Abstract
In addition to neuronal vacuolation and astrocytic hypertrophy, dendritic atrophy is a prominent feature of prion disease. Because increased Notch-1 expression and cleavage releasing its intracellular domain (NICD) inhibit both dendrite growth and maturation, we measured their levels in brains from mice inoculated with Rocky Mountain Laboratory (RML) prions. The level of NICD was elevated in the neocortex, whereas the level of β-catenin, which stimulates dendritic growth, was unchanged. During the incubation period, levels of the disease-causing prion protein isoform, PrPSc, and NICD increased concomitantly in the neocortex. Additionally, increased levels of Notch-1 mRNA and translocation of NICD to the nucleus correlated well with regressive dendritic changes. In scrapie-infected neuroblastoma (ScN2a) cells, the level of NICD was elevated compared with uninfected control (N2a) cells. Long neurofilament protein-containing processes extended from the surface of N2a cells, whereas ScN2a cells had substantially shorter processes. Transfection of ScN2a cells with a Notch-1 small interfering RNA decreased Notch-1 mRNA levels, diminished NICD concentrations, and rescued the long process phenotype. These results suggest that PrPSc in neurons and in ScN2a cells activates Notch-1 cleavage, resulting in atrophy of dendrites in the CNS and shrinkage of processes on the surface of cultured cells. Whether diminishing Notch-1 activation in vivo can prevent or even reverse neurodegeneration in prion disease remains to be established.
Keywords: neurodegeneration, scrapie, Creutzfeldt-Jakob disease, Alzheimer's disease
Prions are infectious proteins that propagate by recruiting a naturally occurring precursor protein and stimulating its conversion into nascent prions (1). Formation of prions involves a conformational change in the precursor protein (2). In mammals, the accumulation of prions is accompanied by neurodegeneration (3-6), whereas in fungi, the prion state is associated with altered metabolism (7, 8). In both mammals (9, 10) and fungi (11), fragments of the precursor proteins have been refolded under cell-free conditions and shown to be infectious upon introduction into the appropriate host.
In mammals, the prion diseases include Creutzfeldt-Jakob disease of humans, scrapie of sheep, and bovine spongiform encephalopathy. In all these disorders, the sole component of the disease-causing isoform of the prion protein (PrPSc) accumulates in the CNS, resulting in presynaptic bouton degeneration, dendritic atrophy, vacuolation of neurons, and hypertrophy of astrocytes (3-6, 12). The prion diseases are invariably fatal. PrPSc is formed from the precursor protein PrPC by a profound conformational change (2). PrPC contains three α-helices (13), at least one of which, along with some portion of the unstructured region, seems to refold into a β-helix during the transformation to PrPSc (14, 15).
The tertiary structure of PrPSc appears to encipher biological information that defines a particular prion strain (10, 16-19). Strains of prions differ phenotypically in their incubation times, ability to infect animals of another species, and neuroanatomical patterns of PrPSc deposition (5, 20, 21). When PrPSc accumulates within neurons and their processes, neuropathological changes that typify the degenerative process, including vacuolation of synaptic regions and nerve cell death, are triggered. Quantitative morphological, functional, neurochemical, and immunohistochemical studies during the course of prion disease have identified a stereotypical progression of neurodegeneration (6, 12). After initiation of disease in a group of neurons by intracerebral inoculation of prions, disease spreads to other neurons and to other brain regions by anterograde axonal transport of PrPSc to axon terminals. This is followed by presynaptic bouton degeneration and dendritic atrophy and, later, by nerve cell death.
To investigate the molecular events that underlie dendritic atrophy, we studied the expression of genes that regulate dendrite growth and maturation during CNS development. We began by examining β-catenin, which initiates dendritic growth and maturation (22), and then measured Notch-1, which inhibits both dendritic and axonal growth and maturation during neuronal development and causes regression of mature dendrites and axons (23-26). We report here that dendritic atrophy was accompanied by increased levels of the Notch-1 intracellular domain (NICD) in neuronal nuclei of prion-infected mice. Elevated levels of NICD were also found in scrapie-infected neuroblastoma (ScN2a) but not in uninfected control (N2a) cells. Although long processes, also referred to as neurites, extend from the surface of N2a cells, ScN2a cells exhibit much shorter processes. To determine whether Notch-1 activation has a role in the shortening of these processes, we transfected ScN2a cells with a Notch-1 small interfering (si) RNA and observed that the normal long-neurite phenotype was rescued. These findings suggest that Notch-1 activation may mediate dendritic atrophy in the brains of humans and animals dying of prion disease.
Materials and Methods
Animals. All animal experiments were performed according to the Guidelines of the Society of Neuroscience and the Institutional Animal Review Committee of the University of California, San Francisco. Animals were housed in pairs under diurnal lighting conditions (12-h light-dark cycles). The macroenvironment was controlled to provide a temperature of 20 ± 2°C and a relative humidity of 45 ± 5%. Female CD1 mice were inoculated intrathalamically on the right with the RML strain of mouse prions, which produces clinical signs of disease at ≈120 days postinoculation (dpi) and terminal disease at ≈150 dpi. They were killed by decapitation at 30, 60, 90, 120, and 130-140 dpi. Age- and sex-matched controls were inoculated in parallel with PBS.
