Abstract
The rickettsia Anaplasma marginale is the most prevalent tick-borne livestock pathogen worldwide and is a severe constraint to animal health. A. marginale establishes lifelong persistence in infected ruminants and these animals serve as a reservoir for ticks to acquire and transmit the pathogen. Within the mammalian host, A. marginale generates antigenic variants by changing a surface coat composed of numerous proteins. By sequencing and annotating the complete 1,197,687-bp genome of the St. Maries strain of A. marginale, we show that this surface coat is dominated by two families containing immunodominant proteins: the msp2 superfamily and the msp1 superfamily. Of the 949 annotated coding sequences, just 62 are predicted to be outer membrane proteins, and of these, 49 belong to one of these two superfamilies. The genome contains unusual functional pseudogenes that belong to the msp2 superfamily and play an integral role in surface coat antigenic variation, and are thus distinctly different from pseudogenes described as byproducts of reductive evolution in other Rickettsiales.
Keywords: rickettsiales, bacterial artificial chromosome, St. Maries strain
Anaplasma marginale, transmitted by ixodid ticks, is the most prevalent tick-borne pathogen of cattle with a world-wide distribution. Acute disease manifests with anemia, weight loss, and often, death. In animals that survive acute disease, A. marginale establishes life-long persistent infection. Persistently infected animals are clinically healthy but serve as reservoirs for continued transmission of the organism; these reservoirs are required because there is no transovarial transmission of the pathogen by the tick vector. Despite its global impact on animal health, there is currently no widely accepted vaccine for A. marginale (for review, see refs. 1 and 2). Related pathogens in the order Rickettsiales include those causing recently emergent tick transmitted diseases such as human granulocytic anaplasmosis (Anaplasma phagocytophilum) and human monocytic ehrlichiosis (Ehrlichia chaffeensis), as well as established diseases such as African heartwater (Ehrlichia ruminantium) and Mediterranean spotted fever (Rickettsia conorii) (see Fig. 4, which is published as supporting information on the PNAS web site). Members of this order are small, obligate intracellular bacteria (3) that typically have small genomes, attributed to reductive evolution following long term intracellular parasitism (4-6). Many obligate intracellular bacteria are difficult to culture, and the need to be grown in a host cell makes it difficult to obtain large amounts of organism-specific DNA necessary for whole genome sequencing (6, 7). The small genome size of A. marginale (1.2 Mb) allowed us to use a bacterial artificial chromosome (BAC)-based strategy to obtain the genome sequence without substantial purification of the organism from the host cell. We report here the complete genome sequence of the St. Maries strain of A. marginale, originally isolated from an animal with severe acute anaplasmosis and shown to be efficiently transmitted by both Dermacentor andersoni and Boophilus microplus (8, 9). The completion of this sequence and the E. ruminantium sequence (7) allows comparative genomics to identify conserved genes and pathways associated with transmission.
Materials and Methods
The Organism. A blood stabilate of the St. Maries strain of A. marginale was inoculated into splenectomized calf no. 836, shown to be free of A. marginale by competitive ELISA (10). During peak rickettsemia, >19% of the erythrocytes were infected. Erythrocytes were isolated by using Histopaque (Sigma) and used in the construction of a BAC library as described (11).
Sequencing. BACs were arrayed in duplicate on nylon membranes and screened with a digoxigenin (DIG)-labeled (Roche Applied Science) bovine total genomic DNA probe or with known A. marginale genes of interest. Ten genes were used for initial screening including msp1α, msp1β, msp2, msp3, msp4, msp5, sodB, groEL, 16S, and opag2 under the following conditions: prehybridization in DIG-Easy Hyb (Roche Applied Science) at 42°C for 12 h followed by hybridization in fresh DIG-Easy Hyb with 10-50 ng/ml of denatured probe. High stringency wash conditions were as follows: two washes in 2× SSC, 0.1% SDS (wt/vol) at room temperature, one wash in the same buffer at 65°C, and a final wash in 0.2× SSC, 0.1% SDS (wt/vol) at 65°C (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7). All washes lasted 15 min. BACs containing genes of interest were selected and sequenced by using the random shotgun method. Briefly, BAC DNA was sheared to 3 kb by using a Hydroshear (Gene Machines) and cloned into pCRScript. Eight 96-well plates of subclones were sequenced per 100-kb BAC by using Big Dye chemistry (Applied Biosystems). BACs were assembled by using phred and phrap (University of Washington, Seattle; refs. 12-14) in conjunction with sequencher (Genecodes). Sequence gaps were closed by primer walking on subclones or BAC DNA. These initial BACs created nucleation points for walking experiments where a BAC with the shortest overlap was chosen for sequencing, and contigs of sequenced BACs were assembled. Each BAC had an average of 7× coverage. Physical gaps were closed by using long-distance PCR (Herculase; Stratagene) on genomic DNA. The ordering of contigs was confirmed by Southern analysis of pulsed field-separated DNA digested with PacI and PmeI (data not shown). The completed sequence has been deposited in GenBank (accession no. CP000030).
