Abstract
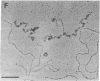
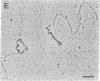
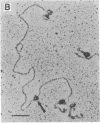
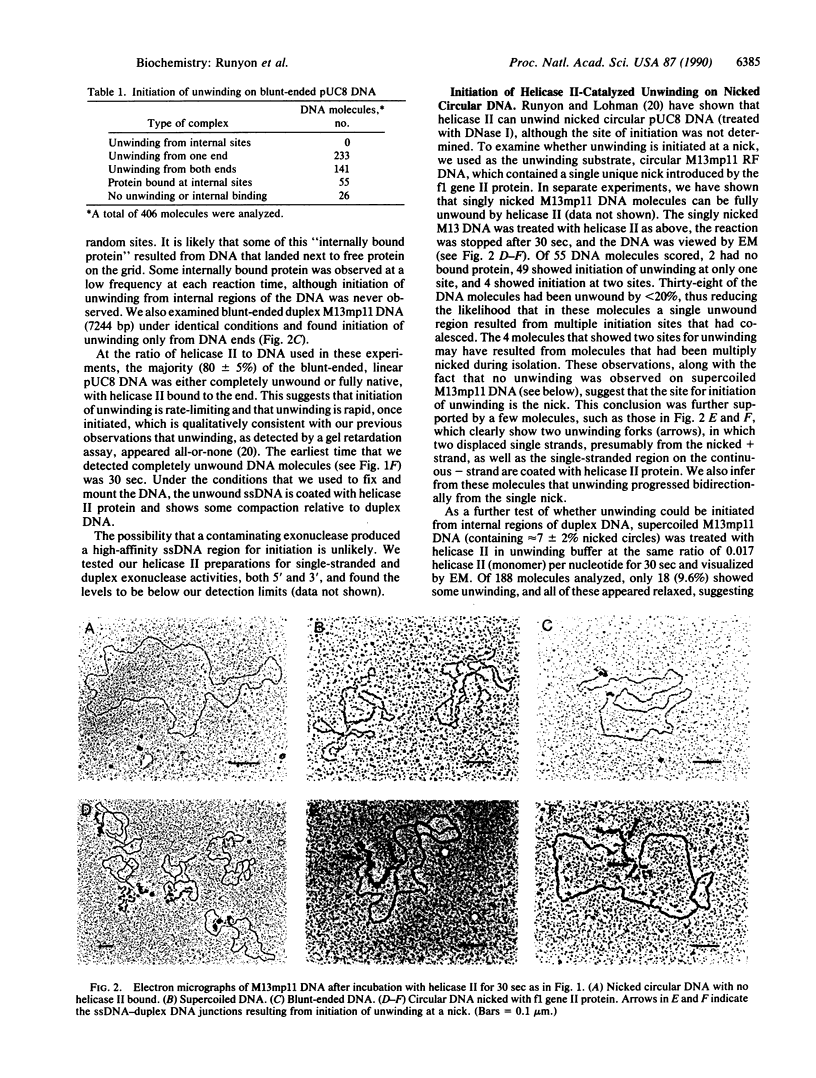
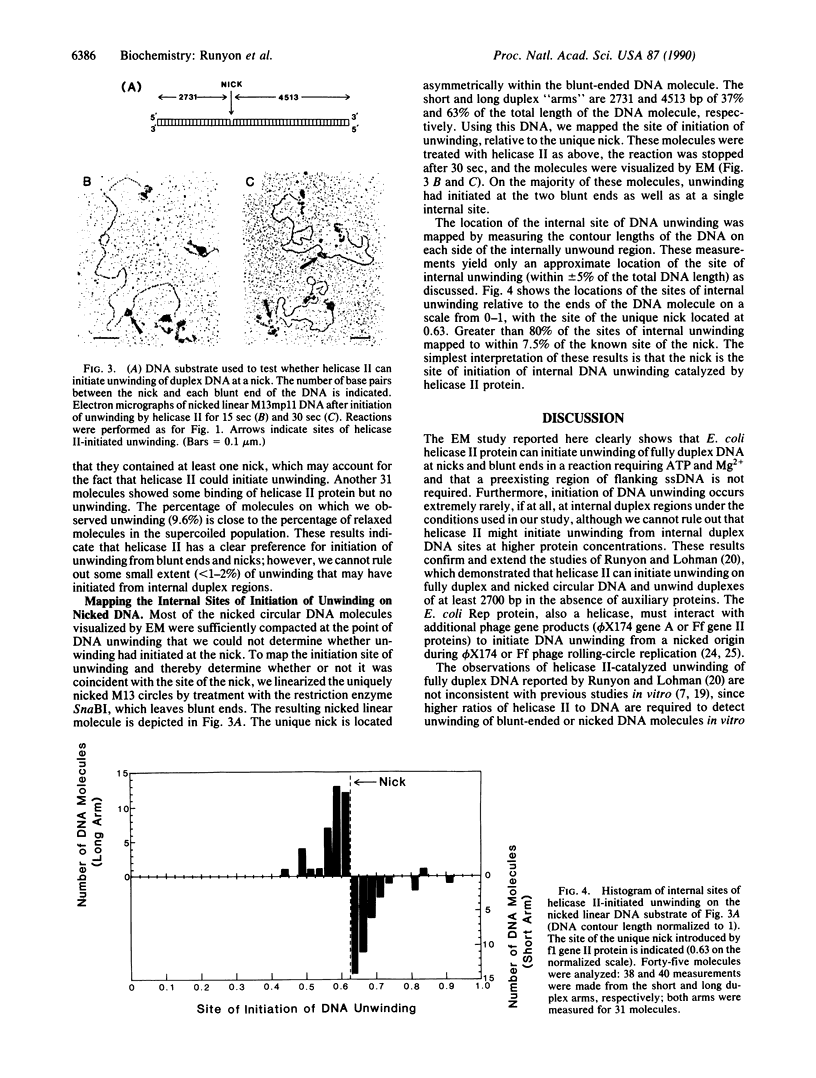
The Escherichia coli uvrD gene product, helicase II, is required for both methyl-directed mismatch and uvrABC excision repair and is believed to function by unwinding duplex DNA. Initiation of unwinding may occur specifically at either a mismatch or a nick, although no direct evidence for this has previously been reported. It has recently been shown that helicase II can unwind fully duplex linear and nicked circular DNA with lengths of at least approximately 2700 base pairs in vitro; hence, a flanking region of single-stranded DNA is not required to initiate DNA unwinding. In studies with uniquely nicked duplex DNA, we present EM evidence that helicase II protein initiates DNA unwinding at the nick, with unwinding proceeding bidirectionally. We also show that helicase II protein initiates DNA unwinding at the blunt ends of linear DNA, rather than in internal regions. These data provide direct evidence that helicase II protein can initiate unwinding of duplex DNA at a nick, in the absence of auxiliary proteins. We propose that helicase II may initiate unwinding from a nick in a number of DNA repair processes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Monem M., Dürwald H., Hoffmann-Berling H. DNA unwinding enzyme II of Escherichia coli. 2. Characterization of the DNA unwinding activity. Eur J Biochem. 1977 Sep 15;79(1):39–45. doi: 10.1111/j.1432-1033.1977.tb11781.x. [DOI] [PubMed] [Google Scholar]
- Arthur H. M., Eastlake P. B. Transcriptional control of the uvrD gene of Escherichia coli. Gene. 1983 Nov;25(2-3):309–316. doi: 10.1016/0378-1119(83)90235-4. [DOI] [PubMed] [Google Scholar]
- Arthur H. M., Lloyd R. G. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet. 1980;180(1):185–191. doi: 10.1007/BF00267368. [DOI] [PubMed] [Google Scholar]
- Caron P. R., Kushner S. R., Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider K., Bäumel I., Meyer T. F. Intermediate stages in enzymatic replication of bacteriophage fd duplex DNA. J Biol Chem. 1982 Jun 10;257(11):6488–6493. [PubMed] [Google Scholar]
