preparation Wizard and LigPrep module from the Schrödinger package (Schrödinger, LLC). Glide XP (extra precision) was used to dock SMAP into the region spanning HEAT domains 5–10 within the A subunit of PP2A. Docking results were visually analyzed using PyMol (https://www.pymol.org/).
PP2A AC protein preparation.
The AC form of the PP2A was purified as described by Xu et al. (40). For PAL experiments, PP2A AC was buffer exchanged 2 times with 20 volumes excess 25 mM Tris, 50 mM sodium chloride, and 5 mM dithiothreitol (DTT), pH 7.8. The final PP2A AC protein concentration was adjusted to 1 μg/μl. For HRF experiments, the purified material was buffer exchanged 2 times with 20 volumes excess of 1× PBS, pH 7.4 (1.1 mM potassium phosphate monobasic, 155.2 mM sodium chloride, 3.0 mM sodium phosphate dibasic, Invitrogen) using a 0.5 ml 10K MWCO filter (Millipore). The PP2A AC-DT complexes for HRF were made by adding various SMAP compounds to PP2A AC at a protein/drug molar ratio of 1:10. For all experiments with and without SMAP compounds, the final PP2A AC protein concentration was adjusted to 3 μM.
PAL and PP2A proteolysis.
PP2A AC protein (1 μg/ μl; 20 μl volume) was exposed to a 350 nm (e.g., long wavelength) UV light source for 15 minutes in the presence of SMAP DT-453 drug at a protein/drug molar ratio of 1:20 and in the absence of DT-453 (positive control). In addition, 20 μl of PP2A AC protein without DT-453 was prepared without any UV light exposure (negative control). Subsequently, the DT-453–PP2A complex and the positive and negative controls were reduced with 20 mM DTT at 37°C for 1 hour and alkylated with 50 mM iodoacetamide at room temperature in the dark for 30 minutes. PAL samples were digested using modified trypsin (Promega) at an enzyme/protein molar ratio of 1:10 at 37°C overnight. PAL experiments were attempted 3× with the same protein preparation.
Synchrotron radiolysis and protein proteolysis.
Radiolysis experiments were performed at beamline X28X of the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, New York, USA) and at beamline 5.3.1 of the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, California, USA). The x-ray beam parameters were optimized by using Alexa Fluor 488 fluorophore assay (41). All samples were exposed for 0 to 20 milliseconds (X28X) and for 0 to 800 μs (5.3.1) at ambient temperature and immediately quenched with methionine amide at a 10 mM final concentration to prevent secondary oxidation (42). Prior to the PP2A AC proteolysis, all protein samples were reduced with 10 mM DTT at 56°C for 45 minutes and alkylated with 25 mM at room temperature in the dark for 45 minutes. For pepsin digestion, formic acid (FA) was added to the radiolyzed samples at a final concentration of 0.5% to adjust the pH of the samples to around 2. For trypsin digestion, the pH of radiolyzed samples was adjusted to 8.0 using 1 M Tris, pH 8.5. Samples then were digested with pepsin and trypsin (Promega) at 37°C for 3 hours and overnight, respectively, at an enzyme/protein molar ratio of 1:10. The digestion reactions were terminated by freezing the samples.
LC-MS analysis.
LC-MS experiments were carried out on an Orbitrap Elite Mass Spectrometer (Thermo Electron) interfaced with a Waters nanoAcquity UPLC System. Peptic peptides were desalted on a trap column (180 μm × 20 mm packed with C18 symmetry, 5 μm, 100 Å; Waters) and subsequently resolved on a reversed phase column (75 μm × 250 mm nano column, packed with C18 BEH130, 1.7 μm, 130 Å, Waters) using a gradient of 2% to 45% mobile phase B (0.1% FA and acetonitrile [ACN]) and mobile phase A (100% water/0.1 % FA) over a period of 60 minutes at 37 °C and a flow rate of 300 nl/min. A total of 2 pmol of digested peptides was loaded on the column. Peptides eluting from the column were introduced into the nanoelectrospray source with a capillary voltage of 2.5 kV. For MS analysis, a full scan was recorded for eluted peptides (m/z range of 360–1600) in the FT mass analyzer at resolution R of 120,000, followed by MS2 of the 20 most intense peptide ion scans in the ion trap (IT) mass analyzer. All MS data were acquired in the positive ion mode. Detected ion currents for peptides in MS1 experiments were used to determine the extent of oxidation for each modified site by separate quantification of the unmodified proteolytic peptides and their radiolytic products. The resulting MS2 data were initially searched against a PP2A AC protein database using Mass Matrix software to identify all specific sites of modification (43). In particular, MS2 spectra were searched for nonspecific peptides of PP2A using mass accuracy values of 8 ppm and 0.7 daltons for MS1 and MS2 scans, respectively, with the allowed variable modifications, including carbamidomethylation for cysteines and all known oxidative modifications for amino acid side chains (44, 45). In addition, all detected MS2 spectra for each site of modification were manually verified.
