Abstract
Many benefits provided by the gut microbiota to the host rely on its intricate interactions with host cells. Perturbations of the gut microbiota, termed gut dysbiosis, affect the interplay between the gut microbiota and host cells, resulting in dysregulation of inflammation that contributes to the pathogenesis of chronic inflammatory diseases, including inflammatory bowel disease, multiple sclerosis, allergic asthma and rheumatoid arthritis. In this review, we provide an overview of how gut bacteria modulates host metabolic and immune functions, summarize studies that examined the roles of gut dysbiosis in chronic inflammatory diseases, and finally discuss measures to correct gut dysbiosis as potential therapeutics for chronic inflammatory diseases.
Introduction
The term ‘microbiota' has been used to define the community of symbiotic microbes including bacteria, archaea, viruses, fungi and protozoa that have established their own niches within a host, and provide supporting roles in host immunity and metabolism.1, 2 It was recently revised that a typical human adult harbors about 1013–1014 of bacteria, which is the same order as the number of total human somatic and germ cells.3 The intestinal tract is the largest reservoir of the microbiota in the human body.3 The intestinal microbiota typically consists of more than 1000 bacterial species whose composition remains relatively stable in each individual throughout adulthood but tremendously diverse between individuals.4 However, several catastrophic changes, such as use of antibiotics, infection, pregnancy and long-term changes of lifestyles, can alter the composition and diversity of the intestinal microbiota.5 Recently, interest in the role of gut microbiota in the human health and disease is being increased as new technologies for meta-analysis of microbial genomes and transcripts as well as metabolites produced by microbiota have become feasible at affordable cost.6, 7 It is now widely recognized that the gut microbiota has essential roles in human metabolism, the development of the immune system, as well as resistance to colonization of enteric pathogens in the gut.1
Given the diverse and critical roles of the gut microbiota in human health, perturbations in the microbial composition, termed dysbiosis, may have a profound negative impact on the human immune system, contributing to various chronic inflammatory disorders. In this article, we review the role of the host–microbe interaction in the host immune system, discuss possible links between intestinal dysbiosis and chronic inflammatory diseases, and finally propose potential treatments for these diseases through correction of intestinal dysbiosis.
Function of intestinal microbiota in the host
The ‘external' metabolic organ
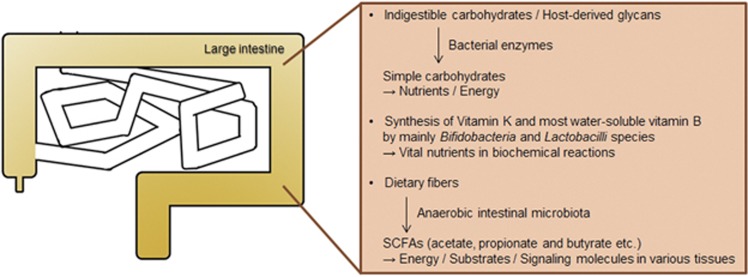
The microbiota assists in the degradation of otherwise indigestible carbohydrates in the human intestine through some digestive enzymes derived from bacteria.8 Most nutrients produced by host enzymes are absorbed in the stomach and small intestine, whereas bacteria residing in the ileum usually utilize only simple carbohydrates as a major energy source.9 By contrast, the indigestible carbohydrates and proteins equivalent to 10–30% of the total ingested energy reach the colon,9, 10 where these otherwise indigestible dietary carbohydrates and host-derived glycans are converted by enzymes produced by strict anaerobic bacteria to simple carbohydrates used as nutrients and energy (Figure 1).8, 11 To do this, for instance, Bacteroides species possess a large number of genes that encode essential enzymes to degrade diverse complex carbohydrates, and members of the phyla Firmicutes, Actinobacteria and Verrucomicrobium produce nutritionally specialized enzymes that have a key role in the degradation of particular substrates, such as plant cell walls, starch particles and mucins.8, 11
Figure 1.
Functions of the microbiota in host metabolism. Some bacterial enzymes are capable of converting food- and host-derived carbohydrates to simple carbohydrates in the colon. Gut symbiotic bacteria such as Bifidobacteria and Lactobacilli are able to synthesize vitamin K and water-soluble vitamin B. SCFAs also are produced from diet fibers by Bacteroides, and used as nutrients, energy source and signaling molecules. SCFA, short-chain fatty acid.
Vitamins are vital nutrients that are necessary for essential biochemical reactions in living cells.12 Intestinal bacteria act as an important supplier of vitamins along with foods.12 Bifidobacteria and Lactobacilli species are recognized as beneficial probiotics capable of converting dietary compounds into vitamin K and most of the water-soluble vitamin B molecules including biotin, folates, nicotinic acid, pyridoxine, riboflavin, cobalamin and pantothenic acid (Figure 1).13 The vitamins synthesized by bacteria are mainly absorbed in the large intestine, whereas uptake of vitamins from dietary sources is achieved in the proximal small intestine.12
Also, anaerobic intestinal bacteria produce short-chain fatty acids (SCFAs) as the end products of fermentation of dietary fibers, among which, acetate, propionate and butyrate are the most abundant.14, 15 SCFAs are transported from the intestinal lumen into the various tissues where they are used as either a source of energy, substrates or signal molecules, to aid in the metabolism of lipids, glucose and cholesterols (Figure 1).16, 17, 18, 19 However, SCFA generation, diet and bacterial composition are delicately interlinked. For example, diets with high fiber-low fat and meat lead to higher amounts of SCFAs than diets with reduced fiber, thereby lowering the pH in the intestine and inhibiting the growth of pH-sensitive microbiota, which affects the production of SCFAs.20, 21 Given the complexity of microbial metabolic pathways and cross-feeding mechanisms involved in SCFA production,15 there is not a simple linear correlation between intestinal SCFA levels and diet or microbial composition. Likewise, administration of a single dietary component or bacterial strain would unlikely result in an appreciable change in the concentration of SCFAs. Taken together, the intestinal microbiota essentially serves as ‘external metabolic organ' that continuously supplies the host with absorbable nutrients and efficient energy.
