Abstract
Recently, we and others have identified two human endogenous retroviruses that entered the primate lineage 25–40 million years ago and that encode highly fusogenic retroviral envelope proteins (syncytin-1 and -2), possibly involved in the formation of the placenta syncytiotrophoblast layer generated by trophoblast cell fusion at the materno–fetal interface. A systematic in silico search throughout mouse genome databases presently identifies two fully coding envelope genes, present as unique copies and unrelated to any known murine endogenous retrovirus, that we named syncytin-A and -B. Quantitative RT-PCR demonstrates placenta-specific expression for both genes, with increasing transcript levels in this organ from 9.5 to 14.5 days postcoitum. In situ hybridization of placenta cryosections further localizes these transcripts in the syncytiotrophoblast-containing labyrinthine zona. Consistently, we show that both genes can trigger cell–cell fusion in ex vivo transfection assays, with distinct cell type specificities suggesting different receptor usage. Genes orthologous to syncytin-A and -B and disclosing a striking conservation of their coding status are found in all Muridae tested (mouse, rat, gerbil, vole, and hamster), dating their entry into the rodent lineage ≈20 million years ago. Together, these data strongly argue for a critical role of syncytin-A and -B in murine syncytiotrophoblast formation, thus unraveling a rather unique situation where two pairs of endogenous retroviruses, independently acquired by the primate and rodent lineages, would have been positively selected for a convergent physiological role.
Keywords: endogenous retrovirus, placenta, syncytiotrophoblast, rodent
Endogenous retroviruses (ERVs) are present in the genome of all vertebrates (1–3). They are most probably the genomic traces of germ-line infections by exogenous retroviruses having taken place during evolution. Over time, increase in their copy number has occurred, at least for some of them, via retrotransposition or germ-cell reinfection. This generated multigenic families containing a few to several hundred elements and now accounting for a substantial fraction of the genome [8% for the human (4) and 10% for the murine (5) genomes]. In mice, eight families of ERVs, with a structure close to the integrated proviral form of exogenous retroviruses (gag-, pol-, and env-related regions bordered by two LTRs), have been described. Those with close exogenous relatives include type B proviruses related to the murine mammary tumor virus (6) and type C proviruses related to the Moloney murine leukemia virus (MLV), with the endogenous MLV, MuRRS, MuRVY, GLN, MuERVC, and MmERV family members (1, 7, 8). A family of sequences with no exogenous relatives, called IAPE (intracisternal A-particle sequences with an envelope gene), was also described (9). Although some of these ERVs have accumulated mutations, deletions, and/or truncations and would therefore require recombination with exogenous counterparts to generate replication-competent retroviruses, others are fully coding (e.g., IAPE and MmERV) and are therefore expected to be replication-competent. Besides this large pool of highly reiterated murine ERVs, few examples exist of coopted ERV genes, which are present at a single copy and can be considered bona fide cellular genes. This is the case of the murine Fv1, Fv4, and Rmcf genes that confer resistance to infection by exogenous MLV retroviruses (10). Fv1 derives from the gag region of an ancient MERV-L element (11), whereas Fv4 and Rmcf consist of intact env genes, amid severely altered proviral structures, whose expression prevents infection via receptor interference. These are unambiguous examples of a contributive role in the host physiology for an ERV-derived gene. Interestingly, in humans, a physiological function has also been hypothesized for two envelope genes of retroviral origin, named syncytin-1 and -2 (12–14). They are expressed almost exclusively in the placenta, at the level of the cytotrophoblast/syncytiotrophoblast (refs. 12–15; S. Blaise, personal communication), and both induce cell–cell fusion in ex vivo assays via an interaction with the type D retrovirus receptor for syncytin-1 (13), thus suggesting they are involved in the formation of the syncytiotrophoblast layer of the placenta (not excluding other possible roles, such as protection against infection and/or immunosuppression of maternal immune response against the fetus; reviewed in refs. 1 and 2). Entry of the two corresponding retroviruses (namely HERV-W and -FRD) into the primate lineage occurred 25–40 million years (Myr) ago, with their env gene being conserved in a functional state in all derived species, consistent with selection for a function of benefit to the host (14, 16). Concerning other species in which placenta trophoblast fusion also takes place, such as rodents, the question of whether they have similarly diverted ERVs for a physiological, syncytin-related function can be raised. Here, taking advantage of the near completion of the mouse genome sequencing and an appropriate screening procedure for retroviral envelopes as performed for the human genome (17), we report the discovery of two previously uncharacterized murine env genes, each present at a single copy and phylogenetically unrelated to human syncytin-1 and -2. These genes are specifically expressed in the placenta, at the level of the syncytiotrophoblast-containing labyrinthine zona, and are highly fusogenic when tested in an ex vivo transfection assay. We further show that orthologs to these murine genes are present in all Muridae tested, with conservation of their coding status, thus suggesting positive selection for a function most probably similar to that of the human syncytin genes. Accordingly, at least in two cases of animal evolution, i.e., in primates and rodents, a pair of ERVs would have been independently acquired and put to work for a common physiological role.
