Abstract
Objective
A memory clinic at an academic medical center has relied on several ad hoc data capture systems including Microsoft Access and Excel for cognitive assessments over the last several years. However these solutions are challenging to maintain and limit the potential of hypothesis-driven or longitudinal research. REDCap, a secure web application based on php and MySQL, is a practical solution for improving data capture and organization. Here, we present a workflow and toolset to facilitate legacy data migration and real-time clinical research data collection into REDCap as well as challenges encountered.
Materials and Methods
Legacy data consisted of neuropsychological tests stored in over 4,000 Excel workbooks. Functions for data extraction, norm scoring, converting to REDCap-compatible formats, accessing the REDCap API, and clinical report generation were developed and executed in Python.
Results
Over 400 unique data points for each workbook were migrated and integrated into our REDCap database. Moving forward, our REDCap-based system replaces the Excel-based data collection method as well as eases the integration to the Electronic Health Record.
Conclusion
In the age of growing data, efficient organization and storage of clinical and research data is critical for advancing research and providing efficient patient care. We believe that the tools and workflow described in this work to promote legacy data integration as well as real time data collection into REDCap ultimately facilitate these goals.
Keywords: Electronic Health Records, Biomedical Informatics, ETL, REDCap, Clinical Informatics, Clinical Research Informatics
1. INTRODUCTION
1.1 Background and Significance
Properly integrating clinical research data from a variety of systems can be a daunting challenge, especially given the specific demands of an academic medical environment. Data collection instruments are often developed to fulfill specific needs, often with little thought placed on future changes or interoperability with other instruments or central management. In our clinical research environment, this has lead to a number of stopgap solutions ranging from old and unmaintainable Microsoft Access databases and Visual Basic applications, paper forms, and Microsoft Excel spreadsheets. The relative inaccessibility to and lack of communication between these data sources presents challenges not only from a clinical research perspective, but also in providing optimal clinical care.
While numerous studies agree that improvements in electronic data capture (EDC) systems can lead to improved efficiency [3,4], accuracy [5,6], cost savings [7], and ultimately improvements in health care delivery, the implementation of these systems is often challenged by common barriers such as user motivation, regulatory requirements, economic and personnel resources, and graphical user interface interaction [8,9]. Various measures can be taken to lower these barriers to transition. For example, adopting online EDCs based on a Software as a Service (SaaS) models or offering no-cost licensing arrangements can save significantly heavy investment in information technology, form editing functions allow systems to stay practical and integrated in a rapidly changing clinical research environment, and precise quantification of costs and benefits can inspire institutions to finally adopt EDCs [10,11].
There exists a wide range of both proprietary and open source solutions in the EDC domain to support organized, flexible data capture and analysis in healthcare or research settings (see reviews [6,12]). Oracle Clinical is an example of a commercial integrated clinical management system that allows remote data capture [13]. While its benefits include enforcing standards and consistency through internal global libraries and allowing a flexible study design that can take into account even the most complex trial scenario, the system is limited by common constraints observed in commercial EDCs, namely prohibitive costs for implementation and management [14]. An example of an open source EDC for management of clinical research studies is OpenClinica [15], which has been approved for every stage of clinical research and has a global community spanning over 100 countries. Some key features are the easily-designed flexible studies through an intuitive GUI (Graphical User Interface), automated visit scheduling, direct access to data, and the ability to integrate with other systems such as EHR and PACS. Seeking to better organize and store our clinical research data, we have adopted REDCap (Research Electronic Data Capture), another open source, web-based EDC backed by a MySQL database engine for data storage and manipulation [16]. REDCap includes a wide range of special features that have accelerated its initial development in 2004 to a multi-national consortium today comprised of users from over 1,650 institutions [17]. Key features include collaboration access across institutions, role-based security restrictions, quality assurance mechanisms, centralized data storage and backup, and data export options for common statistics packages [16,18]. The REDCap user interface allows less-technical users to access and view the project data. In addition, the integrated GUI form designer greatly simplifies form design for clinicians and staff as it does not necessitate a software developer or database administrator. Advanced features and plugins contributed internally and from the external community continue to add to the appeal of REDCap. Available plugins include capabilities to visualize geographical data and generate advanced plots [19], “hooks” to insert custom code and features within standard REDCap environment [20], and pairing with Open Data Kit (ODK, opendatakit.org) software to allow data collection and upload to REDCap directly from mobile devices [21].