Annotation. ORFs likely to encode proteins, coding sequences (CDSs), were predicted by glimmer2 (15) and orpheus (16). All predicted proteins were searched against a nonredundant protein database (nr, National Center for Biotechnology Information) (17). To identify sequencing errors, 300-bp extensions of each ORF >300 bp were compared by blastx to a nr database (17). ORFs with the same blast identity as that of the adjacent sequence were inspected for frameshifts in the sequence and errors corrected where appropriate; adjacent ORFs that did not have a frameshift and had blast identity to different regions of the same protein were considered to be authentic mutated genes and were annotated as a “split domain” by using the convention established for R. conorii (5). The start of each CDS was inspected to define initiation codons by comparing the glimmer2 and orpheus output and by using blast alignments and rbsfinder (The Institute for Genomic Research, Rockville, MD). Putative signal peptides were identified with signalp (version 3; ref. 18). A hidden Markov model was used to determine CDS membership in families and superfamilies by using the Pfam protein families database (19). Guidelines for annotating CDSs were as follows: (i) CDSs with blast scores >100 (<e-20), consistent matches to homologous sequences in the blast output, and sequence similarity throughout the translated product were assigned a gene name and symbol; (ii) CDSs with scores of 50-100 (<e-5) were called conserved hypothetical proteins, or conserved protein family members if a significant pfam score accompanied the blast result; (iii) CDSs identified by glimmer2 with a blast score less than that indicated for conserved hypotheticals were called hypothetical proteins. Each identified CDS was assigned to a category of the Cluster of Orthologous Groups (COG) database (20). Paralogous families were determined by performing an all-versus-all search of the predicted sequences. Repeats were identified by using reputer (14, 21). Transfer RNAs were identified by using trnascan-se (22). Base pair 1 was assigned arbitrarily based on GC nucleotide skew (G - C/G + C) analysis (23). Identified CDSs were assigned to pathways by using the kegg database (24) and ecocyc (25).
Results and Discussion
Sequencing Strategy. A. marginale is an obligate intracellular bacterium that invades and replicates in bovine erythrocytes. The DNA of a single contaminating leukocyte in blood collected from an infected animal is equivalent to ≈3,000 A. marginale genomes; therefore, a small amount of contamination would result in a large percentage of bovine DNA in the sequencing project. Thus, we used a strategy to create a BAC library of pathogen and host cell DNA, with only minimal purification of the organism from the host cell, followed by selection for clones that contained A. marginale genes. The resultant BAC library contained >60% bovine clones, and these were removed from further consideration. This high level of host cell DNA is validation that a whole genome shotgun approach would have been unsuitable for this organism. Four previously cloned and sequenced A. marginale genes (msp2, msp1α, msp1β, and msp4) were used to select clones for sequencing and to establish nucleation points for walking experiments. As walking progressed, two of the BAC contigs collapsed into one; however, the remaining BAC contigs reached endpoints where there were no overlapping BACs. To fill the gaps, additional probes (groEL and sodB) were used to identify BACs for sequencing. Once each contig had no more overlapping BACs in the library, five gaps remained. The gaps, ranging from 1.6 kb to 16 kb, were spanned by long-distance PCR on genomic DNA and the resulting amplicons were sequenced to yield the final finished sequence. The final assembled sequence is composed of 14 complete BACs, four partial BACs, and five gap-spanning PCR fragments. This BAC-based strategy had the additional benefit of separating large repeat units (of up to 4.2 kb) containing the msp2/3 pseudogenes into separate assembly projects.
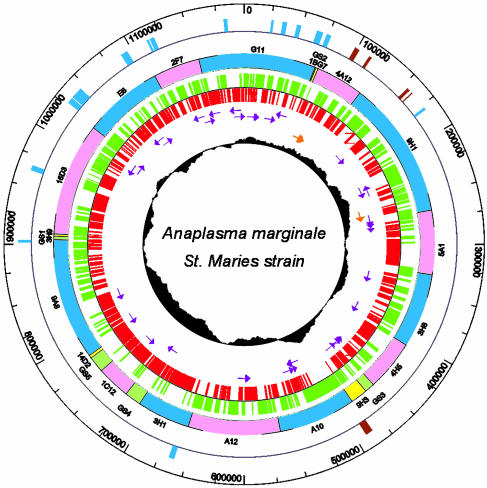
General Features of the Genome. The completed circular genome of the A. marginale St. Maries strain contains 1,197,687 bp and has a G+C content of 49.8%, close to that previously determined by spectral analysis (56 mol%) (26). This G+C content is unusual for obligate intracellular organisms, as many have low G+C contents: the G+C content of the other sequenced Rickettsiales averages 31% (4-7). The origin of replication could not be discerned as the genes (dnaA, gyrA, gyrB, rpmH, dnaN) that are often found clustered near the origin were dispersed throughout the genome and none corresponded with a change in GC or octamer skew (23, 27). Therefore, base pair 1 was set arbitrarily near a change in GC skew. The genome has a high coding density (86%), typical of streamlined intracellular bacteria that have a minimal coding content for maintaining life in particular environmental niches. The A. marginale genome encodes 949 predicted CDSs (Table 1) with a mean size of 1,077 bp. This includes eight CDSs annotated as split domain ORFs, which may be classical pseudogenes. The large mean size of the CDSs is due in part to the presence of several very large CDSs (5-10.5 kb) for which there are no homologs in other closely related bacteria. The genome contains a single split operon of ribosomal RNA genes that seems to be typical of the order Rickettsiales (14). There are 37 tRNA genes representing all 20 amino acids (Fig. 1).