Cell Culture. N2a and ScN2a cells were maintained in Eagle's minimum essential media with Earle's salts containing 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 units of penicillin G per ml, 0.1 mg of streptomycin sulfate/ml, and 10% FCS at 37°C in 5% CO2. To induce differentiation, N2a and ScN2a cells were plated onto glass coverslips that had been sequentially coated with 1 mg/ml poly-l-lysine in 0.1 M borate buffer, pH 8.5, and 0.1 mg of mouse laminin per ml in PBS, respectively. Alternatively, cells were grown in 6-well or 10-cm dishes coated with polylysine/laminin purchased from B-D Labware (Biocoat Cellware, Bedford, MA). To stimulate neural process formation, cells were then cultured in neurobasal medium containing the above-described antibiotics, 2 mM l-glutamine, N2 supplement, and 10 ng/ml murine 2.5S nerve growth factor (Invitrogen).
PrPSc in Synaptosomal Preparations. Synaptosomes were purified from the neocortex as described (6). In brief, the neocortex was dissected and pooled from two mouse brains. Three pools were obtained at each time point for statistical analysis. All steps of the isolation procedure were carried out quickly, and all solutions were chilled on ice. The pools were suspended in 10 volumes (wt/vol) of an ice-cold solution containing 0.32 M sucrose in 5 mM Tris, pH 7.4, and homogenized in a Potter-Elvehjem glass homogenizer with 14 strokes of a Teflon pestle. The crude homogenates were transferred to 50-ml conical tubes and centrifuged at 1,500 × g for 10 min in a Beckman fixed rotor centrifuge to pellet cell debris and nuclei. The supernatants were removed and the pellet resuspended for a second slow-speed clarification process to obtain additional synaptosomes. The supernatants from the two slow-speed centrifugations were pooled and centrifuged at 15,000 × g for 25 min in a fixed-rotor KA-30 centrifuge (Optima LE-8OK, Beckman Coulter) to obtain a pellet enriched in synaptosomes. That pellet was resuspended in 10 volumes of RIPA lysis buffer (1 × PBS, pH 7.4/1% igepal CA-630/0.5% sodium deoxycholate/0.1% SDS), vortexed at 4°C for 20 min and at low speed in an Eppendorf tabletop centrifuge for 5 min to remove additional cellular debris from the synaptosomes.
For additional enrichment of PrPSc, the final synaptosomal supernatants were subjected to two rounds of incubation with sodium phosphotungstate (NaPTA) to selectively precipitate PrPSc (18, 27, 28). Protein concentrations in the supernatants were determined by bichromic acid assay. For the first precipitation, a volume of solution containing 2 mg of total protein was obtained from each sample, and a stock solution of Sarkosyl was added to a final concentration of 2% (wt/vol). These samples were mixed with a stock solution containing 4% NaPTA and 170 mM MgCl2, pH 7.4, to obtain a final concentration of 0.32% NaPTA. After 1-h incubation at 37°C on a rocking platform, the samples were centrifuged for 30 min at 14,000 × g in a microcentrifuge at room temperature (RT). For the second precipitation, the pellets were resuspended in 0.25 ml of PBS with 0.1% Sarkosyl containing 50 μg/ml proteinase K (PK) to eliminate PrPC and protease-sensitive PrPSc. After 1 h at 37°C on a rocking platform, PK digestion was stopped by the addition of 2 mM phenylmethylsulfonyl fluoride. To that solution, NaPTA was added to a final concentration of 0.32% followed by incubation at 37°C for 1 h on a rocking platform and then centrifugation at 14,000 × g for 30 min at RT to precipitate protease-resistant PrPSc. The pellets were resuspended in equal volumes of SDS sample buffer and their proteins separated on 4-12% Bis-Tris polyacrylamide gels and transferred to poly(vinylidene difluoride) membranes for Western blot analysis recommended by the manufacturer (NuPage gel system, Invitrogen). The gels were loaded with aliquots containing equal amounts of protein based on the protein measurements made just before the NaPTA precipitation steps.
Western Blot Analysis of Neocortex and N2a/ScN2a Cells. Neocortex dissected from mouse brains was homogenized in RIPA buffer. For cultured cell lines, cells were scraped from plates, pelleted at low speed, and lysed in 10 mM Tris·HCl/100 mM NaCl/10 mM EDTA/0.5% Triton X-100/0.5% deoxycholate, pH 7.5. Protein concentrations were determined by using the bichromic acid assay protein assay by using BSA as a standard (Pierce). After SDS gel electrophoresis and transfer of proteins to poly-(vinylidene difluoride) membranes, the blots were probed by using the following antibodies: Notch-1 C-20 and β-catenin E-5 from Santa Cruz Biotechnology (Santa Cruz, CA), cleaved Notch-1 Val-1744 from Cell Signaling Technology (Beverly, MA), GAPDH from Chemicon International (Temecula, CA), and PrP-specific HuM-D13 (19). Proteins were detected with an enhanced chemiluminescence system (Amersham Pharmacia). Densitometry of autoradiograms was used to estimate the relative concentrations of target proteins.
