Abstract
Functional characterization of human genes is one of the most challenging tasks in current genomics. Owing to a large number of newly discovered genes, high-throughput methodologies are greatly needed to express in parallel each gene in living cells. To develop a method that allows efficient transfection of plasmids into adherent cells in spatial- and temporal-specific manners, we studied electric pulse-triggered gene transfer using a plasmid-loaded electrode. A plasmid was loaded on a gold electrode surface having an adsorbed layer of poly(ethyleneimine), and cells were then plated directly onto this modified surface. The plasmid was detached from the electrode by applying a short electric pulse and introduced into the cells cultured on the electrode, resulting in efficient gene expression, even in primary cultured cells. The location of transfected cells could be restricted within a small area on a micropatterned electrode, showing the versatility of the method for spatially controlled transfection. Plasmid transfection could also be performed in a temporally controlled manner without a marked loss of the efficiency when an electric pulse was applied within 3 days after cell plating. The method described here will provide an efficient means to transfer multiple genes, in parallel, into cultured mammalian cells for high-throughput reverse genetics research.
INTRODUCTION
With the human genome sequences in hand, a major challenge in current genomics research is to analyze the functions of a large number of genes at the cellular level (1–3). Since newly discovered genes have reached large numbers, high-throughput strategies are absolutely required to simultaneously express each gene in living cells (4,5). To conduct such high-throughput reverse genetics research, we (6) and others (7–10) developed transfectional microarrays that allow parallel transfer of multiple genes into cultured mammalian cells. These methods employ the microarray, onto which plasmids are adsorbed in an array format together with cationic lipids or polymers, as the transfection enhancer. Recently, similar techniques were also used to transfer short-interfering RNAs to knock-down the expression of specific genes (11–13).
Further improvements are considered necessary to extend the utility of the microarray-based methods. One of the most important issues includes transfection efficiency, especially for primary cultured cells. Efficient expression of introduced genes is crucial for the microarray-based functional screening, since low efficiency would make experimental data difficult to be interpreted. The precise restriction of transfected areas in a 2D cell culture is also important for integrated microarrays onto which a large number of different expression constructs are microspotted separately. In addition, temporally controlled transfection will be needed in many cases that cells must adhere, differentiate, or proliferate prior to transfection. The cationic lipid- or polymer-based methods described above (6–13) do not meet this requirement.
Electroporation is a widespread technique for the efficient introduction of exogeneous nucleotides into various types of cells in vitro and in vivo (14–16). To overcome these limitations mentioned above, we attempted to develop a novel method in which electric pulses are used to detach plasmids from the microarray surface and then introduce them into cells cultured on the microarray. In conventional electroporation, cells are exposed to plasmids dissolved in a medium, whereas in our approach, plasmids are adsorbed onto the electrode surface. To precisely control the regions of transfection, plasmids were adsorbed through polyelectrolyte complex formation onto the specific area on the electrode surface that had been modified with a cationic polymer. It would be expected that this configuration facilitates in preventing release of plasmids during cell culture before electroporation and to protect plasmids from nuclease-mediated degradation. These effects are also likely to provide the possibility of transfecting plasmids into cells at a preferred moment after cell seeding.
In this study, we demonstrate that gene transfer can be efficiently performed using our method for a cell line, HEK293, and for primary cultured cells including the mouse embryonic fibroblast and the rat hippocampal neuron. It is further demonstrated that the method can be applied for the spatially and temporally controlled transfection. By spatially restricting transfected areas, we could prepare microarrays that display live cells expressing exogeneous genes. This study was also devoted to examine several conditions of plasmid loading and electric pulsing to achieve higher transfection efficiency and gain an insight into the factors that govern transfection at surfaces.
MATERIALS AND METHODS
Cell culture and plasmids
Human embryonic kidney cells, HEK293, were obtained from Health Science Research Resources Bank, Tokyo, Japan. The cells were routinely maintained in a minimal essential medium (MEM; Invitrogen Corp., Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 0.1 mg/ml streptomycin at 37°C under a 5% CO2 atmosphere. Primary rat hippocampal neurons were obtained from fetal Wistar rats (E18) and cultured in Neurobasal™ medium supplemented with 2% B-27 (Invitrogen), 25 μM glutamic acid and 0.5 mM l-glutamine (17,18). Primary mouse embryonic fibroblasts (PMEF) were prepared from day-13.5 embryos (19) and cultured in DMEM (Sigma Chemical Co., St Louis, MO) containing 10% FBS and 10 μM sodium pyruvate (Sigma) at 37°C under a 5% CO2 atmosphere.
The plasmids used in this study were pEGFP-C1 and pDsRed-C1 (4.7 kb; both from Clontech Laboratories, Palo Alto, CA) coding an enhanced green fluorescent protein (EGFP) and Discosoma sp. red fluorescent protein (DsRed), respectively, under the control of a cytomegalovirus promoter. The plasmids were expanded in Escherichia coli and purified using the EndoFree Plasmid Maxi Kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction. The plasmids were dissolved in 10 mM Tris–HCl buffer (pH 8.0) containing 0.1 mM ethylenediamine-N,N,N′N′-tetraacetic acid (TE buffer) and stored at −20°C until use.