Table 1. General features of the A. marginale genome.
| Genome size, bp | 1,197,687 |
| G + C, % | 49 |
| Protein coding, % | 86 |
| Protein coding genes | 949 |
| Functional assignment | 567 |
| Conserved family assignment | 107 |
| Conserved hypothetical | 126 |
| Hypothetical | 151 |
| Functional pseudogenes | 14 |
| Split domain ORFs | 8 |
| Gene density | 0.79 |
| Mean gene length | 1,077 |
| Ribosomal RNAs | 3 |
| Transfer RNAs | 37 |
Fig. 1.
Genome map of A. marginale. The inner-most circle depicts the GC skew (G - C/G + C). The second and third circles show the position and orientation of rRNA (orange arrows) and tRNA (purple arrows) genes. The fourth and fifth circles show the positions of the predicted CDSs in the reverse (red) and forward (green) orientations. The sixth circle shows the positions of the BACs (full BACs in blue and pink; partial BACs in yellow) and gap-spanning PCR fragments (green) that were sequenced. The seventh circle shows the positions of the msp2 (blue) and msp1 (brown) superfamily genes.
Pseudogenes. Analysis of complete genome sequences of Rickettsia prowazekii, R. conorii, and Wolbachia pipientis wMel has indicated that these obligate intracellular bacteria in the order Rickettsiales have undergone reductive evolution toward highly streamlined genomes containing many pseudogenes (4-6). Although A. marginale has a small genome typical of members of this order, it has relatively few classical pseudogenes, defined as inactive copies of functional genes. Only four genes (murC, aspS, mutL, and aatA) were found with interrupted coding regions and, although these are probably pseudogenes, these were annotated as split domains because functionality remains to be determined. These four genes have a different codon usage than the presumed functional CDSs, but this may be biased because of the small number. Notably, A. marginale has genes defined as functional pseudogenes: truncated copies of genes that are only expressed as part of a functional full-length protein after recombination into a unique expression site (11, 28, 29). Similar functional pseudogenes are also present in A. phagocytophilum (30), but have not yet been described for other genera in the order Rickettsiales.
Membrane Proteins. signalp (version 3; ref. 18) predicted 163 CDSs to contain signal peptides, and all but three contained at least one transmembrane-spanning domain predicted by tmpred (http://ch.embnet.org/software/TMPRED_form.html). Further discrimination of protein location was not computationally possible because psort (http://psort.nibb.ac.jp/form.html) and psortb (http://psort.org/psortb/index) predicted only 43 and 13 outer membrane proteins (OMPs) respectively, missing many of the known A. marginale OMPs. By sequence identity to previously defined OMPs, 13 CDSs were assigned as OMPs. In addition to these 13 OMPs, we describe two previously undefined superfamilies consisting of previously known OMPs and recently identified CDSs: the msp2 superfamily contains 56 members, including 16 pseudogenes, and the msp1 superfamily contains nine members. This brings the total number of predicted OMPs to 62 (not including pseudogenes), consistent with the number expected for a genome of this size (31, 32).
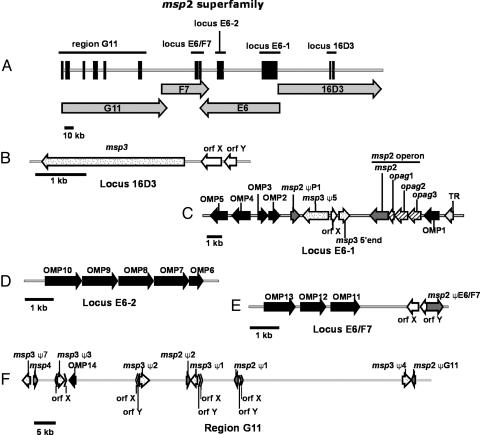
The msp2 Superfamily. MSP2, -3, and -4 reside in the outer membrane with surface exposed domains (33), with MSP2 and -3 being immunodominant proteins that are antigenically variable and serve to evade the host immune response (29, 34-37). The msp2 superfamily (Fig. 2) is built around msp2, msp3, and msp4; the latter two molecules reported to have a low level of sequence identity to msp2 (29, 38). Each of these protein sequences match to pfam01617, a family of surface antigens. The genome contains one full-length expression site gene for msp2, msp3, and msp4. In addition, there are seven functional pseudogenes for msp2 and seven functional pseudogenes for msp3. Four of the msp2 and msp3 functional pseudogenes are closely linked in a tail to tail arrangement known as the pseudogene complex (11). In addition to the functional pseudogenes, there are two remnant sequences of msp3 in the genome, one corresponding to the msp3-specific 5′ end, and another very short sequence corresponding to the conserved 5′ end of a pseudogene. Msp2 is transcribed as part of an operon of four genes, the remaining three genes have been called operon associated genes (opags) 1-3 (39, 40), and have been included in the family. Opags 2 and 3 are also members of pfam01617. The members of the operon display an unusual pattern of differential expression: opag1 does not seem to be translated, OpAG2 and MSP2 are expressed by A. marginale in the bovine erythrocyte and in the tick midgut and salivary gland, whereas OpAG3 is expressed only in the erythrocyte (40). There are 15 previously unidentified genes with sequence identity to the core members of the superfamily (msp2-4) that correspond to pfam01617, and these have been designated OMP1-15. Twelve of these OMP genes are arranged in three clusters representing four putative operons, with the remaining three genes occurring singly (Fig. 2). The remaining members of the superfamily correspond to small genes called orfX (12 copies) and orfY (seven copies) (29). These genes have a signal peptide with sequence identity to MSP3, but otherwise do not correspond to members of pfam01617. They are included as members of the superfamily because of their positional relationship to msp2 and msp3: they are often found flanking an msp2 or an msp3 pseudogene, and are part of the msp3 expression site. When found in conjunction with a pseudogene orfX and -Y are on the strand opposite the pseudogene; however, in the msp3 expression site both are oriented in the same direction as msp3, and are transcribed as part of the msp3 operon (29). Interestingly, orfX and -Y are found in an ≈600-bp repeat (containing two repeat units) that is found in conjunction with the msp2 and msp3 pseudogenes; however, this structure is not absolute, because there are three instances of the repeat that do not contain orfX or -Y (11). This repeat has been hypothesized to function in the recombination of the pseudogenes into the expression site (11).
