Abstract
Stem cells including cancer stem cells (CSCs) require niches to maintain stemness, yet it is unclear how CSCs maintain stemness in the suboptimal environment outside their niches during invasion. Postnatal codeletion of Pten and Trp53 in mouse neural stem cells (NSCs) leads to their expansion in the subventricular zone (SVZ) niches but fails to maintain the stemness outside the SVZs. We discovered that QKI is a major regulator of NSC stemness. Qki deletion against Pten−/−Trp53−/− backdrop helps NSCs maintain their stemness outside the SVZs in Nestin-CreERT2 QkiL/LPtenL/LTrp53L/L mice, which develop glioblastoma with a penetrance of 92% and a median survival of 105 days. Mechanistically, Qki deletion decreases endolysosome-mediated degradation and enriches receptors essential for maintaining self-renewal on the cytoplasmic membrane to cope with low ligand levels outside the niches. Thus, downregulating endolysosome level by QKI loss helps glioma stem cells (GSCs) maintain their stemness in suboptimal environments outside the niches.
INTRODUCTION
Self-renewal is a unique feature of all stem cells that creates identical daughter stem cells without differentiating into other cell types1. To maintain stemness, normal somatic stem cells need to be in a suitable environment, namely a niche, which provides proper cues to instruct them to self-renew and prevent them from differentiating2–5. Niches for cancer stem cells (CSCs) have been characterized in multiple different cancers, and they were shown to often resemble the structures and/or the signals of niches from the original tissues6–9. However, left unanswered is how CSCs still manage to maintain the stemness when they invade and migrate from their niches to other areas, where optimal niches are less likely to be available10.
Glioblastoma (GBM, grade IV glioma) is the most common and lethal type of primary brain tumor in adults, with over 10,000 cases diagnosed annually in the U.S.11,12. Evidence is emerging that a population of glioblastoma cells called glioma stem cells (GSCs) possesses an inexhaustible ability to self-renew and produce intracranial tumors that retain the features of original tumors13–15. Several studies have shown GSC niches to be hypoxic and perivascular, which resemble NSC niches such as the SVZs6,7. However, niche-dependent stemness maintenance fails to explain how GSC stemness is maintained when GSCs invade and migrate to new territories. Glioblastoma is a highly invasive disease16–18, and the ability of glioblastoma cells to use distinct anatomic structures to invade and migrate raises the question of how invading GSCs maintain their stemness when they face various compositions of matrix components in their environment. Therefore, we hypothesize that GSCs must acquire ability to maintain stemness independently of their niches during invasion and migration. In this study, we used premalignant NSCs (PM-NSCs) with deletion of major tumor suppressors and the SVZs as model systems to study the interaction between GSCs and their niches.
To look for genetic alterations that could help PM-NSCs/GSCs maintain the stemness outside their niches, we previously screened a series of candidate genes and identified five tumor suppressors potentially affecting GSC stemness, one of which was Quaking (QKI)19. QKI is a STAR family RNA-binding protein that is involved in RNA homeostasis from various aspects: RNA stability, splicing, translation, miRNA processing, and circular RNA biogenesis20–23. We discovered that knockout of the Quaking (Qki) gene decreased proliferation of Pten−/−Trp53−/− PM-NSCs in the niches but could help maintain their self-renewal outside the niches in the Nestin-CreERT2 QkiL/LPtenL/LTrp53L/L mice. This unique model system allowed us to discover a novel underlying mechanism whereby GSCs maintain their high self-renewal capacity in the suboptimal microenvironment, helping them invade and migrate without losing their stemness.
RESULTS
Deletion of Qki enhances NSC self-renewal and decreases NSC differentiation
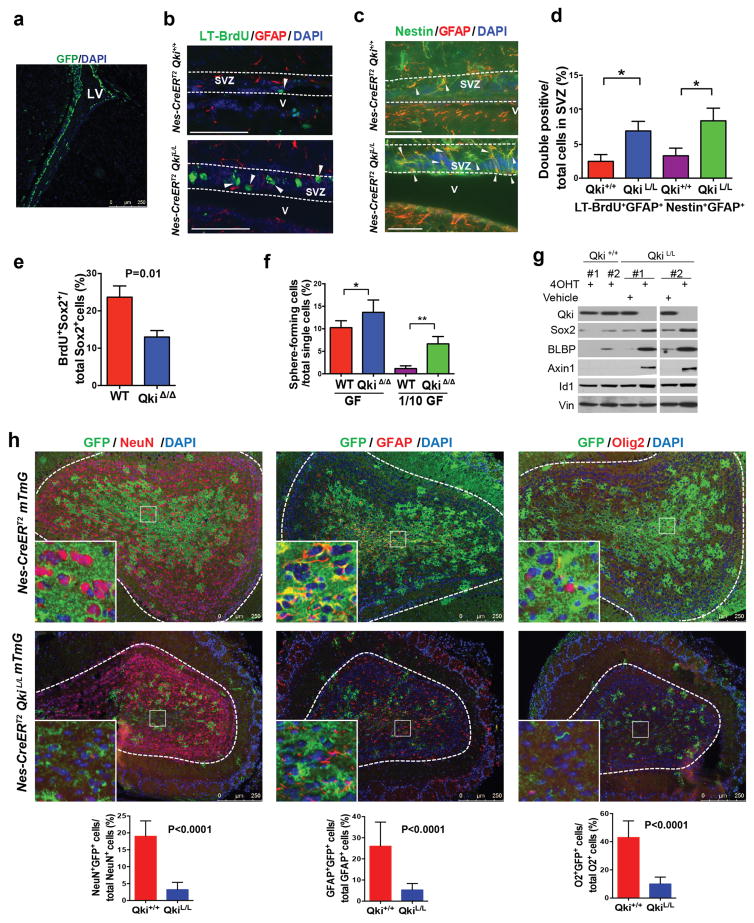
Our previous studies showed that QKI is a tumor suppressor gene that can potentially regulate the stemness of GSCs19,20, and we confirmed that it inhibited the capacity of Pten−/−Trp53−/− PM-NSCs to form secondary neurospheres in vitro (Supplementary Fig. 1a, 1b). Supporting QKI’s function in NSCs, we found that it was highly expressed not only in oligodendrocytes but also colocalized with Nestin in both the lateral ventricle SVZ (LV-SVZ) and the third ventricle SVZ (TV-SVZ) (Supplementary Fig. 1c). To check the impact of Qki deletion on NSCs in vivo, we generated a conditional knockout allele Qki-LoxP, in which Qki exon 2 is flanked by two LoxP sites (Supplementary Fig. 1d, 1e) and crossed it to the inducible Cre line Nestin-CreERT2 and the reporter allele Rosa26-LoxP-mTRed-Stop-LoxP-mGFP (mTmG)24. Consistent with the original characterization of the Nestin-CreERT2 allele25, we found that the GFP signal is exclusively localized in the SVZs of Nestin-CreERT2 mTmG mice shortly after tamoxifen injection (Fig. 1a). Over 85% of GFP+ cells coexpressed NSC markers GFAP and Nestin, confirming the specificity and efficiency of the activity of the Nestin-CreERT2 allele (Supplementary Fig. 1f). To determine the impact of Qki deletion on NSC stemness, we first measured the numbers of NSCs in the SVZs of the Nestin-CreERT2 Qki+/+ and Nestin-CreERT2 QkiL/L mice with a long-term BrdU labeling assay2,3. We labeled all dividing cells of postnatal day (P)1 Nestin-CreERT2 Qki+/+ and Nestin-CreERT2 QkiL/L mice (treated with tamoxifen at P0.5) with BrdU (Supplementary Fig. 2a) and waited for 10 days to determine the number of long-term BrdU-retaining cells in SVZs. Double staining of long-term BrdU (LT-BrdU) and GFAP (a NSC marker in SVZ) was utilized to specifically examine NSCs. We found that the Qki-null SVZs exhibited significantly higher numbers of LT-BrdU+GFAP+ double positive cells than the Qki-wt SVZs (7% vs 2.5%, respectively, P=0.01) (Fig. 1b, 1d). Consistently, the Qki-null SVZs also contained significantly more Nestin+GFAP+ NSCs than the Qki-wt SVZs (8.5% vs 3.3%, respectively, P=0.01) (Fig. 1c–d).
Figure 1. Deletion of Qki enhances NSC self-renewal and decreases NSC differentiation.
(a) Representative image of GFP staining of SVZ of P10 Nestin-CreERT2 mTmG mouse that was injected with tamoxifen at P8. (b–c) Representative IF images of NSCs that are labeled with long-term Brdu+ (LT-BrdU+) GFAP+ (b) or Nestin+ GFAP+ (c) in the SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice injected with tamoxifen at P1. (d) Quantitative analyses of (b–c) (n=6/group; Student’s t test; *P=0.01). (e) Percentages of BrdU+Sox2+ double positive cells among all the Sox2+ early passage Qki-wt and Qki-null NSCs that were cultured with BrdU for 12 hours (6 lines/genotype; Student’s t test). (f) Percentages of early passage (P5) Qki-wt and Qki-null NSCs that can form neurospheres from single cells cultured in either full-strength growth factors (GF) (20ng/ml EGF+10ng/ml FGF) or 1/10 dilution of GF (2ng/ml EGF+1ng/ml FGF); (n=6/group; Student’s t test; *P=0.02; **P<0.0001). (g) Immunoblotting of stemness markers of Qki+/+ and QkiL/L NSCs treated with 4-hydroxytamoxifen (4OHT) or vehicle alone. (h) Representative images and quantification of IF double staining of olfactory bulbs of P40 Nestin-CreERT2 mTmG and Nestin-CreERT2 mTmG QkiL/L mice that were injected with tamoxifen at P8 (n=5/group; Student’s t test). Scale bars represent 250 μm in (a), (h), 100 μm in (b), and 50 μm in (c). Error bars indicate s.d. All the experiments were replicated three times in the lab.
To determine whether the increase of NSC numbers in the SVZ by Qki deletion was associated with enhanced proliferation rate or enhanced self-renewal capacity, we first checked how Qki deletion would impact proliferation rates of NSCs. The short-term BrdU incorporation assay showed that Qki deletion decreased proliferation rates of NSCs in vitro (P=0.01) (Fig. 1e), which is consistent with the observation that the Qki-null SVZs contain more Id1+GFAP+ quiescent NSCs than the Qki-wt SVZs (P=0.03) (Supplementary Fig. 2b). The decreased proliferation rate of NSCs by Qki deletion was not due to the enhancement of cell death, as there was no difference in apoptosis between the Qki-wt and Qki-null NSCs, either in vivo or in vitro (Supplementary Fig. 2c).
To determine whether Qki deletion could enhance the self-renewal capability of NSCs, we first surveyed the sphere-forming ability of single cells, showing that Qki-null NSCs had significantly higher capability to form single cell-derived neurospheres (P=0.02 and P<0.0001 for high and low concentrations of EGF/FGF, respectively) (Fig. 1f), which were subsequently subjected to differentiation medium for determining multipotency. This single cell-based assay showed that one in 25 Qki-null NSCs can form secondary spheres when cultured with low concentrations of EGF/FGF, compared with one in 86 Qki-wt NSCs (passage 5; P<0.0001), indicating that Qki deletion significantly enhances self-renewal of NSCs. Immunoblotting showed that Qki deletion also enhances various self-renewal markers including Sox2, Id1, BLBP, and Wnt-signaling component Axin1 (Fig. 1g). In addition, most of the Qki-wt NSCs at late passages (e.g., passage 25) showed flattened cell morphology, indicating loss of stemness features, while at the same passages, most of the Qki-null NSCs still maintained their spherical morphology (Supplementary Fig. 2d).
Furthermore, we reasoned that if self-renewal were enhanced, the differentiation of NSCs would also be repressed by Qki deletion. Immunofluorescent (IF) costaining of lineage markers and GFP showed that differentiation of adult NSCs into neurons (indicated by NeuN staining), astrocytes (indicated by GFAP staining) and oligodendrocytes (indicated by Olig2 staining) in olfactory bulbs of P40 Nestin-CreERT2 QkiL/L mice was significantly compromised by Qki deletion (Fig. 1h, P<0.0001 for all). Consistently, differentiation of embryonic and neonatal NSCs was also compromised by Qki deletion in the Nestin-CreERT2 QkiL/L mice that were injected with tamoxifen at embryonic day (E)9.5 and P2, as shown by lineage marker staining (Mbp and Olig2 for oligodendrocytes, Tuj1 for neurons, and GFAP for astrocytes; P<0.0003 for all) (Supplementary Fig. 2e–h). Collectively, these data suggest that although Qki deletion directs NSCs toward a more quiescent state, meanwhile it enhances NSC self-renewal capacity and blocks NSC differentiation.