Apart from the features described above, our choice of REDCap for this project was driven by two practical considerations. Our umbrella institution has an enterprise REDCap installation that is maintained and supported by the information technology group for a modest yearly fee (< $250 a year/project). Given the typical complexities of installation, backup and maintenance of a server that houses protected health information, this nominal fee obviates many of the administrative challenges we would have faced using a separate database system. In addition, REDCap provides a robust API that allows the import, export, and modification of data that can significantly aid in automating the data migration process. While not an absolute dependency, the existence of a python based wrapper (pyCAP) that simplified interaction with the REST-based API was also a key advantage.
1.2 Objective
In this work, we describe the workflow, challenges, and tools we have developed (REDLetr, or REDCap Load, Extract, Translate, Revise) to facilitate the migration of legacy clinical data and real-time data collection into a REDCap environment with added features to facilitate integration in standard clinical workflow. This will increase the ability to take advantage of previous data as well as to facilitate future research. The case study we use to describe this process is the migration of neuropsychological testing data from the NIH-funded Alzheimer’s Disease Research Center (ADRC) and affiliated memory clinics at Emory University. In addition, as the applicability of tools to transfer valuable legacy clinical data to centrally organized and flexible research databases is widespread, we have provided a resource containing code and explanations on an accompanying site (https://github.com/dgutman/RedLETR) and believe that it will be of use to other organizations seeking to standardize data capture and improve efficiency.
2. MATERIALS AND METHODS
2.1 Data Description
Legacy data consisted of approximately 4,000 Excel spreadsheets corresponding to patients seen in the ADRC memory clinics at Emory University between 2011 and 2014. This research-oriented clinic is staffed by approximately 15 nurses and physicians in addition to social workers, researchers, and other support staff and sees approximately 1500 patients per year through primary patient encounters, follow up visits, and research studies. As part of each standard visit, patients are administered a comprehensive neuropsychological battery containing tests that measure a wide variety of functions (Supplemental Table 1). These data are later stored in the institutional electronic medical record system (EMR, Cerner Millennium, www.cerner.com). Each spreadsheet consists of at least nine individual forms/tests evaluating the patient’s cognitive function across a number of domains and a summary sheet containing the overall raw scores as well as corresponding normative scores adjusted for a patient’s age, race, education, and/or gender.
2.2 Software
Functions for extracting data, converting to formats compatible with REDCap integration, and normative scoring were developed in Python (Anaconda Python distribution [22], Python version 2.72). The main prerequisite for this project was a working REDCap server with the ability to upload data using the API. The PyCAP toolkit was used to communicate between Python and the REDCap API [23]. We have included a set of package dependencies for the application itself in the requirements.txt file on our GitHub page, along with instructions on how to setup a Python “virtualenv” for this work. Due to the iterative nature of this process, we found iPython Notebook [24], which provides a web-based GUI for Python development, especially useful. With the exception of the Excel password removal using a 64-bit Windows 7 machine due to its use of a Windows DLL, all of our code was executed on a Linux system running Ubuntu 12.04 and 14.04 64-bit as well as MAC OS-X 10.1.
2.3 Data Migration of legacy neurology data to REDCap
Our general workflow for migrating legacy data to the REDCap environment is similar to a standard Extract-Translate-Load (ETL)[25] process, and we describe our procedure in this framework. Because this data migration to an improved system was part of quality improvement and regular operations, Institutional Review Board (IRB) approval was not required for this project.
Pre-Extraction
In order to leverage existing Python tools, we first used the Microsoft Office Migration Manager [26] to convert any XLS (a legacy Excel file format) files to XLSX (the current Excel format; see Code Snippet 1 in the supplemental section). In addition, to allow our tools access to password-protected sheets, we used the win32com.client Python library to unlock/unprotect and re-save each XLSX file (see Code Snippet 2 in the supplemental section).
Extraction: Extracting Data from Excel Worksheets
We leveraged the openpyxl Python library [27] to extract content from individual cells by location from each XLSX spreadsheet. In most cases, such as the score on the “Trails” exercise from the MOCA test (see code Snippet 3, supplemental section), there was a simple one-to-one mapping between variables of interest and cells, which we encoded within its own embedded dictionary. In several tests, such as the FAS or CERAD Animal Vegetable Fluency, where participants are asked to spontaneously generate as many words as possible within a specified time frame, one variable was associated with several cells in what we internally labeled a “complex map”. Here, a one-to-many mapping, where information from multiple cells is binned through concatenation into variables representing a time interval, was more appropriate (see Code Snippet 4, supplemental section). Our pipeline could handle this case separately by enumerating the list of individual cells that comprised a single variable, and we simply concatenated the cells into a single comma-delimited string.
We also encountered a number of cases where different versions of the same test/form existed with identical names. These differences were often driven by a minor update in the instruction section that inadvertently shifted cell mappings by several rows, making them invalid. We resolved this situation by first matching specific text at specific cell locations to identify the appropriate cell-mapping schema for data extraction for each record.