- Geider K., Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- Greenstein D., Horiuchi K. Interaction between the replication origin and the initiator protein of the filamentous phage f1. Binding occurs in two steps. J Mol Biol. 1987 Sep 20;197(2):157–174. doi: 10.1016/0022-2836(87)90115-x. [DOI] [PubMed] [Google Scholar]
- Griffith J., Formosa T. The uvsX protein of bacteriophage T4 arranges single-stranded and double-stranded DNA into similar helical nucleoprotein filaments. J Biol Chem. 1985 Apr 10;260(7):4484–4491. [PubMed] [Google Scholar]
- Grossman L., Caron P. R., Mazur S. J., Oh E. Y. Repair of DNA-containing pyrimidine dimers. FASEB J. 1988 Aug;2(11):2696–2701. doi: 10.1096/fasebj.2.11.3294078. [DOI] [PubMed] [Google Scholar]
- Hickson I. D., Arthur H. M., Bramhill D., Emmerson P. T. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190(2):265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., Bardwell E. Effects of recB21, recF143, and uvrD152 on recombination in lambda bacteriophage-prophage and Hfr by F- crosses. J Bacteriol. 1981 Nov;148(2):739–743. doi: 10.1128/jb.148.2.739-743.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert M. Q., Klein A., Abdel-Monem M. Studies on the functions of DNA helicase I and DNA helicase II of Escherichia coli. J Biol Chem. 1980 Oct 25;255(20):9746–9752. [PubMed] [Google Scholar]
- Kuhn B., Abdel-Monem M., Krell H., Hoffmann-Berling H. Evidence for two mechanisms for DNA unwinding catalyzed by DNA helicases. J Biol Chem. 1979 Nov 25;254(22):11343–11350. [PubMed] [Google Scholar]
- Kumura K., Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J Biol Chem. 1984 Feb 10;259(3):1560–1565. [PubMed] [Google Scholar]
- Kumura K., Sekiguchi M., Steinum A. L., Seeberg E. Stimulation of the UvrABC enzyme-catalyzed repair reactions by the UvrD protein (DNA helicase II). Nucleic Acids Res. 1985 Mar 11;13(5):1483–1492. doi: 10.1093/nar/13.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue R. S., Au K. G., Modrich P. DNA mismatch correction in a defined system. Science. 1989 Jul 14;245(4914):160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G. lexA dependent recombination in uvrD strains of Escherichia coli. Mol Gen Genet. 1983;189(1):157–161. doi: 10.1007/BF00326069. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längle-Rouault F., Maenhaut-Michel G., Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 1987 Apr;6(4):1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples V. F., Kushner S. R. DNA repair in Escherichia coli: identification of the uvrD gene product. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5616–5620. doi: 10.1073/pnas.79.18.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson S. W. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3' to 5' direction. J Biol Chem. 1986 Aug 5;261(22):10169–10175. [PubMed] [Google Scholar]
- Matson S. W., George J. W. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987 Feb 15;262(5):2066–2076. [PubMed] [Google Scholar]
- Matson S. W., Kaiser-Rogers K. A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Geider K. Bacteriophage fd gene II-protein. II. Specific cleavage and relaxation of supercoiled RF from filamentous phages. J Biol Chem. 1979 Dec 25;254(24):12642–12646. [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Myles G. M., Sancar A. DNA repair. Chem Res Toxicol. 1989 Jul-Aug;2(4):197–226. doi: 10.1021/tx00010a001. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Irino N., Nakayama H. recA+ gene-dependent regulation of a uvrD::lacZ fusion in Escherichia coli K12. Mol Gen Genet. 1983;192(3):391–394. doi: 10.1007/BF00392180. [DOI] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Runyon G. T., Lohman T. M. Escherichia coli helicase II (uvrD) protein can completely unwind fully duplex linear and nicked circular DNA. J Biol Chem. 1989 Oct 15;264(29):17502–17512. [PubMed] [Google Scholar]
- Siegel E. C. The Escherichia coli uvrD gene is inducible by DNA damage. Mol Gen Genet. 1983;191(3):397–400. doi: 10.1007/BF00425753. [DOI] [PubMed] [Google Scholar]
- Su S. S., Grilley M., Thresher R., Griffith J., Modrich P. Gap formation is associated with methyl-directed mismatch correction under conditions of restricted DNA synthesis. Genome. 1989;31(1):104–111. doi: 10.1139/g89-020. [DOI] [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taucher-Scholz G., Hoffmann-Berling H. Identification of the gene for DNA helicase II of Escherichia coli. Eur J Biochem. 1983 Dec 15;137(3):573–580. doi: 10.1111/j.1432-1033.1983.tb07864.x. [DOI] [PubMed] [Google Scholar]
- Welsh K. M., Lu A. L., Clark S., Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987 Nov 15;262(32):15624–15629. [PubMed] [Google Scholar]