Immunofluorescence Microscopy. Ten-micrometer-thick coronal cryostat sections were mounted on glass slides and fixed in methanol containing 1% hydrogen peroxide. For triple fluorescence detection in mouse brain, goat anti-Notch-1 C-20 antibody (Santa Cruz Biotechnology) binding was detected by using biotinylated horse anti-goat IgG (heavy and light chain) followed by incubation with fluorescein-containing reagents of the tyramide signal amplification Fluorescent System (PerkinElmer). To detect neuron-specific mouse anti-NeuN antibodies (Chemicon), anti-mouse Alexa Fluor 568 (Molecular Probes) was used as the secondary antibody. Nonspecific binding during this mouse-on-mouse immunohistochemical step was blocked with the Vector M.O.M. kit (Vector Laboratories). Vectashield Mounting Media containing DAPI (Vector Laboratories) was used to identify nuclei. N2a and ScN2a cells grown on coverslips were fixed for 10 min with 4% formaldehyde in PBS and then in 1:1 methanol/acetone for 10 min before being stained for mouse antineurofilament protein (NF200, NovoCastra, Newcastle, UK), which was detected with anti-mouse Alexa Fluor 568. Fluorescence images were recorded with Zeiss LSM 510 and Leica TCS SP confocal microscopes.
Dendrite Length and Spine Density. Dendritic length and spine density were measured as described (29). Golgi silver impregnation was performed according to the instructions with the FD Rapid GolgiStain Kit (FD NeuroTechnologies, Ellicott City, MD). Silver-impregnated dendrites in 100-μm-thick sections were analyzed. Ten pyramidal neurons whose dendritic trees were relatively isolated from other silver-impregnated neurons and that did not have discontinuities or breaks in their dendritic trees were chosen for quantification in each of three ill and three age-matched control animals at several time points during the course of the disease. The primary apical and total basal dendrite lengths of neurons in layer 4 of the somatosensory neocortex overlying the hippocampus were measured at ×200. scion image for windows, release β4.0.2 (Scion, Frederick, MD) was the morphometric program used to make the measurements of dendrite parameters.
Quantitative RT-PCR. Total RNA was isolated from normal and ill mouse brains by homogenizing the tissue in Trizol (Invitrogen) and extracting with chloroform. RNeasy columns (Qiagen, Valencia, CA) were used to purify further the total RNA post-DNase-I treatment (Ambion, Austin, TX, DNA-free kit). RT reactions were performed on total RNA with Taqman core RT reagents (Applied Biosystems) by using random hexamers as primers. Amounts ranging from 250 pg to 2 μg of total RNA were used to create standard curves, and 100 ng of experimental total RNA was used per 100-μl RT reaction. A total of 10 μl of cDNA was used for each PCR reaction. Taqman probes and primers were designed by using primer 3 software (Whitehead Institute Center for Genome Research, Cambridge, MA) and were purchased through Integrated DNA Technologies (Coralville, IA). The PCR reactions were carried out by using Taqman core PCR reagents (Applied Biosystems) with 200 nM concentration of primers and 100 nM of fluorescent probe. For signal detection, the Applied Biosystems HT 9700 sequence detector was programmed to an initial step at 50°C for 2 min and by 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Once the PCR was completed, each sample was given a threshold cycle (CT) value, which is defined as the number of PCR cycles needed to exceed a minimum fluorescent detection threshold. The analysis of the Taqman data was done by using the comparative CT method (Applied Biosystems, Sequence Detection System User Bulletin no. 2). In each case, GAPDH was used as an endogenous reference.
RNA Interference. Three chemically synthesized siRNAs, Notch-1-1 (nucleotides 473-491), Notch-1-2 (nucleotides 482-500), and Notch-1-3 (nucleotides 1529-1547), against murine Notch-1 were purchased from Ambion. These siRNAs were transiently transfected into N2a and ScN2a cells by using Lipofectamine 2000 per the manufacturer's instructions (Invitrogen). An siRNA targeted to GFP (GPF-22) was purchased from Qiagen as a negative control in transfection experiments. Briefly, cells were plated the day before transfection at the desired density in maintenance media without antibiotics, typically 5,000 cells per cm2. Cells were exposed to the siRNA-Lipofectamine mixture for 5 h before the maintenance medium was replaced by differentiation medium for the times indicated.