Translate: Building a REDCap Data Dictionary and project structure
The resulting mapping produced over 400 unique variables to be organized in a REDCap data dictionary that would become the backbone for the project. The data dictionary contained the REDCap variable names corresponding to each variable identified from the Excel sheets above organized by test/form name. This was an iterative process where individual testing instruments were created as separate instruments within REDCap, and variable names were either adapted from pre-existing REDCap versions (if available locally and/or from the REDCap Shared Instrument library [17]) or generated de novo.
During the REDCap data dictionary development process, we developed several data quality tools though Python to check for the fidelity between the variables in the Excel sheets and in the growing REDCap dictionary. For example, we applied constraints to individual variables by limiting input to an appropriate data type (integers, True/False, Yes/No, etc.).
Loading Data into REDCap
Following development of the data dictionary, we created a function to loop through all available XLSX files and then through each corresponding worksheet. For each patient, specific values for each of the 400 locations in the standardized data dictionary were collected based on cell-location mapping. Extracted data were individually loaded into our REDCap project and organized into the appropriate REDCap forms and variables for each of the 4,000 patients. Of note, we did not load the results from the summary sheet of the Excel document due to concerns of the accuracy of the norm scoring criteria of the Excel-based implementation. We later compute these scores for each patient and repopulate them on the remaining REDCap Summary Sheet form (see Instrument Scoring section below).
2.4 Graphical User Interface
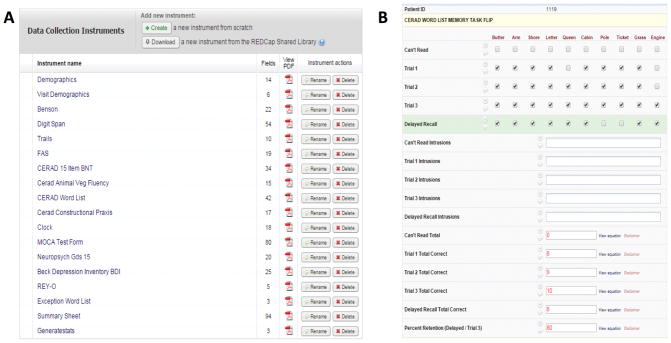
Next, on the GUI side of REDCap, we used the Edit Instrument and Graphical Form Builder tools to modify the basic structure created above. For example, stylistic section headers or pictures for certain tests were added to assist test administration. In addition, calculated fields were implemented where possible to save test administrator’s time and avoid manual errors (Figure 1).
FIGURE 1.
REDCap graphical user interface corresponding to tests administered from the Excel version. Panel A displays all tests included in the battery and Panel B displays the GUI where test administrators enter responses for CERAD Word List Memory task. Red values are those automatically calculated in real time by REDCap.
This design stage allowed healthcare administrators to review the current testing battery implementation and offer suggestions of what questions or tests should be modified in the new version.
2.5 Instrument Scoring
A key functionality required after raw scores had been migrated to REDCap was the need to perform scoring operations. For a given test/instrument, a patient’s raw score had been transformed into standardized scores (such as percentiles and Z-scores) based on the patient’s age, gender, education and/or race. This allowed the clinician to interpret the patient’s performance relative to population norms. Scoring algorithms previously implemented in Excel used a set of functions making references to several hundred cells scattered across various worksheets and were too complicated to be reproduced using REDCap syntax (see code Snippet 5, supplemental section).
To facilitate scoring in our new platform, we loaded norm-scoring tables consisting of means and standard deviations for patient performance on a given test based on their demographics, to a PostgreSQL database. A set of basic Python functions uses the patient’s raw score and demographic information as input to calculate the necessary normative statistics for each test. The resulting raw scores and normalized scores are then sent using the API to the remaining Summary Sheet form in the REDCap Project.
2.6 Clinical Documentation
A requirement for our project was the generation of a single page summary sheet that summarized a visit and contained the raw and normalized scores for each test. This was an integral part of the previous Excel-based clinical workflow and was used by the health professionals to summarize the current cognitive state of the patient directly after the test, as well as to monitor progress during follow up visits. This report serves as the primary documentation for the testing, and is directly uploaded to our Electronic Medical Record (EMR) system by the clinical team.
To develop similar capabilities using the REDCap interface, we first generated a template in MS Word that reflected the Excel-based summary sheet used in the previous workflow. We placed temporary variable names in each area where patient specific demographics, raw test scores, and normalized test scores values would be eventually inserted. Then, by calling the Python library python-docx [28], a function would search for each variable in the template, replace it with the corresponding value from the patient-specific demographics and summary sheet in REDCap, and generate a new Word document that was then uploaded back to REDCap.