Preparation of a plasmid-loaded electrode
A glass plate (24 mm × 26 mm × 1.5 mm, Matsunami Glass Ind., Ltd, Osaka, Japan) was treated with a Piranha solution (concentrated sulfuric acid:hydrogen peroxide = 7:3 by volume) at room temperature for 5 min to remove impurities. The cleaned glass plate was thoroughly washed with deionized water and 2-propanol, and dried under a stream of nitrogen gas. A thin primer layer of chromium (thickness: 1 nm) was deposited onto the glass surface, and then a gold layer (thickness: 19 nm) was further deposited on top of the chromium layer in a continuous process using a thermal evaporator (V-KS200, Osaka Vacuum Instruments, Osaka, Japan) operated at a pressure of 3–4 × 10−4 Pa. Immediately after deposition, the plate was immersed in 1 mM 11-mercaptoundecanoic acid (Aldrich Chemical Co., Milwaukee, WI) solution in ethanol at room temperature for 24 h to form a self-assembled monolayer (SAM) of the alkanethiol on the gold-evaporated glass plate. The plate was then washed with ethanol and water, and dried under a stream of nitrogen gas.
Branched poly(ethyleneimine) (PEI; Aldrich) with an average molecular weight of 800, 25 000 and 750 000 (PEI 800, PEI 25000 and PEI 750000, respectively) was dissolved in phosphate-buffered saline (PBS) to a concentration of 1%. The solution pH was adjusted to 7.4 with HCl solution. The glass-based gold electrode having a SAM of carboxylic acid-terminated alkanethiol was exposed to the PEI solution and kept for 30 min at room temperature to electrostatically adsorb PEI onto the surface. Then, the plate was washed with water to remove weakly adsorbed PEI, sterilized with 70% ethanol and air-dried in a sterile laminar flow hood. The following procedures for DNA loading were carried out under sterile conditions.
The cationic polymer-modified plate was then exposed to a plasmid solution (50 μg/ml) in PBS (pH 7.4) for 2 h at room temperature to adsorb plasmid DNA onto its surface. The resulting plasmid-loaded glass plate was extensively washed with PBS to remove any weakly bound plasmid from the surface, and used for later experiments without drying.
To determine the amount of plasmid loaded on the electrode surface, rhodamine-labeled plasmid DNA (pGeneGrip™ Rhodamine/GFP; 5.7 kb; Gene Therapy Systems, Inc., San Diego, CA) was loaded by the same procedure as described above but in the dark and visualized with a fluorescence microscope (MZ FLIII fluorescence stereomicroscope; Leica, Germany) equipped with a cooled-CCD camera (ORCA-ER; Hamamatsu Photonics K.K., Shizuoka, Japan). The average fluorescence intensity was measured on the plate surface analyzing an area of 13 mm × 13 mm using an AQUA-Lite digital imaging software (Hamamatsu Photonics) and converted to the density of loaded plasmid using a calibration curve that was obtained by depositing the plasmid DNA of known amounts on the PEI-adsorbed gold surface.
Electroporation of cells on the plasmid-loaded electrode
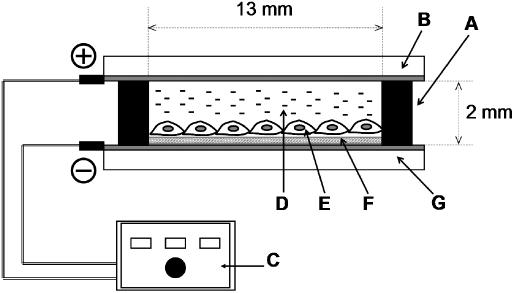
An electroporation setup is schematically depicted in Figure 1. A sterilized silicone frame (inner area, 13 mm × 13 mm; Figure 1, A) was used to confine the cell culture region. A suspension of HEK293 cells was transferred onto the plasmid-loaded surface within the silicone frame at a density of 4.5 × 104 cells/cm2. The plate was placed in a 35 mm polystyrene cell culture dish, and cultured at 37°C under 5% CO2 atmosphere normally for 24 h to allow attachment of cells to the plasmid-loaded electrode surface. When the effect of pre-culture was studied, the culture period was prolonged up to 168 h. The attached cells were washed with PBS, and the inner space of the silicone frame was filled with cold PBS. A counter electrode (gold-evaporated glass plate; Figure 1, B) was placed on the frame with the gold surface down at a distance of 2 mm above the plasmid-loaded electrode. Lower and upper electrodes were then connected to a pulse generator (ElectroSquarePorator T820, BTX, San Diego, CA; Figure 1, C). Cells at 70–80% confluency were treated with a single or multiple electric pulse at a field strength of 50–250 V/cm for a duration of 10 ms. After a 10-min incubation at room temperature, PBS was replaced with a growth medium, and the cells were further cultured.
Figure 1.
A setup for electroporation on a plasmid-loaded electrode. Silicone frame (A), counter electrode (B), electric pulse generator (C), PBS (D), cell (E), plasmid (F) and PEI-adsorbed electrode (G).
Electroporation of primary cells was carried out as described above, but the cell seeding density and the period of pre-culture before electric pulsing were 4.5 × 104 cells/cm2 and 24 h for PMEF, and 2.0 × 105 cells/cm2 and 72 h for hippocampal neurons. A single electric pulse was applied to these cells at a field strength of 100 V/cm for a 10 ms pulse duration.