Materials and Methods
Database Screening and Sequence Analyses. Retroviral envelopes were searched by using the biomotif program (www.lpta.univmontp2.fr/software.html) and the degenerate universal CKS17u consensus motif in ref. 17. Multiple alignments were carried out by using the clustalw program (www.infobiogen.fr). Phylogenetic trees were constructed from alignments by using the neighbor-joining program within clustalw and were viewed with the njplot program. Repeat sequences were identified by using the repeat-masker server (www.repeatmasker.org). Homologous sequences were searched by using the blast programs at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST) and at the Ensembl (www.ensembl.org) sites. Nonsynonymous vs. synonymous substitution ratios (Ka/Ks) were calculated by using the snap.pl program at www.hiv.lanl.gov.
Real-Time Quantitative PCR, RT-PCR, and Northern Blot Analyses. C57BL/6 mice, including pregnant females at different gestational stages [morning of vaginal plug considered as 0.5-day postcoitum (dpc)], were from Janvier (Le Genest-St-Isle, France). Total RNA was extracted from frozen organs (pooled from at least two mice) with the RNeasy RNA isolation kit (Qiagen, Valencia, CA). Reverse transcription and quantitative PCR were performed as in ref. 18, with primers TACTCCTGCCCGATAGATGA and CCGTTTTTCTTAACAGTGGGT for syncytin-A and CCACCACCCATACGTTCAAA and GGTTATAGCAGGTGCCGAAG for syncytin-B. Structure of the syncytin transcripts was determined by RT-PCR (primer sequences available upon request). Northern blot analysis was performed as in ref. 19, using syncytin-A or -B 32P-labeled full-length env gene probes.
In Situ Hybridization. Placenta tissues (14.5 dpc) were processed for serial cryosections (10 μm thick). The antisense and sense riboprobes were in vitro transcribed with T7 RNA polymerase by using digoxigenin-11-UTP (Roche Applied Science, Indianapolis) and the syncytin-A and -B full-length env genes cloned in the two orientations into PGEM-T (Promega) as templates. Cryosections were postfixed at 4°C in 1% paraformaldehyde for 5 min, hybridized at 65°C overnight with the riboprobes, and further incubated with antidigoxigenin antibodies overnight at room temperature. The signal was amplified by using the Vectastain ABC kit (Vector Laboratories) coupled to the Tyramide Signal Amplification system (NEN Life Sciences). Histological examination was performed after DAPI nuclear staining by using a Zeiss Axiophot microscope.
Envelope Gene Expression Vectors. The A-Rless hyperfusogenic mutant amphotropic MLV envelope expression vector and the phCMV-G plasmid are in ref. 14. The syncytin-A and -B envelope expression vectors were constructed as follows: syncytin-A was PCR amplified from C57BL/6 genomic DNA, using the proofreading Platinium Pfx DNA polymerase (Invitrogen) for 30 cycles, and syncytin-B was PCR-amplified from the corresponding C57BL/6 bacterial artificial chromosome (AC134575) DNA, using the proofreading PfuTurbo Hotstart polymerase (Stratagene) for 15 cycles (primer sequences available upon request); the PCR products were cloned into the phCMV-G vector cut with EcoRI and blunt-ended.
Cell Lines and Fusion Assay. The murine MCA205 fibrosarcoma, WOP fibroblast, and LOK kidney cells (gift from S. Fichelson, Institut Cochin de Génétique Moléculaire, Paris), the rat 208F fibroblast cells, hamster BHK-21 kidney cells, canine MDCK kidney cells, feline G355-5 astrocyte cells, monkey VERO kidney cells, the human TE671 rhabdomyosarcoma, 293T embryonal kidney, and HeLa carcinoma cells, were grown in DMEM with 10% FCS; the Chinese hamster ovary cells were grown in DMEM/F12K (1:1), with 7% FCS. For fusion assays, cells were transfected by using Lipofectamine (Invitrogen) and 1–2 μg of DNA for 5 × 105 cells, except for 208F, 293T, and TE671 cells, which were transfected by using calcium phosphate precipitation (Invitrogen, 5–20 μg of DNA for 5 × 105 cells). Plates were inspected for cell fusion 24–48 h after transfection. Syncytia were visualized by using May–Grünwald and Giemsa staining (Sigma) and the fusion index calculated as [(N - S)/T] × 100, where N is the number of nuclei in the syncytia, S is the number of syncytia, and T is the total number of nuclei counted.
DNAs and Southern Blot Analyses. Bacterial artificial chromosome clones were from BACPAC Resources (Oakland, CA). Genomic DNAs were extracted from animal tissues or blood cells. C57BL/6 mouse (Mus musculus), rat (Rattus norvegicus), gerbil (Meriones unguiculatus), and hamster (Mesocricetus auratus) were from Janvier. Guinea pig (Cavia porcellus) DNA was extracted from peripheral blood leukocytes (Charles River Laboratories). Vole (Clethrionomys glareolus) tissue was a gift from F. Catzeflis (Institut des Sciences de l'Evolution, Montpellier, France). Squirrel (Spermophilus beecheyi) and woodchuck (Marmotta monax) tissues were a gift from M.-A. Buendia (Institut Pasteur, Paris). For Southern blot analyses, membranes were hybridized overnight at 65°C in 7% SDS/1 mM EDTA/0.5 M Na2HPO4, using 32P-labeled syncytin-A or -B full-length env probes, and washed at 65°C under low-stringency conditions (2× SSC/0.1% SDS for 15 min and 1× SSC/0.1% SDS for 10 min) before x-ray film exposure.