Deletion of Qki maintains stemness of Pten−/−Trp53−/− PM-NSCs outside SVZs
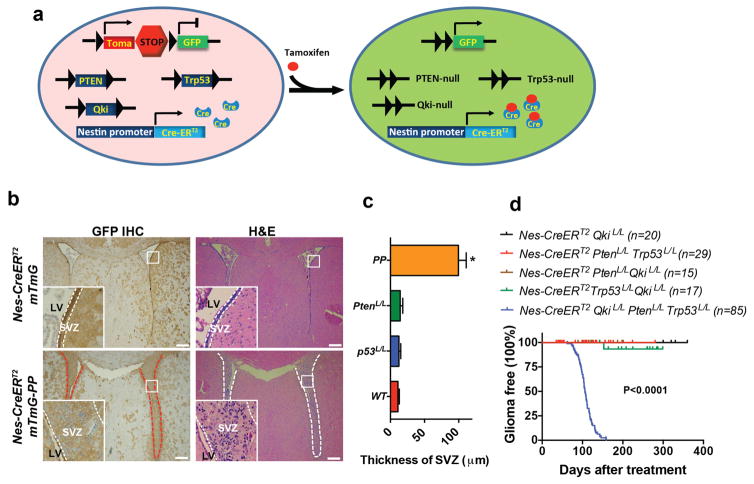
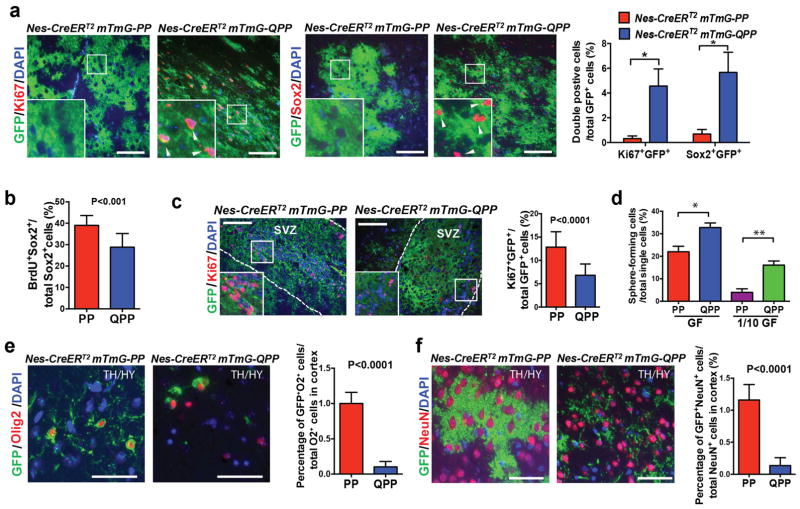
Numerous studies have shown that the NSC in the SVZ is one of the possible cells-of-origin of malignant gliomas26–28. To investigate how Qki deletion would impact the interaction between PM-NSCs and their niches in vivo during gliomagenesis, we crossed the reporter allele mTmG24 into the Nestin-CreERT2 PtenL/LTrp53L/L (denoted PP hereafter) and Nestin-CreERT2 QkiL/LPtenL/LTrp53L/L (denoted QPP hereafter) cohorts to trace the fates of PP and QPP PM-NSCs with GFP labeling (Fig. 2a). The P38 SVZs of the Nestin-CreERT2 mTmG-PP mice that were injected with tamoxifen at P8 were significantly enlarged relative to those of the Nestin-CreERT2 mTmG mice (P<0.0001) (Fig. 2b–c, Supplementary Fig. 3a), indicating that deletion of Pten and Trp53 can significantly expand NSCs in the niches. However, PP PM-NSCs lost their stemness (indicated by loss of staining of stem cell marker Sox2 and proliferating cell marker Ki67) (Supplementary Fig. 3a) when they migrated beyond their niches (e.g., to the thalamus/hypothalamus), where they fully differentiated into oligodendrocytes (Olig2+GFP+ and Mbp+GFP+), neurons (NeuN+GFP+), and astrocytes (S100β+GFP+) with morphologies identical to the wild type cells (Supplementary Fig. 3b). Further supporting the dependency of PP PM-NSCs on their microenvironment, we found that PP PM-NSCs could grow and maintain their stemness only when they were orthotopically injected into the lateral ventricle but not in the cortex (Supplementary Fig. 4a). The PP PM-NSCs that were injected in the cortex would lose their stemness, as evidenced by the higher level of differentiation marker Numb and the lower levels of receptors EGFR and FGFR that are essential for maintaining stemness, compared with those injected into the lateral ventricle (Supplementary Fig. 4b–d). Because no gliomas formed in the Nestin-CreERT2 mTmG-PP mice for up to 12 months (Fig. 2d), we concluded that although deletion of Pten and Trp53 can promote NSC expansion within their niches, it is insufficient to maintain NSC stemness beyond the niches, thereby preventing NSCs from developing into fully malignant gliomas. In contrast, 4.5% and 5.8% of GFP+ cells found outside the SVZs (e.g., in the thalamus/hypothalamus) of the Nestin-CreERT2 mTmG-QPP mice were Ki67+ and Sox2+, respectively (P<0.0001) (Fig. 3a). Similar observations were made in other regions of the brain, including the striatum and cortex (data not shown). Among the Ki67+GFP+ proliferating cells, 70% were Sox2+ stem cells (Supplementary Fig. 4e). These data suggest that unlike PP PM-NSCs, QPP PM-NSCs can still maintain their stemness outside the SVZs.
Figure 2. Deletion of Pten and p53 promotes PM-NSC proliferation inside but not outside of the SVZ.
(a) Scheme of cohort Nestin-CreERT2 mTmG Qki L/LPtenL/LTrp53L/L (or Nestin-CreERT2 mTmG-QPP). GFP is used to label knockout NSCs. (b) Representative images of H&E and GFP IHC staining of SVZs of P38 Nestin-CreERT2 mTmG, and Nestin-CreERT2 mTmG PtenL/LTrp53L/L (or Nestin-CreERT2 mTmG-PP) mice that were injected with tamoxifen at P8. (c) Quantification of the maximal diameters of the SVZs in control, single, or double knockout mice (n=6/group; Student’s t test). (d) Kaplan-Meier survival curves (log-rank test) of Nestin-CreERT2 QkiL/L, Nestin-CreERT2 PtenL/LQkiL/L, Nestin-CreERT2 Trp53L/LQkiL/L, Nestin-CreERT2-PP, and Nestin-CreERT2-QPP mice treated with tamoxifen at P8–10. P<0.0001 means that the differences between Nestin-CreERT2-QPP and any other cohorts are significant. Scale bars represent 100 μm. Error bars indicate s.d. The experiments in (b) were replicated three times in the lab.
Figure 3. Deletion of Qki maintains stemness of PM-NSCs outside the SVZs.
(a) Representative IF images and quantification of Ki67+GFP and Sox2+GFP costaining in thalamus/hypothalamus (TH/HY) of P38 Nestin-CreERT2-PP and Nestin-CreERT2-QPP mice that were injected on P8 (n=6/group; Student’s t test; *P<0.0001). (b) Percentages of BrdU+Sox2+ double positive cells among all the Sox2+ early passage PP and QPP PM-NSCs that were cultured with BrdU for 12 hours (6 lines/genotype; Student’s t test). (c) Representative IF images and quantification of Ki67+GFP costaining of SVZs in P38 Nestin-CreERT2 mTmG-PP and Nestin-CreERT2 mTmG-QPP mice that were injected with tamoxifen at P8 (n=6/group; Student’s t test). (d) Percentages of early passage (P5) PP and QPP PM-NSCs that can form neurospheres from single cells cultured in either full-strength growth factors (GF) (20ng/ml EGF+10ng/ml FGF) or 1/10 dilution of GF (2ng/ml EGF+1ng/ml FGF); (n=6/group; Student’s t test; *P=0.02; **P<0.0001). (e)–(f) Representative IF images and quantification of Olig2+GFP and NeuN+GFP costaining of thalamus/hypothalamus (TH/HY) in P38 Nestin-CreERT2 mTmG-PP and Nestin-CreERT2 mTmG-QPP mice injected with tamoxifen at P8 (n=5/group; Student’s t test). Scale bars represent 100 μm in (a), (c), and 50 μm in (e), (f). Error bars indicate s.d. All the experiments were replicated three times in the lab.
Because we showed that Qki deletion decreased NSC proliferation rate, enhanced NSC self-renewal capacity, and blocked NSC differentiation (Fig. 1, Supplementary Fig. 2), we wanted to determine whether Qki deletion could have similar effects on PM-NSCs. BrdU incorporation assay showed that Qki deletion decreased proliferation rates of PM-NSCs in vitro (P<0.01) (Fig. 3b). Consistently, 30 days after tamoxifen injection, the SVZs of the Nestin-CreERT2 mTmG-QPP mice contained fewer Ki67+GFP+ proliferating cells than the SVZs of the Nestin-CreERT2 mTmG-PP mice (P<0.0001) (Fig. 3c). To determine whether Qki deletion could enhance the self-renewal capability of PM-NSCs, we surveyed the sphere-forming ability of single cells, which showed that Qki-null PM-NSCs had significantly higher capability to form single-cell-derived neurospheres (P=0.02 and P<0.0001 for low and high concentrations of EGF/FGF, respectively) (Fig. 3d). Consistent with the observation that Qki deletion reduced the differentiation capacity of NSCs, QPP PM-NSCs in the thalamus/hypothalamus of P38 mice also exhibited compromised differentiation compared with PP PM-NSCs in the same location (P<0.0001 for all) (Fig. 3e–f, Supplementary Fig. 4f). Collectively, these data showed that similar to NSCs, loss of QKI in PM-NSCs also enhances their self-renewal and reduces their differentiation tendency.
Deletion of Qki under the backdrop of Pten−/−Trp53−/− promotes gliomagenesis
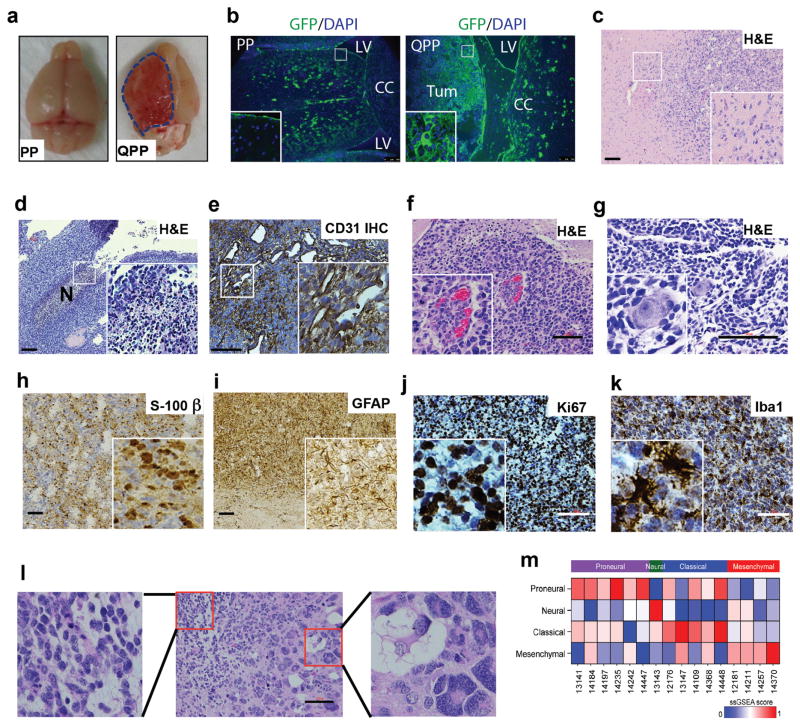
Supporting the essential role of maintaining the PM-NSC stemness outside the niches in gliomagenesis, 92% of the Nestin-CreERT2-QPP mice eventually developed gliomas, with a median survival time of 105 days (Fig. 2d); 44% of the gliomas were macroscopically evident at the cortex (Fig. 4a–b), and the rest occurred in the parenchyma. Immunohistochemistry (IHC), immunoblotting, and PCR confirmed that most tumor cells do not express Qki, Pten and p53, indicating that all three tumor suppressors need to be deleted to permit gliomagenesis (Supplementary Fig. 4g–j). These gliomas were all characterized as grade IV (glioblastoma) because they exhibited typical human glioblastoma features, including an invasive leading edge (Fig. 4c), pseudopalisading necrosis (Fig. 4d), hypervascularity (Fig. 4e), microvascular proliferation (Fig. 4f), perineuronal satellitosis (Fig. 4g), positive S100-β and GFAP staining (Fig. 4h–i), a high proliferative index (Ki67) (Fig. 4j), positive stem-cell-marker staining (e.g., Notch1 and Nestin) (Supplementary Fig. 5a–b), and high Iba1+ microglia/macrophage infiltration (Fig. 4k). They also showed high intra- and intertumoral heterogeneity, manifesting as morphologically different cell types or growth patterns within the same tumors (Fig. 4l) and among different tumors (Supplementary Fig. 5c). Glioblastoma that developed in the Nestin-CreERT2-QPP mice represented all four subtypes, including 37.5% proneural, 6% neural, 31% classical, and 25% mesenchymal, based on transcriptomic profiling (n=16)29 (Fig. 4m), which indicates the plasticity of NSC-derived glioblastoma. Thus, maintaining the stemness of PM-NSCs outside their niches by Qki deletion can promote development of glioblastoma that recapitulates features of human glioblastoma. As with the Nestin-CreERT2 mTmG-QPP mice injected with tamoxifen at P8, the Nestin-CreERT2 mTmG-QPP mice injected with tamoxifen at P30 also developed glioblastoma, with a penetrance of 73% and a median survival time of 200 days, suggesting that like early postnatal NSCs, adult NSCs with deletion of Qki, Pten and Trp53 also have great potential to develop into glioblastoma (Supplementary Fig. 5d). Moreover, supporting the idea that QKI can function as a tumor suppressor under different genetic backgrounds, we found that the Nestin-CreERT2 QkiL/LPtenL/LInk4a/ArfL/L mice injected with tamoxifen at P8 developed glioblastoma with a penetrance of 76% and a median survival time of 190 days, whereas the Nestin-CreERT2 PtenL/LInk4a/ArfL/L mice never developed gliomas (Supplementary Fig. 5e).
