Abstract
Steady-state and transient conformational changes upon the thermal unfolding of ubiquitin were investigated with nonlinear IR spectroscopy of the amide I vibrations. Equilibrium temperature-dependent 2D IR spectroscopy reveals the unfolding of the β-sheet of ubiquitin through the loss of cross peaks formed between transitions arising from delocalized vibrations of the β-sheet. Transient unfolding after a nanosecond temperature jump is monitored with dispersed vibrational echo spectroscopy, a projection of the 2D IR spectrum. Whereas the equilibrium study follows a simple two-state unfolding, the transient experiments observe complex relaxation behavior that differs for various spectral components and spans 6 decades in time. The transient behavior can be separated into fast and slow time scales. From 100 ns to 0.5 ms, the spectral features associated with β-sheet unfolding relax in a sequential, nonexponential manner, with time constants of 3 μs and 80 μs. By modeling the amide I vibrations of ubiquitin, this observation is explained as unfolding of the less stable strands III–V of the β-sheet before unfolding of the hairpin that forms part of the hydrophobic core. This downhill unfolding is followed by exponential barrier-crossing kinetics on a 3-ms time scale.
Keywords: protein-folding dynamics, temperature jump, nonlinear IR spectroscopy
Describing the conformational changes of proteins as they fold from a disordered denatured state to a compact native state remains an important experimental objective. Studies that examine this subject provide a molecular interpretation to the conceptual framework of the energy landscape picture (1–3) and allow more direct comparison of experiment and simulation (4, 5). Viewed as a problem in molecular dynamics, characterizing protein folding poses considerable challenges because it calls for a statistical yet structurally sensitive description of a heterogeneous ensemble in solution evolving over many decades in time.
Most protein-folding experiments measure kinetics: the rate of appearance or disappearance of an experimental signature for a particular species, typically on millisecond or longer time scales. These results give information on the height of energetic barriers much higher than thermal energy but say little about how structure changed along the path. A number of fast folding experiments of proteins and peptides have shown that downhill folding, in which evolution is governed by energy barriers ≲kT, can be initiated with a nanosecond temperature jump (T-jump). Such experiments work in a diffusive regime that allows a freer exploration of available structures and provide evidence that the relevant molecular time scales for folding is nanoseconds to microseconds (6–9). If downhill folding can be initiated and followed with a structure-sensitive probe, then meaningful information can be obtained on the underlying molecular dynamics of folding. We report here on such an experiment, a conformationally sensitive probing of downhill unfolding of ubiquitin over nanosecond-to-millisecond time scales after a T-jump.
The vast range of length and time scales involved in protein folding and unfolding ensures that no single experimental technique can capture all relevant structural changes at high spatial resolution in solution. Optical and IR spectroscopies such as fluorescence-based methods (10, 11), time-resolved circular dichroism (12), Raman (13), and IR absorption (14, 15) are desirable in fast folding experiments because they have the intrinsic time resolution to follow the fastest processes. Nevertheless, relating these experiments to nuclear coordinates that characterize a protein structure or conformation is not trivial, particularly in the presence of disorder.
Of these probing methods, we have chosen to use vibrational spectroscopy of the amide transitions of the protein backbone, because it is intrinsic to all proteins and has perhaps the most direct connection to protein conformation. IR spectroscopy on amide I transitions (primarily CO stretching; 1,600–1,700 cm-1) has been widely used for folding studies of proteins (7, 14–17) and peptides (9, 18–20), primarily because amide I spectra have peak positions that depend on secondary structure. The frequency–structure correlation is a result of through-space electrostatic couplings that depend on the distance and orientation between the many amide I vibrations of each of the protein's peptide units (21, 22). These couplings, which are spectroscopically significant for separations on the order of 3–8 Å, lead to vibrational excitations that extend over large regions of the protein and have IR transition frequencies that are sensitive to the configuration of peptide units with respect to one another and hydrogen bonding to the peptide carbonyls (21, 23).