Results
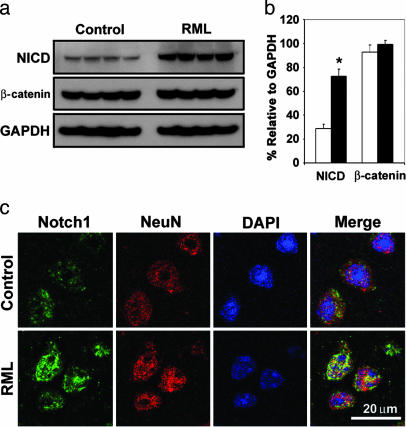
NICD Levels Increase During Prion Infection. Homogenates were prepared from neocortex of prion-inoculated CD1 mice that were killed ≈130 dpi. Western blots were used to assess the levels of β-catenin and NICD. We observed a statistically significant 2- to 3-fold increase in the level of NICD in prion-infected mice but no change in β-catenin (Fig. 1 a and b). Confocal microscopy showed accumulation of NICD in the nuclei of many neocortical neurons; in contrast, little or no NICD was located in the nuclei of neurons in uninfected control cortex (Fig. 1c).
Fig. 1.
Increased concentrations of NICD but not β-catenin occur during RML infection in CD1 mice. (a) Western blot analysis of NICD (Notch-1 C20 antibody), β-catenin, and GAPDH in neocortical homogenates from four ill mice at 130 dpi and four age-matched control mice. Identical results were obtained with the cleaved Notch-1 Val-1744 antibody. (b) Densitometry estimates of the concentrations of NICD and β-catenin relative to GAPDH show a statistically significant 2.5-fold increase in NICD concentration (Student's t test; *, P < 0.0001) but no significant change in β-catenin during RML infection (filled bars) compared with controls (open bars). (c) Confocal microscopy shows accumulation of NICD in the nuclei of layer IV neurons in RML-infected mice at 140 dpi. In uninfected control neurons, merge shows small amounts of NICD mostly in the cytoplasm. A C-terminal Notch-1 antibody was used to localize NICD. A NeuN antibody was used to identify neurons. DAPI identifies nuclei.
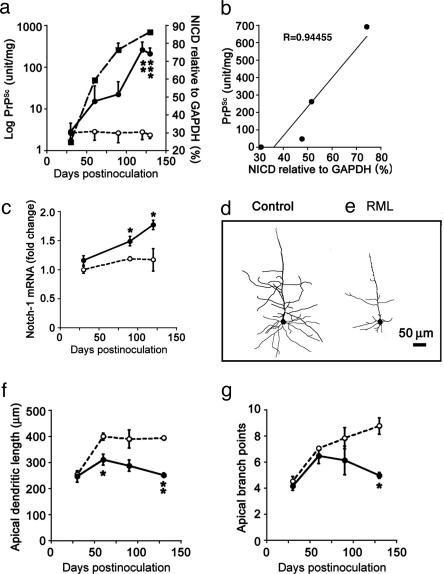
Increased NICD Levels Correlate with PrPSc Accumulation. We found that increases in NICD levels correlated with PrPSc accumulation in prion-infected mice. Because previous studies indicated that the initial accumulation of PrPSc in the neocortex is within presynaptic boutons after its anterograde transport along thalamocortical pathways (6), we measured PrPSc concentration by using synaptosomal preparations. Further enrichment of PrPSc from synaptosomes was accomplished by selective precipitation with NaPTA (18). Compared with uninoculated controls, the level of NICD in prion-inoculated mice increased progressively throughout the incubation period, reaching statistically significant increases at 120 and 130 dpi (Fig. 2a). The levels of PrPSc rose ≈700-fold during the incubation period, whereas the levels of NICD increased ≈2.5-fold compared with controls. The progressive increase of NICD concentration correlated well with the accumulation of PrPSc (R = 0.94) (Fig. 2b). A progressive increase in neocortical Notch-1 mRNA expression, measured by quantitative RT-PCR also followed accumulation of PrPSc (Fig. 2c). At 30 dpi, the levels of Notch-1 mRNA were similar in control and prion-infected mice (n = 3). By 90 dpi, a ≈20% increase in Notch-1 mRNA was detected, which rose to ≈50% (P < 0.05) at 120 dpi. These in vivo results suggest a close temporal relationship between PrPSc accumulation and elevated levels of NICD and raise the possibility that PrPSc directly or indirectly triggers increased Notch-1 expression and cleavage.
Fig. 2.
PrPSc accumulation in neocortical synaptosomes coincides with increased amounts of NICD, increased expression of Notch-1 mRNA, and regressive changes in dendrites. (a) Kinetics of the log of neocortical PrPSc accumulation in synaptosomes (filled squares) relative to NICD concentrations (filled circles) during the course of prion disease. NICD levels in age-matched PBS-inoculated mice are shown as controls (open circles). (b) A plot of synaptosomal PrPSc vs. NICD shows a high degree of correlation. (c) Quantitative RT-PCR measurements of Notch-1 mRNA levels at three time points during the course of prion disease (filled circles) relative to PBS-inoculated controls (open circles). (d and e) Camera lucida drawings of Golgi silver-stained dendritic trees show one type of regressive change; compare PBS-inoculated age-matched control cerebral cortex (d) with RML-infected cerebral cortex at 90 dpi (e). Note that the Golgi method stains only a small percentage of neurons. (f and g) Apical dendrite lengths (f) and numbers of apical dendrite branch points (g) during RML infection (filled circles) compared with uninoculated agematched controls (open circles) are shown. Data points and bars represent means and SEM, respectively, calculated from three independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0001 by Student's t test.