Fig. 2.
msp2 superfamily schematic. (A) The distribution of many superfamily members in a 360-kb region of the genome that is comprised of four BAC sequences (G11, F7, E6, and 16D3). The loci identified in A are shown in more detail in B-F.(B) The msp3 operon containing orfX, orfY, and msp3 (C). The locus shown contains the msp2 operon, several OMPs and pseudogenes as well as orfX and a putative transcriptional regulator (TR) that is found in the syntenic region in E. ruminantium, E. chaffensis, and A. phagocytophilum.(D) A putative operon of five OMP genes. (E) Another putative operon of OMP genes that is situated near a msp2 pseudogene. The 95-kb region depicted in F illustrates the positional relationship of the msp2/3 pseudogenes with orfX/Y. OMP14 and msp4 also occur in this region of the genome.
The orthologs of msp2 in members of the genus Ehrlichia, E. ruminantium, E. canis and E. chaffeensis (41-43) are arranged as tandemly repeated full-length genes in one (E. ruminantium, E. chaffeensis) or two (E. canis) loci containing 16-25 paralogs. There is synteny between the arrangement of these ehrlichial genes and part of the msp2 superfamily in the region of the msp2 operon in both A. marginale and A. phagocytophilum (30, 44). Interestingly, the mechanism for generating antigenic variation in these immunodominant OMPs is very different between these two genera: Erhlichial species use multiple genes from the tandem array of OMPs, whereas A. marginale and A. phagocytophilum use a recombination mechanism (41-43). One possible explanation for the evolution of these different mechanisms may be mutL, an enzyme involved in mismatch repair. In A. marginale, this gene contains a variable stretch of G residues (9-13) sometimes resulting in a frameshift, and thus an inactive molecule. Genomes with defective mismatch repair have elevated rates of mutation and recombination (45), which is a necessary event for the antigenic variation system used by A. marginale and A. phagocytophilum.
The msp4 gene is known to be difficult to clone in E. coli (46), an observation that may be clarified by the genome sequence. Msp4 is flanked by two msp3 pseudogenes: one 336 bp upstream and the other 4,687 bp downstream from msp4. Additionally, the recA gene is located between the two msp3 pseudogenes. The close proximity of these repeat units coupled with an additional recA likely makes this region of the genome unstable when cloned in prokaryotic vectors.
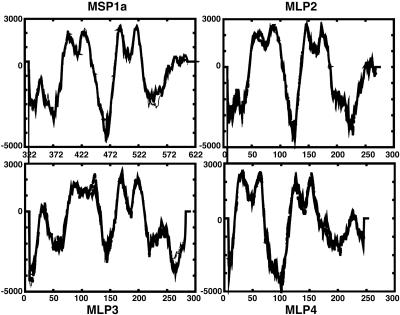
The Msp1 Superfamily. MSP1 is a surface exposed heteromeric complex consisting of MSP1a and MSP1b. Msp1α is a single copy gene and exhibits strain differences caused by a variable number and sequence of tandem repeats units of 86-89 bp in length (47). These repeats have been designated by letters, and the St. Maries strain msp1α contains three repeats with the designation JBB (47). Interestingly, MSP1a has no canonical signal peptide, although it has been demonstrated to be surface exposed (48). We have identified three CDSs immediately downstream from msp1α with structural similarity to the C-terminal half of MSP1a as shown by transmembrane protein predictions (Fig. 3), and designate these as MSP1a-like proteins (MLPs) 2-4. MLP2 and -4 have 30% and 37% sequence identity, respectively, to the C-terminal end of MSP1a, whereas MLP3 has no appreciable sequence identity to MSP1a. Msp1β is encoded by a small multigene family of five genes with two full-length and three partial genes (pg). The arrangement of the genes in the St. Maries strain is similar to the arrangement in the Florida strain (49); however, the gene sequences are not identical between strains. Percent identities between the two strains for each gene are as follows: msp1β-1, 95%; msp1β-2, 77%; msp1β pg1, 99%; msp1β pg2, 28%; and msp1β pg3, 91%. The partial genes may be functional pseudogenes that allow for antigenic variation of MSP1b, but this has not yet been demonstrated.