Modification rate calculation.
The integrated peak areas of the unmodified peptide (Au), and of a peptide in which a residue is modified (Am) derived from selected ion chromatograms, were used to calculate the fraction unmodified (Fu) for each specific modified species according to the following formula: Fu = 1 – (Am/(Au + SAm)), where ΣAm is the sum of all modified products for a particular peptide. Dose-response curves were generated using unmodified fractions for each specific site of modification plotted versus x-ray exposure time. The fraction unmodified for each site of modification was fit to the following first-order equation: Fu = (Fu)0e-kt, where Fu and (Fu)0 are the fractions of unmodified values at each site of modification at time t and time 0 (seconds), respectively, and k is a first order rate constant as previously described (46, 47). Dose-response curves were presented as unmodified fractions for each specific site of modification plotted versus x-ray exposure time. The modification rates were obtained from the slope of the dose-response curves. The protection factors (PFs) were calculated by dividing the modification rates for each modified site derived from the drug-free PP2A AC protein by the modification rates for the same sites of modification derived from the PP2A AC protein bound to the various SMAP compounds. The peptide segments and the amino acid side chains in each segment for which PFs were determined are provided as supplementary information (Supplemental Table 4).
Statistics.
Enhanced chemiluminescent images of immunoblots were analyzed by scanning densitometry and quantified with ImageJ (NIH) software. All values were normalized to GAPDH expression and expressed as fold change relative to control. Analysis was performed using GraphPad Prism. Statistical significance was assumed for a 2-tailed P value of less than 0.05 using Student’s t test or ANOVA with Tukey’s post-hoc test (presented as means; error bars indicate SD). Except where otherwise noted, box boundaries of all box-and-whisker plots represent the range of values obtained in the experiment and whiskers represent mean ± SD.
Study approval.
All animal experiments were approved by the IACUCs at Icahn School of Medicine at Mount Sinai and Case Western Reserve University. Animal use and care were in strict compliance with institutional guidelines, and all experiments conformed to the relevant regulatory standards established by Icahn School of Medicine at Mount Sinai and Case Western Reserve University.
Author contributions
JS, MO, and GN designed the research studies. DBK, NZ, and MO created the compounds. JS, AP, RT, JK, SI, SM, DDW, CMO, DH, NSD, DS, SY, DL, ACB, GG, LW, ES, CCF, EY, RAA, AS, BS, VG, HMG, DM, and KM conducted experiments and interpreted the data. ZW, ACL, RCS, EYC, QD, MD, SH, AM, AD, NS, MDG, DLB, YAI, WX, MRC, MO, and GN provided observations and scientific interpretations. All authors discussed the results and provided input on the manuscript.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the contributions of Amy and George Trainor and David U’Prichard for insight and critical evaluation of the work. We also thank Suresh Y. Ramanathan, Christopher I. Shaari, and Ariana Shaari for assistance with manuscript. We would like to acknowledge the Young Scientist Foundation (www.ysf.org). We thank Dr. Sayan Gupta for assistance with sample irradiation at the Advanced Light Source (ALS) of Lawrence Berkeley National Laboratory (Berkeley, CA) which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We also thank Dr. Jennifer Bohon for assistance with sample irradiation at the National Synchrotron Light Source (NSLS) of Brookhaven National Laboratory (Upton, NY) which is supported by NIH grant P30EB009998 from the National Health Institute and by the National Science Foundation grant, DBI-1228549. G. Narla and M.D. Galsky would like to acknowledge NIH/NCI R01CA181654. G. Narla is supported by the Pardee-Gerstacker Professorship in Cancer Research. A. DiFeo is supported by the The Norma C. and Albert I. Geller Designated Professor in Ovarian Cancer Research.
Footnotes
Conflict of interest: The Icahn School of Medicine at Mount Sinai, on behalf of G. Narla, M. Ohlmeyer, N.S. Dhawan, and D.B. Kastrinsky, has filed patents covering composition of matter on the small molecules disclosed herein for the treatment of human cancer and other diseases (International Application Numbers: PCT/US15/19770, PCT/US15/19764; and US Patent: US 9,540,358 B2). Dual Therapeutics LLC has licensed this intellectual property for the clinical and commercial development of this series of small molecule PP2A activators. G. Narla, M. Ohlmeyer, Y.A. Ioannou, M.D. Galsky, N.S. Dhawan, and D.B. Kastrinsky have an ownership interest in Dual Therapeutics LLC. G. Narla and M. Ohlmeyer are consultants for Dual Therapeutics LLC.