PrPSc Accumulation Results in Dendritic Atrophy. Loss and shortening of dendritic branches during prion disease in hamster neocortex has been reported (30), as has progressive loss of dendritic spines after local accumulation of PrPSc in mouse hippocampus (12). Here, we examined 10 Golgi-silver-impregnated pyramidal neurons in layer 4 from each of three control (Fig. 2d) and prion-infected mice (Fig. 2e) at four time points during the incubation period. Lengths of apical and basal dendrites, the numbers of dendritic branch points, and the numbers of dendritic spines were quantified as a function of PrPSc accumulation in the neocortex. Only changes in the lengths of apical dendrites (Fig. 2f) and the numbers of apical dendrite branch points are shown (Fig. 2g), because all of the regressive changes produced similar curves when plotted as a function of the incubation period. These degenerative changes appear to follow the accumulation of PrPSc in neocortex and worsen as PrPSc accumulation continues to increase (Fig. 2a).
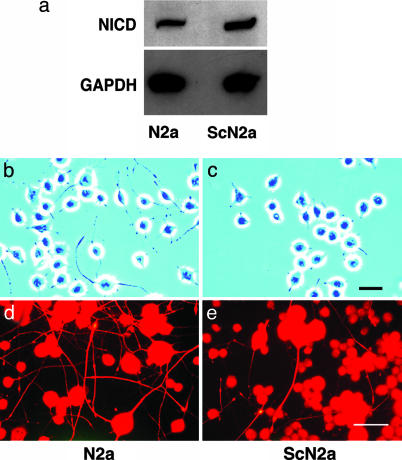
Elevated NICD Levels Interfere with Growth of Neuritic Processes. Similar to prion-infected brains, ScN2a cells had elevated levels of NICD compared with N2a cells (Fig. 3a). Both the ScN2a and control N2a cells were grown in a defined neurobasal medium containing N2 supplement and 10 ng/ml nerve growth factor that promotes neuronal differentiation and growth of neuritic processes. By convention, processes with a length of less than twice the cell diameter were designated “processes,” and those with lengths more than twice the cell diameter were designated “neurites.” Uninfected N2a cells grew numerous long neurites under these culture conditions (Fig. 3b), whereas most ScN2a cells grew short processes (Fig. 3c). Immunohistochemistry shows that the cell bodies and neurites of N2a and ScN2a cells contain high molecular-weight neurofilament protein (NFP200), which also highlights the difference in numbers of neurites (Fig. 3 d and e).
Fig. 3.
ScN2a cells have a higher concentration of NICD and fewer cells with neurites compared with N2a cells. (a) Western blots show ≈2× as much NICD in ScN2a cells (cleaved Notch-1 Val-1744 antibody). GADPH was used to normalize the data. (b and c) Phase-contrast microscopy shows that many N2a cells grew neurites that are more than two cell diameters in length (b); in contrast, most processes of ScN2a cells are less than two cell diameters in length (c). (d and e) Fluorescence immunohistochemistry shows that the cell bodies and long neurites of both N2a (d) and ScN2a cells (e) are immunopositive for the high molecular-weight neurofilament protein, NF200. N2a cells grew far more NF200-immunopositive neurites than ScN2a cells. (Bars = 90 μm; bar in c applies to b; bar in e applies to d.)
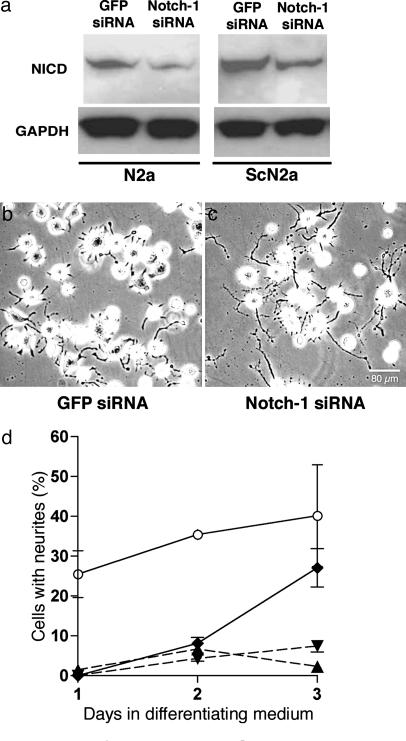
Inhibition of Notch-1 Activation Enables Normal Neurite Growth. To test the possibility that the short-process phenotype is related to elevated NICD levels, ScN2a cells were transfected with Notch-1 siRNA or GFP siRNA as a negative control. Three different commercially available Notch-1 siRNAs were obtained and tested for knockdown of gene expression in N2a and ScN2a cells. One of those siRNAs that produced a consistent reduction of NICD in both N2a and ScN2a cells was used in all subsequent experiments. Subconfluent cultures were transfected for 5 h with 5 nM or 50 nM Notch-1 siRNA in a standard growth medium containing FCS. The medium was replaced with the differentiating growth medium, and the effects on NICD levels and neurite outgrowth were measured over 72 h. Because both 5 and 50 nM Notch-1 siRNA produced ≈40% decreases in Notch-1 mRNA by 48 h relative to untreated cells, as measured by quantitative RT-PCR, all subsequent transfections were performed by using 5 nM siRNA. In both N2a and ScN2a cells, 5 nM Notch-1 siRNA decreased NICD levels by ≈30% at 72 h (Fig. 4a), whereas 5 nM GFP siRNA had no detectable effect. The levels of NICD were quantified by densitometry of Western blots from three separate experiments.