The number of cells expressing fluorescence proteins (EGFP and DsRed) was counted 48 h after electroporation at five randomly chosen areas under an epifluorescence microscope (BX-51; Olympus, Tokyo, Japan). In a separate experiment, viable cells were determined from the number of trypan blue-excluding cells 48 h after electric pulsing. To assess viability, cells were cultured on the plasmid-loaded electrode for 72 h without electric pulsing, and the number of viable cells was determined as a control in the same manner as just described. The transfection efficiency and cell viability were determined according to the following equations:

All experiments were performed at least five times, and values are expressed as a mean ± SD.
DNA release assay
An electric pulse was applied to the plasmid-loaded electrode on which no cells were cultured. Then the amount of released DNA into the medium (PBS) was determined with the intercalating fluorescent dye, PicoGreen (Molecular Probes, Inc., Eugene, OR). The fluorescence intensity was measured with a fluorescence spectrophotometer (F-2500, Hitachi, Ltd., Tokyo, Japan) at excitation and emission wavelengths of 480 and 525 nm, respectively. All experiments were performed three times, and values are expressed as mean ± SD.
Spatially controlled gene transfer
A plasmid-arrayed electrode was prepared in a similar fashion to that described previously (6). In brief, a SAM was prepared on a gold-coated glass surface using 1-hexadecanthiol (Tokyo Kasei Kogyo Co., Tokyo, Japan) or methoxypoly(ethylene glycol) thiol (SUNBRIGHT ME-050SH, Mw = 5000, NOF Corporation, Tokyo, Japan). These monolayers were irradiated with an ultraviolet light through a photomask having an array of transparent, square or circular dots. The plate was then immersed in an 11-mercaptoundecanoic acid solution to form a carboxylic acid-terminated SAM within the irradiated regions. PEI and plasmid DNA were sequentially adsorbed by manually pipetting their solutions onto the carboxylic acid-terminated SAM. The electroporation of cells on the plasmid-loaded electrode was performed in the same manner as described above to examine the possibility of spatially controlled gene transfer.
Conventional lipofection and electroporation
Control experiments of cell transfection were performed by conventional lipofection and electroporation. In lipofection, Lipofectamine™ 2000 (Invitrogen) was used according to the manufacturer's instructions. Briefly, a plasmid (pEGFP-C1; 1 μg) and Lipofectamine™ 2000 (3 μl) were added to 0.1 ml of Opti-MEM (Invitrogen) to form lipid–plasmid complexes. Hippocampus neurons and PMEF cultured in a 24-well cell culture plate were incubated in Opti-MEM (0.5 ml) containing the complexes for 4 h at 37°C and 5% CO2, and further incubated for 48 h in 0.5 ml of a fresh complete medium without the lipid–plasmid complexes (20). Electroporation of the plasmid into neurons was performed according to the standard procedure (21) in which a cell suspension (0.5 ml, 3.0 × 106 cells/ml) in PBS containing 15 μg of a plasmid (pEGFP-C1) was treated with three pulses of 10 ms duration each at a field strength of 250 V/cm in an electroporation cuvette (0.4 mm gap between electrodes; BTX). After 48 h of cell culture, the transfection efficiency and cell viability were evaluated in the same manner as described above.
RESULTS
Electric pulse-triggered transfection on a DNA-loaded electrode
EGFP was successfully expressed by HEK293 cells electroporated on the electrode to which plasmid DNA and PEI 800 were electrostatically adsorbed (Figure 2A). The transfection efficiency was ∼90%. A control experiment in which cells were cultured on the plasmid-loaded electrode with no exposure to electric pulses resulted in no expression of EGFP (Figure 2B). EGFP expression could be observed 5–6 h after electroporation and reached a maximum 48 h after electroporation, followed by a gradual decrease most likely due to the proliferation of cells and plasmid degradation. Other water-soluble cationic polymers, such as poly(allylamine) and poly-L-lysine, were used instead of PEI which resulted in a slightly lower transfection efficiency, though conditions of electrode preparation and pulsing for these polymers was not optimized. In addition, reverse polarity of the electric field where the plasmid-loaded electrode was used as an anode, produced a much lower transfection efficiency compared to the reverse set-up.
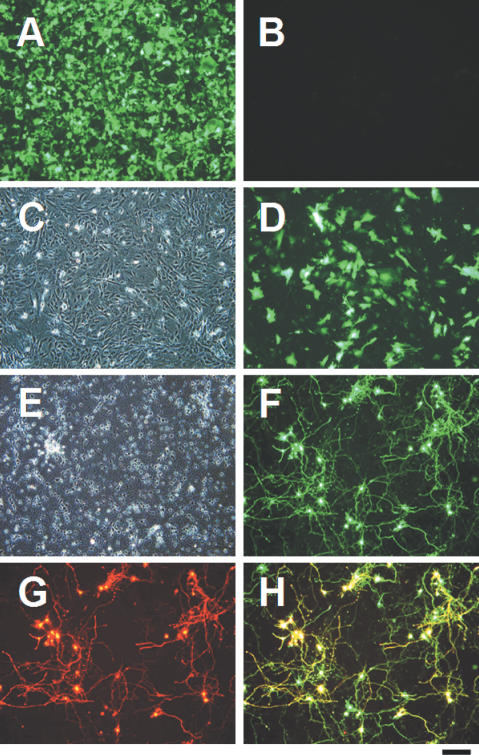
Figure 2.