Characterization of the Syncytin-A and -B Genes from Rodents. The orthologous genes and flanking sequences were sorted out step by step by a series of low-stringency direct PCRs carried out for 30–45 cycles (10 sec at 94°C, 30 sec at 50°C, 1–6 min at 68°C), using 100 ng of genomic DNA, the Expand long-template enzyme mix (Roche Applied Science); in some cases, inverted PCRs were performed as described in ref. 20 by using genomic DNA first digested by appropriate restriction enzymes and then self-ligated, and pairs of primers in opposite orientation. For unambiguous sequence determination, the entire syncytin-A and -B orthologous genes were finally reamplified from 100 ng of genomic DNA for 25 cycles, using the Expand long-template enzyme mix (Roche Applied Science) and primers designed from the characterized flanking sequences, and sequencing was performed on the bulk of the PCR products without cloning (sequences have been deposited in the GenBank database; accession nos. AY849973–AY849981).
Results
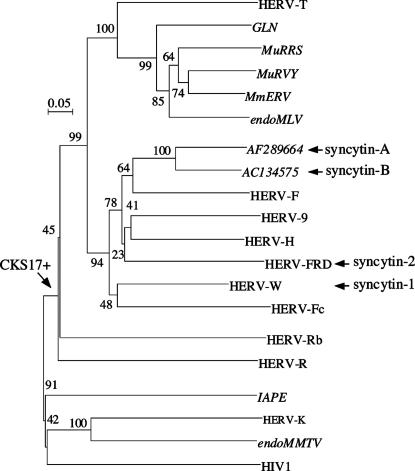
Identification of Two Previously Uncharacterized Retroviral Envelope Genes in the Mouse Genome. To identify env genes within the mouse genome, we used a method previously devised to screen the whole human genome for such genes (17). Basically, it makes use of the degenerate CKS17u consensus motif, associated with the immunosuppressive domain of retroviral envelopes and designed to match the majority of envelope genes of exogenous and endogenous origin (17). Murine sequences from the GoldenPath database and from the nr (nonredundant) and HTGS (high-throughput genomic sequences) divisions of the GenBank database (release 132) were screened with this motif by using the biomotif program. A 2-kb region encompassing 1,500 bp upstream and 500 bp downstream of the CKS17u motif was extracted from positive sequences, and only those regions containing an ORF >1.6 kb were considered further. Selected ORFs (≈200), plus representatives of known endogenous and exogenous retroviral env genes, were then aligned and phylogenetic trees constructed to identify and classify the sequences that were sorted out. All of them (except two; see below) were found to belong to multicopy families of type C origin, with about one-half as MmERV (8), one-quarter as endogenous MLV (21), and one-quarter as GLN sequences (providing, by the way, the first description of GLN fully coding env genes in the mouse genome; ref. 22). Note that IAPE and endogenous MMTV env genes were not sorted out by the screen, because they are devoid of the CKS17 motif. Interestingly, two previously uncharacterized env sequences (AF289664 and AC134575), not belonging to any known murine ERV family and each present as a unique copy, were identified with that screen. A phylogenetic analysis (Fig. 1) places them within the enlarged HERV-F/H family, also containing the human syncytin-1 and -2 env genes, but in branches distinct from the latter. These two newly identified murine env sequences were subsequently named syncytin-A and -B because of their functional properties closely related to those of the human syncytin genes (see below).
Fig. 1.
Envelope-based phylogenetic tree with positions of syncytin-A and -B. The tree was determined by the neighbor-joining method by using TM envelope sequences (see ref. 17), essentially from murine ERVs (in italics) and human HERVs. The horizontal branch length and scale indicate the percentage of amino acid substitutions from the node, and the arrow on the left points to the node of the CKS17-positive sequence group. Percent bootstrap values obtained from 1,000 replicates are indicated. Accession numbers for each representative element together with the sequence alignment used to generate the tree are given in Fig. 8, which is published as supporting information on the PNAS web site.