Fig. 4.
An siRNA against Notch-1 decreases NICD concentration by a mean of ≈30% in both N2a and ScN2a cells and rescues the long-neurite phenotype in ScN2a cells. (a) NICD levels in N2a and ScN2a cells are reduced 3 d after transfection with 5 nM Notch-1 siRNA (Notch-1-1, Ambion); in contrast, 5 nM GFP siRNA had no apparent effect. (b and c) Phase-contrast microscopy shows that ScN2a cells treated with Notch-1 siRNA (c) have longer processes than ScN2a cells treated with GFP siRNA (b) 3 d posttransfection and growth in differentiating medium. [Bar (c) = 80 μm and also applies to b.] (d) Cells with neurites measuring more than or equal to two cell diameters at different times points in differentiating medium expressed as a percentage of total cells. Data points and bars represent mean percentages and SEM, respectively, from at least three independent experiments. For transfection experiments, ≥1,000 cells were examined at each time point. Open circles, control N2a cells; filled diamonds, ScN2a cells treated with 5 nM Notch-1 siRNA; filled inverted triangles, ScN2a cells treated with 5 nM GFP siRNA; filled triangles, untreated ScN2a cells.
At 72 h in differentiation medium, <8% of nontransfected ScN2a cells or ScN2a cells transfected with GFP siRNA had surface neurites (Fig. 4 b and d); in contrast, ≈27% of ScN2a cells treated with Notch-1 siRNA grew long neurites (Fig. 4 c and d). The number of N2a cells with neurites ranged from ≈26% at 24 h (day 1) to ≈40% at 72 h (Fig. 4d). The difference between Notch-1 siRNA-transfected ScN2a cells and nontransfected N2a cells was not statistically significant at 72 h (Fig. 4d). These results argue that decreasing NICD concentration in ScN2a cells by selectively knocking down Notch-1 mRNA expression resulted in recovery of the normal neurite phenotype.
Discussion
The results described above show a temporal association between PrPSc accumulation and increased levels of NICD in the neocortex of mice infected with RML prions. Elevated levels of Notch-1 mRNA; elevated levels of the Notch-1 cleavage product, NICD; as well as translocation of the NICD to the nucleus were found in the brains of these mice. As reported earlier (12, 30) and verified here, regressive dendritic changes were prominent in the brains of these prion-infected mice (Fig. 2). Because dendritic growth in the developing nervous system is inhibited by NICD, our results raise the possibility that increased levels of NICD and its translocation to the nucleus may be responsible for dendritic atrophy in prion diseases. To explore this possibility, ScN2a cells were studied to determine whether the relationship between PrPSc and Notch-1 in brains of prion-infected mice was recapitulated in cultured cells. Compared with uninfected controls, NICD was elevated ≈50%, and the number of cells with long neurites was significantly reduced in ScN2a cells.
To test the possibility that elevated NICD levels in ScN2a cells are responsible for the shortened processes on the surface of these cells, we used siRNA to diminish the level of NICD. Under conditions in which NICD levels were decreased by using siRNA, recovery of the normal neurite-length phenotype was observed. These findings argue that the short-process phenotype in ScN2a cells and, by inference, regressive dendritic changes in prioninfected mice are due to increased levels of NICD.
It will be important to inhibit Notch-1 activation in mice inoculated with prions and to determine whether the incubation time is prolonged. Might mice continue to produce PrPSc but not exhibit neurological dysfunction under such circumstances? Although the results of the studies reported here show that inhibition of Notch-1 activation restores dendritic processes on the surface of cultured cells, it is unknown whether these events occur in animals. If inhibition of Notch-1 activation does prevent dendritic atrophy in vivo, it will be of interest to learn whether neuronal vacuolation and astrocytic gliosis, which are generally present in prion disease, are diminished concomitantly.
Because only the RML prion strain was used in both the mouse and cultured cell studies described here, we must determine whether Notch-1 activation occurs with other strains. Much evidence indicates that different strains of prions reflect distinct conformers of PrPSc (10, 16-19). It is also critical to establish whether Notch-1 activation features in the pathogenesis of human prion disease, including the inherited forms of these disorders.