Electric pulse-triggered transfection on a plasmid-loaded electrode. HEK293 cells and PMEF were cultured on the pEGFP-loaded electrode for 24 h, and then treated with a single electric pulse for 10 ms duration at the field strengths of 75 and 100 V/cm, respectively. Primary hippocampal neurons were cultured on the pEGFP/pDsRed-loaded electrode for 72 h, and then treated with a single electric pulse for 10 ms duration at 100 V/cm. Two days after electroporation, cells were observed for assessing EGFP and DsRed expression. (A) Fluorescence image of EGFP-expressing HEK293 cells treated with an electric pulse. (B) No EGFP was expressed by HEK293 on a plasmid-loaded electrode when an electric pulse had not been applied. (C) Phase-contrast image of transfected PMEF. (D) Fluorescence image of the same sight as in (C). (E) Phase-contrast image of transfected hippocampal neurons. (F and G) Fluorescence images of the same sight as in (E) acquired using an interference filter for (F) EGFP and (G) DsRed. (H) Overlay image of (F) and (G). Scale bar = 200 μm.
We also succeeded in gene transfer into PMEF cells (Figure 2C and D). Furthermore, electroporation of two independent plasmids encoding EGFP and DsRed into hippocampal neurons also resulted in the co-expression of both fluorescent proteins in many cells (Figure 2E–H), indicating that two different plasmids could be co-introduced into individual cells. Transfection efficiency was approximately 40% and 25% for PMEF and hippocampus neurons, respectively, with 90% cell viability for both cases. On the other hand, transfection of these cells by the conventional electroporation or lipofection methods under optimal conditions resulted in an efficiency of 10–20% with only 50–70% cell viability (data not shown).
Influence of the molecular weight of PEI
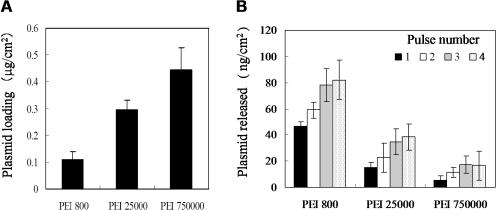
To study the influence of the molecular weight of PEI on plasmid loading, a rhodamine-labeled plasmid DNA was electrostatically adsorbed to the electrode surface which had been modified with PEI 800, PEI 25000 or PEI 750000. The fluorescence intensity measured on the plasmid-loaded surface was converted to the surface density of the plasmid using a calibration curve obtained separately. As shown in Figure 3A, an increase in the molecular weight of PEI gave rise to a larger amount of plasmid loading onto the electrode surface. This result may be explained from the molecular weight dependence on the extent of surface charge reversal by PEI adsorption.
Figure 3.
Influence of the molecular weight of PEI on the amount of loaded and released plasmids. (A) Amount of plasmid DNA loaded on the PEI-adsorbed electrode surface. (B) Amount of plasmid DNA released into PBS after the electrode was pulsed 1 to 4 times (each 10 ms duration) at the field strength of 75 V/cm. The data are expressed as a mean ± SD for three independent experiments.
In Figure 3B, the amount of plasmid DNA released from the electrode upon application of electric pulses is plotted as a function of the molecular weight of PEI and the number of pulses applied. As seen clearly, an increase in the molecular weight of PEI resulted in a decrease in the amount of released DNA when comparing data for the same pulse number. Furthermore, no release of DNA was observed from any surface unless an electric pulse was applied. In addition, the amount of released plasmid increased with increasing pulse numbers, regardless of the molecular weight of PEI. These results indicate that PEI of a lower molecular weight is more effective in releasing plasmid upon electric pulsing compared to those of higher molecular weight. In a preliminary study using surface plasmon imaging, results suggested that PEI is also released in the medium together with plasmid DNA.
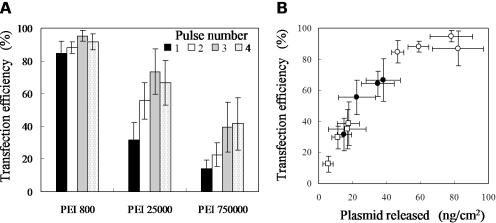
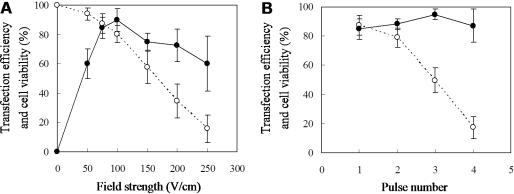
As shown in Figure 4A, transfection efficiency increased with a decreasing molecular weight of PEI. The highest efficiency was achieved when PEI 800 was adsorbed to the electrode surface. The influence of pulse number was not detected when PEI 800 was used, whereas PEI 25000 and PEI 750000 resulted in enhanced transfection by increasing the pulse number up to 3. When 4 electric pulses were applied to the electrode prepared with PEI 25000 and PEI 750000, transfection efficiency was similar to that obtained with 3 pulses; however, severe damage of the cells was observed with 4 pulses compared to the application of 3 pulses.
Figure 4.
Influence of the molecular weight of PEI on the transfection efficiency. Transfection efficiency is plotted as a function of (A) the molecular weight of PEI and (B) the amount of plasmid DNA released upon electric pulsing at the field strength of 75 V/cm with a pulse number of 1–4 and a pulse duration of 10 ms. Molecular weight of PEI: (open circles) 800, (solid circles) 25 000, and (open squares) 750 000. The data are expressed as a mean ± SD for five independent experiments.