Although the IR spectrum encodes information on the 3D structure of the protein, traditional IR spectroscopy has been limited in its use as a structural probe because spectral congestion among many overlapping features washes out the information of interest. 13C isotope labeling, which red-shifts the labeled amide I frequency by 35–40 cm-1 and is thought to localize the vibration, is one approach to spectral simplification that has been used for small helical peptides (9, 18–20). To probe conformationally sensitive delocalized states and defeat spectral congestion, we choose to probe protein unfolding with nonlinear IR spectroscopy. Our methods use sequences of femtosecond IR pulses to interrogate vibrational couplings, dissect congested spectra, and reveal transient structural information with picosecond time resolution. One such method is 2D IR spectroscopy, in which a vibrational spectrum is spread over two frequency axes, revealing couplings through the formation of cross peaks and isolating inhomogeneous broadening on the diagonal axis (24). As a particular strength, we have found that 2D IR spectroscopy is a sensitive indicator of antiparallel β-sheet conformation in proteins, as a result of cross peaks that form between two characteristic IR-active vibrational modes of the sheet (25–27). The shapes of these spectra are indicators of the number of strands in the sheet and its disorder. A closely related but technically simpler nonlinear IR experiment is dispersed vibrational echo (DVE) spectroscopy, which is related to a projection of the 2D IR spectrum onto one frequency axis. As a result of this projection, DVE spectra are not as detailed as 2D IR spectra, yet they still retain a unique sensitivity to vibrational couplings and secondary structure that is not present in conventional IR experiments (27).
We report on conformational changes during the thermal unfolding of ubiquitin studied both at equilibrium, with temperature-dependent 2D IR and DVE spectroscopy, and transiently, with DVE spectroscopy after a nanosecond T-jump. Equilibrium experiments suggest that the system obeys simple two-state folding kinetics, but transient experiments reveal a more complex relaxation behavior spanning 6 decades in time. From 100 ns to 0.5 ms, we observe a nonexponential, sequential loss of two β-sheet transitions that originate in delocalized β-sheet vibrations. On millisecond time scales, we cross into a kinetic regime, characterized by a concerted 3-ms exponential unfolding. An energy landscape represented in terms of native contacts between different strands of the β-sheet is proposed to explain the experiments.
Methods
Nonlinear IR Spectroscopy. Detailed descriptions of the nonlinear IR experiments can be found in refs. 24, 26, and 27, including the methods of acquiring 2D IR spectra and the relationship between 2D IR spectra and DVE spectra. Briefly, the mid-IR experiments are performed at a 1-kHz repetition rate by using 90-fs, 6-μm pulses. These pulses have a 160-cm-1 bandwidth (full width at half maximum), which covers the fundamental transitions of the amide I band (v = 0→1) and anharmonically shifted v = 1→2 transitions. Three 150-nJ pulses are focused onto the sample in a box geometry, giving rise to a background-free signal that is radiated into a phase-matched direction (ks = -kα + kβ + kχ). For the DVE measurement, the pulses are time-coincident. The signal is dispersed by a monochromator onto a 64-channel MCT array detector (see ref. 24 for details) with 4-cm-1 resolution, and the intensity as a function of detection frequency S̃DVE(ω3) is observed. For the 2D IR experiment, the timing between the first and second pulses (τ1) is varied with a 4-fs step. The signal is overlapped after the sample with a fourth local oscillator pulse for phase-sensitive detection and then dispersed in the monochromator. The 2D IR correlation spectrum S̃C(ω1,ω3) is obtained from the numerical Fourier transform of the interference signal along τ1 for all detection frequencies ω3. We plot the real part of the complex correlation spectrum obtained by summing rephasing (kR = -k1 + k2 + k3) and nonrephasing (kNR = k1 - k2 + k3) spectra. 2D IR and DVE spectra are obtained in crossed (ZZYY) and parallel (ZZZZ) polarization geometries, respectively.
T-Jump Experiments. The T-jump before transient DVE probing is created by nanosecond excitation of the OD stretch overtone of the D2O buffer solution at λ = 2 μm. Picosecond vibrational relaxation raises the temperature of the solution on the time scale of the excitation pulse (15). The 6-mJ, 8-ns T-jump pulse is obtained from a beta barium borate (BBO)-based optical parametric oscillator pumped by the output of a 20-Hz, frequency-doubled, Q-switched Nd:yttrium/aluminum-garnet laser and focused to a 500-μm diameter at the sample. This setup leads to uniform T-jumps of ≈12°C in the 100-μm-diameter spot probed by the mid-IR pulses.