The data presented here suggest that PrPSc accumulation in plasma membranes (6, 31) directly or indirectly activates Notch-1 cleavage, releasing NICD. In turn, elevated levels of NICD produce dendritic atrophy. It is notable that Notch-1 and the β-amyloid precursor protein (APP) are both cleaved by γ-secretase (32). Cleavage of APP by γ-secretase is a critical step in the formation of Aβ42 peptide that comprises amyloid plaques in Alzheimer's disease (AD). Additionally, elevated levels of Notch-1 protein (33, 34) and dendritic atrophy (35) are features of AD.
From the studies reported here, it is important to ask whether inhibitors of Notch-1 expression or cleavage might be used as pharmacotherapeutics for prion disease. Indeed, inhibitors of γ-secretase are being developed in an effort to find therapeutics for AD. Whether inhibition of γ-secretase activity might also delay cognitive decline secondary to synaptic degeneration in Creutzfeldt-Jakob disease might be worth investigating.
Acknowledgments
We thank Peter Nelken and Amy Tang for additional technical support and Hang Nguyen for editing the manuscript. This work was funded by grants from the National Institutes of Health (AG10770, AG02132, and AG021601). E.B.-B. was supported in part by the John Douglas French Foundation for Alzheimer's disease.
Author contributions: N.I., J.L.C., S.B.P., E.J.H., and S.J.D. designed research; N.I., J.L.C., E.B.-B., E.S., E.J.H., and S.J.D. performed research; S.B.P. contributed new reagents/analytic tools; N.I., J.L.C., E.B.-B., E.S., E.J.H., and S.J.D. analyzed data; and N.I., J.L.C., E.J.H., and S.J.D. wrote the paper.
Abbreviations: NICD, Notch-1 intracellular domain; PrPSc, disease-causing isoform of the prion protein; ScN2a, scrapie-infected neuroblastoma cells; N2a, uninfected control cells; siRNA, small interfering RNA; RML, Rocky Mountain Laboratory; dpi, days postinoculation; NaPTA, sodium phosphotungstate.
References
- 1.Prusiner, S. B., Scott, M. R., DeArmond, S. J. & Cohen, F. E. (1998) Cell 93, 337-348. [DOI] [PubMed] [Google Scholar]
- 2.Cohen, F. E., Pan, K.-M., Huang, Z., Baldwin, M., Fletterick, R. J. & Prusiner, S. B. (1994) Science 264, 530-531. [DOI] [PubMed] [Google Scholar]
- 3.DeArmond, S. J., Mobley, W. C., DeMott, D. L., Barry, R. A., Beckstead, J. H. & Prusiner, S. B. (1987) Neurology 37, 1271-1280. [DOI] [PubMed] [Google Scholar]
- 4.Jendroska, K., Heinzel, F. P., Torchia, M., Stowring, L., Kretzschmar, H. A., Kon, A., Stern, A., Prusiner, S. B. & DeArmond, S. J. (1991) Neurology 41, 1482-1490. [DOI] [PubMed] [Google Scholar]
- 5.Hecker, R., Taraboulos, A., Scott, M., Pan, K.-M., Torchia, M., Jendroska, K., DeArmond, S. J. & Prusiner, S. B. (1992) Genes Dev. 6, 1213-1228. [DOI] [PubMed] [Google Scholar]
- 6.Bouzamondo-Bernstein, E., Hopkins, S. D., Spillman, P., Uyehara-Lock, J., Deering, C., Safar, J., Prusiner, S. B., Ralston, H. J. & DeArmond, S. J. (2004) J. Neuropathol. Exp. Neurol. 63, 882-899. [DOI] [PubMed] [Google Scholar]
- 7.Wickner, R. B. (1994) Science 264, 566-569. [DOI] [PubMed] [Google Scholar]
- 8.Kajava, A. V., Baxa, U., Wickner, R. B. & Steven, A. C. (2004) Proc. Natl. Acad. Sci. USA 101, 7885-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneko, K., Ball, H. L., Wille, H., Zhang, H., Groth, D., Torchia, M., Tremblay, P., Safar, J., Prusiner, S. B., DeArmond, S. J., et al. (2000) J. Mol. Biol. 295, 997-1007. [DOI] [PubMed] [Google Scholar]
- 10.Legname, G., Baskakov, I. V., Nguyen, H. O., Riesner, D., Cohen, F. E., DeArmond, S. J. & Prusiner, S. B. (2004) Science 305, 673-676. [DOI] [PubMed] [Google Scholar]
- 11.Osherovich, L. Z. & Weissman, J. S. (2001) Cell 106, 183-194. [DOI] [PubMed] [Google Scholar]
- 12.Jeffrey, M., Halliday, W. G., Bell, J., Johnston, A. R., Macleod, N. K., Ingham, C., Sayers, A. R., Brown, D. A. & Fraser, J. R. (2000) Neuropathol. Appl. Neurobiol. 26, 41-54. [DOI] [PubMed] [Google Scholar]