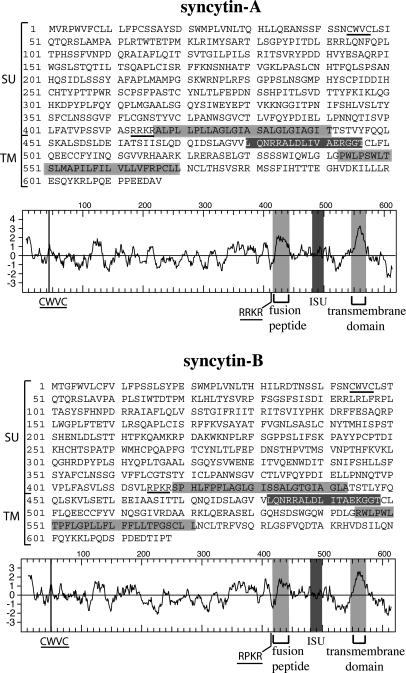
Amino Acid Sequence and Genomic Organization of the Syncytin-A and -B Genes. Analysis of syncytin-A and -B amino acid sequences together with their hydrophobic profiles (Fig. 2), discloses all of the characteristic features of retroviral envelopes (reviewed in ref. 23), with a canonical cleavage site (consensus: R/K-X-R/K-R; ref. 24) between the surface (SU) and transmembrane (TM) moieties of the proteins and the presence of hydrophobic domains corresponding to the fusion peptide and the TM domain. A CWVC domain, involved in the interaction between the SU and TM moieties in retroviral envelopes (25), and an immunosuppressive domain (26) can also be identified. Amino acid sequence alignment shows 67% identity between the two genes, with maximum similarity in the TM region (data not shown). Syncytin-A and -B loci can be assigned to chromosomal positions 5qG2 and 14qD1, respectively. Analysis of the genomic sequences surrounding the env ORFs discloses extremely degenerate proviral structures (Fig. 3), with only short regions with homology to pol and the presence of solitary ERV and MaLR LTRs, which could serve for initiation and termination of syncytin transcription (see below), as well as of repetitive B1, B2, and LINE elements. Finally, the regions flanking the syncytin-A and -B proviral structures are not orthologous (no sequence similarity), consistent with distinct retroviral integration events.
Fig. 2.
Primary sequence, hydrophobicity profile, and predicted features of the syncytin-A and -B envelopes. The SU and TM moieties of the envelopes are delineated, with the canonical furin cleavage site (R/K-N-R/K-R) between the two subunits and the CWVC domain involved in SU–TM interaction underlined; the hydrophobic fusion peptide and TM domains are shaded in light gray, and the putative immunosuppressive domain (ISU) in dark gray.
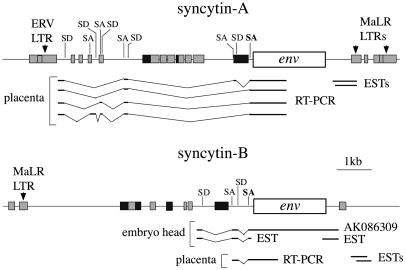
Fig. 3.
Genomic organization of the syncytin-A and -B chromosomal locus. The envelope ORF (open box), the pol-related sequences (dark gray boxes), and murine repetitive elements (light-gray boxes) are indicated. Partial structure of envelope transcripts, as determined experimentally by RT-PCR (with reverse primers in the env ORF) or by an in silico search throughout EST and cDNA databases, are schematized, together with the related splice donor (SD) and splice acceptor (SA) sites positioned on the DNA sequences (with the canonical retroviral envelope SA site in bold).
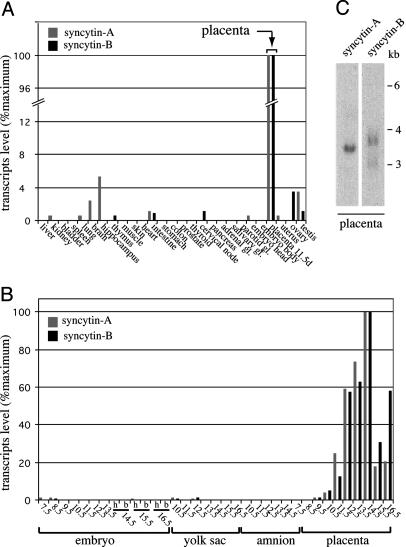
Placenta-Specific Expression of the Syncytin-A and -B Genes. Syncytin-A and -B transcript levels in mouse tissues were investigated by real-time quantitative RT-PCR, by using primer pairs designed and demonstrated to be specific for each gene. As illustrated in Fig. 4A, syncytin-A and -B are expressed essentially in the placenta, with only few organs above background level. Analysis of embryos and embryonic annexes (yolk sac, amnion, and placenta) at different developmental stages (from 7.5 to 16.5 dpc) further supports placenta-specific expression of syncytin-A and -B and reveals strong induction of both genes in this organ between 9.5 and 14.5 dpc (Fig. 4B). Northern blot analysis of placenta RNAs discloses a single transcript of 3.5 kb for syncytin-A and two transcripts of 3.7 and 3 kb for syncytin-B (Fig. 4C). Their structure was partially characterized by RT-PCR and in silico searches throughout EST and cDNA databases (Fig. 3). It revealed multiple alternative splicing for both env transcripts, reminiscent of the spliced subgenomic env mRNAs of retroviruses and consistent with a retroviral origin of these genes, and two alternative polyadenylation signals for syncytin-B, all features compatible with the observed size of the transcripts.
Fig. 4.
Real-time quantitative RT-PCR and Northern blot analysis of syncytin-A and -B transcripts in mouse tissues. (A and B) syncytin-A (gray bars) and -B (dark bars) transcript levels in tissues of adult male (same results for the corresponding female tissues, when tested, namely thyroid, cervical node, pancreas, adrenal, salivary, and parotid glands; data not shown), adult female (uterus and ovaries), and 11.5-day-pregnant female (embryos and placenta) mice (A), and embryonic tissues (embryo, yolk sac, amnion, and placenta) at different times postcoitum (from day 7.5 to 16.5) (B). The embryo head (h) and body (b) were analyzed separately whenever possible. (C) Northern blot analysis of 11.5 dpc mouse placenta total RNA, probed with a syncytin-A or -B full-length env probe. Exposure time, 4 days.