The initial temperature (T0) is 58°C, and the final temperature attained as a result of the interaction of the sample with the near-IR laser is designated T. To detect small spectral changes in the DVE signal, we measure the difference signal relative to a reference spectrum, ΔS = [SDVE(T,τ) - SDVE(T0)], where τ is the delay from the T-jump. The DVE reference spectrum SDVE(T0) is the average DVE spectrum at T0 from the last two IR pulses from the 1-kHz train preceding the T-jump.
More detailed descriptions of the T-jump experiment, sample preparation method, and calculation of IR spectra are available in the supporting information, which is published on the PNAS web site.
Results
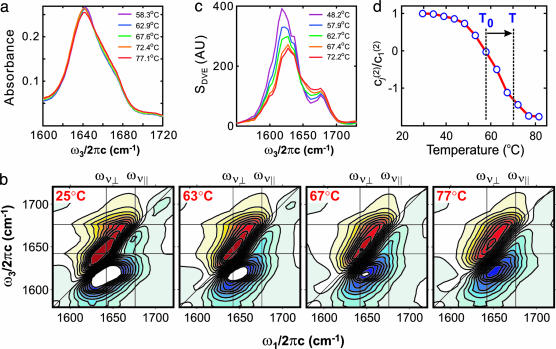
Equilibrium Thermal Unfolding. The conformational changes upon the thermal unfolding of ubiquitin as monitored by temperature-dependent Fourier transform IR (FTIR), 2D IR, and DVE spectra are shown in Fig. 1. FTIR absorption spectra, shown in Fig. 1a, are asymmetric, with a peak at 1,642 cm-1 and a shoulder at 1,676 cm-1. Although broad and quite featureless, the asymmetry is suggestive of the two-peak structure that arises from amide I vibrational couplings in antiparallel β-sheets (21, 28). The strong band (1,630–1,650 cm-1) has a transition moment roughly perpendicular to the β-strands, and we label it ν⊥, whereas the transition moment of the weak band (≈1,680 cm-1) is near parallel to the strands and is designated ν∥. As the temperature is raised from 58°C to 77°C, a very small decrease in the peak intensity and increase in the 1,650–1,660 cm-1 range is observed.
Fig. 1.
Temperature-dependent IR spectroscopy of ubiquitin unfolding. (a) FTIR spectra from 58°C to 77°C. (b) 2D IR spectra from 25°C to 77°C. Twenty-one equally spaced contours are drawn from -60% to 60% of the maximum amplitude of the 25°C spectrum. (c) DVE spectra from 48°C to 72°C. (d) Thermal melting curve constructed from the normalized singular value decomposition second-component coefficients c2(T)/c2(T1) of temperature-dependent DVE spectra. Vertical dashed lines indicate the T-jump window.
The vibrational transitions within the congested amide I line shape are more apparent in 2D IR spectra of ubiquitin, shown from 25°C to 77°C in Fig. 1b. In these spectra, the ω1 dimension marks the frequency of the initially excited vibration, and ω3 is the detection frequency for vibrational transitions induced by the initial excitation. Red contours originate from fundamental transitions (v = 1→0), and blue contours are anharmonically shifted, out-of-phase induced absorptions to two quantum states (v = 1→2). The characteristic ν⊥ and ν∥ transitions underlying the congested FTIR spectrum are apparent in the 2D IR spectrum from the stretched ridges at ω3 = ων∥ and ων⊥. The ν⊥ diagonal peak is diagonally elongated because of the inhomogeneous broadening that results from structural disorder within the β-sheet and overlap with resonances from other secondary structures. The ν∥ diagonal peak does not appear because of its weak transition dipole moment, but it is apparent through a cross peak between the ν⊥ transition and ν∥ transition that is elongated into a ridge along ω3 = ων∥. The cross peak is elongated parallel to the ω1 axis because the ν⊥ transition is sensitive to the size and disorder of the β-sheet, whereas the ν∥ transition is not (25, 26). Another cross peak at (ω1, ω3) ≈ (ων∥, ων⊥) is present as an elongation of the negative peak along ω3 ≈ ων⊥ because of interference with a negative peak anharmonically shifted below the diagonal axis. The features of diagonal elongation and ω1-stretching of cross peaks leads to a characteristic Z shape in the 2D IR spectra of β-sheet proteins (26).