Figure 4. Deletion of Qki against the backdrop of Pten−/−Trp53−/− promotes gliomagenesis.
(a) Representative images of brains of Nestin-CreERT2-PP and Nestin-CreERT2-QPP mice at 3 months after tamoxifen injection. (b) Representative GFP staining of Nestin-CreERT2 mTmG-PP and Nestin-CreERT2 mTmG-QPP mice at 3 months after tamoxifen injection. (c) Representative H&E staining of invasive front of QPP gliomas. (d) Representative H&E staining of pseudopalisading necrosis in QPP gliomas. (e) Representative IHC staining of CD31 in QPP gliomas. (f) Representative H&E staining of microvascular proliferation in QPP gliomas. (g) Representative H&E staining of perineuronal satellitosis in QPP gliomas. (h–k) Representative IHC staining of S-100β, GFAP, Ki67, and Iba1 in QPP gliomas. (l) Representative H&E images of a QPP tumor showing intratumoral cellular heterogeneity. Biphasic-like pattern consisting of cells with eosinophilic fibrillary cytoplasm and round or oval nuclei (left inset) and multinucleated giant cells (right inset) are shown. (m) QPP glioma subtype classification. Each row represents a gene signature, and each column represents a tumor. Top color bar indicates the predicted subtype. Colors from blue to red represent the activation level of the signature. Scale bars represent 100 μm.
RNA binding protein QKI is a novel regulator of endolysosome
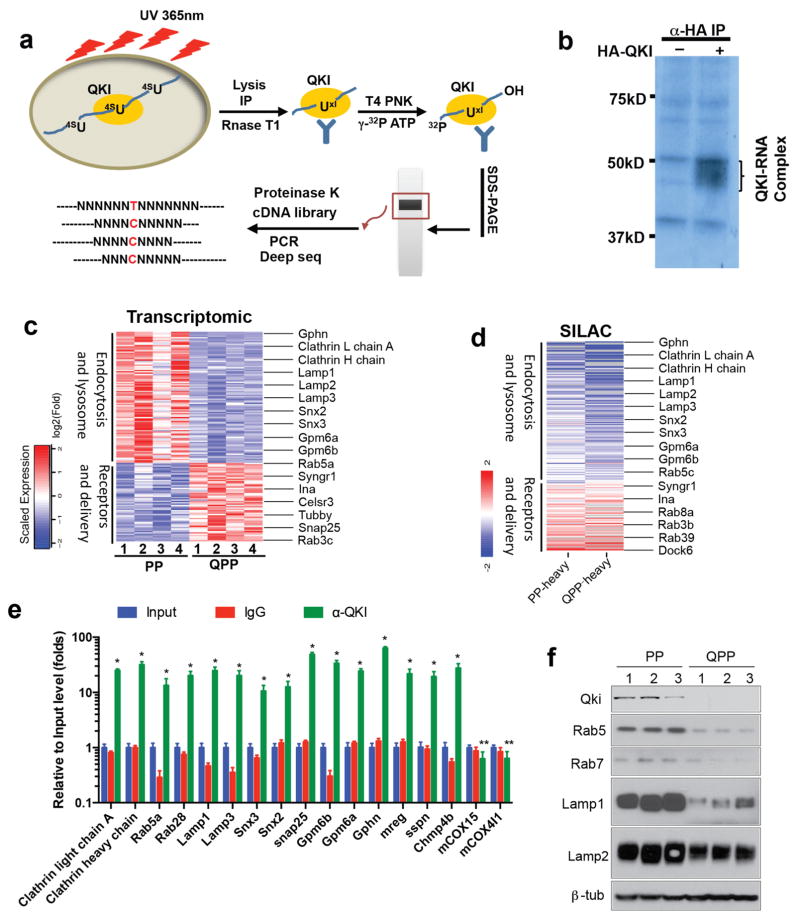
To understand how Qki deletion can enhance self-renewal capacity of Pten−/−Trp53−/− PM-NSCs, especially outside their niches, we sought to identify the genes that are changed by Qki deletion at the total mRNA level, alternative splicing level, and protein level, because QKI has been shown to regulate RNA stability, alternative splicing and translation23. We first performed transcriptomic profiling with RNA-seq on Qki-wt and Qki-null NSCs as well as on PP and QPP PM-NSCs. We then integrated these two datasets and identified 388 overlapped genes (P<0.01, fold change>1.5) (Supplementary Table 1). Analyzing the same RNA-seq dataset with SpliceSeq Analyzer also found that alternative splicing of 217 genes is altered by Qki deletion in both NSCs and PM-NSCs (P<0.05) (Supplementary Table 2). To identify genes changed by Qki deletion at the protein level, we performed Stable Isotope Labeling by Amino acid in Cell culture (SILAC)-coupled Mass Spectrometry-based quantitative proteomics on PP and QPP PM-NSCs (Supplementary Fig. 6a). We discovered that 290 proteins are upregulated (Supplementary Table 3) and 343 proteins are downregulated (Supplementary Table 4) in QPP PM-NSCs compared with PP PM-NSCs (fold change >1.2). To determine which genes that were changed by Qki deletion at different levels are the direct targets of the QKI protein, we performed Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP)-coupled RNA-seq analyses on PM-NSCs to identify QKI-associated RNAs, with known QKI-binding motif AYUAAY30 (Fig. 5a). P32-labeling-coupled SDS-PAGE revealed that this approach could specifically pull down QKI-associated RNAs (Fig. 5b), which were subsequently characterized with Illumina RNA-sequencing and computational analyses. We then integrated the transcriptomic profiles, proteomic profiles, and PAR-CLIP data, and found that 104 genes that changed at the mRNA level, 73 genes that changed at the splicing level, and 148 genes that changed at the protein level showed high confidence in being direct targets of QKI because the RNAs they encode also physically interacted with the QKI protein (Supplementary Table 5). Pathway analysis on these QKI targets by Ingenuity Pathway Analysis (IPA) identified 15 pathways that are highly enriched (P<0.001) (Supplementary Fig. 6b). Twelve of these 15 pathways (80%) are involved in receptor signaling and trafficking, including cytoskeleton regulation, endocytosis, and lipid biosynthesis (which is critical for vesicle formation). In order to further break down which pathways are upregulated or downregulated by Qki deletion, we found that 45% of the genes that are upregulated at the mRNA level and 18% of the genes that are upregulated at the protein level encode either receptors (especially GPCR) or proteins that are involved in delivering receptors to the cytoplasmic membranes (Fig 5c–d, Supplementary Table 2). We also found that 39% of the genes are downregulated by Qki deletion at the mRNA level, and 40% of the genes that are downregulated at the protein level are involved in regulating endocytosis-mediated receptor degradation, including multiple caveolin and clathrin subunits and various components of endosomes and lysosomes (Fig. 5c–d, Supplementary Table 2). Finally, we also discovered that 37% of the genes that are changed at the splicing level encode proteins that are involved in receptor trafficking (Supplementary Table 2). We found that Qki deletion doesn’t affect the expression of endolysosome biogenesis regulators TFE3/TFEB/MITF, suggesting that QKI provides another level of regulation by regulating RNA homeostasis of the endolysosome components. To verify the direct interaction of QKI and its substrates, we picked the top 15 QKI substrates in the endolysosomal pathway and were able to confirm the enrichment of these RNAs in the anti-QKI immunocomplex by PAR-CLIP analysis followed by quantitative PCR (qPCR) (Fig. 5e).
Figure 5. QKI is a novel regulator of endolysosomes.
(a) Scheme of Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP) experimental procedures to identify QKI targets. (b) Autoradiography showing HA-QKI5-associated RNAs pulled down from Pten−/−Trp53−/− (PP) PM-NSCs with PAR-CLIP and separated by SDS-PAGE. (c) Heat map of transcriptomic profiles of PP and QPP PM-NSCs showing genes involved in receptor delivery and endolysosome. Some classical endolysosome genes are highlighted. (d) Heat map of Stable Isotope Labeling by Amino Acid (SILAC)-based proteomic profiles of PP and QPP PM-NSCs showing genes involved in receptor delivery and endocytosis. PP-heavy and QPP-heavy indicate that PP and QPP PM-NSCs were labeled with heavy arginine/lysine, respectively. Some classical endolysosome genes are highlighted. (e) QKI target RNAs were immunoprecipitated (IP’ed) by IgG and QKI antibody with the PAR-CLIP method. Input and IP’ed RNA levels were measured by qPCR. * indicates P<0.0001 compared with input and α-IgG, and ** indicates not significant. (f) Immunoblotting of endosome/lysosome markers in PP and QPP PM-NSCs. The experiments in (b), (e), and (f) were replicated three times in the lab.
Qki deletion decreases endolysosome level
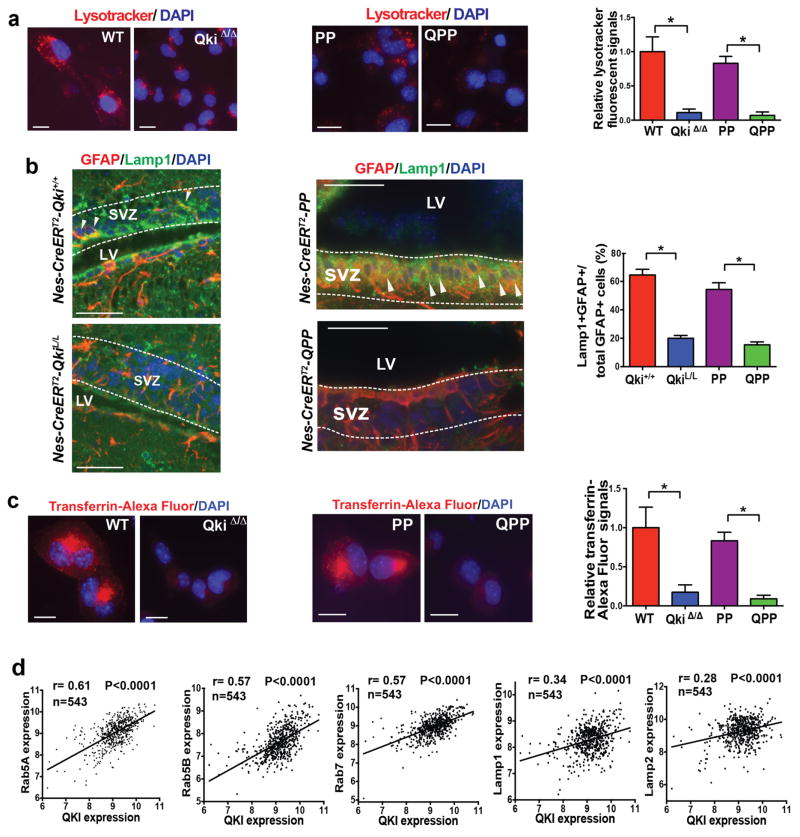
The enrichment of endolysosome components in the QKI targets made us hypothesize that Qki deletion maintains self-renewal of stem cells in suboptimal environments by downregulating endolysosome-dependent degradation. Downregulation of endolysosome levels was confirmed by immunoblotting of key components such as endosome components Rab5 and Rab7 and lysosome components Lamp1 and Lamp2 (Fig. 5f, Supplementary Fig. 7a). Downregulation of endolysosome levels by Qki deletion was also confirmed by the Lysotracker staining (Fig. 6a) as well as by GFAP/Lamp1 (Fig. 6b) and LT-BrdU/Lamp1 (Supplementary Fig. 7b) double staining in SVZs in vivo. Furthermore, uptake of Alexa Fluor-labeled transferrin and low pH-sensitive probe PHrodo Red EGF from the culture medium was greatly decreased in Qki-null NSCs and PM-NSCs compared with Qki-wt NSCs and PM-NSCs (Fig. 6c, Supplementary Fig. 7c, 7d), suggesting that Qki deletion greatly compromises the endocytosis of receptors. We also found that the mRNA levels of endolysosomal components/regulators CLTC, Rab5A/5B/5C, Rab7, Rab28, Lamp1/2, Snx2/3, and Gphn were all positively correlated with QKI mRNA levels in the TCGA dataset (Pearson correlation; P<0.0001 for all; Fig. 6d, Supplementary Fig. 7e), suggesting that downregulation of endolysosome levels by low QKI also occurs in human glioblastoma samples. Altogether, these data suggest that QKI positively regulates multiple components of endolysosome-dependent degradation machinery through various RNA regulating mechanisms.