Role of intestinal microbiota in the development and function of host immune system
Studies using germ-free (GF) and gnotobiotic animals, colonized with defined bacteria, have provided direct evidence that the microbiota has a crucial role in the development and maintenance of the host immune system.2, 22 Notably, GF mice show remarkable defects in the development of the gut-associated lymphoid tissues (GALT).2, 22 For example, GF mice exhibit relatively fewer and smaller Peyer's patches and mesenteric lymph nodes, and impaired development and maturation of isolated lymphoid follicles that serve as an inductive site for IgA responses, compared with the mice housed under specific pathogen free (SPF) conditions (Figure 2).22, 23 However, the impaired development of these structures can be reversed by exposing GF mice to intestinal bacteria or peptidoglycans isolated from Gram-negative bacteria,22, 23 showing that the microbiota has a dynamic relationship with the formation of GALT. The development and maturation of IgA-producing B cells are defective in the intestine of GF mice and sustained exposure to bacteria is required for the continued production of commensal-specific IgA.24 In addition, the intestinal epithelial cells in GF animals are regenerated at a lower rate and form structurally altered microvilli compared with conventionally raised animals.25 The intestinal mucus layer, which functions as a lubricant and protects the epithelium from irritations by luminal contents including certain microbes, is thinner in GF animals, indicating that resident bacteria are involved in the normal formation of the mucus layer.26, 27 These defects in the intestinal physical barriers in GF animals seem to be associated with more susceptible phenotypes to pathogen infections and excessively heavy and prolonged bleeding during colitis induction.28 Moreover, typical properties of mucus layer could be changed by certain bacteria and their community. For example, an increase in the abundance of Proteobacteria and TM7 bacteria in the gut drastically enhanced the permeability of the normally sterile mucus inner layer to more penetrable region, resulting in bacterial infiltration into intestinal inner layer close to epithelium.27
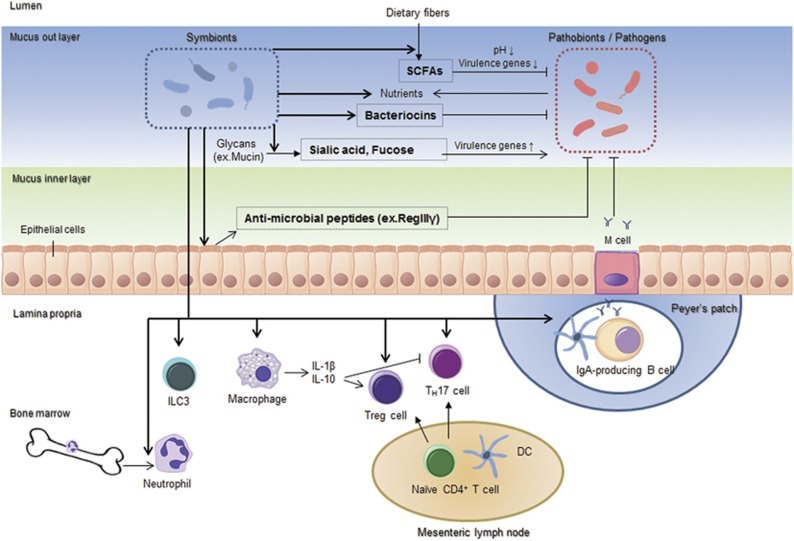
Figure 2.
Symbionts directly and indirectly regulate the growth and colonization of pathobionts and pathogens. Symbionts outcompete pathobionts and pathogens by consuming limited nutrients, producing bacteriocins and lowering luminal pH through SCFA production. SCFAs and byproducts produced by symbionts regulate the expression of virulence genes in the pathobionts and pathogens. Moreover, symbionts elicit the host immune responses, such as recruitment of neutrophils, cytokine production of macrophages, differentiation of TH17, ILC3 and Treg cells, IgA production by B cells, and antimicrobial peptides production from epithelial cells. SCFA, short-chain fatty acid.
Symbiotic intestinal bacteria have been well known to be essential for the development and function of specific lymphocyte subsets. GF mice and antibiotic-treated mice show reduced numbers of mucosal T helper (TH)17 cells and type 3 innate lymphoid cells (Figure 2), which are involved in the resistance to extracellular microbes and the pathogenesis of autoimmune diseases by secreting interleukin (IL)-17 and IL-22.4, 29 Moreover, mice lacking segmented filamentous bacteria (SFB) in their gut microbiota have fewer TH17 cells in the small intestine than in mice with abundant SFB.29 Normal developments of mucosal TH17 cells can be restored following reconstitution with conventional microbiota or SFB alone (Figure 2).29 However, SFB are not the only bacteria that possess Th17-inducing capacities. GF mice colonized with Altered Schaedler Flora (ASF), a cocktail of 8 defined commensals including Clostridia, also showed significantly increased numbers of Th17 cells, albeit to a lesser extent as observed in SFB-colonized GF mice.30 Likewise, the number of regulatory T cells (Tregs), which suppress immunopathology mediated by effector T cells and induce tolerance toward self-components, is reduced in the intestinal lamina propria of both GF and antibiotic-treated mice, whereas recolonization with conventional microbiota or selective species of bacteria restore the Treg cell population (Figure 2).4 For example, the administration with Bacteroides fragilis or its product, polysaccharide A, induces the development of Tregs and production of IL-10, anti-inflammatory cytokine in the intestine.31 Furthermore, Kenya Honda and colleagues have reported unique Treg-inducing activities in indigenous Clostridia species isolated from mice and healthy humans (Figure 2).32
Important roles for the gut microbiota in innate immunity in the gut have been documented as well. Phagocytes, such as macrophages and neutrophils, are tightly regulated to maintain intestinal homeostasis.4 Intestinal bacteria continuously stimulate resident macrophages to release large amounts of IL-10 that promotes induction of Treg cells and prevents excessive development of TH17 cells, which is critical for maintaining tolerogenic immune responses in the intestine (Figure 2).33 The stimulation by commensals also upregulates the expression of pro-IL-1β (precursor form of IL-1β) in intestinal phagocytes without eliciting inflammasome responses and harmful release of mature IL-1β.34 During infection of pathogens, such as Salmonella and Pseudomonas aeruginosa, however, maturation and secretion of IL-1β immediately occurs by the activation of inflammasome through Nod-like receptors, NLRC4, that are expressed in intestinal phagocytes to protect the host from pathogens.34 The numbers of circulating and bone marrow neutrophils are also affected by the microbiota as well.35 For example, intestinal microbiota augments host response against pathogens, such as Streptococcus pneumoniae and Staphylococcus aureus, through constitutive stimulation of neutrophils derived from the bone marrow (Figure 2).36