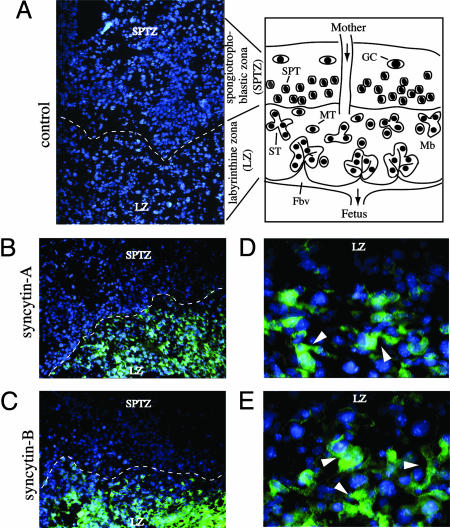
Localization of Syncytin-A and -B Expression in the Placenta by in Situ Hybridization. In situ hybridizations (Fig. 5) were performed on 14.5 dpc placenta cryosections (see scheme of the placenta tissue in Fig. 5 and ref. 27), using digoxigenin-labeled probes. Syncytin-A and -B expression is observed all over the labyrinthine zona (Fig. 5 B and C), with labeling detected neither in the spongiotrophoblastic and giant cell zona nor in the maternal decidua (not shown); no labeling can be detected in control hybridizations using a sense syncytin-A probe (Fig. 5A). The murine labyrinthine zona mainly contains syncytiotrophoblasts formed by fusion of mononuclear trophoblastic cells, arranged as barriers between the maternal and fetal blood circulation (see scheme). Higher magnification of the labeled labyrinthine zona (Fig. 5 D and E) provides indication for syncytin-A and -B expression at the level of syncytiotrophoblasts, with the labeling encompassing structures with several nuclei. Labyrinth-specific expression of both genes is consistent with the kinetics of accumulation of syncytin-A and -B transcripts as revealed by quantitative RT-PCR (Fig. 4B), which follow the kinetics of development of the placenta labyrinthine layer, taking place from 9.5 to 15.5 dpc (27, 28).
Fig. 5.
In situ hybridization of 14.5 dpc mouse placenta for syncytin-A and -B expression. Scheme of a mouse mature placenta cross section: the labyrinthine zona (LZ) is the site of exchange between the maternal blood (Mb) and the fetal blood vessels (Fbv) and contains mononuclear trophoblast cells (MT) that terminally differentiate, by cell fusion, into syncytiotrophoblasts (ST) directly lining the fetal blood vessels; in Muridae, both cell types are arranged as a three-layered barrier (two syncytial and one mononuclear layer, not represented) between the maternal and fetal blood circulation; the spongiotrophoblast zona with the spongiotrophoblast cells (SPT) and the outermost layer with the trophoblast giant cells (GC) (producing hormone/growth factors) overlie the labyrinthine zona. (A) Negative control: tissue section probed with a digoxigenin-labeled sense syncytin-A riboprobe. (B and C) Tissue sections probed with digoxigenin-labeled antisense syncytin-A and -B riboprobes, respectively, disclosing hybridization signals only within the labyrinthine zona (the dotted lines delineate the separation between the labyrinthine and spongiotrophoblastic zona). (D and E) Magnification of the labyrinthine zona in B and C, with the arrowheads pointing to labeled structures with several nuclei. Nuclei were stained in blue by using DAPI. A–C, ×100; D and E, ×400.
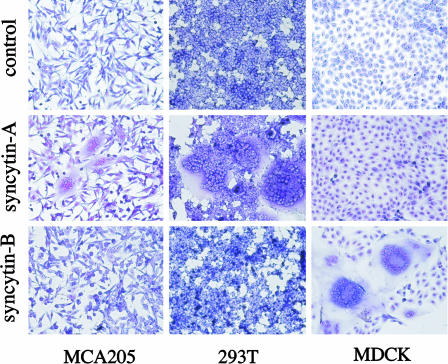
Syncytin-A and -B Are Fusogenic, with Different Receptor Usage. To determine whether the syncytin-A and -B gene products have fusogenic properties, expression vectors for the corresponding proteins were used to transfect a series of cells of distinct origins, which were thereafter screened for the presence of syncytia (Fig. 6). The highly fusogenic A-Rless MLV mutant envelope (29) and syncytin-A in antisense orientation were used as controls. As illustrated in Table 1, cell–cell fusion can be observed for several murine, human, and monkey cell lines with syncytin-A, whereas syncytin-B is found to be fusogenic only with the canine MDCK kidney cells, suggesting a rather rarely distributed receptor for the latter. That the fusogenic potencies of syncytin-A and -B proteins are different is strongly in favor of different receptor usage for these two envelope proteins. Yet, the observed cell specificity patterns for fusion are distinct from those of the human syncytin-1 and -2 proteins (14) and do not allow any prediction as to the nature of the receptors involved.
Fig. 6.
Syncytin-mediated cell–cell fusion. Shown is syncytia formation by the syncytin-A and -B envelope genes. The indicated cells were transfected with expression vectors for syncytin-A or -B, or a negative control (syncytin-A in antisense orientation) and were stained with May–Grünwald and Giemsa 24–48 h after transfection.