Figure 6. Qki deletion downregulates endolysosomes.
(a) Representative images and quantification of Lysotracker staining of Qki-wt and Qki-null NSCs and Pten−/−Trp53−/− (PP) and Qki−/−Pten−/−Trp53−/− (QPP) PM-NSCs (n=7/group; Student’s t test). (b) Representative images and quantification of the percentages of GFAP+Lamp1+ double positive cells among all the GFAP+ cells in SVZs of P12 Nestin-CreERT2Qki+/+, Nestin-CreERT2QkiL/L, Nestin-CreERT2-PP, and Nestin-CreERT2-QPP mice treated with tamoxifen at P1 (n=3–5/group; Student’s t test). Arrowheads indicate costaining. (c) Representative images and quantification of Transferrin-Alexa-Fluor staining of Qki-wt and Qki-null NSCs and PP and QPP PM-NSCs (n=6/group; Student’s t test). (d) Expression correlation between QKI and Rab5A, Rab5B, Rab7, Lamp1, and Lamp2 in TCGA microarray dataset (n=543, Pearson correlation). Scale bars represent 50 μm. *P<0.0001. Error bars indicate s.d. The experiments in (a)-(c) were replicated three times in the lab.
Downregulation of endolysosome-dependent receptor degradation caused by Qki deletion enhances PM-NSC stemness
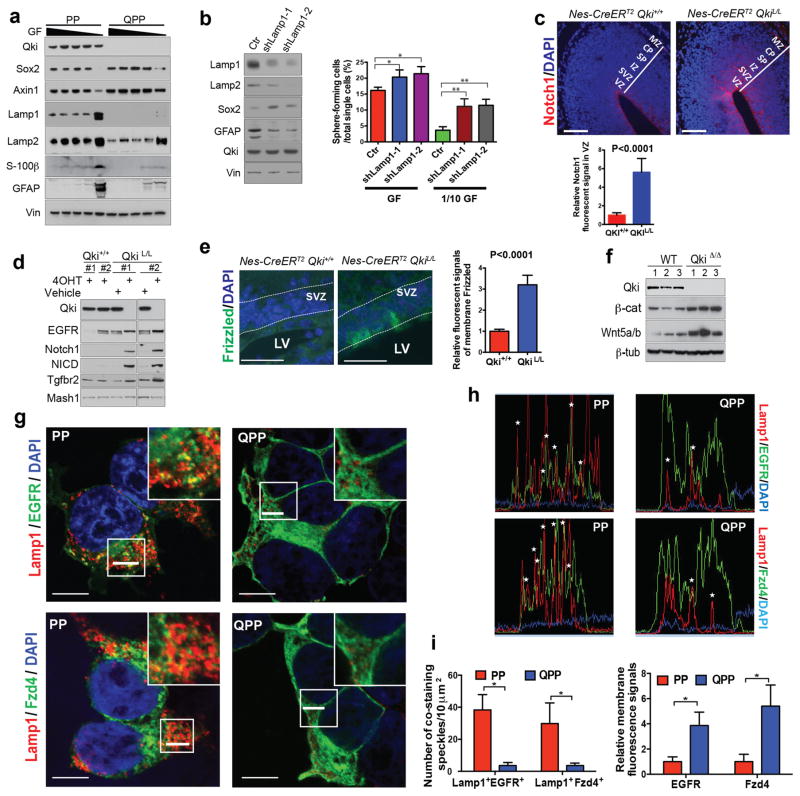
One of the main functions of niches is to provide various ligands for receptors that are essential for maintaining stem cell self-renewal1. Because Qki deletion could enhance receptor/ligand activity by downregulating endolysosome-dependent receptor/ligand degradation, we hypothesized that this mechanism would help maintain NSC self-renewal when the concentrations of exogenous ligands were lower. Supporting the role of decreased endolysosome-dependent degradation in regulating stemness, we found that Lamp1/2 levels in PM-NSCs were much lower than those of the differentiated PM-NSCs that were induced by low concentrations of EGF/FGF (Fig. 7a). Compared with PP PM-NSCs, when cultured with low concentrations of EGF/FGF, QPP PM-NSCs still expressed low levels of lysosomes, maintained high levels of stem cell markers (such as Sox2 and Axin1), and expressed low levels of differentiation markers, such as GFAP and S-100β (Fig. 7a). To determine whether low levels of endolysosomes play a causal role in determining stem cell state, we inhibited endolysosomal degradation by Lamp1 knockdown and discovered that it significantly enhanced the self-renewal of PP PM-NSCs, as indicated by the enhanced serial sphere-forming ability, particularly with low concentrations of EGF/FGF (P<0.0001 for both assays) (Fig. 7b). Reinforcing the function of endolysosomes in tumorigenesis, we discovered that knockdown of Lamp1 greatly promoted xenograft tumor growth of PP PM-NSCs, whereas the control PP PM-NSCs did not form any tumors during the observation period (over 3 months, Supplementary Fig. 7f). Supporting the idea that restoration of endolysosomes can reverse the low differentiation capability and high self-renewal ability caused by Qki deletion, we found that endolysosome biogenesis induced by ectopic TFE3 in QPP PM-NSCs could significantly decrease the expression of stemness marker Sox2 and serial sphere-forming ability, and could also increase the expression of differentiation marker GFAP in QPP PM-NSCs (Supplementary Fig. 7g).
Figure 7. Qki deletion promotes self-renewal by decreasing endolysosome-dependent receptor degradation.
(a) Immunoblotting of self-renewal and differentiation markers and lysosome markers in PP and QPP PM-NSCs that were cultured with various dilutions of EGF (20ng/ml)/FGF (10ng/ml) mixture (100%, 75%, 50%, 25%, 0%). GF: growth factors. (b) Percentages of PP PM-NSCs with Lamp1 knockdown or control knockdown that can form neurospheres from single cells cultured in either full-strength growth factors (GF) (20ng/ml EGF+10ng/ml FGF) or 1/10 dilution of GF (2ng/ml EGF+1ng/ml FGF). (Student’s t test; *P=0.02; **P<0.0001). (c) Representative images and quantification of IF staining of Notch1 in E14.5 cortex of Nestin-CreERT2QK+/+ and Nestin-CreERT2QKL/L embryos treated with tamoxifen at day E9.5 (n=5/group; Student’s t test; scale bars indicate 100 μm). (d) Immunoblotting of EGFR, Notch1, NICD, and Tgfbr2 in Qki+/+ and QkiL/L NSCs treated with 4OHT or vehicle alone. (e) Representative images and quantification of IF staining of Frizzled in SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice treated with tamoxifen at P1 (n=5/group, 40 cells measured/animal; Student’s t test; scale bars indicate 50 μm). (f) Immunoblotting of Wnt5a/b and β-catenin in Qki-wt and Qki-null NSCs. (g) Representative images of costaining of Lamp1 with EGFR or Fzd4 15 mins after EGF or Wnt5a/b ligand withdrawal in PP and QPP PM-NSCs (scale bars indicate 10 μm). The white bars in the boxes will be analyzed in (h). (h) IF colocalizing profiles (performed by the FV10-ASW 3.0 Reviewer) of the white bars in the square boxes shown in (g). Asterisks indicate colocalization. (i) Quantitative analyses of costaining of speckles and membrane fluorescence signals shown in (g) (n=6; Student’s t test). Error bars indicate s.e.m. in (e) and s.d. in (b) and (i). MZ: marginal zone; CP: cortical plate; SP: subplate; IZ: intermediate zone; SVZ: subventricular zone; VZ: ventricular zone. The experiments shown in all figures were replicated three times in the lab.
We next sought to check whether the receptors that are known to be important for maintaining NSC self-renewal (such as Notch1, Frizzled, and RTKs)31,32 can be enriched by downregulation of endolysosomes caused by Qki deletion. IF staining showed that the VZs and SVZs of the E14.5 Nestin-CreERT2 QkiL/L embryos that were treated with tamoxifen at E9.5 expressed a significantly higher level of Notch1 than those of control Nestin-CreERT2 Qki+/+ embryos (P<0.0001) (Fig. 7c). Consistently, immunoblotting also showed that both Notch1 and cleaved Notch1 (NICD) were greatly upregulated in Qki-null NSCs compared with Qki-wt NSCs in vitro (Fig. 7d). IF staining also revealed that membrane Frizzled levels were much higher in the SVZs of the P12 Nestin-CreERT2 QkiL/L mice that were treated with tamoxifen at P1 compared with control Nestin-CreERT2 Qki+/+ mice (P<0.0001) (Fig. 7e). Immunoblotting showed that autocrine Wnt ligands Wnt5a/b are also upregulated in Qki-null NSCs compared with Qki-wt NSCs, and as a consequence of higher levels of Wnt receptors and ligands, Wnt’s downstream target (activated β-catenin) level is also upregulated in Qki-null NSCs compared with Qki-wt NSCs (Fig. 7f). Similar to cNotch1 and Frizzled, the EGFR protein level in NSCs was also significantly upregulated by Qki deletion in vitro (Fig. 7d). Finally, we found that transient expression of Qki in QPP PM-NSCs could rescue Lamp1 level and downregulate Notch1 and FGFR protein levels (Supplementary Fig. 8a). Supporting the notion that the receptors are enriched by Qki deletion through the skewed degradation rather than transcription and/or translation, costaining of ectopically expressed EGFR and Frizzled with Lamp1 showed that the receptors undergoing endocytosis are greatly downregulated while the receptors remaining on the cytoplasmic membranes are greatly upregulated in Qki-null PM-NSCs (P<0.0001) (Fig. 7g–i).
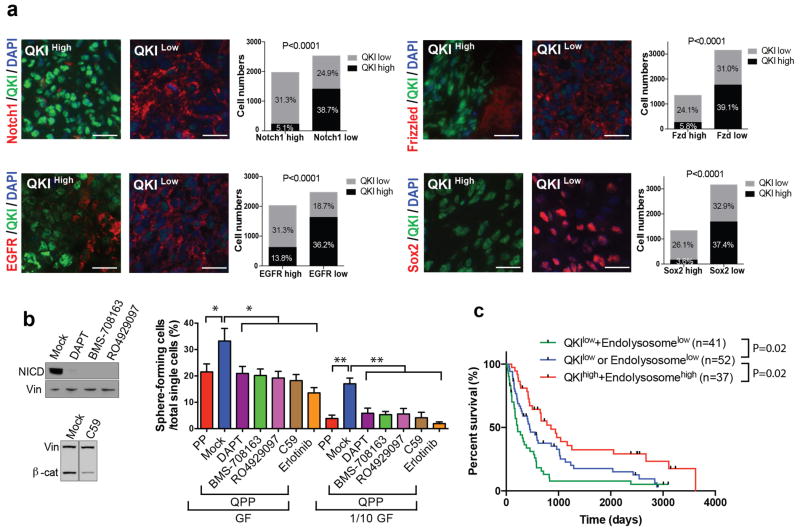
Supporting the idea that decreased receptor degradation caused by low QKI levels also contributes to enhanced receptor levels and GSC populations in human glioblastoma, we found that positivity of QKI staining negatively correlated with the positivity of Frizzled, Notch1, and EGFR as well as stem cell marker Sox2 staining in human glioblastoma tissue microarray (TMA) samples (chi-square test; P<0.0001 for all; Fig. 8a). Reinforcing the notion that Qki deletion enhances self-renewal by downregulating endolysosome-dependent degradation of receptors, we found that pharmacological inhibition of Notch1, Wnt, and EGFR activities could diminish higher self-renewal capacity caused by Qki deletion (Fig. 8b, Supplementary Fig. 8b–c). Finally, to evaluate whether QKI and endolysosome status can predict patient survival, IHC was used to measure the QKI protein levels and endolysosome levels (indicated by punctate Lamp1 staining) in human glioblastoma TMA samples with known patient survival data (Supplementary Table 6). We found that over 63% and 50% of the cancer cells in half of the glioblastoma samples expressed QKI and endolysosome levels below the detection limit, respectively (Supplementary Fig. 8d–g). Consistent with the importance of QKI and endolysosomes in human gliomagenesis, higher percentages of the QKI-low or endolysosome-low cells correlated significantly with poorer prognosis in human glioblastoma patients (Fig. 8c). Overall, the above data demonstrate that loss of QKI enables PM-NSCs to maintain their self-renewal capacity when subjected to low concentrations of ligands (as found in suboptimal environments outside of niches) by downregulating endolysosome-mediated degradation of various receptors (Notch1, Frizzled, RTKs, etc.) that are critical for maintaining stemness.
Figure 8. QKI protein levels negatively correlate with protein levels of self-renewal receptors and patient survivals.