The results shown in Figures 3 and 4 led us to suppose that transfection efficiency was primarily governed by the amount of plasmid DNA released upon electric pulsing. To examine this hypothesis, the transfection efficiency was plotted against the amount of released DNA for all the cases with different molecular weights of PEI. As clearly demonstrated in Figure 4B, transfection efficiency was directly proportional to the amount of released DNA, though a large amount of released DNA (>60 ng/cm2) resulted in saturation of the efficiency. Based on these results, all of the subsequent experiments were conducted using PEI 800.
Influence of electric field strength
A single electric pulse of different strengths with constant pulse duration of 10 ms was applied to HEK293 cells adhered on the plasmid-loaded electrode. Figure 5A shows the resulting transfection efficiency as a function of the field strength. No EGFP expression was detected when an electric pulse was not applied (0 V/cm). The transfection efficiency was increased with increasing field strength and reached a maximum level at the field strength of 100 V/cm. At the field strength of 75 and 100 V/cm, ∼80% of the cells survived, and >85% of the live cells expressed EGFP. A further increase in the field strength caused a considerable decrease in cell viability, which was associated with slight reduction in transfection efficiency. This is most likely due to severe cell damage under the stronger electric fields. In Figure 5B, the effect of pulse number is shown for electroporation at the field strength of 75 V/cm. As can be seen, multiple pulsing had no appreciable effect on the transfection efficiency, but affected cell viability considerably. In addition, the prolonged duration of pulsing up to 90 ms had little effect on the transfection efficiency but reduced cell viability, when a single pulse was applied at the field strength of 75 V/cm (data not shown).
Figure 5.
Effects of field strength (A) and pulse number (B) on transfection efficiency and cell viability. HEK293 cells on the pEGFP-loaded electrode were treated with electric pulses at 10 ms duration. (A) A single pulse was applied at various field strengths between 0 and 250 V/cm. (B) The number of pulses was varied between 1 and 4 at the field strength of 75 V/cm. Transfection efficiency (solid circle) and percent cell viability (open circle) are expressed as a mean ± SD for five independent experiments.
Based on the results shown above, we optimized electric conditions for HEK293 cells: a single pulse for 10 ms duration at a field strength of 75 V/cm.
Temporally controlled electroporation
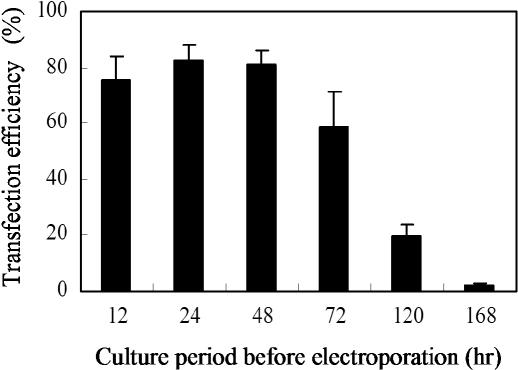
To examine whether our method can be used for controlling the time of transfection, an electric pulse was applied 12–168 h after cell seeding, and transfection efficiency was determined 48 h after electric pulsing, with the results shown in Figure 6. Transfection efficiency was maintained more than 60% when cells were cultured for 12–72 h before applying an electric pulse. These results indicate that the loaded plasmids retained their gene expression activity during at least 3 days under a cell culture environment, showing the versatility of our method for transfecting cells in a temporally controlled manner. Cells cultured for more than 120 h before electric pulsing, however, exhibited a transfection efficiency less than 20%. This is probably due to the gradual degradation of plasmids in a serum-containing culture medium.
Figure 6.
Temporally controlled gene transfer into HEK293 cells on the plasmid-loaded electrode. HEK293 cells were treated with a single pulse at 75 V/cm at different time points after cell seeding. The transfection efficiency was determined 48 h after electroporation.
Spatially controlled electroporation
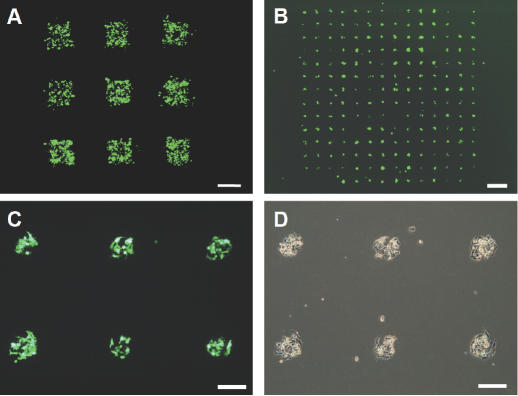
The plasmid-arrayed electrode was prepared by the photo-assisted patterning technique and used for transfecting cells at defined regions in the 2D culture of HEK293 cells. Figure 7A shows representative results of electroporation on the electrode onto which PEI 800 and the EGFP-coding plasmid were adsorbed to 9 square islands (1 mm × 1 mm) presenting a carboxylic acid-terminated SAM. EGFP-expressing cells can be seen only on the plasmid-loaded square islands but not on the surrounding regions, demonstrating that electroporation could be spatially controlled on the millimeter length scale. As shown in Figure 7B–D, the size of the islands could be reduced down to 200 μm in diameter. These photographs also demonstrate that cell adhesion can be prevented effectively by tethering poly(ethylene glycol) chains to the surface of surrounding areas between islands.