Fig. 3.
Structural similarity of MSP1a and MLPs. tmpred profiles of MSP1a and the MLPs are shown. Only the C-terminal 300 aa of MSP1a were used for analysis. Peaks >500 are transmembrane-spanning domains.
The proteins in the msp2 and msp1 superfamilies represent a significant proportion of the molecules expected on the surface of the organism. MSP1, MSP2, and MSP3 are immunodominant molecules to which most of the host immune response is targeted (37, 38, 50-57). Persistence of A. marginale in the bovine host allows ticks to acquire and subsequently transmit the pathogen, which is a requirement because ticks do not maintain A. marginale between generations. Persistent infection is maintained through antigenic variation of MSP2 and MSP3, permitting the organism to evade the host immune response. The genome sequence has allowed elucidation of the combinatorial segmental gene conversion mechanism used by A. marginale to effect this antigenic variation (28). Interestingly, although there are seven functional pseudogenes for msp2, two of these are duplicated in the St. Maries strain genome, thereby restricting the number of possible combinatorial variants that can be made as compared to the South Idaho strain, which has at least nine different msp2 pseudogenes (11, 28). We have demonstrated that there is simultaneous switching of msp2 and msp3 variants during infection (36), and the ability of these two molecules to work in concert may serve to amplify antigenic diversity of the surface coat and increase the repertoire used to evade the host immune response.
Paralogous Gene Families. The largest repeat family, both in number and in length, corresponds to the msp2 superfamily. The transcription terminator rho is usually single copy, but has been duplicated in the A. marginale genome and separated by 333 kb (Fig. 5, which is published as supporting information on the PNAS web site). This repeat appears to have occurred during an inversion event around the origin of replication, as seen by comparison with Ehrlichia ruminantium (7), and flanks the inverted element. There are three tandemly occurring CDSs that have a low level of sequence identity to orfX of the msp2 superfamily; however, they are not initiated with the characteristic start sequence that defines the orfX paralogs (MLLK). The recently described aaap gene (58) involved in actin filament formation has two paralogs immediately upstream. Transporter proteins account for four families including eight ATP-binding cassette transport proteins, four major facilitator superfamily proteins, three virB6-like proteins, and two putative symporter proteins. There are two small families of putative cell surface proteins containing two or three members, and a family of four exported protein genes. The remaining 12 families of paralogous genes contain two to four members and range from different enzymes containing shared domains to undefined products. There are no insertion elements present in the genome.
Metabolism. Metabolic reconstruction shows that most of the glycolytic enzymes are present, but neither glucokinase nor a sugar transport system was detected, indicating that A. marginale may primarily use gluconeogenesis. In addition, key enzymes for the Entner-Doudoroff pathway were not found. Very few genes for enzymes involved in amino acid biosynthesis were found: no complete pathways were detected, and enzymes involved in the terminal biosynthetic step were present for just eight amino acids: serine, glycine, proline, tyrosine, cysteine, phenylalanine, glutamine, and glutamate. Aerobic respiration is achieved through the TCA cycle for which a complete set of enzymes is present. Enzymes for the nonoxidative pentose phosphate pathway are present, although transaldolase could not be definitively identified. All enzymes necessary for fatty acid synthesis were found. Complete pathways for de novo purine and pyrimidine biosynthesis were detected.
Transporters. Only a single amino acid transporter (for proline) could be unambiguously assigned. Another transporter was putatively identified for alanine. Given that very few amino acids can be synthesized in A. marginale, it is surprising to find so few transporters for amino acids. However, there are numerous ATP-binding cassette-type transporters with no assigned function, and perhaps some of these may perform this role. Several transport systems were present for cations, anions, and oligopeptides. Two multidrug resistance pumps were identified. Transport systems for ribonucleotides and phosphate were present. The sec pathway for the secretion of polypeptides is present, with a putatively assigned secE, and missing the nonessential component secG. The tat transport system does not appear to be intact, as only tatC was found. A type IV secretion system was identified, arranged as previously reported in A. phagocytophilum and E. chaffeensis (59), and also the same as in E. ruminantium (7): with one operon containing sodB, virB3, virB4, and virB6 and a second, distantly spaced operon containing virB8, virB9, virB10, virB11, and virD4. Unlike A. phagocytophilum and E. chaffeensis, there is no linkage of these operons with antigenic OMP genes (59). In addition to the previously described operons, the virB6 gene is followed by three virB6 paralogs, and there is one additional copy each for virB8 and virB4 that occur distantly in the genome and are unlinked to other type IV secretory system genes.