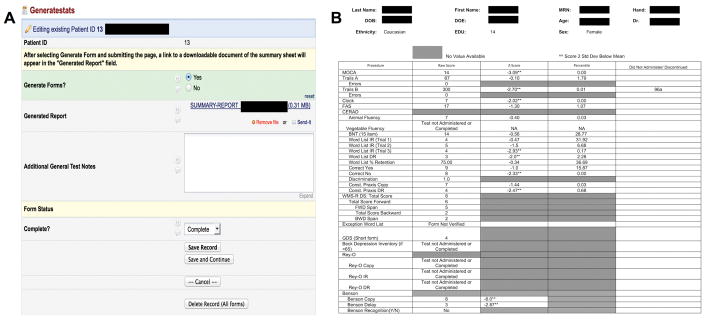
Generation of the report itself was supported by the “data trigger” functionality available through REDCap. By navigating to a “Generate Stats” form and submitting (Figure 2A), the test administrator sends an HTTP POST message to a separate application (either on the same server or remote) that responds to generate and upload the resulting Word document to the REDCap interface. This sheet could then be downloaded, printed, edited, and uploaded to the EMR.
FIGURE 2.
User interface to generate testing report at end of testing battery administration (A). Test administrators select “Generate Report”, submit the page, and reload. Upon reloading, a downloadable Microsoft Word version of the testing summary sheet is available as a link. Panel B displays an example of a testing report displayed after neuropsychological battery administration. The first column indicates name of test, second column provides raw scores, and third and fourth columns provide statistic scores calculated according to the procedure described in the Instrument Scoring section. The fifth column shows selected notes section for each test. Patient sensitive information has been blocked out in black.
2.7 Evaluating User Satisfaction
In order to evaluate our new REDCap-based research database in a quantitative aspect, we asked each neuropsychological test administrator to respond to a user satisfaction survey. The survey used in this case was the Computer System Usability Questionnaire (CSUQ) which was developed by IBM and validated for reliability and validity/sensitivity as well as used in other projects for points of care technology in healthcare settings [29,30]. This questionnaire contains 19 questions, each with seven possible responses ranging from Strongly Disagree (1) to Strongly Agree (7) broken up into sub scores that measure overall system usage, quality of documentation, and interface quality.
3. RESULTS
3.1 Migration of legacy data
The REDCap project created from the data dictionary generated in the extraction/translate steps and from REDCap’s instrument editing tools faithfully reflects the neuropsychology testing battery from the original Excel workbook as well as provides added functionality. 400 unique data elements from 4,000 patients were migrated into the updated database system.
3.2 Development of integrated REDCap-based research database for future data collection
In addition, in late January 2015 our department successfully transitioned to our REDCap-based data management system and we are currently planning similar implementations in other departments. Inherent features in REDCap to automatically calculate fields in real time along with our custom implementation to generate a summary report with normed scores recapitulating a visit (Figure 2B) are some of the new program’s most distinctive features. Currently, the summary sheet is simply uploaded into the EMR as a Word document. We have also developed a feature where the patient’s summary sheet information coded into a QR code can be generated by clicking on a link in REDCap. A clinician can then simply use a 2D barcode reader to read the summary sheet directly into a text field of the patient’s EMR.
3.3 User evaluation of new REDCap-based testing battery administration
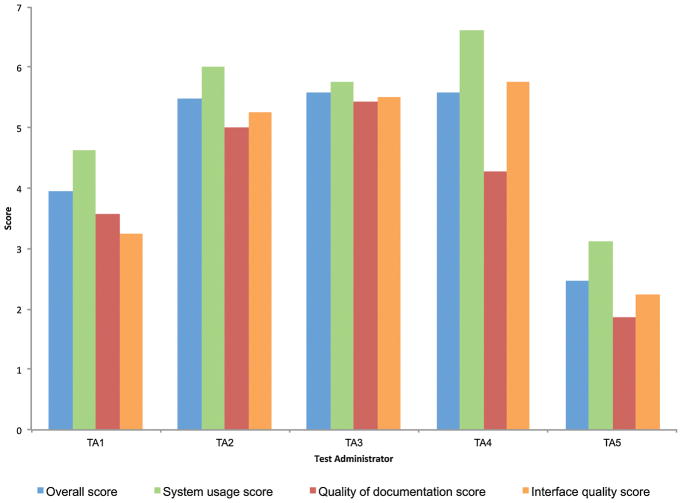
In terms of user experience, the average across test administrators was 4.61 for overall score, 5.23 for system usage score, 4.03 for documentation quality score, and 4.20 for interface quality score. Figure 3 shows overall and subscores for each individual rater. Most of the users reported medium to high scores in all four areas. It is possible that the variability in scores results from an initial learning curve that is quicker for some raters than others. In addition, some users noted that inherent features in the REDCap interface, compared to our other version, made it more difficult to quickly find old/recent patients or avoid accidently clicking form reset buttons. Compared to using the previous Excel-based system, most users found the main advantages also stemmed from inherent features of REDCap’s intuitive GUI that makes it easy to edit data and create reports displaying subsets of specific data of interest.