(a) Representative images and quantification of IF costaining of QKI and Notch1, Frizzled, EGFR, or Sox2 in human glioblastoma Tissue Microarray (TMA) (chi-square test; 45 samples/staining; 10 independent fields of 104 μm2 /sample; 10 individual cells/field; P<0.0001 for all; scale bars indicate 20 μm). (b) Percentages of PP PM-NSCs and QPP PM-NSCs treated with 10 μM Notch1 inhibitor (DAPT, BMS-708163, RO4929097), 250 μM Wnt inhibitor (C59), or 1 μM EGFR inhibitor (Erlotinib), which can form neurospheres from single cells cultured in either full-strength growth factors (GF) (20ng/ml EGF+10ng/ml FGF) or 1/10 dilution of GF (2ng/ml EGF+1ng/ml FGF) (n=6/group; Student’s t test; *P<0.0005; **P<0.0003). Immunoblotting of NICD and β-catenin is shown on the left. (c) Kaplan-Meier survival curves of glioblastoma patients based on QKI protein levels and punctate Lamp1 staining levels (log-rank test). The median level was used as the cutoff. Representative QKI-high, QKI-low, endolysosome-high and endolysosome-low samples are shown in Supplementary Fig. 8d–g. The experiments in (b) were replicated three times in the lab.
DISCUSSION
The niche is a special environment where stem cells are maintained in an undifferentiated state and propagated through self-renewal1. As seen for somatic stem cells, numerous studies have identified niche structures for CSCs that are important for maintaining their self-renewal and promoting tumorigenesis6–9. However, these seemingly specialized CSC niches are absent when CSCs invade other areas or tissues by various routes and metastasize to distant organs via the circulation. This raises the question as to how CSCs maintain their self-renewal capability outside of their niches. In this study, we found that although postnatal deletion of Pten and Trp53 could expand the NSC population in the SVZs, it was insufficient to maintain NSC self-renewal outside the SVZs, indicating that the inability of NSCs to maintain self-renewal outside their niches prevents Pten−/−Trp53−/− PM-NSCs from developing into gliomas. We discovered that deletion of Qki in Pten−/−Trp53−/− PM-NSCs enhanced self-renewal, especially outside of their niches, thereby promoting gliomagenesis. Previously, Zheng et al showed that deletion of Pten and Trp53 in embryonic NSCs could enhance sphere-forming ability/stemness by upregulating Myc, leading to the development of both low-grade and high-grade gliomas33, indicating that embryonic NSCs are easier to be transformed than postnatal NSCs. Nevertheless, compared with our postnatal QPP model, in Zheng’s model system (with deletion of Pten and Trp53 in embryonic NSCs) glioblastoma developed with a much longer latency period (105 days vs. 300 days) and a lower penetrance (92% vs. 25%), suggesting that additional genetic/epigenetic alterations still must be acquired to allow embryonic PP PM-NSCs to maintain self-renewal outside their niches, so as to permit full progression of gliomas.
Niches provide various factors that are essential for maintaining stem cell characteristics, among which are extracellular matrix components and ligands for various pathways that are important for self-renewal such as Wnt, Notch, RTKs, Shh, etc.1 Upon ligand activation, receptor-ligand complexes are internalized through endocytosis and are directed to and degraded within the lysosomes. This serves as a major pathway to desensitize the signaling that is mediated by ligand-receptor complexes so that cells can be poised to receive fresh signals for fate determination34. Mutations of cancer genes that uncouple receptors from endocytosis-mediated downregulation have been found to play critical roles in tumorigenesis. One example is c-Cbl, an E3 ubiquitin ligase that conjugates ubiquitin onto the receptors to poise them for endocytosis35, 36. Ying et al discovered that a novel tumor suppressor, Mig-6, recruits EGFR to endocytic vesicles and drives EGFR into lysosome-mediated degradation by binding with a SNARE protein STX837. In our study, we have established the first link between downregulation of endolysosomes and enhanced self-renewal of PM-NSCs and GSCs, which provides a novel mechanism by which PM-NSCs and GSCs cope with suboptimal environments during gliomagenesis.
QKI has long been studied as a gene critical for oligodendrocyte differentiation and myelin formation23,38,39, and our results confirmed that QKI is required for oligodendrocyte differentiation in vivo. QKI has also been shown to be important for the development of smooth muscle, endothelial cells, and monocytes/macrophages23. In addition, we discovered that QKI is also highly expressed in NSCs and is a major regulator of self-renewal and differentiation. QKI has been shown to regulate RNA homeostasis in various aspects: RNA stability, splicing, translation, miRNA processing, and circular RNA biogenesis20–23,39. Here we found that QKI can regulate endocytosis by at least modulating RNA stability and splicing. It is likely that QKI also regulates endocytosis indirectly by affecting miRNA processing and circular RNA biogenesis.
Supporting the importance of QKI’s function as a tumor suppressor, a previous analysis of the TCGA database of glioblastoma defined QKI as the sole gene within the minimal common region of the 6q26 deletions (32% deletion rate and 1.7% mutation rate)40. Furthermore, QKI downregulation by methylation of the QKI locus (chromosome 6, base 163,755,107) was also reported in 50 of 250 (20%) glioblastoma samples20. In angiocentric glioma, nearly 90% of tumors have Myb-QKI translocation, which results in disruption of QKI function41,42. Other than in gliomas, QKI has been shown to be a tumor suppressor in other malignancies, including gastric43, breast44, colon45, prostate46, and oral cancers47.
In conclusion, the downregulation of endolysosome-dependent degradation of receptor-ligand complexes caused by Qki deletion is a novel mechanism for PM-NSCs and GSCs to maintain stemness in suboptimal environments outside their niches. This study may lead to cancer therapeutic opportunities by targeting the mechanisms involved in maintaining CSC stemness, such as receptor trafficking, endocytosis, and autophagy/lysosome-dependent protein degradation.
EXPERIMENTAL PROCEDURES
Mice
Pten-LoxP, Trp53-LoxP, and Ink4a/Arf-LoxP alleles have been described previously 33,48. Nestin-CreERT2 was a gift from Dr. Ryoichiro Kageyama at Kyoto University 25. The Qki-LoxP allele was generated based on the scheme depicted in Supplementary Fig. 1d. Specifically, two LoxP sites were inserted flanking exon2, which will generate an out-of-frame transcript when deleted. All the alleles were maintained in FvB/C57Bl6 hybrid mice interbred in pathogen-free conditions at the facility at The University of Texas M.D. Anderson Cancer Center (M.D. Anderson). The mice were monitored for signs of ill health every other day, and euthanized and necropsied when moribund. Tamoxifen was dissolved in corn oil at the concentration of 10 mg/ml and injected subcutaneously into mice of various ages based on the following doses (where P = postnatal day): P1, 10 μl, one time; P7, 20 μl, two times for two consecutive days; P12–P14, 40 μl, two times for two consecutive days; P21, 80 μl, two times for two consecutive days; P30, 100 μl, two times for two consecutive days. To induce glioma development in the Nestin-CreERT2 QkiL/L PtenL/L Trp53L/L and Nestin-CreERT2 QkiL/L PtenL/L Ink4a/ArfL/L cohorts, tamoxifen was injected subcutaneously into P7-P10 or P30 mice two times for two consecutive days. All manipulations were performed with Institutional Animal Care and Use Committee (IACUC) approval. For the sample sizes of Nestin-CreERT2 QkiL/L PtenL/L Trp53L/L and Nestin-CreERT2 QkiL/L PtenL/L Ink4a/ArfL/L cohorts, because our previous studies showed that Nestin-CreERT2 PtenL/L Trp53L/L and Nestin-CreERT2 PtenL/L Ink4a/ArfL/L cohorts with postnatal tamoxifen treatment never developed brain tumors, we estimated that for a pilot experiment, if Nestin-CreERT2 QkiL/L PtenL/L Trp53L/L and Nestin-CreERT2 QkiL/L PtenL/L Ink4a/ArfL/L animals generated 50% tumor penetrance (which justify good mouse models), 10 mice from each cohort would be enough to generate significant statistical power (P<0.01, with Kaplan-Meier survival analysis) to distinguish the survival times. We kept including more animals in the follow-up studies after the pilot experiments because we needed more tumor tissue for histopathological analyses, molecular profiling, and generating cell lines. There were no randomization and blinding events in the animal studies.
Tumor dissection, sample preparation, histology, and tumor characterization
Mice were overdosed with anesthetic or carbon dioxide, and their brains were removed with or without transcardial perfusion with 4% paraformaldehyde (PFA). The brains were fixed in formalin or post-fixed in 4% PFA at room temperature. Serial sections were prepared at 5 μm thickness for paraffin sections or 8 μm for cryostat sections, and subjected to staining with hematoxylin and eosin or to immunofluorescence (IF)/immunohistochemical (IHC) studies. Histopathological diagnosis of the tumors was made by neuropathologist Dr. Greg Fuller and veterinary pathologist Dr. Mark McArthur from M.D. Anderson.
Antibodies
Antibodies were obtained and used as follows; active beta-catenin (05-665, mouse, 1:1000 for western blot (WB)), MAP2 (AB5622, rabbit, 1:200 for IF), MSI1 (MABE268, mouse, 1:10000 for WB), Nestin (MAB353, mouse, 1:500 for WB), NeuN (MAB377, mouse, 1:200 for IF), Olig2 (MABN50, mouse, 1:100 for IF, IHC), phospho-H3 (06-570, rabbit, 1:200 for IF), Sox2 (AB5603, rabbit, 1:100 for IF, 1:1000 for WB) from EMD Millipore (Billerica, MA), Axin1 (2087S, rabbit, 1:500 for WB), beta-tubulin (2146S, mouse, 1:50000 for WB), cleaved Notch1 (4147S, rabbit, 1:500 for WB), GAPDH (2118S, rabbit, 1:10000 for WB), Notch1 (3608S, rabbit, 1:500 for WB), p62 (5114S, rabbit, 1:1000 for WB), Pten (9559, rabbit, 1:000 for WB), Rab5 (3547P, rabbit, 1:3000 for WB), Rab7 (9367S, rabbit, 1:3000 for WB), TGFbR2 (2518, rabbit), Wnt5a/b (2530P, rabbit, 1:1000 for WB), BIP (3177, rabbit, 1:1000 for WB), GFP (2037, rabbit, 1:500 for IHC and IF), IRE1alpha (3294, rabbit, 1:1000 for WB), PDI (3501, rabbit, 1:1000 for WB) from Cell Signaling Technology (Danvers, MA), beta-actin (A5441, mouse, 1:50000 for WB), vinculin (V9131, mouse, 1:50000 for WB) from Sigma-Aldrich (St. Louis, MO), BLBP (ab32423, mouse, rabbit, 1:200 for IF), BrdU (ab6326, rat, 1:100 for IF), Dcx (ab18723, rabbit, 1:500 for IF), Lamp1 (ab24170, rabbit, 1:5000 for WB), Lamp2 (ab13524, rat, 1:5000 for WB), synaptophysin (ab32127, rabbit, 1:500 for IHC), Iba1 (ab107159, 1:500 for IHC), GFP (ab13970, chicken, 1:200 for IF and IHC), transferrin receptor (ab84036, rabbit, 1:2000 for WB) from Abcam (Cabridge, MA), BrdU (M0744, mouse, 1:500 for IF), GFAP (Z0334, rabbit, 1:1000 for IF, IHC) from DAKO, Agilent Technologies (Carpinteria, CA), FGFR (sc-123, rabbit, 1:1000 for WB, 1:100 for IF), Frizzled (SC-9169, rabbit, 1:100 for IF) and EGFR (SC-03, rabbit 1:1000 for WB) from Santa Cruz Biotechnology (Santa Cruz, CA), GFAP (556330, mouse, 1:200 for IF, IHC), nestin (556309, mouse, 1:100 for IF, IHC, 1:1000 for WB) from Becton, Dickinson and Company, BD Biosciences (San Diego, CA), Id1 (BCH-1/#37-2, rabbit, 1:3000 for WB, 1:200 for IF) from BioCheck (Foster City, CA), Ki67 (VP-RM04, rabbit, 1:200 for IF, IHC) from Vector Laboratories (Burlingame, CA), LC3B (NB100-2331, rabbit, 1:5000 for WB) from Novus Biologicals (Littleton, CO), Mash1 (10R-M106B, mouse) from Fitzgerald Industries (Acton, MA), Mbp (SMI-94R, mouse, 1:1000 for IF, IHC), Tuj1 (MMS-435P-250, mouse, 1:500 for IF, 1:5000 for WB) from Covance (Princeton, NJ), O4 (MAB1326, mouse IgM, 1:200 for IF) from R&D Systems (Minneapolis, MN), Anti-QKI (QK IHC) (IHC-00574, rabbit, 1:500 IHC/IF) from Bethyl lab (Montgomery,TX), CD31 (AC-0083, rabbit, 1:500 for IHC) from Epitomics (Cabridge, MA), S100beta (RB-044-A0, rabbit, 1:300 for IHC) from Labsource/Fisher (Rockford, IL).