Reference information:J Clin Invest. 2017;127(6):2081–2090.https://doi.org/10.1172/JCI89548.
See the related Commentary at A SMAP in the face for cancer.
Contributor Information
Jaya Sangodkar, Email: jaya.sangodkar@mssm.edu.
Abbey Perl, Email: alp80@case.edu.
Rita Tohme, Email: rita.g.tohme@gmail.com.
Janna Kiselar, Email: jxk217@case.edu.
David B. Kastrinsky, Email: david.kastrinsky@mssm.edu.
Nilesh Zaware, Email: nilesh.zaware@mssm.edu.
Sudeh Izadmehr, Email: Sudeh.Izadmehr@mssm.edu.
Sahar Mazhar, Email: saharmazhar@hotmail.com.
Divya Hoon, Email: divya.hoon@gmail.com.
Neil S. Dhawan, Email: neil.dhawan@mssm.edu.
Daniela Schlatzer, Email: dms73@case.edu.
Shen Yao, Email: Shen.yao@mssm.edu.
Daniel Leonard, Email: dleonard03@mac.com.
Giridharan Gokulrangan, Email: Giridharan.Gokulrangan@bms.com.
Lifu Wang, Email: lifu1026@gmail.com.
Elena Svenson, Email: ecs83@case.edu.
Eric Yuan, Email: eric.yuan@me.com.
Rita A. Avelar, Email: raa103@case.edu.
Agnes Stachnik, Email: agnes.stachnik@gmail.com.
Blake Smith, Email: e.smith.blake@gmail.com.
Vickram Gidwani, Email: vickgidwani@gmail.com.
Daniel McQuaid, Email: danielcmcquaid@gmail.com.
Kimberly McClinch, Email: kmcclinch@gmail.com.
Zhizhi Wang, Email: wxu@u.washington.edu.
Alice C. Levine, Email: alice.levine@mountsinai.org.
Rosalie C. Sears, Email: searsr@ohsu.edu.
Qiaonan Duan, Email: geonann@gmail.com.
Manish Datt, Email: manishdatt@gmail.com.
Shozeb Haider, Email: shozeb.Haider@ucl.ac.uk.
Analisa DiFeo, Email: avd11@case.edu.
Neelesh Sharma, Email: neelesh.sharma@novartis.com.
Wenqing Xu, Email: zzwang@u.washington.edu.
Michael Ohlmeyer, Email: michael.ohlmeyer@mssm.edu.
Goutham Narla, Email: gxn71@case.edu.
References
- 1.Gross S, Rahal R, Stransky N, Lengauer C, Hoeflich KP. Targeting cancer with kinase inhibitors. J Clin Invest. 2015;125(5):1780–1789. doi: 10.1172/JCI76094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9(1):28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139(3):468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Perrotti D, Neviani P. Protein phosphatase 2A: a target for anticancer therapy. Lancet Oncol. 2013;14(6):e229–e238. doi: 10.1016/S1470-2045(12)70558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sangodkar J, Farrington CC, McClinch K, Galsky MD, Kastrinsky DB, Narla G. All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. FEBS J. 2016;283(6):1004–1024. doi: 10.1111/febs.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353(Pt 3):417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445(7123):53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 8.Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795(1):1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Khew-Goodall Y, Hemmings BA. Tissue-specific expression of mRNAs encoding alpha- and beta-catalytic subunits of protein phosphatase 2A. FEBS Lett. 1988;238(2):265–268. doi: 10.1016/0014-5793(88)80493-9. [DOI] [PubMed] [Google Scholar]
- 10.Stone SR, Hofsteenge J, Hemmings BA. Molecular cloning of cDNAs encoding two isoforms of the catalytic subunit of protein phosphatase 2A. Biochemistry. 1987;26(23):7215–7220. doi: 10.1021/bi00397a003. [DOI] [PubMed] [Google Scholar]
- 11.Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, Chiang CW. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283(4):1882–1892. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- 12.Letourneux C, Rocher G, Porteu F. B56-containing PP2A dephosphorylate ERK and their activity is controlled by the early gene IEX-1 and ERK. EMBO J. 2006;25(4):727–738. doi: 10.1038/sj.emboj.7600980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang SS, et al. Alterations of the PPP2R1B gene in human lung and colon cancer. Science. 1998;282(5387):284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 14.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junttila MR, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130(1):51–62. doi: 10.1016/j.cell.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 16.Kastrinsky DB, et al. Reengineered tricyclic anti-cancer agents. Bioorg Med Chem. 2015;23(19):6528–6534. doi: 10.1016/j.bmc.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez A, et al. Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J Clin Invest. 2014;124(2):644–655. doi: 10.1172/JCI65093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH, et al. Haloperidol regulates the phosphorylation level of the MEK-ERK-p90RSK signal pathway via protein phosphatase 2A in the rat frontal cortex. Int J Neuropsychopharmacol. 2008;11(4):509–517. doi: 10.1017/S1461145707008292. [DOI] [PubMed] [Google Scholar]
- 19.Murren JR, et al. Trifluoperazine as a modulator of multidrug resistance in refractory breast cancer. Cancer Chemother Pharmacol. 1996;38(1):65–70. doi: 10.1007/s002800050449. [DOI] [PubMed] [Google Scholar]