Figure 7.
Spatially restricted gene transfer. HEK293 cells were seeded on the plasmid-arrayed electrode, cultured for 24 h, and then treated with a single electric pulse at the field strength of 75 V/cm for 10 ms duration. Fluorescence images were acquired 48 h after electric pulsing. (A) Fluorescence image showing EGFP expression in the restricted regions of HEK293 cells 48 h after electric pulsing. Note that cells are adhered to the entire surface of the electrode, while fluorescently active cells are observed only in the plasmid-loaded square islands. (B) Fluorescence image of HEK293 cells transfected on the electrode with a plasmid-loaded island array prepared using the poly(ethylene glycol)-tethering alkanethiol. (C) Higher magnification image of (B). (D) Phase-contrast image of (C). Note that cells adhere only on the islands but not on the surrounding regions. Scale bar: (A and B) 1 mm; (C and D) 200 μm.
DISCUSSION
Electroporation is one of the most efficient methods for introducing nucleotides into cells (14–16). However, one cannot precisely restrict the regions of transfection in a 2D cell culture using the conventional procedures. Furthermore, since electroporation is usually carried out for cells suspended in a DNA-containing medium, gene transfer is difficult for cells in the adherent state. The present study demonstrates that electroporation using the plasmid-loaded electrode provides a means to transfect various types of cells, including primary cells, in a spatially and temporally controlled manner and thus, a useful way to prepare a transfectional microarray.
Other approaches were reported for spatially and temporally controlling electroporation in vitro and in vivo (22–26). In a typical case, a plasmid solution is locally administered to adhering cells with an aid of a micropipette followed by treatment with electric pulses using miniature electrodes. However, it seems that site specificity is limited in these methods because a DNA solution was immediately spread over the medium. More importantly, these methods are not practical when multiple genes are introduced in parallel, as required for transfection on a microarray.
In this study, we demonstrated that spatially and temporally controlled transfection was achieved by using the plasmid-loaded gold electrode. In this method, the basement gold layer has dual roles: the sulfur atom of the alkanethiol binds to the gold substrate, forming a SAM on the surface, while a conductive gold layer is also necessary for applying electric pulses. The plasmid-loaded electrode was prepared by adsorbing plasmid DNA onto the gold surface that had been modified with a carboxylic acid-terminated SAM and PEI. Since PEI is a cationic polymer, it is expected that the anionic SAM surface attracts PEI molecules by electrostatic interactions. As a result, the electrode surface is modified with an adsorbed layer of PEI. In such a system, the outermost surface of the electrode is likely to carry a net positive charge due to excess cationic moieties (27), and thus attracts DNA molecules bearing net negative charges due to the dissociation of backbone phosphate groups. Eventually, plasmid DNA forms stable polyelectrolyte complexes with the PEI loaded on the electrode surface.
We showed that EGFP coded by the plasmid vector was efficiently expressed in the cells when electric pulses were applied to the cells typically pre-cultured for 24 h on the plasmid-loaded electrode. It is likely that the electric pulse-triggered gene expression involves two crucial steps. First, the loaded plasmid detaches from the electrode surface upon pulsing at the extremely high electric field. This is evidenced by the results of the release tests (Figure 3B). The second step may include electropermeabilization by which transient pores are generated in the cellular membranes upon application of electric pulses, as in the case with the conventional electroporation (28). It is thought that nucleotides are transported into the cytoplasm by electrophoresis across the cell membrane through the pores (29).
The transfection efficiency was dependent to a great extent on the molecular weight of the PEI (Figure 4A): The efficiency increased in the order of PEI 750000 < PEI 25000 < PEI 800, when compared under the same pulsing conditions. This trend was well correlated to the amount of plasmids released upon electric pulsing from the surfaces with the adsorbed PEI of different molecular weights (Figure 4B), while an opposite tendency was observed for the loading amounts. These findings suggest that availability of plasmids for adjacent cells is the predominant factor that governs transfection efficiency. It is probable that a PEI of a lower molecular weight forms an adsorbed layer more unstable against electric pulses than those of higher molecular weights. Accordingly, we concluded that the surface with adsorbed PEI 800 was the most effective surface studied.
The transfection efficiency also varied depending on the field strength of the electric pulse (Figure 5A). This effect can be explained in part from the enhancement of plasmid release by the increasing field strength. In addition, intensive pulsing may promote the creation of transient pores in a cell membrane and the internalization of DNA into the cytoplasm (28). The results that the number of pulses and the duration of each pulse had little effect on transfection efficiency (Figure 5B) may be attributed to the fact that rather mild conditions are sufficient for the release of the largest part of the loaded plasmid. Conversely, the total energy, which is proportional to the field strength and the pulse number, was shown to greatly affect the viability of cells after pulsing (Figure 5A and B). Therefore, pulse conditions should be balanced in order to achieve the optimal efficiency of transfection.
The spatial control of transfection was accomplished by utilizing the micropatterning of a SAM formed on a gold electrode (Figure 7). The size, shape and density of DNA-loaded islands can be readily designed using an appropriate photomask. A single island created in this study had an area of 0.03 (circular) and 1 mm2 (square), to which approximately 30 and 2000 cells adhered, respectively, in the typical case with HEK293 cells. In an integrated transfectional microarray, cross-contamination between different plasmids spotted to the juxtapose islands should be avoided. The results shown in Figure 7 indicate that transfection is sufficiently restricted within the islands on the SAM-based plasmid microarray, illustrating that this method will be advantageous in expressing multiple genes in parallel for the high-throughput functional analysis at a cellular level.