Cell Wall Components. Several genes for lipopolysaccharide (LPS) biosynthesis were absent. All of the genes for lipid A biosynthesis were missing. A complete pathway for peptidoglycan synthesis was not present: although all genes for diaminopimelate (DAP) synthesis were found, only some of the genes for the synthesis of murein sacculus were present (ddlB, glmU, mraY1, murA,-B,-D,-E,-F, and -G, and slt). The murC gene was present but contains a frameshift and therefore appears to be a pseudogene. The presence of the genes for DAP synthesis was puzzling because these genes are normally associated with lysine biosynthesis, but the gene (lysA)for the final step in the lysine biosynthetic pathway was missing. The lack of a traditional cell wall seems to be a common feature for the family Anaplasmataceae (60), but not the order Rickettsiales because members of the family Rickettsiaceae are capable of synthesizing LPS and peptidoglycan (4, 5). Unlike other members of the family, A. marginale does not seem to be particularly fragile, and may be able to strengthen its cell wall in an alternative way. We have demonstrated that many of the MSPs are covalently and noncovalently linked in homeric and heteromeric complexes on the cell surface that may serve to strengthen the cell wall (33).
Conclusions and Perspectives
The BAC-based approach used to sequence A. marginale avoided problems associated with host DNA contamination that occurs when isolating infected cells directly from the mammalian host. The A. marginale, E. ruminantium (7), and W. pipientis (6) genomes are compact and streamlined, indicating that genome structure for organisms in the family Anaplasmataceae is similar to that of organisms in the family Rickettsiaceae (4, 5). Although tick-transmitted pathogens in these two families have similarities in their infection biology, there is a large gap in current knowledge regarding the microbial determinants of transmission. The completion of sequences of multiple tick transmitted bacterial species in the families Anaplasmataceae and Rickettsiaceae allows comparative genomic approaches to detect genes and pathways unique to tick-transmitted species. Importantly, comparative approaches are unbiased to the location or function of a protein and will detect surface proteins, regulators, and transporters that may be required for replication in the tick as well as novel enzymes and proteins of unknown function. To illustrate this approach, we compared three tick transmitted genomes (A. marginale, E. ruminantium, and R. conorii) with the non-tick-transmitted W. pipientis genome. Table 2 contains a list of orthologs that are shared between the tick-transmitted genomes and not found in W. pipientis. The majority of genes on the list had pfam matches, but a gene name or function could not be definitively assigned, providing candidates that may be involved in transmission. The expected completion of whole genome sequences for additional species in these two families will markedly increase the resolution of this type of comparative approach.
Table 2. Orthologs common to tick-transmitted Rickettsiales.
| A. marginale locus ID/gene name | Annotated product | Homolog E. ruminantium | Homolog R. conorii | pfam match |
|---|---|---|---|---|
| AM102 | Conserved family | ER7510 | RC0617 | Pf00561 |
| AM166 | Conserved family | ER6830 | RC0443/cyaY | Pf01491 |
| AM220 | Conserved family | ER1470 | RC1342 | Pf00753 |
| AM524/truB | tRNA pseudouridine 55 synthase | ER3520 | RC0665 | Pf01509 |
| AM527 | Conserved hypothetical | ER3550 | RC0692/bioC | |
| AM560 | Conserved family cell surface protein | ER4050 | RC0259 | Pf00497 |
| AM619/folE | GTP cyclohydrolase I | ER4000 | RC0527 | |
| AM847 | Conserved hypothetical | ER5530 | RC0355 | Pf04039 |
| AM848 | Conserved family | ER5540 | RC0355 | |
| AM875 | Conserved hypothetical | ER5780 | RC0191 | Pf01613 |
| AM916/cmk | Cytidylate kinase | ER6110/cmk | RC0748/cmk | |
| AM923 | Conserved family | ER6190/ATPase | RC0282/n2B | Pf03969 |
| AM975 | Conserved family pyrophosphokinase | ER6520 | RC0037 | Pf01288 |
| RC0038 | ||||
| AM1275 | Conserved hypothetical | ER8910 | RC0013 | |
| AM1327/xseA | Exodeoxyribonuclease large subunit | ER0370 | RC1026 |
Finally, as persistent infection in the mammalian host is required for ticks to continuously acquire and transmit A. marginale, knowledge of the complete composition of the antigenically variable surface coat is directly applicable to both understanding immune evasion and vaccine development. The genome sequence definitively shows that the A. marginale surface is dominated by two families of OMPs: the msp2 superfamily and the msp1 family, each containing immunodominant members. These two families comprise more than half of the molecules predicted to be on the surface of this organism and thus are a primary focus of ongoing studies.
Supplementary Material
Acknowledgments
We thank Anthony F. Barbet for helpful discussion, and Pete Hetrick for excellent technical assistance. BAC library construction and BAC sequencing were provided by Amplicon Express (Pullman, WA). This work was supported by U.S. Department of Agriculture Microbial Genome Sequencing Program Grant 2001-5210011342, U.S. Department of Agriculture Grants USDA-ARS-CRIS 5248-32000-012-00D and USDA-SCA 58-5348-8-044, and National Institutes of Health Grants RO1 AI44005, RO1 AI45580, and T32-AI07025.
Author contributions: K.A.B., L.S.K., D.R.H., G.H.P., T.C.M., and D.P.K.J. designed research; K.A.B., L.S.K., D.R.H., M.J.D., and D.P.K.J. performed research; K.A.B., L.S.K., D.R.H., M.J.D., D.L.T., G.H.P., and D.P.K.J. analyzed data; and K.A.B., G.H.P., and D.P.K.J. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BAC, bacterial artificial chromosome; OMP, outer membrane protein; MLP, MSP1a-like protein; CDS, coding sequence.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. CP000030).