FIGURE 3.
Summary of Computer System Usability Questionnaire (CSUQ) received from each of the five test administrators in our neurology clinic. Responses for each of the 19 questions in this questionnaire range from 1 (strongly disagree) to 7 (strongly agree). Overall scores average responses from all 19 questions for each administrator. System usage (Q1–Q8), Quality of documentation (Q9–Q15), and Interface quality (Q16–Q19) scores are subscores derived from averages of indicated questions.
4. DISCUSSION
Using a case study in our ADRC’s memory clinics, we present a workflow for the data migration of valuable legacy data stored in various formats such as Excel or .csv files to the REDCap environment for research data management. In addition, we describe the development and implementation of a redesigned interface with added features to improve its usability and seamless integration to the clinical workflow.
Specific novel features that we have added to the generic REDCap functionality are quality-checked conversion from data stored in Excel sheets to REDCap-formatted data-dictionaries, calculation of normed score from raw scores, clinical documentation report generation, and barcode generation for facilitating report upload to EMR. As informal discussions and literature suggests, we believe that other institutions are facing similar challenges, wishing to upgrade from spreadsheet-based collection instruments. The tools and workflow developed in this project may benefit those unable to transition due to typical barriers in the process [6] or fear of losing potentially valuable legacy data [31].
4.1 Benefits
A driving motivation behind this work was to create a comprehensive, flexible data management system in our neurology clinic integrated with the clinical workflow, but also to be able to take advantage of the availability and utility of legacy data sources for both clinic and research purposes. Indeed, access to this legacy information is particularly important given the nature of our clinic, which focuses on neurodegenerative diseases. While the “raw” Excel files continue to be available if needed, realistically this information would have been rarely, if ever, accessed once we transitioned our testing to REDCap.
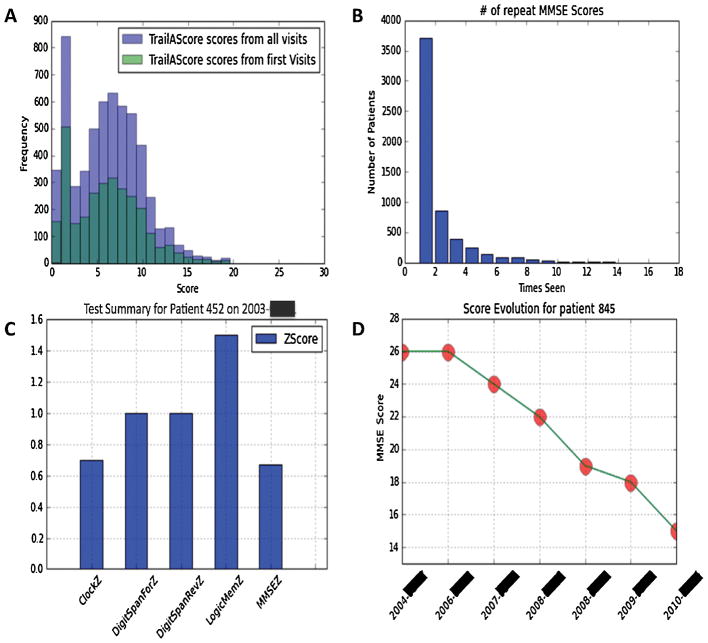
On the clinical side, we are beginning to add these data to some of the de novo reports we are generating for new tests, which the clinicians have found helpful. Apart from facilitating this migration of legacy data, our tools developed can leverage advanced Python libraries through PyCap to plot graphics that aid doctors in measuring population level statistics or patient level progress (Figure 4). This of course would not have been realistically feasible had staff needed to search, combine, and manually integrate data for data housed in disparate locations.
FIGURE 4.
Example of visualizations produced through a combination of PyCap and Python plotting functions. Notice data can be visualized at a population level (A,B) as well as at a patient level (C,D) for visualizing patient evolution over follow up visits. Patient IDs have been randomly generated and exact dates blocked out to maintain confidentiality.