Table 1. Fusion host range of the syncytin-A and -B envelopes genes.
| Fusion efficiency
|
|||||
|---|---|---|---|---|---|
| Species | Cell | Syncytin-A | Syncytin-B | A-Rless | Control |
| Mouse | MCA205 | ++ | – | +++ | – |
| WOP | – | – | +++ | – | |
| LOK | ++ | – | +++ | – | |
| Rat | 208F | – | – | ++ | – |
| Hamster | BHK-21 | – | – | + | – |
| CHO | – | – | – | – | |
| Dog | MDCK | – | +++ | – | – |
| Cat | G355-5 | – | – | +++ | – |
| Monkey | VERO | ++ | – | +++ | – |
| Human | TE671 | + | – | ++ | – |
| 293T | +++ | – | + | – | |
| HeLa | – | – | + | – | |
Experimental conditions were the same as in Fig. 6, with the hyperfusogenic mutant amphotropic MLV envelope (A-Rless) used as a reference. Fusion indices were determined as indicated in Materials and Methods, with the following scale: –, <5; +, 5–20; ++, 20–70; +++, >70.
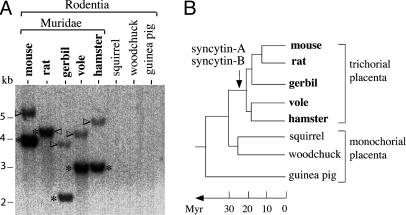
Syncytin-A and -B env Gene Acquisition and Conservation During Rodent Evolution. To tentatively identify syncytin-A and -B orthologs in other species, DNAs from eight rodents were digested with DraI, which does not cut in the murine syncytin-A and -B env genes, and examined by Southern blot with syncytin-A or -B probes under low-stringency conditions. As illustrated in Fig. 7, hybridizing fragments are detected for both genes in Muridae only (mouse, rat, gerbil, vole, and hamster), with no signal for the squirrel, woodchuck, and guinea pig. In the rat, the DraI-restricted syncytin-A and -B bands display identical sizes, consistent with sequencing data (see below). According to the zoo-blot, the date of entry of the syncytin-A and -B genes into the rodent lineage should be before speciation of Muridae, ≈20 Myr ago (30). blast searches performed using rat genome databases provided the sequence of the syncytin-A (GenBank accession no. AC091000) and the syncytin-B (contig RNOR03237248 in Ensembl and XM_224362 in GenBank) orthologs, as fully coding genes. To identify the other Muridae orthologs, sets of primers based on regions conserved between the rat and mouse sequences were first used to amplify portions of the orthologous loci, under low-stringency PCR conditions. Step-by-step series of low-stringency PCRs, sequencing, and design of new primers then allowed (except for gerbil syncytin-A, with only part of the gene identified) a complete characterization of the orthologous genes and flanking sequences for the Muridae in Fig. 7 (see Materials and Methods). Remarkably, all syncytin-A and -B orthologs have conserved a fully coding status. Pairwise comparison of the sequences between the mouse, rat, gerbil, vole, and hamster disclosed, for both the syncytin-A and -B genes, a percentage of amino acid identity comprised between 77% and 91% (Table 2, which is published as supporting information on the PNAS web site), compatible with the rate of nucleotide substitution observed in rodents (31). It also reveals a high prevalence of synonymous vs. nonsynonymous substitutions (average Ka/Ks values for all pairwise comparisons: 0.21 and 0.15 for syncytin-A and -B, respectively), consistent with purifying selection on these genes and a beneficial function for the host.
Fig. 7.
Conservation of the syncytin-A and -B genes during rodent evolution. (A) Southern blot of DraI-restricted genomic DNA from the indicated rodent species, with the syncytin-B fragments (asterisk) and the syncytin-A fragments (arrowhead) both found in Muridae only (the syncytin-A bands have a lower intensity due to the use of a syncytin-B probe, with the opposite result when using a syncytin-A probe; not shown). (B) Schematized rodent phylogenetic tree, with the arrow indicating the date of entry of the two syncytin-A and -B envelope genes; the placenta type, in terms of structure of the syncytiotrophoblast materno–fetal barrier, is indicated (see Discussion).
Discussion
The present search for complete env genes in mouse genome databases has identified numerous sequences of known highly reiterated ERV families, together with two fully coding env genes, present at a single copy, and that we named syncytin-A and -B. These mouse genes display fusogenic activities when expressed in transfected cells, with differences in the cell types prone to fusion, indicating differences in receptor usage. Both genes are specifically expressed in the placenta and, more precisely, in the syncytiotrophoblast-containing labyrinthine zona. These features are closely related to those of the human syncytin-1 and -2 genes, suggesting a role in trophoblast fusion and placenta physiology. Yet the murine and human genes are not orthologs, with entry dates ≈20 Myr ago for the murine genes and 25–40 Myr ago for the human genes, i.e., after the speciation of rodents and primates. Phylogenetic analyses (Fig. 1), disclosing distinct branching of the human and murine syncytin genes, as well as evidence that they are not syntenic, further support these conclusions. Analysis of the syncytin-A and -B genomic loci finally indicates that the two genes were acquired from two ancient retroviral integration events, with recognizable vestiges of proviral structures (conservation of short pol-related sequences and generation of multiple spliced env transcripts). These degenerate structures, much more conserved for the human syncytin-1 and -2 loci, are reminiscent of the severely altered Fv4 murine locus (10), a feature that may be explained by the high rate of nucleotide substitution in rodents (31).