Immunohistochemistry
After deparaffinization, antigen retrieval, quenching of endogenous peroxidase, and blocking, brain sections were incubated with primary antibodies for 2 h at room temperature or overnight at 4°C. The sections were incubated with horseradish peroxidase (HRP)-conjugated polymer (Biocare Medical, Concord, CA) for 45 minutes and then diaminobenzidine using the Ultravision DAB Plus Substrate Detection System (Thermo Fischer Scientific, Waltham, MA) for 2–10 minutes at room temperature, followed by hematoxylin staining.
Immunofluorescence
For IF study of tissues, brain sections were treated by deparaffinization and antigen retrieval. For immunocytochemical study, cells on fibronectin-coated cover slips were fixed with 4% PFA and treated with 0.1% Triton X-100. After blocking with serum of goat or horse or bovine serum albumin, the samples were incubated with primary antibodies for 2 h at room temperature or overnight at 4°C. The samples were then incubated with species-appropriate secondary antibodies from goat or donkey coupled to AlexaFluor dyes (488, 555, 568 or 594, Thermo Fischer Scientific) for 45 minutes at room temperature at a dilution of 1:1000. VECTASHIELD with DAPI (Vector Laboratories) was used to mount cover slips. Most IF images (except Fig. 7g–i) were taken with a Nikon Upright Eclipse Ni-E microscope. The confocal images in Fig. 7g–i were taken with an Olympus FluoView FV1000 confocal microscope. The colocalization analyses were performed with a FV10-ASW 3.0 Reviewer.
NSC isolation, neurosphere culture, and self-renewal assay
Mouse NSCs, PM-NSCs, and GSCs were isolated by using Neural Tissue Dissociation Kits (Miltenyi Biotec, San Diego, CA) according to the manufacturer’s instructions. All the cell lines were found to test negative for mycoplasma. Briefly, mouse brains or tumors were cut into small pieces with a scalpel, and then extracellular adhesion proteins were enzymatically degraded. Isolated cells were maintained in NeuroCultTM Basal Medium (Stemcell Technologies, Vancouver, Canada) containing NeuroCultTM Proliferation Supplement (Stemcell Technologies), 20 ng/ml EGF (EMD Millipore), 10 ng/ml basic FGF (EMD Millipore), 50 units/ml penicillin G and 50 μg/ml streptomycin (Thermo Fischer Scientific) in a humidified atmosphere containing 5% CO2 at 37°C. For knockout of Qki, Pten, and Trp53, cells were treated with 100 nM 4-hydroxytamoxifen (Sigma-Aldrich) every two days from two to four times. For immunocytochemical assay, glass cover slips were coated with 2 μg/ml fibronectin (R&D Systems) in PBS overnight, and cells were seeded on the cover slips. For induction of differentiation, cells were cultured with DMEM/F12 (Thermo Fischer Scientific) containing either 2% FBS or 100 ng/ml IGF-1, 60 ng/ml T3 (Thermo Fischer Scientific), and 50 units/ml penicillin G and 50 μg/ml streptomycin. For self-renewal/proliferation assays, NSCs and PM-NSCs were dissociated and seeded in 96-well plates as single cells. The formation of neurospheres was monitored every day. All the neurospheres bigger than 50 μm in diameter were considered as true neurospheres, and the sizes of these spheres were monitored and measured.
Long-term BrdU incorporation assay in vivo
Given that the NSC population in SVZs is relatively quiescent, utilizing the long-term BrdU labeling assay will capture the true NSCs while the transit-amplifying progenitor cells (TAPs) will continue to proliferate and dilute the BrdU. We injected P1 Nestin-CreERT2 Qki+/+ and Nestin-CreERT2 QkiL/L mice with tamoxifen followed by BrdU injection for 1 day (with an interval of 6 hours) and waited for 1 day and for 10 days to check short-term and long-term BrdU retention, respectively. IF staining of BrdU at 1 day after BrdU injection showed that most of the proliferating cells in the SVZs were labeled. Because at this stage cells in the SVZs grow very rapidly, at 10 days, most of the BrdU staining had been diluted out, with only relatively quiescent NSCs still retaining BrdU staining.
Endocytosis and cell fluorescence measurement
The endocytosis assay was performed using pHrodoTM Red EGF conjugates (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cell fluorescence was measured using ImageJ. Briefly, cells on fibronectin-coated 96-well flat bottom plates were kept on ice for 10 min, washed with cold Live Cell Imaging Solution (LCIS, Thermo Fisher Scientific) and treated with 2 μg/ml EGF conjugate for 5 min. Then cells were washed with LCIS and images were obtained using a fluorescence microscope (Leica DMi8). The images were analyzed using ImageJ to measure corrected total cell fluorescence.
RNA isolation
RNA was isolated from mouse brains, tumors, NSCs, PM-NSCs, and GSCs using the RNeasy Mini Kit (Qiagen, Venlo, Netherlands), according to the manufacturer’s instructions.
Protein isolation
Cells were harvested, washed with phosphate buffered saline (PBS), lysed in RIPA buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], Sigma-Aldrich) with a protease inhibitor cocktail tablet cOmplete mini (Roche Diagnostics, Indianapolis, IN), phosphatase inhibitor cocktail 2 (Sigma-Aldrich) and 1 μM DTT, and centrifuged at 10,000 × g at 4°C for 15 min. The supernatant was used as the cell lysate in standard western blotting. Briefly, 20–50 μg of protein were subjected to SDS-PAGE on a 4–12% gradient polyacrylamide gel (NuPage, Thermo Fischer Scientific), transferred to a nitrocellulose membrane (Trans Blot Turbo, Bio-Rad Laboratories, Hercules, CA), and incubated with the indicated primary antibodies followed by incubation with species-appropriate HRP-conjugated secondary antibodies. Bound antibodies were visualized using Super Signal, the enhanced chemiluminescence system (Thermo Fischer Scientific).
Subcutaneous and intracranial cell injections
For s.c. injection, female nude mice (NCr, Taconic) aged 6–8 wks were anesthetized, and 1 million control PP NSCs and Lamp1 knockdown PP NSCs were injected into each flank of the nude mice. Tumor volumes were calculated based on ½(length X width2) of the tumor. For intracranial injections, female SCID mice (Taconic) aged 6–8 wk were anesthetized and placed into a stereotactic apparatus equipped with a z-axis (Stoelting). A small hole was bored in the skull 0.5 mm anterior and 3.0 mm lateral to the bregma using a dental drill; 1 × 105 cells in NeuroCult were injected into the right lateral ventricle using a 10-μl Hamilton syringe with an unbeveled 30-gauge needle. The scalp was closed using a 9-mm Autoclip Applier. Animals were monitored for the development of neurological deficits every other day. All manipulations were performed with IACUC approval.
RNA-sequencing and alternative splicing analyses
Total RNA sequencing was performed by the Illumina Next Generation Sequencing Service at the Sequencing and Microarray Facility of M.D. Anderson. RNA sequence data analyses were performed with the typical RNA data analysis pipeline. We used the Tophat program to align RNA sequence data against the mouse genome mm10 version, allowing two mismatches. After applying the easyRNA R package to extract the raw count tables based on the aligned bam files, we used edgR to perform normalization and differential gene expression analysis. Alternative splicing analyses were performed on the SpliceSeq platform. The detailed protocol is listed on the website http://bioinformatics.mdanderson.org/main/SpliceSeqV2:Methods.
SILAC quantification
Pairs of Qki-wt and Qki-null PM-NSCs were cultured with DMEM:F12 (1:1) medium minus L-Lysine and L-Arginine (Pierce), supplemented with B27, EGF/FGF, regular L-Arginine-HCl and L-Leucine-HCl, or with heavy L-Arginine-HCl (13C6) and L-Leucine-HCl (13C6), for more than six passages. Cells were then lysed with RIPA buffer and mixed 1:1, accurately. The mixed lysates were fractionated by SDS-PAGE until markers had migrated 5 cm toward the anode. The gel was then stained with Coomassie Blue, and cut into five 1-cm pieces. Each piece was further cut into 1-mm cubes, which were combined in one Eppendorf tube and sent to the Proteomics Core Facility at The University of Texas Southwestern Medical School for quantification.
Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP)
The PAR-CLIP assay followed procedures described previously30. In brief, Qki−/−Pten−/−Trp53−/− PM-NSCs were transduced with HA-QKI5. Cells were cultured in 0.5 mM 4-thiouridine for 14 hours followed by crosslinking with 0.6 J/cm2 (around 2 mins) of 365 nm UV light in a Stratalinker 2400 (Stratagene). Crosslinked cells were lysed in NP40 buffer with a Dounce homogenizer and filtered through a 0.2 μm-membrane syringe filter. The cell lysate was treated with RnaseT (1 U/μl) at 22°C. QKI-RNA complexes were immunoprecipitated with anti-HA antibody. Beads were washed and treated with RNaseT1 (100 U/μl) for 15 min at 22 °C and then were dephosphorylated using intestinal alkaline phosphatase (0.5 U/μl, New England Biolabs [NEB]) for 10 min at 37°C. RNAs on the beads were labeled with gamma-32P-ATP to a final concentration of 0.5 μCi/μl and T4 PNK (NEB) to 1 U/μl in one original bead volume for 30 min at 37°C. QKI-RNA complexes on the beads were then denatured for 5 min in a heat block at 95°C and separated on a Novex Bis-Tris 4–12% (Invitrogen) precast polyacrylamide gel. QKI-RNA complexes were then transferred to a nitrocellulose membrane, and bands that corresponded to the expected size (40–50 kDa) were cut out. The bands were treated with Proteinase K for 30 min at 55°C. The RNAs were recovered by acidic phenol/chloroform/IAA extraction (25:24:1, pH 4.0), followed by a chloroform extraction and sent out for illumina RNA-sequencing. qPCR primers for verifying QKI targets are listed in Supplementary Table 7.
Electron microscopy
Cells were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.3, overnight. Samples were processed by Kenneth Dunner Jr. at the High Resolution Electron Microscopy Facility at M.D. Anderson.
IF and IHC analyses on human tissue microarray (TMA)
The TMAs consisted of resected glioma tissues from glioblastoma (n = 130) and normal brain (cortex; n = 19). The human tumor sample collection has been described previously49. In brief, tumor tissues were resected from patients and collected in accordance with the Institutional Review Board of M.D. Anderson. For TMA construction, two 1-mm cores were obtained per tumor sample. The rationale for using a TMA was to facilitate an analysis of the largest number of tumor samples possible. The neuropathologists gathered the tissue sections from the archived paraffin blocks and confirmed the tumor’s pathologic type. For IHC staining of QKI and IF staining of QKI, EGFR, Notch1, Frizzled, and Sox2, the procedures were described in the sections “Immunohistochemistry” and “Immunofluorescence.” For analyzing the costaining of QKI with EGFR, Notch1, Frizzled, and Sox2, 45 individual samples were used. 10 randomly picked independent fields of 104 μm2 were selected for each sample, and 10 individual cells were picked for each field. For individual cells, if the staining was obviously higher than the background, they were labeled as “high”; otherwise, they were labeled as “low.” The correlations between QKI staining and EGFR, Notch1, Frizzled, or Sox2 staining were determined with the chi-square test. P<0.0001 indicates that the costaining of QKI with EGFR, Notch1, Frizzled, or Sox2 is negatively correlated, but not randomly distributed. IHC staining of Lamp1 was used to determine the endolysosome levels. 10 independent fields of 104 μm2 were picked for each sample and 10 individual cells were selected for each field. For individual cells, if the Lamp1 staining exhibited obvious punctate speckles, they were labeled as “endolysosome-high”; otherwise, they were labeled as “endolysosome-low.”
Subtype classification
To predict expression subtypes, we calculated reads per kilobase per million (RPKM) for each gene. We filtered out genes with low expression (RPKM sum less than 1 in the cohort), resulting in 13,806 genes in the data matrix. We used single sample gene sent enrichment analysis (ssGSEA) to calculate an activation score for each subtype gene signature, and the scores were subsequently normalized across the cohort (PMID: 20129251). Subtypes were assigned according to the maximum score.
Statistics
For all statistics, the numbers used to calculate the statistics are indicated in the figure legends. The replicates are either individual cell lines or individual animals. All the t tests are two-tailed and un-paired.
Supplementary Material
Supplementary Figure 1. QKI is expressed in NSCs and inhibits self-renewal of Pten−/−Trp53−/− PM-NSCs
(a) Strategy to identify QKI as a potential tumor suppressor whose deletion might enhance self-renewal of NSCs. (b) Percentage of Pten−/−Trp53−/− (PP) PM-NSCs that are able to form secondary spheres when overexpressed with Qki-6 or mock (control) (n=7/group; Student’s t test). (c) Representative IF image of costaining of Qki and Nestin in the LV-SVZ and TV-SVZ of P7 mice. (d) Scheme of generation of Qki-LoxP conditional knockout allele. (e) Immunoblotting of Qki in NSCs derived from the SVZs of wild type, Nestin-CreERT2Qki+/L, and Nestin-CreERT2QkiL/L mice. (f) Representative images of GFP+Nestin and GFP+GFAP staining of SVZ of P10 Nestin-CreERT2 mTmG mouse that was injected with tamoxifen at P8. Scale bars represent 100 μm. The experiments shown in (b), (c), (e), and (f) were replicated three times in the lab.