On the research side, our implementation can aid researchers in longitudinal or hypothesis-driven studies. For example, over the past fifteen months since we have rolled out this system, we have captured over 1700 patient encounters. Besides the advantage of having an improved process to capture and centrally-store the data, inherent features in REDCap allow us to leverage this data for research purposes. For example, a parallel project involves using data stored in these visit records and accessed through REDCap data export functionalities to identify initial cognitive variables or demographic data that are associated with marked cognitive decline.
4.2 Limitations
As discussed above, REDCap was chosen because of its features that make it an increasingly popular data collection system all around the world. However, REDCap has some inherent limitations that could be addressed to make it a more ideal system. For example, REDCap will accept data out of range with just a warning message (for example, character values in a numeric section), which could cause errors in downstream statistical analyses. In addition, it can be difficult to edit the forms once in production mode - this requires coordination between administrators and the IT department and once approved, could modify data collected in other areas of the battery. Finally, the most significant disadvantage of REDCap is its “flat” data model, which makes it more difficult to join between projects or be able to store or mine data as can be performed in typical relational database models. This also poses some difficulty for our case study where patients can have any number of follow-up visits.
Besides limitations due to REDCap itself, we noted some limitations in our workflow and tools developed for migrating legacy data to a REDCap environment. For example, we found the data curation and clean up involved to be particularly time consuming and challenging due to the domain expertise required to understand tests/versions in designing our data dictionary, implementing appropriate quality checks for data, and learning meanings for internally-used codes (i.e., 997: Did not administer due to patient behavior). This is perhaps an unavoidable aspect for any data migration project, and although we have attempted to streamline this process as much as possible with our available tools and built-in checks, a successful implementation typically requires significant back and forth exchange between the developers and end users. Finally, our project requires an understanding of Python and add-on packages, especially for implementing functionalities such as norm scoring or report generation, which may not be intuitive for people without strong programming backgrounds. While we have attempted to provide sufficient documentation, this coding barrier may still pose problems if norms are updated or tests in the battery are changed.
4.3 Future directions
Due to the successful implementation of our workflow in our Memory Clinic, we have begun planning similar implementations in other departments with clinical trials based on Excel sheets or other stopgap solutions faced with similar problems we initially faced. Future implementations will improve the standardization of our workflow and improve quality checking. In addition, while our barcode method serves as a convenient tool to tie our system back to the EMR, future implementations will take advantage of tools and capabilities that allow secure direct integration results with the EMR or i2b2 translational research platforms [32,33].
Finally, the use of defined data elements has been another important output to this project. Our neurology department has been in the process of generating a federated Brain Health DataMart, which attempts to integrate clinical and research data from a variety of sources including the Clinical Data Warehouse, the LIMS system, as well as REDCap. The process of data-mapping to other local sources of information has been significantly simplified by the use of these elements in our REDCap project, allowing the integration team to do the bulk of the work without much direct input from our team. Compared to other research and clinical groups participating in the data mart, our integration process was much simpler. Since our naming conventions were common between our different areas under the same overall institution, semantic interoperability between heterogeneous databases was not a challenge in this initial case study. However, as our workflow grows internally and externally, future iterations could facilitate semantic interoperability across projects through the development of standards defined by archetypes according to, for example, the openEHR archetype methodology [1,2].
5. CONCLUSION
The numerous data points that clinical centers or hospitals collect across time though standard practices offer tremendous opportunities for hypothesis-driven research to further understand disease and uncover trends not otherwise noticed. As computational processing and storage becomes less expensive and data more easily available, it is critical that clinical research data management systems be efficiently integrated and structured to accelerate this type of large data research. The transfer of legacy data from ad hoc data collection tools to a flexible, web-based database system is a challenging, but well rewarded process and we hope that our work will provide valuable insight into a seamless transition for similar institutions.
Supplementary Material
8. SUMMARY TABLE.
What was already known
Many institutions are transferring to advanced research databases such as REDCap to save costs, become more efficient, and advance discoveries in research and healthcare.
Legacy data collected from ad hoc spreadsheet-based research studies contain valuable data that could be better leveraged if integrated in a centralized database.
Advanced databases alone such as REDCap typically do not sufficiently meet clinical research demands.
What this study added
A set of open-sourced tools to facilitate migrating spreadsheet-based legacy data to the REDCap research database environment
A set of open-sourced tools that increase the basic functionality of REDCap, such as norm scoring and report generation features that help integrate REDCap into standard operating procedures of healthcare centers
Acknowledgments
We would like to especially thank all of the personnel at the Emory University Neurology Clinic and ADRC for their input and assistance during the transition process described in this manuscript. Funding was provided through NIH Grants P30 NS055077 and P50 AG025688.