The syncytin-A and -B genes display remarkably similar patterns of expression, both quantitatively, with identical kinetics of transcript accumulation during placenta development, and qualitatively, with a similar localization of the transcripts in the syncytiotrophoblast-containing labyrinthine zona. The presence of two syncytin genes with closely related and possibly redundant properties, as also observed for the human syncytin genes, is intriguing. One possibility is that conservation of env genes recognizing two distinct receptors is a safeguard for the host. Another hypothesis could be that cooperation between the two genes is required for an efficient fusion process. Additional studies are required to clarify this point. Among factors known to be critical for in vivo syncytiotrophoblast formation (32, 33), the glial cells missing-1 (Gcm1) transcription factor is of special interest, because its pattern of expression is remarkably similar to that of syncytin-A and -B (28). In addition, Gcm1-deficient mice display a failure in labyrinth formation and lack syncytiotrophoblasts (34, 35). Interestingly, it was shown that human syncytin-1 expression can be activated by the human Gcm1 factor (36), and several putative Gcm1-binding sites can be identified in the murine syncytin-A and -B loci (A.D., unpublished data). One could therefore hypothesize that syncytin-A and -B expression in the placenta is induced by Gcm1 and that both the human and murine syncytin genes are similarly regulated.
An interesting result of the present study is the conservation of the syncytin-A and -B genes with a fully coding status over >20 Myr of rodent evolution, since their entry into the Muridae branch. Consistently, Muridae and non-Muridae rodents display well established differences regarding the structure and number of syncytiotrophoblast layers at the materno–fetal interface: although most rodents have a one-layered syncytiotrophoblast barrier (monochorial placenta), a three-layered structure, with one cellular and two syncytiotrophoblast layers (trichorial placenta), has emerged in Muridae (ref. 37 and Fig. 7). One could therefore hypothesize, provided that all syncytin orthologs are fusogenic, that these phenotypic changes result from the retrovirus-mediated acquisition of the syncytin-A and -B genes in Muridae, which would have given a selective advantage to the host. Yet, the presence of placenta syncytial cells in most other rodents suggests that fusiogenic gene(s) did exist before the capture of the syncytin genes, but we know nothing about their nature or subsequent evolution in rodents. Indeed, placenta syncytial cells have originated on multiple independent occasions during mammalian evolution, being observed for example in Artiodactyla, but not in pigs or camels, in Carnivora, Rodentia, and Primata (38). Our work and other reports (reviewed in refs. 39–41) therefore lead to the proposal that several independent retroviral infections may have contributed to the emergence of a common syncytial barrier in different species and played a pivotal convergent role in placenta morphogenesis and physiology.
Finally, knockout mouse mutants for the syncytin genes should now allow a definite appraisal of the role of ERVs in the reproduction of rodents. Taking into account the remarkable similarities between human and mouse placentation (27), the possible involvement of human syncytin genes in placenta physiology and dysfunctions, such as intrauterine growth restriction or Down's syndrome, might also be addressed via the mouse model.
Supplementary Material
Acknowledgments
We thank S. Fichelson, F. Catzeflis, and M.-A. Buendia for the gift of cells and tissues; P. Dessen at the Institut Gustave Roussy Bioinformatic Center for help with the computing work; and S. Blaise and M. Dewannieux for discussions. We thank C. Lavialle for comments and critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique and the Ligue Nationale Contre le Cancer (Equipe Labellisée).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Myr, million years; ERV, endogenous retrovirus; MLV, murine leukeumia virus; dpc, days postcoitum; SU, surface; TM, transmembrane.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY849973–AY849976 for the mouse, rat, vole, and hamster syncytin-A, respectively; and AY849977–AY849981 for the mouse, rat, gerbil, vole, and hamster syncytin-B, respectively).