Another important feature of our method is that electroporation can be performed on adherent cells pre-cultured for a certain period of time. No transfection occurred during pre-culture (Figure 2B), and gene expression could be initiated by electric pulsing even after a 3-day pre-culture without a marked loss of transfection efficiency (Figure 6). These results suggest that loaded plasmid remains adsorbed to the electrode during the pre-culture period. It is also likely that the polyelectrolyte complex formation with PEI serves to protect plasmids from nuclease-mediated degradation. These results clearly demonstrate the possibility of transferring plasmids into cells at a preferred time after cell seeding.
The possibility of temporal control will be of great advantage in the studies of genes whose functions are required to synchronize closely with the specific state of cells or their organization. For example, as shown in Figure 2D, our method could be used to express exogeneous genes in neurons which had developed cellular networks. Another potential application of our method will be the transfection into polarized cells or organized cellular structures. For these applications, conventional electroporation is not appropriate, because cellular structure, such as cell–cell junction structure, would be disrupted during a required harvesting procedure. For instance, cell detachment by trypsin treatment can cause damage to the membrane proteins, plasma membrane and cytoskeleton, resulting in low transfection efficiency and also reduced cell viability (30,31). To overcome this limitation, several approaches have been reported for in situ electroporation of adherent cells (31–38). However, as mentioned earlier, these methods are ineffective for specifically restricting the area of transfection because of the immediate spreading of a DNA solution over the medium.
In conclusion, we developed a novel method for controlling the location and the time of gene transfection. By the application of a short electric pulse to the plasmid-loaded electrode, cells directly cultured on the electrode surface expressed the encoded genes. It was found that the pulse condition and electrode preparation had great effects on the transfection efficiency. The efficiency was primarily governed by the availability of DNA released from the electrode surface and the physical effects such as membrane permeabilization and cellular damage. Using the plasmid-loaded electrode, the time of transfection could also be controlled when the period of pre-culture was within 3 days. Moreover, it was shown that the micropatterning technique provided a way to precisely control the location of gene transfection, demonstrating the potential of the method for creating transfectional microarrays.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Dr Tracy Richey for her valuable comments on the manuscript. This study was partly supported by Kobe Cluster, the Knowledge-Based Cluster Creation Project, Ministry of Education, Culture, Sports, Science and Technology (MEXT).
REFERENCES
- 1.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Tyers M. and Mann,M. (2003) From genomics to proteomics. Nature, 422, 193–197. [DOI] [PubMed] [Google Scholar]
- 4.Blagoev B. and Pandey,A. (2001) Microarrays go live–new prospects for proteomics. Trends Biochem. Sci., 26, 639–641. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter A.E. and Sabatini,D.M. (2004) Systematic genome-wide screens of gene function. Nature Rev. Genet., 5, 11–22. [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi F., Kato,K. and Iwata,H. (2004) Micropatterned, self-assembled monolayers for fabrication of transfected cell microarrays. Biochim. Biophys. Acta, 1672, 138–147. [DOI] [PubMed] [Google Scholar]
- 7.Ziauddin J. and Sabatini,D.M. (2001) Microarrays of cells expressing defined cDNAs. Nature, 411, 107–110. [DOI] [PubMed] [Google Scholar]
- 8.Yoshikawa T., Uchimura,E., Kishi,M., Funeriu,D.P., Miyake,M. and Miyake,J. (2004) Transfection microarray of human mesenchymal stem cells and on-chip siRNA gene knockdown. J. Control. Release, 96, 227–232. [DOI] [PubMed] [Google Scholar]
- 9.Chang F.H., Lee,C.H., Chen,M.T., Kuo,C.C., Chiang,Y.L., Hang,C.Y. and Roffler,S. (2004) Surfection: a new platform for transfected cell arrays. Nucleic Acids Res., 32, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baghdoyan S., Roupioz,Y., Pitaval,A., Castel,D., Khomyakova,E., Papine,A., Soussaline,F. and Gidrol,X. (2004) Quantitative analysis of highly parallel transfection in cell microarrays. Nucleic Acids Res., 32, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva J.M., Mizuno,H., Brady,A., Lucito,R. and Hannon,G.J. (2004) RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc. Natl Acad. Sci. USA, 101, 6548–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mousses S., Caplen,N.J., Cornelison,R., Weaver,D., Basik,M., Hautaniemi,S., Elkahloun,A.G., Lotufo,R.A., Choudary,A., Dougherty,E.R. et al. (2003) RNAi microarray analysis in cultured mammalian cells. Genome Res., 13, 2341–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R., Conklin,D.S. and Mittal,V. (2003) High-throughput selection of effective RNAi probes for gene silencing. Genome Res., 13, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann E., Schaefer-Ridder,M., Wang,Y. and Hofschneider,P.H. (1982) Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J., 1, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gehl J. (2003) Electroporation: theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol. Scand., 177, 437–447. [DOI] [PubMed] [Google Scholar]