Supplementary Figure 2. Qki deletion increases NSC self-renewal capacity and decreases NSC differentiation
(a) Representative images and quantification of IF staining of short-term BrdU (ST-BrdU) and Qki of P2 SVZs in Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice treated with tamoxifen at P0.5 and labeled with BrdU at P1 (Student’s t test, n=7 for each group). (b) Representative images and quantification of IF costaining of Id1 and GFAP of the SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice injected with tamoxifen at P1 (Student’s t test, n=3 for each group). (c) Percentage of apoptotic cells is indicated by the positive cleaved (Cle’d) caspase-3 positive IF staining in SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice in vivo (left panel) and Qki+/+ and Qki−/− NSCs (passage 5) cultured in vitro (right panel) (Student’s t test, n=7 for each group; n.s., no significance). (d) Representative images of Qki-wt and Qki-null NSCs at passage 25 that are stained with Nestin and Qki. (e–f) Representative images and quantification of IHC staining of Mbp and Olig2 of P12 corpus callosum (CC) of Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice treated with tamoxifen at P2 (n=5–6/group; Student’s t test). (g) Representative images and quantification of Tuj1 and Qki IF staining in E14.5 cortex of Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L embryos treated with tamoxifen at day E9.5 (n=5/group; Student’s t test). (h) Representative images and quantification of IF staining of GFAP in E17.5 cortex of Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L embryos treated with tamoxifen at day E9.5 (n=6/group; Student’s t test). Scale bars represent 100 μm in (a), (d), and (e)–(f), and 50 μm in (b). Error bars indicate s.d. CC: corpus callosum; MZ: marginal zone; CP: cortical plate; SP: subplate; IZ: intermediate zone; SVZ: subventricular zone; VZ: ventricular zone. The experiments shown in all figures were replicated three times in the lab.
Supplementary Figure 3. Deletion of Pten and Trp53 expands NSCs in SVZ niches, yet fails to maintain NSC stemness outside the niches
(a) Representative IF images and quantification of Ki67+GFP costaining or Sox2+GFP costaining of the SVZs or thalamus/hypothalamus (TH/HY) in P38 Nestin-CreERT2mTmG and Nestin-CreERT2 mTmG-PP mice injected at P8 (n=6/group; Student’s t test). (b) Representative IF images of Olig2+GFP costaining, NeuN+GFP costaining or S100β+GFP costaining of thalamus/hypothalamus (TH/HY) in P38 Nestin-CreERT2 mTmG and Nestin-CreERT2 mTmG-PP mice injected with tamoxifen at P8. Scale bars represent 100 μm. *P<0.0001; n.s. indicates no significance. Error bars indicate s.d. The experiments shown in all figures were replicated three times in the lab.
Supplementary Figure 4. Qki deletion maintains stemness of PM-NSCs outside of their niches
(a) Representative H&E images of the SCID mouse brains with injection of PP PM-NSCs in lateral ventricle (LV) and cortex (CTX). (b) Representative IF images of Numb staining the SCID mouse brains with injection of PP PM-NSCs in LV and CTX. (c) Representative IF images of EGFR staining the SCID mouse brains with injection of PP PM-NSCs in LV and CTX. (d) Representative IF images of FGFR staining of the SCID mouse brains with injection of PP PM-NSCs in LV and CTX. (e) Representative IF image of Ki67+Sox2+GFP costaining of TH/HY in P38 Nestin-CreERT2 mTmG-QPP mice that were injected at P8. (f) Representative IF images and quantification of Mbp+GFP costaining of thalamus/hypothalamus (TH/HY) in P38 Nestin-CreERT2 mTmG-PP and Nestin-CreERT2 mTmG-QPP mice injected with tamoxifen at P8 (n=5/group; Student’s t test). (g) Representative IHC images of Qki staining of P38 Nestin-CreERT2-PP and Nestin-CreERT2-QPP mice injected with tamoxifen at P8. (h) Representative IHC images of Pten staining of P38 Nestin-CreERT2-PP and Nestin-CreERT2-QPP mice injected with tamoxifen at P8. (i) Immunoblotting of Qki and Pten in neurosphere cultures derived from SVZs of Nestin-CreERT2 Qki+/+, Nestin-CreERT2 QkiL/L, Nestin-CreERT2-PP, and Nestin-CreERT2-QPP mice. (i) PCR of Trp53 locus in neurosphere cultures derived from SVZs of Nestin-CreERT2 QkiL/L, Nestin-CreERT2-PP, and Nestin-CreERT2-QPP mice. Scale bars represent 100 μm in (a), (b), (g) and (h) and 50 μm in (c)-(f). Error bars indicate s.d. The experiments in all figures were replicated three times in the lab.
Supplementary Figure 5. Nestin-CreERT2mTmG QkiL/LPtenL/LTrp53L/L (QPP) gliomas recapitulate human glioblastoma
(a–b) Representative IHC staining of Notch1 (a) and Nestin (b) in QPP gliomas. (c) Representative QPP glioma H&E images of: a tumor consisting of spindle-shape cells with oval or round nuclei with dense chromatin (upper left); a tumor with abundant multinucleated giant cells (upper right); a tumor consisting of cells with eosinophilic fibrillary cytoplasm and round or polygonal nuclei with prominent nucleoli (lower left); and a tumor with structures reminiscent of Homer-Wright rosettes (lower right). Scale bars represent 100 μm. (d) Kaplan-Meier survival curves (log-rank test) of Nestin-CreERT2 QkiL/L, Nestin-CreERT2 PtenL/LTrp53L/L (PP) and Nestin-CreERT2 QkiL/LPtenL/LTrp53L/L (QPP) mice treated with tamoxifen at P30. P<0.0001 means that the differences between Nestin-CreERT2 QkiL/LPtenL/LTrp53L/L and any other cohorts are significant. (e) Kaplan-Meier survival curves (log-rank test) of Nestin-CreERT2 PtenL/LInk4a/ArfL/L and Nestin-CreERT2 QkiL/LPtenL/LInk4a/ArfL/L mice treated with tamoxifen at P8.
Supplementary Figure 6. QKI is a novel regulator of endolysosomes
(a) Scheme of stable isotope labeling by amino acids in cell culture (SILAC)-based proteomic analysis. LC-MS/MS: liquid chromatography-tandem mass spectrometry; m/z: mass-to-charge ratio. (b) Top pathways (P<0.001) that are enriched by Qki deletion as detected by Ingenuity Pathway Analysis (IPA).
Supplementary Figure 7. Qki deletion promotes self-renewal by decreasing endolysosome-dependent receptor degradation
(a) Immunoblotting of QKI and Lamp1 in human GSCs with shRNAs against QKI and control shRNA. (b) Representative images and quantification of IF costaining of long-term BrdU and Lamp1 in the SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice treated with tamoxifen at P1 (n=3/group; Student’s t test). (c) Immunoblotting of transferrin receptor (TfR) in the Qki+/+ and Qki−/− NSCs and in PP and QPP PM-NSCs. (d) Endocytosis of EGFR was measured by the low pH-sensitive Red EGF probe in the Qki+/+ and Qki−/− NSCs and in PP and QPP PM-NSCs after EGF depletion for 1 hour. (Student’s t test; n=83 for Qki+/+ NSCs, n=93 for Qki−/− NSCs, n=89 for PP PM-NSCs, and n=110 for QPP PM-NSCs). (e) Expression correlation between QKI and CLTC, Rab5C, Rab28, SNX2, SNX3 and GPHN in the TCGA microarray dataset (n=543, Pearson correlation). (f) Subcutaneous xenograft tumor formation of PP PM-NSCs with Lamp1 knockdown (18 individual animals) or control knockdown (9 individual animals). Tumor volumes (mm3) were calculated based on ½(length X width2). (g) Left panel: immunoblotting of Lamp1, Sox2, and GFAP in QPP PM-NSCs with TFE3 overexpression or mock (control). Right panel: percentages of QPP PM-NSCs with TFE3 overexpression or mock that can form neurospheres from single cells cultured in either full-strength growth factors (GF) or 1/10 dilution of GF. (Student’s t test; *P<0.0001). Scale bar represents 50 μm in (b) and 20 μm in (d). Error bars indicate s.d. in (b) and (g) and s.e.m in (d). The experiments in (a), (b), (d) and (g) were replicated three times in the lab.
Supplementary Figure 8. QKI level inversely correlates with receptor levels and patient survival
(a) Immunoblotting of Lamp1, Notch1, and FGFR in QPP PM-NSCs after overexpression of QKI-6. (b) Immunoblotting of self-renewal and differentiation markers in Pten−/−Trp53−/− (PP) and Qki−/−Pten−/−Trp53−/− (QPP) PM-NSCs treated with EGFR inhibitor. (c) Percentages of PP PM-NSCs treated with 10 μM Notch1 inhibitor (DAPT, BMS-708163, RO4929097), 250 μM Wnt inhibitor (C59), and 1 μM EGFR inhibitor (Erlotinib), which can form neurospheres from single cells cultured in either full-strength growth factors (GF) (20ng/ml EGF+10ng/ml FGF) or 1/10 dilution of GF (2ng/ml EGF+1ng/ml FGF). (Student’s t test; *P<0.001; **P<0.01). (d) Human TMA glioblastoma samples were plotted according to the percentages of QKI-positive cells that were detected by IHC. (e) Representative IHC images of a normal human brain sample and human glioblastoma samples with high and low percentages of QKI-positive cells. (f) Human TMA glioblastoma samples were plotted according to the percentages of punctate Lamp1-positive cells that were detected by IHC. (g) Representative IHC images of a normal human brain sample and human glioblastoma samples with high and low percentages of punctate Lamp1-positive cells. White scale bars indicate 100 μm; black scale bars indicate 50 μm. The experiments in (a), (b), and (c) were replicated three times in the lab.
Acknowledgments
We thank Shan Jiang, Zhuangna Fang, and Kun Zhao for mouse husbandry and care, and all members of the Hu laboratory for helpful discussions. We thank Yuan Qi for performing alternative splicing analyses. We thank Konrad Ryszard Gabrusiewicz for helping us analyze the glioblastoma TMA. We thank Kenneth Dunner for EM studies. We also thank David M. Wildrick for editorial assistance. J.H. is supported by NIH K99/R00 Pathway to Independence Award (R00 CA172700), NCI Brain Cancer SPORE Career Development Award (2P50CA127001), and Sidney Kimmel Scholar Award. A.H. was supported by the HHMI Medical Research Fellows Program. A.B.H. is supported by NIH R01 (CA120813).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Data availability
All the dataset have been deposited in the Gene Expression Omnibus (GEO) under accession GSE84134. Series record GSE84134 provides access to all of our data and is the accession that should be quoted in any manuscript discussing the data.
Contributions
J.H and T.S. designed the study. T.S., A.L.H., L.Y., X.Z., C.D. and B.D.L. performed the experiments. S.Z., Q.W., Y.Z., Y.C. and R.G.W.V. performed bioinformatics analysis. G.N.F. provided pathological analyses for the QPP gliomas. Q.C. helped to analyze TMA samples. J.W.H., Y.Y., B.H. and A.B.H. contributed animal and clinical samples.