Direct and indirect regulation of pathogens by commensals
Some of commensals beneficial to the host are referred to as symbionts, whereas others that post harmful effect to the host under certain situations are called pathobionts.2, 37 The commensal microbiota regulates the colonization of pathobionts and invading pathogens in both a direct and indirect manner. Symbionts produce bacteriocins, proteinaceous toxins, that most directly target and kill similar or closely related bacterial strains (Figure 2).38 Also, symbionts and metabolically related pathobionts or pathogens compete for limited nutrients, which gives symbionts the unique ability to resist colonization of related pathobionts or pathogens in the gut (Figure 2). For example, Citrobacter rodentium, an enterohaemorrhagic Escherichia coli (EHEC) mouse strain, could be outcompeted by commensal E. coli strains, but not commensal Bacteroides, through competitions for simple sugars.39
Certain commensal bacteria can indirectly regulate the colonization of pathogens through their byproducts (Figure 2). Bacteroides thetaiotaomicron has sialidase and fucosidase activities, which mediate the production of sialic acid and fucose from host mucus, respectively.40 Sialic acid can be catabolized by both Salmonella enterica serovar Typhimurium and Clostridium difficile, to promote the growth of these two pathogens.41 Fucose induces the expression of virulence genes in EHEC through the fucose receptor signaling cascade, which in turn facilitate vigorous colonization of EHEC.42 The intestinal microbiota also produces several SCFAs as the end products of fermentation of dietary fibers.14, 15 The low pH from the ileum to the cecum is attributed to high concentrations of SCFAs, which inhibits the growth of pH-sensitive pathogenic bacteria, such as E. coli pathogenic strain and Salmonella (Figure 2).20, 21 Moreover, SCFAs regulate the expression of virulence gene in some pathogens, such as Salmonella, E. coli, Campylobacter jejuni, and Campylobacter jejuni.43
In addition, the microbiota indirectly exerts protective effects on pathogens by stimulating the host immune system. The epithelium stimulated by the microbiota produces mucus and antimicrobial peptides, which are critical to the maintenance of a sterile mucus inner layer against both commensals and pathogens (Figure 2).27 Although several antimicrobial proteins, such as α-defensins, are constitutively expressed without bacterial signals, Gram-negative bacteria, like B. thetaiotaomicron, but not Gram-positive microbes, induce the expression of RegIIIγ, a key subset of antimicrobial peptides, which has a specific microbicidal effect on Gram-positive bacteria.44, 45 Direct and indirect regulation of pathogens by commensals prevents the attachment of pathogens to the surface of intestinal epithelium at initial stage of enteric infection.
Role of intestinal microbiota in chronic inflammatory diseases
Inflammatory bowel disease
Inflammatory bowel disease (IBD), in the form of ulcerative colitis (UC) or Crohn's disease (CD), is a chronic inflammatory disease caused by a combination of genetic, environmental and microbial factors.46 The first evidence that intestinal bacteria are involved in the pathogenesis of IBD came from a clinical study showing that fecal stream diversion can improve the symptoms of CD.47 Since then, many studies have supported that IBD is directly and indirectly associated with alterations of the intestinal microbiota. Several metagenomic analyses revealed the reduced diversity of fecal microbiota in patients with colitis compared with that in healthy controls (Table 1).48, 49 In addition, patients with CD or UC have reduced numbers of members of the phyla Bacteroidetes and Firmicutes, whereas species belonging to the phyla Actinobacteria and Proteobacteria, such as E. coli, are increased in mucosa-associated microbiota (Table 1).6, 50, 51, 52 It has been speculated that intestinal dysbiosis might not be a mere consequence but rather a cause of IBD pathogenesis. For instance, more bacteria penetrate the intestinal mucus layer in patients with IBD than that observed in healthy controls, indicating possibly more virulent intestinal bacteria in patients with IBD that possess a heightened accessibility to the epithelial mucus layer.53 Consistently, increase in the relative proportion of Bacteroidales and Clostridiales is likely to trigger inflammation in high-fat-fed rat models.54 However, many bacterial species, such as Enterobacteriaceae, appear to have the unique ability to bloom in an inflammatory environment,55 suggesting that it is likely that many or most of the changes in microbiota composition observed in IBD are secondary to inflammation rather than the cause of colitis.
Table 1. Studies investigating intestinal microbiota in patients.