References
- 1.Palmer, G. H., Rurangirwa, F. R., Kocan, K. M. & Brown, W. C. (1999) Parasitol. Today 15, 281-286. [DOI] [PubMed] [Google Scholar]
- 2.Kocan, K. M., de la Fuente, J., Guglielmone, A. A. & Melendez, R. D. (2003) Clin. Microbiol. Rev. 16, 698-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumler, J. S., Barbet, A. F., Bekker, C. P., Dasch, G. A., Palmer, G. H., Ray, S. C., Rikihisa, Y. & Rurangirwa, F. R. (2001) Int. J. Syst. Evol. Microbiol. 51, 2145-2165. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, S. G., Zomorodipour, A., Andersson, J. O., Sicheritz-Ponten, T., Alsmark, U. C., Podowski, R. M., Naslund, A. K., Eriksson, A. S., Winkler, H. H. & Kurland, C. G. (1998) Nature 396, 133-140. [DOI] [PubMed] [Google Scholar]
- 5.Ogata, H., Audic, S., Renesto-Audiffren, P., Fournier, P. E., Barbe, V., Samson, D., Roux, V., Cossart, P., Weissenbach, J., Claverie, J. M. & Raoult, D. (2001) Science 293, 2093-2098. [DOI] [PubMed] [Google Scholar]
- 6.Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., McGraw, E. A., Martin, W., Esser, C., Ahmadinejad, N., et al. (2004) PLoS Biol. 2, E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, N. E., Liebenberg, J., de Villiers, E. P., Brayton, K. A., Louw, E., Pretorius, A., Faber, F. E., van Heerden, H., Josemans, A., van Kleef, M., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 838-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriks, I. S., Stiller, D., Goff, W. L., Panton, M., Parish, S. M., McElwain, T. F. & Palmer, G. H. (1994) J. Vet. Diagn. Invest. 6, 435-441. [DOI] [PubMed] [Google Scholar]
- 9.Futse, J. E., Ueti, M. W., Knowles, D. P., Jr., & Palmer, G. H. (2003) J. Clin. Microbiol. 41, 3829-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torioni de Echaide, S., Knowles, D. P., McGuire, T. C., Palmer, G. H., Suarez, C. E. & McElwain, T. F. (1998) J. Clin. Microbiol. 36, 777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brayton, K. A., Knowles, D. P., McGuire, T. C. & Palmer, G. H. (2001) Proc. Natl. Acad. Sci. USA 98, 4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 13.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 14.Rurangirwa, F. R., Brayton, K. A., McGuire, T. C., Knowles, D. P. & Palmer, G. H. (2002) Int. J. Syst. Evol. Microbiol. 52, 1405-1409. [DOI] [PubMed] [Google Scholar]
- 15.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frishman, D., Mironov, A., Mewes, H. W. & Gelfand, M. (1998) Nucleic Acids Res. 26, 2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 18.Dyrlov Bendtsen, J., Nielsen, H., Von Heijne, G. & Brunak, S. (2004) J. Mol. Biol. 340, 783-795. [DOI] [PubMed] [Google Scholar]
- 19.Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E. L., et al. (2004) Nucleic Acids Res. 32, D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatusov, R. L., Koonin, E. V. & Lipman, D. J. (1997) Science 278, 631-637. [DOI] [PubMed] [Google Scholar]
- 21.Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J. & Giegerich, R. (2001) Nucleic Acids Res. 29, 4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobry, J. R. (1996) Mol. Biol. Evol. 13, 660-665. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa, M. & Goto, S. (2000) Nucleic Acids Res. 28, 27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karp, P. D., Riley, M., Saier, M., Paulsen, I. T., Collado-Vides, J., Paley, S. M., Pellegrini-Toole, A., Bonavides, C. & Gama-Castro, S. (2002) Nucleic Acids Res. 30, 56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alleman, A. R., Kamper, S. M., Viseshakul, N. & Barbet, A. F. (1993) J. Gen. Microbiol. 139, 2439-2444. [DOI] [PubMed] [Google Scholar]
- 27.Salzberg, S. L., Salzberg, A. J., Kerlavage, A. R. & Tomb, J. F. (1998) Gene 217, 57-67. [DOI] [PubMed] [Google Scholar]
- 28.Brayton, K. A., Palmer, G. H., Lundgren, A., Yi, J. & Barbet, A. F. (2002) Mol. Microbiol. 43, 1151-1159. [DOI] [PubMed] [Google Scholar]
- 29.Meeus, P. F., Brayton, K. A., Palmer, G. H. & Barbet, A. F. (2003) Mol. Microbiol. 47, 633-643. [DOI] [PubMed] [Google Scholar]
- 30.Barbet, A. F., Meeus, P. F., Belanger, M., Bowie, M. V., Yi, J., Lundgren, A. M., Alleman, A. R., Wong, S. J., Chu, F. K., Munderloh, U. G. & Jauron, S. D. (2003) Infect. Immun. 71, 1706-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tettelin, H., Saunders, N. J., Heidelberg, J., Jeffries, A. C., Nelson, K. E., Eisen, J. A., Ketchum, K. A., Hood, D. W., Peden, J. F., Dodson, R. J., et al. (2000) Science 287, 1809-1815. [DOI] [PubMed] [Google Scholar]