- 13.Donne, D. G., Viles, J. H., Groth, D., Mehlhorn, I., James, T. L., Cohen, F. E., Prusiner, S. B., Wright, P. E. & Dyson, H. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13452-13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wille, H., Michelitsch, M. D., Guénebaut, V., Supattapone, S., Serban, A., Cohen, F. E., Agard, D. A. & Prusiner, S. B. (2002) Proc. Natl. Acad. Sci. USA 99, 3563-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govaerts, C., Wille, H., Prusiner, S. B. & Cohen, F. E. (2004) Proc. Natl. Acad. Sci. USA 101, 8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bessen, R. A. & Marsh, R. F. (1994) J. Virol. 68, 7859-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telling, G. C., Parchi, P., DeArmond, S. J., Cortelli, P., Montagna, P., Gabizon, R., Lugaresi, E., Gambetti, P. & Prusiner, S. (1996) Science 274, 2079-2082. [DOI] [PubMed] [Google Scholar]
- 18.Safar, J., Wille, H., Itri, V., Groth, D., Serban, H., Torchia, M., Cohen, F. E. & Prusiner, S. B. (1998) Nat. Med. 4, 1157-1165. [DOI] [PubMed] [Google Scholar]
- 19.Peretz, D., Scott, M., Groth, D., Williamson, R. A., Burton, D. R., Cohen, F. E. & Prusiner, S. B. (2001) Protein Sci. 10, 854-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruce, M. E., McBride, P. A. & Farquhar, C. F. (1989) Neurosci. Lett. 102, 1-6. [DOI] [PubMed] [Google Scholar]
- 21.DeArmond, S. J., Sanchez, H., Qiu, Y., Ninchak-Casey, A., Daggett, V., Paminiano-Camerino, A., Cayetano, J., Yehiely, F., Rogers, M., Groth, D., et al. (1997) Neuron 19, 1337-1348. [DOI] [PubMed] [Google Scholar]
- 22.Yu, X. & Malenka, R. (2003) Nat. Neurosci. 6, 1169-1177. [DOI] [PubMed] [Google Scholar]
- 23.Berezovska, O., McLean, P., Knowles, R., Frosh, M., Lu, F. M., Lux, S. E. & Hyman, B. T. (1999) Neuroscience 93, 433-439. [DOI] [PubMed] [Google Scholar]
- 24.Huang, E. J., Li, H., Tang, A. T., Wiggins, A. K., Neve, R. L., Zhong, W., Jan, L. Y. & Jan, Y. N. (2005) Genes Dev. 19, 138-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redmond, L., Oh, S. R., Hicks, C., Weinmaster, G. & Ghosh, A. (2000) Nat. Neurosci. 3, 30-40. [DOI] [PubMed] [Google Scholar]
- 26.Sestan, N., Artavanis-Tsakonas, S. & Rakic, P. (1999) Science 286, 741-746. [DOI] [PubMed] [Google Scholar]
- 27.Safar, J. G., Scott, M., Monaghan, J., Deering, C., Didorenko, S., Vergara, J., Ball, H., Legname, G., Leclerc, E., Solforosi, L., et al. (2002) Nat. Biotechnol. 20, 1147-1150. [DOI] [PubMed] [Google Scholar]
- 28.Tremblay, P., Ball, H. L., Kaneko, K., Groth, D., Hegde, R. S., Cohen, F. E., DeArmond, S. J., Prusiner, S. B. & Safar, J. G. (2004) J. Virol. 78, 2088-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leuner, B., Falduto, J. & Shors, T. J. (2003) J. Neurosci. 23, 659-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan, R. N., Baringer, J. R. & Prusiner, S. B. (1987) J. Neuropathol. Exp. Neurol. 46, 461-473. [DOI] [PubMed] [Google Scholar]
- 31.Vey, M., Pilkuhn, S., Wille, H., Nixon, R., DeArmond, S. J., Smart, E. J., Anderson, R. G. W., Taraboulos, A. & Prusiner, S. B. (1996) Proc. Natl. Acad. Sci. USA 93, 14945-14949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lleo, A., Berezovska, O., Ramdya, P., Fukumoto, H., Raju, S., Shah, T. & Hyman, B. T. (2003) J. Biol. Chem. 278, 47370-47375. [DOI] [PubMed] [Google Scholar]
- 33.Berezovska, O., Xia, M. Q. & Hyman, B. T. (1998) J. Neuropathol. Exp. Neurol. 57, 738-745. [DOI] [PubMed] [Google Scholar]
- 34.Sestan, N. & Rakic, P. (2002) in Notch from Neurodevelopment to Neurodegeneration: Keeping the Fate, eds. Israel, A., De Strooper, B., Checler, F. & Christen, Y. (Springer, Berlin), pp. 19-40.
- 35.Scheibel, A. B. (1979) in Congenital and Acquired Cognitive Disorders, ed. Katzman, R. (Raven, New York), pp. 107-124.