| Disease subjects | Methodology | Alteration in microbiota | References |
|---|---|---|---|
| Intestinal bowel disease | |||
| UC patients | Terminal restriction fragment length polymorphism | Diversity of mucosa-associated microbiota ↓ | 49 |
| CD patients | 16S rRNA gene sequencing/fluorescent in situ hybridization | Diversity of phylum Firmicutes ↓ | 48 |
| CD patients | 16S rRNA gene sequencing | Faecalibacterium and Roseburia ↓ Enterobacteriaceae and Ruminococcus gnavus ↑ | 52 |
| CD patients | 16S rRNA gene sequencing/fluorescent in situ hybridization | Faecalibacterium prausnitzii, Ruminococcus bromii, Oscillibacter valericigenes, Bifidobacterium bifidum, and Eubacterium rectale ↓ Escherichia coli, Enterococcus faecium and species from the Proteobacteria ↑ | 51 |
| UC and CD patients | 16S rRNA gene sequencing | Members of the phyla Firmicutes and Bacteroidetes↓ | 50 |
| Multiple sclerosis | |||
| MS patients | Fluorescent in situ hybridization | Faecalibacterium ↓ | 71 |
| MS patients | 16S rRNA gene sequencing | Faecalibacterium, Prevotella, and Anaerostipes ↓ | 72 |
| Relapsing-remitting MS patients | 16S rRNA gene sequencing | Parabacteroides, Adlercreutzia and Prevotella ↓ Psuedomonas, Mycoplana, Haemophilus, Blautia, and Dorea ↑ | 73 |
| Allergic diseases | |||
| Atopic patients (newborn ~2 years old) | Bacterial cultivation | Bacteroides, Bifidobacteria ↓ Enterococci, Clostridia, Staphylococcus aureus ↑ | 83 |
| Atopic patients (2 years old) | Quantitative real-time PCR | E. coli and Clostridium difficile ↑ | 84 |
| Atopic patients (3 weeks ~3 months old) | Bacterial cultivation/ gas–liquid chromatography/fluorescence in situ hybridization | Bifidobacteria ↓ Clostridia ↑ | 85 |
| Asthma patient (school age) | 16S rRNA gene sequencing /barcoded 16S rDNA 454 pyrosequencing | Diversity of total microbiota ↓ | 91 |
| Rheumatoid arthritis | |||
| RA patients | Metagenomic sequencing and assembly | Haemophilus spp. ↓ Lactobacillus salivarius ↑ | 96 |
| RA patients (new-onset) | 16S rRNA gene sequencing | Prevotella (P. copri)↑ | 97 |
| RA patients (early) | Quantitative real-time PCR | Lactobacillus ↑ | 99 |
| RA patients | 16S rRNA gene sequencing | Faecalibacterium ↓ Eggerthella and Collinsella ↑ | 100 |
Abbreviations: CD, Crohn's disease; MS, multiple sclerosis; RA, rheumatoid arthritis; UC, ulcerative colitis.
↓ Decreased, ↑ increased, when compared with the healthy subjects.
Preclinical studies in animals provide evidence to support that altered microbial composition of the gastrointestinal tract contributes to the development of IBD. Animals with spontaneous or DSS-induced colitis develop only minor inflammation under GF conditions.56, 57, 58 Moreover, a number of studies have shown that certain pathobionts can contribute to the induction of colitis in genetically IBD-prone animals, and other bacteria have an opposing role. For example, E. coli or Enterococcus faecalis promote the occurrence of colitis in Il-10- or Il2-deficient mice, which could be reversed by colonization with Bacteroides vulgatus.59, 60, 61 In addition, the reduction of Bacteroides fragilis and Faecalibacterium prausnitzii, which correlates with higher risks for CD in humans, might promote intestinal inflammation through downregulation of Treg cells.62, 63, 64 In addition, Enterobacteriaceae species Klebsiella pneumoniae and Proteus mirabilis were shown to correlate with spontaneous development of colitis in T-bet−/− × Rag2−/−mice, perhaps in part because of their abilities to potently induce TNFα in innate immune cells.65 These observations demonstrate immunomodulatory effects of gut symbiotic bacteria in the pathogenesis of IBD, and also shed light on potential therapeutic approaches directed at specific pathobionts for the treatment of IBD.
Multiple sclerosis
Multiple sclerosis (MS) is a chronic inflammatory, demyelinating and neurodegenerative disorder of the central nervous system (CNS) that causes neurological dysfunctions such as blindness, paresis and sensory disturbances.66 The importance of intestinal microbiota in the occurrence of MS was first suggested by several preclinical studies using an experimental autoimmune encephalomyelitis (EAE) model, a murine model of MS.66 GF mice fail to develop spontaneous and inducible EAE, which is attributed to impaired generation of TH17 cells.67, 68 Likewise, antibiotic treatments that induced Treg cells and IL-10-producing B cells were shown to suppress clinical severity in EAE model.69 Moreover, mono-colonization with SFB, known to induce the TH17 population in the intestine, can induce EAE in the GF mice, whereas colonization of B. fragilis or administration of B. fragilis-derived polysaccharide A, maintains the resistance to EAE through induction of Treg cells.68, 70 These results indicate potential roles for certain gut symbiotic bacteria in the pathogenesis of the EAE model through their abilities to modulate immune responses.
Up to date, few but highly informative metagenomics studies have been performed to investigate a clinical correlation between gut microbiota and MS. Patients with MS have significantly altered microbial profiles in the intestine compared with the healthy people, in spite of insignificant differences in bacterial richness and diversity between these two groups. For instance, individuals with MS have less Faecalibacterium that produce the anti-inflammatory SCFA butyrate (Table 1),71 as well as decreased levels of the bacterial genus Prevotella (Bacteroidetes phylum) and the genera Faecalibacterium and Anaerostipes (both of Firmicutes phylum) (Table 1).72 A very recent study reported reduced abundance in the genera Parabacteroides, Prevotella (both of Bacteroidetes phylum) and Adlercreutzia (Actinobacteria phylum) in the feces from patients with relapsing-remitting MS, whereas the general Pseudomonas, Mycoplana, Haemophilus (above Proteobacteria phylum), Blautia and Dorea were more abundant in these patients (Table 1).73 Thus far, studies have suggested intestinal dysbiosis characterized by overall decreases in Bacteroidetes and Firmicutes phyla correlates with the development of MS. However, additional studies are required to delineate the mechanisms underlying intestinal dysbiosis-mediated immune dysregulation that is associated with the development of MS.