- 32.Fraser, C. M., Casjens, S., Huang, W. M., Sutton, G. G., Clayton, R., Lathigra, R., White, O., Ketchum, K. A., Dodson, R., Hickey, E. K., et al. (1997) Nature 390, 580-586. [DOI] [PubMed] [Google Scholar]
- 33.Vidotto, M. C., McGuire, T. C., McElwain, T. F., Palmer, G. H. & Knowles, D. P., Jr. (1994) Infect. Immun. 62, 2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.French, D. M., Brown, W. C. & Palmer, G. H. (1999) Infect. Immun. 67, 5834-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.French, D. M., McElwain, T. F., McGuire, T. C. & Palmer, G. H. (1998) Infect. Immun. 66, 1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brayton, K. A., Meeus, P. F., Barbet, A. F. & Palmer, G. H. (2003) Infect. Immun. 71, 6627-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown, W. C., Brayton, K. A., Styer, C. M. & Palmer, G. H. (2003) J. Immunol. 170, 3790-3798. [DOI] [PubMed] [Google Scholar]
- 38.Palmer, G. H., Eid, G., Barbet, A. F., McGuire, T. C. & McElwain, T. F. (1994) Infect. Immun. 62, 3808-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbet, A. F., Lundgren, A., Yi, J., Rurangirwa, F. R. & Palmer, G. H. (2000) Infect. Immun. 68, 6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohr, C. V., Brayton, K. A., Shkap, V., Molad, T., Barbet, A. F., Brown, W. C. & Palmer, G. H. (2002) Infect. Immun. 70, 6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Heerden, H., Collins, N. E., Brayton, K. A., Rademeyer, C. & Allsopp, B. A. (2004) Gene 330, 159-168. [DOI] [PubMed] [Google Scholar]
- 42.Ohashi, N., Unver, A., Zhi, N. & Rikihisa, Y. (1998) J. Clin. Microbiol. 36, 2671-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohashi, N., Zhi, N., Zhang, Y. & Rikihisa, Y. (1998) Infect. Immun. 66, 132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lohr, C. V., Brayton, K. A., Barbet, A. F. & Palmer, G. H. (2004) Gene 325, 115-121. [DOI] [PubMed] [Google Scholar]
- 45.Schofield, M. J. & Hsieh, P. (2003) Annu. Rev. Microbiol. 57, 579-608. [DOI] [PubMed] [Google Scholar]
- 46.Oberle, S. M. & Barbet, A. F. (1993) Gene 136, 291-294. [DOI] [PubMed] [Google Scholar]
- 47.Palmer, G. H., Rurangirwa, F. R. & McElwain, T. F. (2001) J. Clin. Microbiol. 39, 631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palmer, G. H., Barbet, A. F., Davis, W. C. & McGuire, T. C. (1986) Science 231, 1299-1302. [DOI] [PubMed] [Google Scholar]
- 49.Viseshakul, N., Kamper, S., Bowie, M. V. & Barbet, A. F. (2000) Gene 253, 45-53. [DOI] [PubMed] [Google Scholar]
- 50.Brown, W. C., McGuire, T. C., Zhu, D., Lewin, H. A., Sosnow, J. & Palmer, G. H. (2001) J. Immunol. 166, 1114-1124. [DOI] [PubMed] [Google Scholar]
- 51.Alleman, A. R., Palmer, G. H., McGuire, T. C., McElwain, T. F., Perryman, L. E. & Barbet, A. F. (1997) Infect. Immun. 65, 156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oberle, S. M., Palmer, G. H., Barbet, A. F. & McGuire, T. C. (1988) Infect. Immun. 56, 1567-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blouin, E. F., Saliki, J. T., de la Fuente, J., Garcia-Garcia, J. C. & Kocan, K. M. (2003) Vet. Parasitol. 111, 247-260. [DOI] [PubMed] [Google Scholar]
- 54.Kocan, K. M., Halbur, T., Blouin, E. F., Onet, V., de la Fuente, J., Garcia-Garcia, J. C. & Saliki, J. T. (2001) Vet. Parasitol. 102, 151-161. [DOI] [PubMed] [Google Scholar]
- 55.Brown, W. C., Palmer, G. H., Lewin, H. A. & McGuire, T. C. (2001) Infect. Immun. 69, 6853-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown, W. C., Shkap, V., Zhu, D., McGuire, T. C., Tuo, W., McElwain, T. F. & Palmer, G. H. (1998) Infect. Immun. 66, 5406-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbet, A. F., Palmer, G. H., Myler, P. J. & McGuire, T. C. (1987) Infect. Immun. 55, 2428-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stich, R. W., Olah, G. A., Brayton, K. A., Brown, W. C., Fecheimer, M., Green-Church, K., Jittapalapong, S., Kocan, K. M., McGuire, T. C., Rurangirwa, F. R. & Palmer, G. H. (2004) Infect. Immun. 72, 7257-7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohashi, N., Zhi, N., Lin, Q. & Rikihisa, Y. (2002) Infect. Immun. 70, 2128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin, M. & Rikihisa, Y. (2003) Infect. Immun. 71, 5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.