References
- 1.Boeke, J. D. & Stoye, J. P. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 343-435. [PubMed]
- 2.Bannert, N. & Kurth, R. (2004) Proc. Natl. Acad. Sci. USA 13, 14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Parseval, N. & Heidmann, T., Cytogenet. Genome Res., in press. [DOI] [PubMed]
- 4.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 5.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 6.Imai, S., Okumoto, M., Iwai, M., Haga, S., Mori, N., Miyashita, N., Moriwaki, K., Hilgers, J. & Sarkar, N. H. (1994) J. Virol. 68, 3437-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao, Y., Jacobs, C. P., Wang, L. & Hardies, S. C. (1999) Mamm. Genome 10, 477-481. [DOI] [PubMed] [Google Scholar]
- 8.Bromham, L., Clark, F. & McKee, J. J. (2001) J. Virol. 75, 3053-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuss, F. U. & Schaller, H. C. (1991) J. Virol. 65, 5702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Best, S., Le Tissier, P. R. & Stoye, J. P. (1997) Trends Microbiol. 5, 313-318. [DOI] [PubMed] [Google Scholar]
- 11.Benit, L., de Parseval, N., Casella, J.-F., Callebaut, I., Cordonnier, A. & Heidmann, T. (1997) J. Virol. 71, 5652-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mi, S., Lee, X., Li, X., Veldman, G., Finnerty, H., Racie, L., LaVallie, E., Tang, X., Edouard, P., Howes, S., Keith, J. J. & McCoy, J. (2000) Nature 17, 785-789. [DOI] [PubMed] [Google Scholar]
- 13.Blond, J. L., Lavillette, D., Cheynet, V., Bouton, O., Oriol, G., Chapel-Fernandes, S., Mandrand, B., Mallet, F. & Cosset, F. L. (2000) J. Virol. 74, 3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaise, S., de Parseval, N., Bénit, L. & Heidmann, T. (2003) Proc. Natl. Acad. Sci. USA 100, 13013-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frendo, J. L., Olivier, D., Cheynet, V., Blond, J. L., Bouton, O., Vidaud, M., Rabreau, M., Evain-Brion, D. & Mallet, F. (2003) Mol. Cell. Biol. 23, 3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallet, F., Bouton, O., Prudhomme, S., Cheynet, V., Oriol, G., Bonnaud, B., Lucotte, G., Duret, L. & Mandrand, B. (2004) Proc. Natl. Acad. Sci. USA 101, 1731-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benit, L., Dessen, P. & Heidmann, T. (2001) J. Virol. 75, 11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Parseval, N., Lazar, V., Bénit, L., Casella, J. & Heidmann, T. (2003) J. Virol. 77, 10414-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbot, W., Wasowicz, M., Dupressoir, A., Botteri, C. & Heidmann, T. (2002) Biochim. Biophys. Acta 1576, 81-91. [DOI] [PubMed] [Google Scholar]
- 20.Heidmann, O. & Heidmann, T. (1991) Cell 64, 159-170. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, N. A., Copeland, N. G., Taylor, B. A. & Lee, B. K. (1982) J. Virol. 43, 26-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obata, M. M. & Khan, A. S. (1988) J. Virol. 62, 4381-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter, E. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 71-119.
- 24.Coffin, J. M. (1986) Cell 4, 1-4. [Google Scholar]
- 25.Pinter, A., Kopelman, R., Li, Z., Kayman, S. C. & Sanders, D. A. (1997) J. Virol. 71, 8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cianciolo, G., Copeland, T. D., Orozlan, S. & Snyderman, R. (1985) Science 230, 453-455. [DOI] [PubMed] [Google Scholar]
- 27.Georgiades, P., Ferguson-Smith, A. C. & Burton, G. J. (2002) Placenta 23, 3-19. [DOI] [PubMed] [Google Scholar]
- 28.Basyuk, E., Cross, J., Corbin, J., Nakayama, H., Hunter, P., Nait-Oumesmar, B. & Lazzarini, R. (1999) Dev. Dyn. 214, 303-311. [DOI] [PubMed] [Google Scholar]
- 29.Rein, A., Mirro, J., Gordon-Haynes, J., Ernst, S. M. & Nagashima, K. (1994) J. Virol. 68, 1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson, M., Catzeflis, F., Briolay, J. & Mouchiroud, D. (1997) Mol. Phylogenet. Evol. 8, 423-434. [DOI] [PubMed] [Google Scholar]
- 31.Wu, C. I. & Li, W. H. (1985) Proc. Natl. Acad. Sci. USA 82, 1741-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossant, J. & Cross, J. (2001) Nature 2, 538-548. [DOI] [PubMed] [Google Scholar]
- 33.Sapin, V., Blanchon, L., Serre, A. F., Lemery, D., Dastugue, B. & Ward, S. J. (2001) Transgenic Res. 10, 377-398. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber, J., Riethmacher-Sonnenberg, E., Riethmacher, D., Tuerk, E. E., Enderich, J., Bosl, M. R. & Wegner, M. (2000) Mol. Cell. Biol. 20, 2466-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anson-Cartwright, L., Dawson, K., Holmyard, D., Fisher, S., Lazzarini, R. & Cross, J. (2000) Nat. Genet. 25, 311-314. [DOI] [PubMed] [Google Scholar]
- 36.Yu, C. Y., Shen, K., Lin, M., Chen, P., Lin, C., Chang, G.-D. & Chen, H. (2002) J. Biol. Chem. 277, 50062-50068. [DOI] [PubMed] [Google Scholar]
- 37.Mess, A. (2003) J. Exp. Zoolog. 299A, 78-98. [DOI] [PubMed] [Google Scholar]
- 38.Leiser, R. & Kaufmann, P. (1994) Exp. Clin. Endocrinol. 102, 122-134. [DOI] [PubMed] [Google Scholar]
- 39.Harris, J. R. (1998) BioEssays 20, 307-316. [DOI] [PubMed] [Google Scholar]
- 40.Muir, A., Lever, A. & Moffett, A. (2004) Placenta 25, S16-S25. [DOI] [PubMed] [Google Scholar]
- 41.Palmarini, M., Mura, M. & Spencer, T. E. (2004) J. Gen. Virol. 85, 1-13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.