Asthma and allergic diseases
Allergic asthma is a chronic inflammatory disorder associated with TH2-driven inflammatory responses to inhaled innocuous allergens, resulting in airway hyper-responsiveness and tissue remodeling of airway structures, and bronchial obstruction.74, 75 About 300 million people worldwide are affected by allergic asthma, characterized by recurrent episodes of wheezing as a result of mucus plugging, bronchial mucosal thickening and brochoconstriction.75 The diversity of microbial stimuli exposed to during early childhood, such as breastfeeding, having multiple siblings, contacting farm animals and pets, leads to protective mechanisms against asthma.76, 77, 78, 79 One theory is that microbial products trigger innate immune response through pattern-recognition receptors, such as the Toll-like receptors, which may activate several signaling pathways such as induction of Treg cells, as well as Type 1 helper T (Th1) cells, which can counter-balance the predominance of Th2 cells that mediate asthmatic attacks. In support of this theory, GF mice that have been sensitized and challenged with ovalbumin (OVA) suffer from more exaggerated allergic airway inflammation than SPF mice,80 indicating the importance of microbiota in the prevention of asthma development. Intriguingly, this severe inflammation could be restored by colonization with a conventional microbiota in neonatal GF mice, but not in adult GF mice.81 In support of this hypothesis, Olszak et al.81 revealed that microbial colonization in neonatal mice can contribute to the reduced expression of Cxcl16 mRNA in the colon, ileum and lung by inhibiting hypermethylation of Cxcl16, thereby reducing the recruitment of iNKT cells into these tissues, which alleviates allergic airway inflammation in response to OVA. This observation also implicates that epigenetic modifications of Cxcl16 gene seen during childhood by diverse microbial exposure cannot be achieved later in adulthood even with the same microbial exposure.81
In addition to the exposure to microbes early in life, the microbial composition is also an important factor to dictate development of the allergic diseases. Several studies have reported distinct patterns in the intestinal microbiota infants that developed allergies later in life.82, 83, 84, 85 The intestinal flora of babies that suffered from allergic inflammation, consists of more prevalent Clostridia and Enterococci and less prevalent Lactobacilli, Bifidobacteria and Bacteroidetes compared with healthy subjects (Table 1).83, 84, 85 The childbirth process has a significant impact on the formation of microbial community in neonates. Infants delivered through vaginal tract typically form a microbial composition including abundances of Snethia and Lactobacillus spp., which is substantially similar to that found in the mother's vaginal tract; whereas infants born via cesarean section possess a microbiota enriched with Staphylococcus and Streptococcus spp., similar to that of maternal skin microbiota.86 The latter has been reported at significantly higher risks for allergic diseases.87 Also, use of antibiotics by the mother during pregnancy or the infant affects the microbial communities in the lung and intestine of the infant, which is associated with increased incidence of asthma and other allergic diseases later in life.88, 89
Microbes residing in the respiratory tract are naturally associated with asthma development.90 Surprisingly, intestinal microbiota appears to contribute to allergic airway inflammation. For instance, children developing asthma have a lower diversity in intestinal microbiota than non-asthmatic children do (Table 1).91 Oral administration with Lactobacillus reuteri leads to an increase in Treg cell population in the blood, spleen and mediastinal lymph nodes, resulting in reduction of asthmatic responses in mice with OVA sensitization and challenge.92 Similarly, mice treated with a cocktail of Clostridium strains have reduced IgE production after OVA sensitization, likely because of the induction of Treg cells in the colonic mucosa by these Clostridium strains.32 Moreover, a high-fiber diet increases the ratio of Firmicutes to Bacteroidetes in the intestine, which elevates the levels of circulating SCFA, is associated with protection against airway inflammation.93 These observations suggest that intestinal microbiota, through yet undefined mechanisms, may polarize immune cells such as T helper cells that either promote or favor the development of allergic inflammation.
Rheumatoid arthritis
Rheumatoid arthritis (RA) is a chronic autoimmune disorder that symmetrically affects large and small joints with inflammation and ultimately incurs in joint deformity.94 The relatively higher concordance of 12–15% in monozygotic twins than 2–4% in dizygotic twins suggests the contribution of genetic factors to RA pathogenesis.95 However, the low concordance even in monozygotic twins indicates the importance of environmental factors as well.95 In addition to genetic and environmental factors, a number of studies have provided evidence that microbial factors are involved in the development of RA. A metagenomic analysis of fecal, dental and salivary samples showed that the microbial community of RA patients can be distinguished from that of healthy subjects.96 For instance, individuals with RA have relatively high abundance of Lactobacillus salivarius in their intestine and oral cavity, which correlates with clinical severity.96 In addition, the number of Prevotella copri is increased in patients in the first year after RA onset (Table 1).97 Similarly, other studies also showed a positive correlation between P. copri and early development of RA in untreated patients 97, 98 and increased amounts of Lactobacillus species in early RA patients (Table 1),99 although it remains unclear how/whether higher abundance of these bacterial species have a role in the development of RA. Moreover, patients with RA exhibit reduced microbial diversity in their intestines compared with controls, which is proportional to increased disease duration and autoantibody levels.100 The dysbiosis in RA individuals is characterized by an increase of harmful microbes including Eggerthella and Collinsella and a decrease of beneficial bacteria such as Faecalibacterium, one of the most abundant Firmicutes in the human gut (Table 1).100 Interestingly, treatment with disease modifying anti-rheumatic drugs (DMARDs) can reverse this perturbation of intestinal flora in RA patients closely to that of healthy individuals.96 Hence, the microbial profile in the intestine might be used as a marker of RA development.
Spontaneous arthritis models, such as SKG, K/BxN and IL-1 receptor antagonist knockout (Il1ra−/−) mice and HLA-B27 transgenic rats, fail to induce arthritis under GF conditions.56, 101, 102, 103 In contrast, F344 rats develop severe arthritis induced by adjuvant stimulation when raised under GF conditions.104 Despite these conflicting results, the microbiota may modulate the sensitivity of arthritis models possibly through modulation of the immune response. Furthermore, Collinsella and P. copri that are abundant in RA patients, aggravate disease severity in a humanized arthritis model and the SKG model, respectively,98, 100 suggesting that these two bacteria might indeed have a role in the pathogenesis of RA. However, more work is needed to understand the role of the microbiota in RA development.
Microbiota-target treatment
As described so far, intestinal dysbiosis observed in various chronic inflammatory disorders, such as IBD, MS, allergic asthma and RA, has been suggested to contribute to the pathogenesis of these diseases. Thus, approaches to remove disease-causing pathobionts or boost beneficial symbionts now emerge as promising therapeutics to mitigate dysregulated inflammation underlying these diseases.