Footnotes
5. AUTHORS CONTRIBUTIONS
All authors WD, JC, AL, and DG have made substantial contributions to all of the following: (1) the conception and design of the study and acquisition of data, (2) drafting the article or revising it critically for important intellectual content, (3) and final approval of the version to be submitted.
7. STATEMENT ON CONFLICTS OF INTEREST
Conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen R, Klein GO, Sundvall E, Karlsson D, Ahlfeldt H. Archetype-based conversion of EHR content models: pilot experience with a regional EHR system. BMC Med Inform Decis Mak. 2009;9:33. doi: 10.1186/1472-6947-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalra D, Beale T, Heard S. The openEHR Foundation. Stud Health Technol Inform. 2005;115:153–73. [PubMed] [Google Scholar]
- 3.Helms RW. Data Quality Issues in Electronic Data Capture. Drug Inf J. 2001;35:827–37. [Google Scholar]
- 4.Litchfield J, Freeman J, Schou H, Elsley M, Fuller R, Chubb B. Is the future for clinical trials internet-based? A cluster randomized clinical trial. Clin Trials. 2005;2:72–9. doi: 10.1191/1740774505cn069oa. [DOI] [PubMed] [Google Scholar]
- 5.Velikova G, Wright EP, Smith AB, Cull A, Gould A, Forman D, et al. Automated collection of quality-of-life data: a comparison of paper and computer touch-screen questionnaires. J Clin Oncol. 1999;17:998–1007. doi: 10.1200/JCO.1999.17.3.998. [DOI] [PubMed] [Google Scholar]
- 6.Shah J, Rajgor D, Pradhan S, McCready M, Zaveri A, Pietrobon R. Electronic data capture for registries and clinical trials in orthopaedic surgery: open source versus commercial systems. Clin Orthop Relat Res. 2010;468:2664–71. doi: 10.1007/s11999-010-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prokscha S. Practical guide to clinical data management. CRC Press; 2011. [Google Scholar]
- 8.Welker JA. Implementation of electronic data capture systems: barriers and solutions. Contemp Clin Trials. 2007;28:329–36. doi: 10.1016/j.cct.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 9.de Lusignan S, van Weel C. The use of routinely collected computer data for research in primary care: opportunities and challenges. Fam Pract. 2006;23:253–63. doi: 10.1093/fampra/cmi106. [DOI] [PubMed] [Google Scholar]
- 10.El Emam K, Jonker E, Sampson M, Krleza-Jerić K, Neisa A. The use of electronic data capture tools in clinical trials: Web-survey of 259 Canadian trials. J Med Internet Res. 2009;11:e8. doi: 10.2196/jmir.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masys DR, Harris PA, Fearn PA, Kohane IS. Designing a public square for research computing. Sci Transl Med. 2012;4:149fs32. doi: 10.1126/scitranslmed.3004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroux H, McBride S, Gibson S. On selecting a clinical trial management system for large scale, multi-centre, multi-modal clinical research study. Stud Health Technol Inform. 2011;168:89–95. [PubMed] [Google Scholar]
- 13. [accessed April 9, 2016];Oracle Clinical. - Overview | Oracle. n.d http://www.oracle.com/us/products/applications/health-sciences/e-clinical/clinical/index.html.
- 14.Fegan GW, Lang TA. Could an open-source clinical trial data-management system be what we have all been looking for? PLoS Med. 2008;5:e6. doi: 10.1371/journal.pmed.0050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.OpenClinica Open Source EDC. [accessed April 9, 2016];OpenClinica. n.d https://community.openclinica.com/
- 16.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obeid JS, McGraw CA, Minor BL, Conde JG, Pawluk R, Lin M, et al. Procurement of shared data instruments for Research Electronic Data Capture (REDCap) J Biomed Inform. 2013;46:259–65. doi: 10.1016/j.jbi.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong TC, Captur G, Valeti U, Moon J, Schelbert EB. Feasibility of the REDCap platform for Single Center and Collaborative Multicenter CMR Research. J Cardiovasc Magn Reson. 2014;16:P89. [Google Scholar]
- 19. [accessed April 8, 2016];REDCap Plugins (U of Iowa) - REDCap Documentation - Institute for Clinical and Translational Science. n.d https://www.icts.uiowa.edu/confluence/pages/viewpage.action?pageId=67307660.
- 20.123andy. 123andy/redcap-hook-framework. [accessed April 9, 2016];GitHub. n.d https://github.com/123andy/redcap-hook-framework.