- 16.Jaroszeski M.J., Gilbert,R., Nicolau,C. and Heller,R. (1999) In vivo gene delivery by electroporation. Adv. Drug Deliv. Rev., 35, 131–137. [DOI] [PubMed] [Google Scholar]
- 17.Evans M.S., Collings,M.A. and Brewer,G.J. (1998) Electrophysiology of embryonic, adult and aged rat hippocampal neurons in serum-free culture. J. Neurosci. Meth., 79, 37–46. [DOI] [PubMed] [Google Scholar]
- 18.Brewer G.J. and Price,P.J. (1996) Viable cultured neurons in ambient carbon dioxide and hibernation storage for a month. NeuroReport, 7, 1509–1512. [DOI] [PubMed] [Google Scholar]
- 19.Freshney R.I. (1983) Disaggregation of the tissue and primary culture. In Freshney,R.I. (ed.), Culture of Animal Cells. Alan R. Liss, Inc., NY, pp. 99–l18. [Google Scholar]
- 20.Dalby B., Cates,S., Harris,A., Ohki,E.C., Tilkins,M.L., Price,P.J. and Ciccarone,V.C. (2004) Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods, 33, 95–103. [DOI] [PubMed] [Google Scholar]
- 21.Chang D.C. (1992) Design of protocols for electroporation and electrofusion: selection of electrical parameters. In Chang,D.C., Chassy,B.M., Saunders,J.A. and Sowers,A.E. (eds), Guide to Electroporation and Electrofusion. Academic Press, NY, pp. 429–455. [Google Scholar]
- 22.Lundqvist J.A., Sahlin,F., Aberg,M.A., Stromberg,A., Eriksson,P.S. and Orwar,O. (1998) Altering the biochemical state of individual cultured cells and organelles with ultramicroelectrodes. Proc. Natl Acad. Sci. USA, 95, 10356–10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olofsson J., Nolkrantz,K., Ryttsen,F., Lambie,B.A., Weber,S.G. and Orwar,O. (2003) Single-cell electroporation. Curr. Opin. Biotechnol., 14, 29–34. [DOI] [PubMed] [Google Scholar]
- 24.Haas K., Sin,W.C., Javaherian,A., Li,Z. and Cline,H.T. (2001) Single-cell electroporation for gene transfer in vivo. Neuron, 29, 583–591. [DOI] [PubMed] [Google Scholar]
- 25.Saito T. and Nakatsuji,N. (2001) Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol., 240, 237–246. [DOI] [PubMed] [Google Scholar]
- 26.Pekarik V., Bourikas,D., Miglino,N., Joset,P., Preiswerk,S. and Stoeckli,E.T. (2003) Screening for gene function in chicken embryo using RNAi and electroporation. Nat. Biotechnol., 21, 93–96. [DOI] [PubMed] [Google Scholar]
- 27.Decher G. (1997) Fuzzy nanoassemblies: toward layered polymeric multicomposites. Science, 277, 1232–1237. [Google Scholar]
- 28.Neumann E., Kakorin,S. and Toensing,K. (1999) Fundamentals of electroporative delivery of drugs and genes. Bioelectrochem. Bioenerg., 48, 3–16. [DOI] [PubMed] [Google Scholar]
- 29.Sukharev S.I., Klenchin,V.A., Serov,S.M., Chernomordik,L.V. and Chizmadzhev,Y. (1992) Electroporation and electrophoretic DNA transfer into cells. The effect of DNA interaction with electropores. Biophys. J., 63, 1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nanda G.S. and Mishra,K.P. (1994) Modification of electroporative response of erythrocytes by EDTA in isotonic solutions. Bioelectrochem. Bioenerg., 34, 189–193. [Google Scholar]
- 31.Zheng Q. and Chang,D.C. (1991) High-efficiency gene transfection by in situ electroporation of cultured cells. Biochim. Biophys. Acta, 1088, 104–110. [DOI] [PubMed] [Google Scholar]
- 32.Raptis L. and Firth,K.L. (1990) Electroporation of adherent cells in situ. DNA Cell Biol., 9, 615–621. [DOI] [PubMed] [Google Scholar]
- 33.Wegener J., Keese,C.R. and Giaever,I. (2002) Recovery of adherent cells after in situ electroporation monitored electrically. Biotechniques, 33, 348–357. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y.C., Li,M., Fan,C.S. and Wu,L.W. (2003) A microchip for electroporation of primary endothelial cells. Sens. Actuators, A, 108, 12–19. [Google Scholar]
- 35.Raptis L., Firth,K.L., Tomai,E. and Forkert,P.G. (2000) Improved procedure for examination of gap junctional intercellular communication by in situ electroporation on a partly conductive slide. Biotechniques, 29, 222–226. [DOI] [PubMed] [Google Scholar]
- 36.Raptis L., Balboa,V., Hsu,T., Vultur,A., Turkson,J., Jove,R. and Firth,K.L. (2003) In situ electroporation of large numbers of cells using minimal volumes of material. Anal. Biochem., 317, 124–128. [DOI] [PubMed] [Google Scholar]
- 37.Yang T.A., Heiser,W.C. and Sedivy,J.M. (1995) Efficient in situ electroporation of mammalian cells grown on microporous membrane. Nucleic Acids Res., 23, 2803–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller K.J., Horbaschek,M., Lucas,K., Zimmermann,U. and Sukhorukov,V.L. (2003) Electrotransfection of anchorage-dependent mammalian cells. Exp. Cell Res., 288, 344–353. [DOI] [PubMed] [Google Scholar]