Antibiotics
Antibiotic treatment used in the clinical setting has been mainly used to for the purpose to either treat or prevent infection.105, 106 However, simultaneously, it can cause unintended damage by depleting beneficial commensals105, 106 Even though the microbiota can be restored to the original status following withdrawal of antibiotics, in some cases, full restoration may not be achieved after prolonged treatment.106 In some other cases, blooms of pathobionts, albeit temporary, may pose long-lasting negative impacts on host immune responses or other functions.107 It should be noted that the scale and persistence of such perturbations depends on the class of antibiotics administered. For example, the administration of either a combination of amoxicillin, metronidazole and bismuth, or vancomycin alone, significantly reduces microbial diversity in intestinal microbiota; whereas compositional alterations can be almost fully reversed to baseline about 3 weeks after termination of antibiotic treatment.105 In contrast, following administration with clindamycin, which has a strong and broad activity against almost anaerobes, for 7 days, the diversity of Bacteroides is never restored to its original level until over two years of recovery.107 Antibiotic-induced alterations of the gut microbiota can promote various diseases. For instance, antibiotic treatments lead to an increase in the abundance of host-derived free sialic acid in the intestine, which can then be utilized by certain pathogens and pathobionts, such as S. enterica serovar Typhimurium and C. difficile, resulting in blooms of pathogenic bacteria in the intestinal tract and subsequently symptoms including diarrhea.41
Therefore, the use of narrow-spectrum antibiotics appears as an appealing alternative to minimize the risk of gut dysbiosis and blooms of pathobionts.108 There has been an increasing interest in the use of ampicillin and penicillin, relatively narrow-spectrum antibiotics, for the management of hospitalized infants and children with community-acquired pneumonia.109 A follow-up study of this strategy showed that ampicillin and penicillin have a similar efficacy with empirically used broad-spectrum antibiotics.110 Like this, it remains challenging to determine which narrow-spectrum antibiotics have sufficient efficacy and specificities against particular infectious agents to replace the empirically used broad-spectrum antibiotics. To do that, understanding the mechanisms by which pathogens colonize the intestinal tract, outcompete commensal bacteria, or translocate systemically as well as finding intrinsic factors that can differentiate pathogens from commensals, will provide critical insights into the development of effective and specific narrow-spectrum antibiotics that target the pathogens or selective pathobionts, whereas remaining innocuous to other commensals.
Probiotics
Probiotics are live microorganisms that provide health benefits to the host when consumed in adequate amounts.111 For many years, studies of probiotics have reported prophylactic and therapeutic effects of probiotics in the various disease models associated with intestinal dysbiosis. For instance, a probiotic cocktail composed of four Lactobacillus and Bifidobacterium strains attenuates severity of acute colitis.112 A mixture of five probiotics, consisting of Lactobacillus casei, Lactobacillus acidophilus, Lactobacillus reuteni, Bifidobacterium bifidum and Streptococcus thermophiles, has prophylactic and therapeutic effects in the murine EAE model through decreases in TH1 and TH17 cells and induction of Treg cells.113 Oral administrations of Lactobacillus rhamnosus attenuates OVA-induced airway inflammation.114 Also, L. casei and L. acidophilus diminish arthritic symptoms in the collagen-induced arthritis model by inhibiting proinflammatory cytokines and inducing anti-inflammatory cytokines.115
Compared to preclinical data, available clinical reports on the benefits of probiotics, however, are scarce.108 A randomized trial showed that consumption of a probiotic drink containing Lactobacillus casei, Lactobacillus bulgaricus and Streptococcus thermophilus reduced the risk of antibiotic-associated diarrhea and Clostridium difficile infection (CDI) in a small number of patients over the age of 50.116 A meta-analysis of sixteen randomized, parallel, controlled studies (3432 participants) of the incidence of antibiotic-associated diarrhea in children treated with adjunctive probiotic therapy supported that potential of probiotics to prevent antibiotic-associated diarrhea.117 In addition, a probiotic combination of Lactobacillus acidophilus and Bifidobacterium lactis was shown to reduce birch pollen allergy symptoms including infiltration of eosinophils.118 Despite results from these clinical studies, the administration of probiotics is not strongly recommended, due to insufficient data to support their preventive or therapeutic efficacy and conflicting reports on their benefits.108 Alternatively, consumption of prebiotics, which are mostly plant-derived fibers that selectively enrich beneficial bacteria, may have advantages over probiotic therapy to introduce more persistent and stable compositional changes to a dysbiotic microbial community. Further investigations of the mechanisms of protection by probiotics or prebiotics will advance the design of probiotics-based therapeutics to correct dysbiotic microbiota that could potentially promote development of disease if untreated.
Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) refers to the transfer of the entire fecal microbial community from a healthy donor into a recipient patient with the goal to completely restore healthy microbial community in the patient.118 FMT has been used for patient with recurrent CDI.119 As predicted, CDI patients treated with FMT exhibited the microbial structural changes more similar to the donor profile than bacterial flora prior to transplant, which resulted in functional changes and hence therapeutic effects.120 Promising results were observed in using FMT to treat IBD as well. A meta-analysis of previous nine case studies of IBD patients treated with FMT showed that a pooled proportion achieving remission was 22% in UC patients and 60.5% in CD patients.121 Given that the microbial basis of IBD is far more complex and variable than that of recurrent CDI, the results suggest a positive effect of FMT, albeit at a lower successful rate compared with that achieved by FMT in recurrent CDI (between 80 and 95% efficacy).119 Overall, the efficacy of FMT in IBD treatment needs to be corroborated by additional large-scale clinical studies. The potentials of FMT to improve other chronic inflammatory diseases associated with intestinal dysbiosis, however, have not been extensively explored. Recent studies have begun to investigate the effect of FMT on MS patient treated with FMT.122 Therefore, further studies are required to determine the roles of FMT in the treatment of other inflammatory disorders including MS.
Conclusion
Knowledge related to the interplay between the microbiota and the human immune system is being accumulated at a remarkable rate, as metagenomic and metatranscriptomic approaches to the microbiome and metabolomic analysis of small metabolites at affordable cost have been possible.123 The information reveals us that the microbiota is no longer a by-stander in human health, disease and survival. As described in this review article, the intestinal microbiota is likely to be involved in the pathogenesis of various chronic inflammatory diseases, such as IBD, MS, allergic asthma and RA. Particular bacterial species with the gut microbial community have been identified to directly or indirectly contribute the onset and deterioration of these disorders. Therefore, measures to correct intestinal dysbiosis, such as FMT may offer long-lasting and effective ways to mitigate dysbiosis-induced inflammation that in part promotes the manifestation of the aforementioned diseases. Further identification of the specific symbiotic bacteria capable of inhibiting the colonization of pathogenic bacteria and promoting host immunity and metabolism will aid in developing microbiota-based precision medicine to reverse dysbiosis-induced and disease-associated defects.