- 21.Gutierrez JB, Harb OS, Zheng J, Tisch DJ, Charlebois ED, Stoeckert CJ, Jr, et al. A Framework for Global Collaborative Data Management for Malaria Research. Am J Trop Med Hyg. 2015;93:124–32. doi: 10.4269/ajtmh.15-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anaconda Scientific Python Distribution. [accessed April 24, 2015];n.d https://store.continuum.io/cshop/anaconda/
- 23.Burns SS, Browne A, Davis GN, Rimrodt SL, Cutting LE, Nashville TN. PyCap (Version 1.0) Vanderbilt University and Philadelphia, PA: Childrens Hospital of Philadelphia; GitHub; n.d. [accessed April 24, 2015]. [Computer Software] https://github.com/sburns/PyCap. [Google Scholar]
- 24.Perez F, Granger BE. IPython:;1. A System for Interactive Scientific Computing. Comput Sci Eng. n.d;9:21–9. [Google Scholar]
- 25.Post A, Kurc T, Overcash M, Cantrell D, Morris T, Eckerson K, et al. A Temporal Abstraction-based Extract, Transform and Load Process for Creating Registry Databases for Research. AMIA Jt Summits Transl Sci Proc. 2011;2011:46–50. [PMC free article] [PubMed] [Google Scholar]
- 26.Office Migration Planning Manager (OMPM): Office Compatibility. Microsoft Download Center; n.d. [accessed April 24, 2015]. http://www.microsoft.com/en-us/download/details.aspx?id=11454. [Google Scholar]
- 27. [accessed April 24, 2015];openpyxl - A Python library to read/write Excel 2007 xlsx/xlsm files — openpyxl 2.2.1 documentation. n.d https://openpyxl.readthedocs.org/en/latest/
- 28. [accessed April 29, 2016];python-docx - A Python library for creating and updating Microsoft Word (.docx) files — python-docx 0.8.5 documentation. n.d https://python-docx.readthedocs.io/en/latest/
- 29.Lewis JR. IBM computer usability satisfaction questionnaires: Psychometric evaluation and instructions for use. Int J Hum Comput Interact. 1995;7:57–78. [Google Scholar]
- 30.Dillon TW, Ratcliffe TM. Evaluating point-of-care technology with the IBM computer usability satisfaction questionnaire. Issues in Information Systems. 2004;5:441–6. [Google Scholar]
- 31.Franklin JD, Guidry A, Brinkley JF. A partnership approach for Electronic Data Capture in small-scale clinical trials. J Biomed Inform. 2011;44(Suppl 1):S103–8. doi: 10.1016/j.jbi.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adagarla B, Connolly DW, McMahon TM, Nair M, VanHoose LD, Sharma P, et al. SEINE: Methods for Electronic Data Capture and Integrated Data Repository Synthesis with Patient Registry Use Cases. 2015 [Google Scholar]
- 33.Forrest CB, Margolis PA, Bailey LC, Marsolo K, Del Beccaro MA, Finkelstein JA, et al. PEDSnet: a National Pediatric Learning Health System. J Am Med Inform Assoc. 2014;21:602–6. doi: 10.1136/amiajnl-2014-002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasreddine ZS, Phillips NA, Bédirian V, Charbonneau S, Whitehead V, Collin I, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53:695–9. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- 35.St Clair-Thompson HL, Allen RJ. Are forward and backward recall the same? A dual-task study of digit recall. Mem Cognit. 2013;41:519–32. doi: 10.3758/s13421-012-0277-2. [DOI] [PubMed] [Google Scholar]
- 36.Agrell B, Dehlin O. The clock-drawing test. Age Ageing. 1998;27:399–404. doi: 10.1093/ageing/afs149. [DOI] [PubMed] [Google Scholar]
- 37.Spreen O, Benton AL. Neurosensory Center Comprehensive Examination for Aphasia (NCCEA) 1977 [Google Scholar]
- 38.Morris JC, Heyman A, Mohs RC, Hughes JP, van Belle G, Fillenbaum G, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) Part I. Clinical and neuropsychological assessment of Alzheimer’s disease. Neurology. 1989;39:1159–65. doi: 10.1212/wnl.39.9.1159. [DOI] [PubMed] [Google Scholar]
- 39.Calero MD, Arnedo ML, Navarro E, Ruiz-Pedrosa M, Carnero C. Usefulness of a 15-item version of the Boston Naming Test in neuropsychological assessment of low-educational elders with dementia. J Gerontol B Psychol Sci Soc Sci. 2002;57:P187–91. doi: 10.1093/geronb/57.2.p187. [DOI] [PubMed] [Google Scholar]
- 40.Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer’s disease. Am J Psychiatry. 1984;141:1356–64. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- 41.Sheikh JI, Yesavage JA. Geriatric depression scale (GDS): recent evidence and development of a shorter version. Clinical Gerontology. 1986;5:165–73. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.