References
- 1.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 2.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suh H, Deng W, Gage FH. Signaling in adult neurogenesis. Annu Rev Cell Dev Biol. 2009;25:253–75. doi: 10.1146/annurev.cellbio.042308.113256. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–78. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 5.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–9. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 6.Calabrese C, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood. 2009;114:1150–7. doi: 10.1182/blood-2009-01-202606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merlos-Suarez A, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–24. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Neaves WB. Normal stem cells and cancer stem cells: the niche matters. Cancer research. 2006;66:4553–7. doi: 10.1158/0008-5472.CAN-05-3986. [DOI] [PubMed] [Google Scholar]
- 11.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 12.Dunn GP, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–84. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rich JN, Eyler CE. Cancer stem cells in brain tumor biology. Cold Spring Harbor symposia on quantitative biology. 2008;73:411–20. doi: 10.1101/sqb.2008.73.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nature reviews Cancer. 2006;6:425–36. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 16.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–36. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 17.Scherer HJ. The forms of growth in gliomas and their practical significance. Brain. 1940;63:1–35. [Google Scholar]
- 18.Scherer HJ. Structural development in gliomas. Am J Cancer. 1938:333–351. [Google Scholar]
- 19.Hu J, et al. From the Cover: Neutralization of terminal differentiation in gliomagenesis. Proc Natl Acad Sci U S A. 2013;110:14520–7. doi: 10.1073/pnas.1308610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen AJ, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26:1459–72. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Vogel G, Yu Z, Richard S. The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells. Mol Cell Biol. 2013;33:1233–43. doi: 10.1128/MCB.01604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conn SJ, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Darbelli L, Richard S. Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip Rev RNA. 2016;7:399–412. doi: 10.1002/wrna.1344. [DOI] [PubMed] [Google Scholar]
- 24.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 25.Imayoshi I, et al. Roles of continuous neurogenesis in the structural and functional integrity of the adult forebrain. Nat Neurosci. 2008;11:1153–61. doi: 10.1038/nn.2185. [DOI] [PubMed] [Google Scholar]
- 26.Holland EC, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–7. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 27.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacques TS, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29:222–35. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–17. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–33. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 35.Levkowitz G, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–40. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 36.Levkowitz G, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–74. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying H, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6912–7. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Tian D, Ku L, Osterhout DJ, Feng Y. The selective RNA-binding protein quaking I (QKI) is necessary and sufficient for promoting oligodendroglia differentiation. J Biol Chem. 2007;282:23553–60. doi: 10.1074/jbc.M702045200. [DOI] [PubMed] [Google Scholar]
- 39.Chenard CA, Richard S. New implications for the QUAKING RNA binding protein in human disease. J Neurosci Res. 2008;86:233–42. doi: 10.1002/jnr.21485. [DOI] [PubMed] [Google Scholar]
- 40.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qaddoumi I, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016 doi: 10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandopadhayay P, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016;48:273–82. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bian Y, et al. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem Biophys Res Commun. 2012;422:187–93. doi: 10.1016/j.bbrc.2012.04.138. [DOI] [PubMed] [Google Scholar]
- 44.Yu F, et al. Post-transcriptional repression of FOXO1 by QKI results in low levels of FOXO1 expression in breast cancer cells. Oncol Rep. 2014;31:1459–65. doi: 10.3892/or.2013.2957. [DOI] [PubMed] [Google Scholar]
- 45.Yang G, et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology. 2010;138:231–40. e1–5. doi: 10.1053/j.gastro.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, et al. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic and prognostic protein. Cancer Biol Ther. 2014;15:108–18. doi: 10.4161/cbt.26722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu W, et al. QKI impairs self-renewal and tumorigenicity of oral cancer cells via repression of SOX2. Cancer Biol Ther. 2014;15:1174–84. doi: 10.4161/cbt.29502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhat KP, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–46. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. QKI is expressed in NSCs and inhibits self-renewal of Pten−/−Trp53−/− PM-NSCs
(a) Strategy to identify QKI as a potential tumor suppressor whose deletion might enhance self-renewal of NSCs. (b) Percentage of Pten−/−Trp53−/− (PP) PM-NSCs that are able to form secondary spheres when overexpressed with Qki-6 or mock (control) (n=7/group; Student’s t test). (c) Representative IF image of costaining of Qki and Nestin in the LV-SVZ and TV-SVZ of P7 mice. (d) Scheme of generation of Qki-LoxP conditional knockout allele. (e) Immunoblotting of Qki in NSCs derived from the SVZs of wild type, Nestin-CreERT2Qki+/L, and Nestin-CreERT2QkiL/L mice. (f) Representative images of GFP+Nestin and GFP+GFAP staining of SVZ of P10 Nestin-CreERT2 mTmG mouse that was injected with tamoxifen at P8. Scale bars represent 100 μm. The experiments shown in (b), (c), (e), and (f) were replicated three times in the lab.
Supplementary Figure 2. Qki deletion increases NSC self-renewal capacity and decreases NSC differentiation
(a) Representative images and quantification of IF staining of short-term BrdU (ST-BrdU) and Qki of P2 SVZs in Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice treated with tamoxifen at P0.5 and labeled with BrdU at P1 (Student’s t test, n=7 for each group). (b) Representative images and quantification of IF costaining of Id1 and GFAP of the SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice injected with tamoxifen at P1 (Student’s t test, n=3 for each group). (c) Percentage of apoptotic cells is indicated by the positive cleaved (Cle’d) caspase-3 positive IF staining in SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice in vivo (left panel) and Qki+/+ and Qki−/− NSCs (passage 5) cultured in vitro (right panel) (Student’s t test, n=7 for each group; n.s., no significance). (d) Representative images of Qki-wt and Qki-null NSCs at passage 25 that are stained with Nestin and Qki. (e–f) Representative images and quantification of IHC staining of Mbp and Olig2 of P12 corpus callosum (CC) of Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice treated with tamoxifen at P2 (n=5–6/group; Student’s t test). (g) Representative images and quantification of Tuj1 and Qki IF staining in E14.5 cortex of Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L embryos treated with tamoxifen at day E9.5 (n=5/group; Student’s t test). (h) Representative images and quantification of IF staining of GFAP in E17.5 cortex of Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L embryos treated with tamoxifen at day E9.5 (n=6/group; Student’s t test). Scale bars represent 100 μm in (a), (d), and (e)–(f), and 50 μm in (b). Error bars indicate s.d. CC: corpus callosum; MZ: marginal zone; CP: cortical plate; SP: subplate; IZ: intermediate zone; SVZ: subventricular zone; VZ: ventricular zone. The experiments shown in all figures were replicated three times in the lab.
Supplementary Figure 3. Deletion of Pten and Trp53 expands NSCs in SVZ niches, yet fails to maintain NSC stemness outside the niches
(a) Representative IF images and quantification of Ki67+GFP costaining or Sox2+GFP costaining of the SVZs or thalamus/hypothalamus (TH/HY) in P38 Nestin-CreERT2mTmG and Nestin-CreERT2 mTmG-PP mice injected at P8 (n=6/group; Student’s t test). (b) Representative IF images of Olig2+GFP costaining, NeuN+GFP costaining or S100β+GFP costaining of thalamus/hypothalamus (TH/HY) in P38 Nestin-CreERT2 mTmG and Nestin-CreERT2 mTmG-PP mice injected with tamoxifen at P8. Scale bars represent 100 μm. *P<0.0001; n.s. indicates no significance. Error bars indicate s.d. The experiments shown in all figures were replicated three times in the lab.
Supplementary Figure 4. Qki deletion maintains stemness of PM-NSCs outside of their niches
(a) Representative H&E images of the SCID mouse brains with injection of PP PM-NSCs in lateral ventricle (LV) and cortex (CTX). (b) Representative IF images of Numb staining the SCID mouse brains with injection of PP PM-NSCs in LV and CTX. (c) Representative IF images of EGFR staining the SCID mouse brains with injection of PP PM-NSCs in LV and CTX. (d) Representative IF images of FGFR staining of the SCID mouse brains with injection of PP PM-NSCs in LV and CTX. (e) Representative IF image of Ki67+Sox2+GFP costaining of TH/HY in P38 Nestin-CreERT2 mTmG-QPP mice that were injected at P8. (f) Representative IF images and quantification of Mbp+GFP costaining of thalamus/hypothalamus (TH/HY) in P38 Nestin-CreERT2 mTmG-PP and Nestin-CreERT2 mTmG-QPP mice injected with tamoxifen at P8 (n=5/group; Student’s t test). (g) Representative IHC images of Qki staining of P38 Nestin-CreERT2-PP and Nestin-CreERT2-QPP mice injected with tamoxifen at P8. (h) Representative IHC images of Pten staining of P38 Nestin-CreERT2-PP and Nestin-CreERT2-QPP mice injected with tamoxifen at P8. (i) Immunoblotting of Qki and Pten in neurosphere cultures derived from SVZs of Nestin-CreERT2 Qki+/+, Nestin-CreERT2 QkiL/L, Nestin-CreERT2-PP, and Nestin-CreERT2-QPP mice. (i) PCR of Trp53 locus in neurosphere cultures derived from SVZs of Nestin-CreERT2 QkiL/L, Nestin-CreERT2-PP, and Nestin-CreERT2-QPP mice. Scale bars represent 100 μm in (a), (b), (g) and (h) and 50 μm in (c)-(f). Error bars indicate s.d. The experiments in all figures were replicated three times in the lab.
Supplementary Figure 5. Nestin-CreERT2mTmG QkiL/LPtenL/LTrp53L/L (QPP) gliomas recapitulate human glioblastoma
(a–b) Representative IHC staining of Notch1 (a) and Nestin (b) in QPP gliomas. (c) Representative QPP glioma H&E images of: a tumor consisting of spindle-shape cells with oval or round nuclei with dense chromatin (upper left); a tumor with abundant multinucleated giant cells (upper right); a tumor consisting of cells with eosinophilic fibrillary cytoplasm and round or polygonal nuclei with prominent nucleoli (lower left); and a tumor with structures reminiscent of Homer-Wright rosettes (lower right). Scale bars represent 100 μm. (d) Kaplan-Meier survival curves (log-rank test) of Nestin-CreERT2 QkiL/L, Nestin-CreERT2 PtenL/LTrp53L/L (PP) and Nestin-CreERT2 QkiL/LPtenL/LTrp53L/L (QPP) mice treated with tamoxifen at P30. P<0.0001 means that the differences between Nestin-CreERT2 QkiL/LPtenL/LTrp53L/L and any other cohorts are significant. (e) Kaplan-Meier survival curves (log-rank test) of Nestin-CreERT2 PtenL/LInk4a/ArfL/L and Nestin-CreERT2 QkiL/LPtenL/LInk4a/ArfL/L mice treated with tamoxifen at P8.
Supplementary Figure 6. QKI is a novel regulator of endolysosomes
(a) Scheme of stable isotope labeling by amino acids in cell culture (SILAC)-based proteomic analysis. LC-MS/MS: liquid chromatography-tandem mass spectrometry; m/z: mass-to-charge ratio. (b) Top pathways (P<0.001) that are enriched by Qki deletion as detected by Ingenuity Pathway Analysis (IPA).
Supplementary Figure 7. Qki deletion promotes self-renewal by decreasing endolysosome-dependent receptor degradation
(a) Immunoblotting of QKI and Lamp1 in human GSCs with shRNAs against QKI and control shRNA. (b) Representative images and quantification of IF costaining of long-term BrdU and Lamp1 in the SVZs of P12 Nestin-CreERT2Qki+/+ and Nestin-CreERT2QkiL/L mice treated with tamoxifen at P1 (n=3/group; Student’s t test). (c) Immunoblotting of transferrin receptor (TfR) in the Qki+/+ and Qki−/− NSCs and in PP and QPP PM-NSCs. (d) Endocytosis of EGFR was measured by the low pH-sensitive Red EGF probe in the Qki+/+ and Qki−/− NSCs and in PP and QPP PM-NSCs after EGF depletion for 1 hour. (Student’s t test; n=83 for Qki+/+ NSCs, n=93 for Qki−/− NSCs, n=89 for PP PM-NSCs, and n=110 for QPP PM-NSCs). (e) Expression correlation between QKI and CLTC, Rab5C, Rab28, SNX2, SNX3 and GPHN in the TCGA microarray dataset (n=543, Pearson correlation). (f) Subcutaneous xenograft tumor formation of PP PM-NSCs with Lamp1 knockdown (18 individual animals) or control knockdown (9 individual animals). Tumor volumes (mm3) were calculated based on ½(length X width2). (g) Left panel: immunoblotting of Lamp1, Sox2, and GFAP in QPP PM-NSCs with TFE3 overexpression or mock (control). Right panel: percentages of QPP PM-NSCs with TFE3 overexpression or mock that can form neurospheres from single cells cultured in either full-strength growth factors (GF) or 1/10 dilution of GF. (Student’s t test; *P<0.0001). Scale bar represents 50 μm in (b) and 20 μm in (d). Error bars indicate s.d. in (b) and (g) and s.e.m in (d). The experiments in (a), (b), (d) and (g) were replicated three times in the lab.
Supplementary Figure 8. QKI level inversely correlates with receptor levels and patient survival
(a) Immunoblotting of Lamp1, Notch1, and FGFR in QPP PM-NSCs after overexpression of QKI-6. (b) Immunoblotting of self-renewal and differentiation markers in Pten−/−Trp53−/− (PP) and Qki−/−Pten−/−Trp53−/− (QPP) PM-NSCs treated with EGFR inhibitor. (c) Percentages of PP PM-NSCs treated with 10 μM Notch1 inhibitor (DAPT, BMS-708163, RO4929097), 250 μM Wnt inhibitor (C59), and 1 μM EGFR inhibitor (Erlotinib), which can form neurospheres from single cells cultured in either full-strength growth factors (GF) (20ng/ml EGF+10ng/ml FGF) or 1/10 dilution of GF (2ng/ml EGF+1ng/ml FGF). (Student’s t test; *P<0.001; **P<0.01). (d) Human TMA glioblastoma samples were plotted according to the percentages of QKI-positive cells that were detected by IHC. (e) Representative IHC images of a normal human brain sample and human glioblastoma samples with high and low percentages of QKI-positive cells. (f) Human TMA glioblastoma samples were plotted according to the percentages of punctate Lamp1-positive cells that were detected by IHC. (g) Representative IHC images of a normal human brain sample and human glioblastoma samples with high and low percentages of punctate Lamp1-positive cells. White scale bars indicate 100 μm; black scale bars indicate 50 μm. The experiments in (a), (b), and (c) were replicated three times in the lab.