- 20.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410(6832):1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, et al. Structural and biochemical insights into the regulation of protein phosphatase 2A by small t antigen of SV40. Nat Struct Mol Biol. 2007;14(6):527–534. doi: 10.1038/nsmb1254. [DOI] [PubMed] [Google Scholar]
- 22.Cho US, Morrone S, Sablina AA, Arroyo JD, Hahn WC, Xu W. Structural basis of PP2A inhibition by small t antigen. PLoS Biol. 2007;5(8):e202. doi: 10.1371/journal.pbio.0050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sablina AA, Hahn WC. SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev. 2008;27(2):137–146. doi: 10.1007/s10555-008-9116-0. [DOI] [PubMed] [Google Scholar]
- 24.Kiselar JG, Chance MR. Future directions of structural mass spectrometry using hydroxyl radical footprinting. J Mass Spectrom. 2010;45(12):1373–1382. doi: 10.1002/jms.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Chance MR. Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting. Anal Chem. 2011;83(19):7234–7241. doi: 10.1021/ac200567u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 28.Kim JY, et al. Activity-based proteomics reveals heterogeneous kinome and ATP-binding proteome responses to MEK inhibition in KRAS mutant lung cancer. Proteomes. 2016;4(2):16. doi: 10.3390/proteomes4020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marusiak AA, et al. Mixed lineage kinases activate MEK independently of RAF to mediate resistance to RAF inhibitors. Nat Commun. 2014;5:3901. doi: 10.1038/ncomms4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCubrey JA, et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascade inhibitors: how mutations can result in therapy resistance and how to overcome resistance. Oncotarget. 2012;3(10):1068–1111. doi: 10.18632/oncotarget.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng J, et al. Combination treatment with MEK and AKT inhibitors is more effective than each drug alone in human non-small cell lung cancer in vitro and in vivo. PLoS One. 2010;5(11):e14124. doi: 10.1371/journal.pone.0014124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu T, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res. 2012;18(8):2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 33.Pallas DC, et al. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60(1):167–176. doi: 10.1016/0092-8674(90)90726-U. [DOI] [PubMed] [Google Scholar]
- 34.Janghorban M, et al. Targeting c-MYC by antagonizing PP2A inhibitors in breast cancer. Proc Natl Acad Sci U S A. 2014;111(25):9157–9162. doi: 10.1073/pnas.1317630111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujiki H, Suganuma M. Tumor promotion by inhibitors of protein phosphatases 1 and 2A: the okadaic acid class of compounds. Adv Cancer Res. 1993;61:143–194. doi: 10.1016/s0065-230x(08)60958-6. [DOI] [PubMed] [Google Scholar]
- 36.Côme C, et al. CIP2A is associated with human breast cancer aggressivity. Clin Cancer Res. 2009;15(16):5092–5100. doi: 10.1158/1078-0432.CCR-08-3283. [DOI] [PubMed] [Google Scholar]
- 37.Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saddoughi SA, et al. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol Med. 2013;5(1):105–121. doi: 10.1002/emmm.201201283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sangodkar J, et al. Targeting the FOXO1/KLF6 axis regulates EGFR signaling and treatment response. J Clin Invest. 2012;122(7):2637–2651. doi: 10.1172/JCI62058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, et al. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127(6):1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 41.Gupta S, Sullivan M, Toomey J, Kiselar J, Chance MR. The beamline X28C of the Center for Synchrotron Biosciences: a national resource for biomolecular structure and dynamics experiments using synchrotron footprinting. J Synchrotron Radiat. 2007;14(Pt 3):233–243. doi: 10.1107/S0909049507013118. [DOI] [PubMed] [Google Scholar]
- 42.Xu G, Kiselar J, He Q, Chance MR. Secondary reactions and strategies to improve quantitative protein footprinting. Anal Chem. 2005;77(10):3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- 43.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107(8):3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 45.Xu G, Chance MR. Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal Chem. 2005;77(14):4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 46.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu Rev Biophys Biomol Struct. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 47.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. Int J Radiat Biol. 2002;78(2):101–114. doi: 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.