Acknowledgments
This work was supported by grants from National Institutes of Health grants R01DK61707 and R01DK091191 (GN) and a grant from the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2015R1A3A2032927) (DK).
Footnotes
The authors declare no conflict of interest.
References
- Kamada N, Seo S-U, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Med 2013; 13: 321–335. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016; 14: e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Nunez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 2014; 146: 1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 2015; 26: 26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Arthritis Res Ther 2013; 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012; 3: 289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajmalnik-Brown R, Ilhan Z-E, Kang D-W, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract 2012; 27: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 1990; 70: 567–590. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005; 307: 1955–1959. [DOI] [PubMed] [Google Scholar]
- LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 2013; 24: 160–168. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Vaughan EE. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Ann Rev Microbiol 2009; 63: 269–290. [DOI] [PubMed] [Google Scholar]
- den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 2013; 54: 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de los Reyes-Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 2016; 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009; 58: 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi T, Suruga K, Oshima Y, Fukiharu M, Tsukamoto Y, Goda T. Dietary acetic acid reduces serum cholesterol and triacylglycerols in rats fed a cholesterol-rich diet. Br J Nutr 2006; 95: 916–924. [DOI] [PubMed] [Google Scholar]
- Todesco T, Rao AV, Bosello O, Jenkins DJ. Propionate lowers blood glucose and alters lipid metabolism in healthy subjects. Am J Clin Nutr 1991; 54: 860–865. [DOI] [PubMed] [Google Scholar]
- Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology 1982; 83: 424–429. [PubMed] [Google Scholar]
- Cherrington CA, Hinton M, Pearson GR, Chopra I. Short-chain organic acids at pH 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J Appl Microbiol 1991; 70: 161–165. [DOI] [PubMed] [Google Scholar]
- Prohaszka L, Jayarao BM, Fabian A, Kovacs S. The role of intestinal volatile fatty acids in the Salmonella shedding of pigs. Zentralbl Veterinarmed B 1990; 37: 570–574. [DOI] [PubMed] [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev 1998; 62: 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008; 456: 507–510. [DOI] [PubMed] [Google Scholar]
- Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun 1978; 21: 532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest 1963; 12: 355–364. [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA 2008; 105: 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 2014; 16: 164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Morimoto M, Sagara E, Shimizu C, Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim 2001; 50: 387–395. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking Markus B, Cahenzli J, Lawson Melissa AE, Ng Derek CK, Slack E, Hapfelmeier S et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011; 34: 794–806. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3(+) regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA 2010; 107: 12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med 2012; 209: 139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol 2012; 13: 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'Leary CE et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 2014; 20: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010; 16: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornef M. Pathogens, commensal symbionts, and pathobionts: discovery and functional effects on the host. ILAR J 2015; 56: 159–162. [DOI] [PubMed] [Google Scholar]
- Hammami R, Fernandez B, Lacroix C, Fliss I. Anti-infective properties of bacteriocins: an update. Cell Mol Life Sci 2013; 70: 2947–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 2012; 336: 1325–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem 2006; 281: 36269–36279. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013; 502: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG et al. Fucose sensing regulates bacterial intestinal colonization. Nature 2012; 492: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, O'Riordan MXD. Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv Appl Microbiol 2013; 85: 93–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem 2000; 275: 40478–40482. [DOI] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006; 313: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 2011; 140: 1720–1728. [DOI] [PubMed] [Google Scholar]
- Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet 1991; 338: 771–774. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006; 55: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa J, Kudo T, Sakata S, Benno Y, Sugiyama T. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol 2009; 44: 180–186. [DOI] [PubMed] [Google Scholar]
- Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007; 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M et al. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm Bowel Dis 2011; 17: 185–192. [DOI] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010; 139: 1844–1854.e1841. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 2007; 56: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol 2010; 299: G440–G448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Inohara N, Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol 2016; 10: 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med 1994; 180: 2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianda L, Hanby AM, Wright NA, Sebesteny A, Hayday AC, Owen MJ. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol 1997; 150: 91–97. [PMC free article] [PubMed] [Google Scholar]
- Hudcovic T, Štěpánková R, Cebra J, Tlaskalová-Hogenová H. The role of microflora in the development of intestinal inflammation: acute and chronic colitis induced by dextran sulfate in germ-free and conventionally reared immunocompetent and immunodeficient mice. Folia Microbiol 2001; 46: 565–572. [DOI] [PubMed] [Google Scholar]
- Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 2005; 128: 891–906. [DOI] [PubMed] [Google Scholar]
- Waidmann M, Bechtold O, Frick JS, Lehr HA, Schubert S, Dobrindt U et al. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 2003; 125: 162–177. [DOI] [PubMed] [Google Scholar]
- Bohn E, Bechtold O, Zahir N, Frick JS, Reimann J, Jilge B et al. Host gene expression in the colon of gnotobiotic interleukin-2-deficient mice colonized with commensal colitogenic or noncolitogenic bacterial strains: common patterns and bacteria strain specific signatures. Inflamm Bowel Dis 2006; 12: 853–862. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008; 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005; 122: 107–118. [DOI] [PubMed] [Google Scholar]
- Qiu X, Zhang M, Yang X, Hong N, Yu C. Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating TNBS-induced colitis. J Crohns Colitis 2013; 7: e558–e568. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010; 8: 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wekerle H, Kurschus FC. Animal models of multiple sclerosis. Drug Discov Today Dis Models 2006; 3: 359–367. [Google Scholar]
- Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011; 479: 538–541. [DOI] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 2011; 108: 4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Repáraz J, Mielcarz DW, Haque-Begum S, Kasper LH. Induction of a regulatory B cell population in experimental allergic encephalomyelitis by alteration of the gut commensal microflora. Gut Microbes 2010; 1: 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol 2010; 185: 4101–4108. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J et al. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med 2015; 63: 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS ONE 2015; 10: e0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Soldan MMP et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep 2016; 6: 28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse WW, Lemanske RFJr. Asthma. New Eng J Med 2001; 344: 350–362. [DOI] [PubMed] [Google Scholar]
- Bosnjak B, Stelzmueller B, Erb KJ, Epstein MM. Treatment of allergic asthma: modulation of Th2 cells and their responses. Respir Res 2011; 12: 114–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan DP. Hay fever, hygiene, and household size. BMJ 1989; 299: 1259–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownby DR, Johnson C, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA 2002; 288: 963–972. [DOI] [PubMed] [Google Scholar]
- Riedler J, Braun-Fahrlander C, Eder W, Schreuer M, Waser M, Maisch S et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet 2001; 358: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2012; 2: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst T, Sichelstiel A, Schär C, Yadava K, Bürki K, Cahenzli J et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med 2011; 184: 198–205. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336: 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkstén B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy 1999; 29: 342–346. [DOI] [PubMed] [Google Scholar]
- Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol 2001; 108: 516–520. [DOI] [PubMed] [Google Scholar]
- Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 2007; 56: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol 2001; 107: 129–134. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010; 107: 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz-Polster H, David MR, Buist AS, Vollmer WM, O'Connor EA, Frazier EA et al. Caesarean section delivery and the risk of allergic disorders in childhood. Clin Exp Allergy 2005; 35: 1466–1472. [DOI] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 2012; 13: 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensballe LG, Simonsen J, Jensen SM, Bonnelykke K, Bisgaard H. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. J Pediatr 2013; 162: 832–838.e833. [DOI] [PubMed] [Google Scholar]
- Beigelman A, Weinstock GM, Bacharier LB. The relationships between environmental bacterial exposure, airway bacterial colonization and asthma. Curr Opin Allergy Clin Immunol 2014; 14: 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy 2014; 44: 842–850. [DOI] [PubMed] [Google Scholar]
- Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med 2009; 179: 186–193. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014; 20: 159–166. [DOI] [PubMed] [Google Scholar]
- McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. New Eng J Med 2011; 365: 2205–2219. [DOI] [PubMed] [Google Scholar]
- Silman AJ, MacGregor AJ, Thomson W, Holligan S, Carthy D, Farhan A et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Rheumatology 1993; 32: 903–907. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015; 21: 895–905. [DOI] [PubMed] [Google Scholar]
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013; 2: e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheum 2016; 68: 2646–2661. [DOI] [PubMed] [Google Scholar]
- Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol 2013; 67: 170–176. [DOI] [PubMed] [Google Scholar]
- Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med 2016; 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehaume LM, Mondot S, Aguirre de Carcer D, Velasco J, Benham H, Hasnain SZ et al. ZAP-70 genotype disrupts the relationship between microbiota and host, leading to spondyloarthritis and ileitis in SKG mice. Arthritis Rheum 2014; 66: 2780–2792. [DOI] [PubMed] [Google Scholar]
- Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest 2008; 118: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010; 32: 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohashi O, Kuwata J, Umehara K, Uemura F, Takahashi T, Ozawa A. Susceptibility to adjuvant-induced arthritis among germfree, specific-pathogen-free, and conventional rats. Infect Immun 1979; 26: 791–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal microbiota. Gut Microbes 2010; 1: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 2016; 22: 458–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007; 1: 56–66. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med 2012; 4: 137rv5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley JS, Byington CL, Shah SS, Alverson B, Carter ER, Harrison C et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis 2011; 53: e25–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DJ, Hall M, Shah SS, Parikh K, Tyler A, Neuman MI et al. Narrow vs broad-spectrum antimicrobial therapy for children hospitalized with pneumonia. Pediatrics 2013; 132: e1141–e1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers GT, de Vos WM, Brummer RJ, Morelli L, Corthier G, Marteau P. Health benefits and health claims of probiotics: bridging science and marketing. Br J Nutr 2011; 106: 1291–1296. [DOI] [PubMed] [Google Scholar]
- Toumi R, Soufli I, Rafa H, Belkhelfa M, Biad A, Touil-Boukoffa C. Probiotic bacteria lactobacillus and bifidobacterium attenuate inflammation in dextran sulfate sodium-induced experimental colitis in mice. Int J Immunopathol Pharmacol 2014; 27: 615–627. [DOI] [PubMed] [Google Scholar]
- Kwon H-K, Kim G-C, Kim Y, Hwang W, Jash A, Sahoo A et al. Amelioration of experimental autoimmune encephalomyelitis by probiotic mixture is mediated by a shift in T helper cell immune response. Clin Immunol 2013; 146: 217–227. [DOI] [PubMed] [Google Scholar]
- Wu C-T, Chen P-J, Lee Y-T, Ko J-L, Lue K-H. Effects of immunomodulatory supplementation with Lactobacillus rhamnosus on airway inflammation in a mouse asthma model. J Microbiol Immunol Infect 2016; 49: 625–635. [DOI] [PubMed] [Google Scholar]
- Amdekar S, Singh V, Kumar A, Sharma P, Singh R. Lactobacillus casei and Lactobacillus acidophilus regulate inflammatory pathway and improve antioxidant status in collagen-induced arthritic rats. J Interferon Cytokine Res 2013; 33: 1–8. [DOI] [PubMed] [Google Scholar]
- Hickson M, D'Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C et al. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ 2007; 335: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston BC, Goldenberg JZ, Vandvik PO, Sun X, Guyatt GH. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev 2011; 9: CD004827. [DOI] [PubMed] [Google Scholar]
- Ouwehand AC, Nermes M, Collado MC, Rautonen N, Salminen S, Isolauri E. Specific probiotics alleviate allergic rhinitis during the birch pollen season. World J Gastroenterol 2009; 15: 3261–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. New Eng J Med 2013; 368: 407–415. [DOI] [PubMed] [Google Scholar]
- Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS et al. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 2014; 5: e00893–e00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 2014; 8: 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borody T, Leis S, Campbell J, Torres M, Nowak A. Fecal microbiota transplantation (FMT) in multiple sclerosis (MS). Am J Gastroenterol 2011; 106: S352. [Google Scholar]
- Kinross JM, Darzi AW, Nicholson JK. Gut microbiome–host interactions in health and disease. Genome Med 2011; 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]