As the temperature is raised, we observe a concerted blue-shift of the ν⊥ diagonal peak and shrinking of the cross-peak ridges. The overall shape as the temperature is raised tends toward a diagonally elongated doublet. These observations are similar to those seen for the unfolding of ribonuclease A (27) and are consistent with melting of the β-sheet structure in ubiquitin. At 77°C, the cross peak still persists as a weak protrusion from the diagonal peak, indicating the existence of the residual β-sheet secondary structure.
Temperature-dependent DVE spectra of ubiquitin are shown in Fig. 1c. The 48°C spectrum shows two distinct peaks at 1,621 cm-1 and 1,677 cm-1. These resonances can be interpreted in terms of the features in the 2D IR spectra, because the DVE spectrum is the absolute value squared of the projection of the complex 2D IR spectrum onto the ω3 axis (27). As a result of the projection along the elongated ω3 = ων⊥ negative ridge and the ω3 = ων∥ cross-peak ridge, two distinct peaks are observed in the DVE spectrum, which are distinct signatures of the ubiquitin β-sheet. More detailed features are observed in the DVE spectra, and the spectral changes as the temperature is raised are much more dramatic than those in FTIR spectra. The peak maximum (ν⊥) shifts from 1,621 cm-1 to 1,630 cm-1, and the peak intensity decreases.
A melting curve for ubiquitin can be obtained by singular value decomposition (SVD) of the equilibrium temperature-dependent DVE spectra (27). More than 99% of the spectra can be reconstructed by linear combinations of the first- and second-component SVD spectra, and the scaled second-component coefficient at each temperature is used for the melting curve. This melting curve (Fig. 1d) shows a sigmoid shape with a melting temperature of ≈61°C, suggesting simple two-state unfolding kinetics.
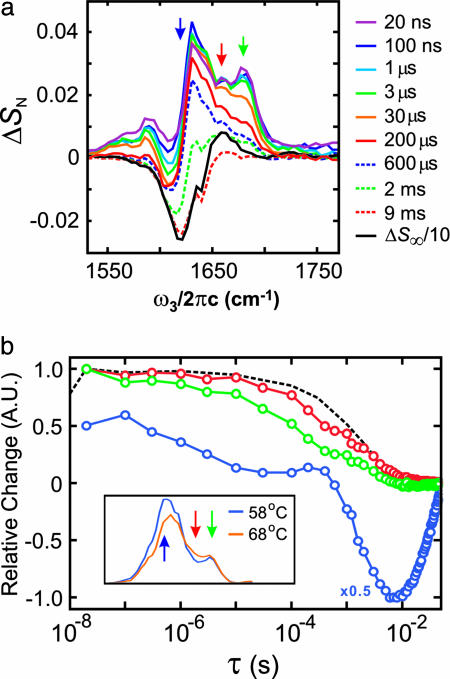
Transient Spectral Change. DVE spectra were used to select a T0 of 58°C, leading to the 12°C T-jump window shown in Fig. 1d. The resulting transient DVE difference spectra are shown in Fig. 2a for nanosecond-to-millisecond delays. Also shown is the equilibrium difference spectrum ΔS∞, which should be recovered for long times after a step T-jump. Fig. 2b shows a semilogarithmic plot of the response of three frequency channels: two at the peaks associated with features originating in the ν⊥ and ν∥ β-sheet transitions and one in the intermediate spectral region commonly associated with random coils and α-helices. The time-dependent spectral changes consist of five roughly sequential stages: (i) the initial rise of the whole spectrum, (ii) a microsecond drop of the signal in the ω3 = ων⊥ region, (iii) the drop of the high-frequency signal, including the ω3 = ων∥ region, (iv) a millisecond unfolding in the β-sheet region resembling the equilibrium difference spectrum, and (v) refolding. The unfolding also can be distinguished by nonexponential relaxation for times before ≈0.5 ms, followed by exponential behavior for longer times.
Fig. 2.
Transient DVE spectral changes after T-jump. (a) DVE difference spectra normalized to the peak signal ΔSN = ΔS/SDVE (T0,ω3 = 1,621 cm-1) at several time delays from the T-jump. The solid black curve shows the equilibrium difference spectrum ΔS∞ = [SDVE(T) - SDVE(T0)]/SDVE (T0, ω3 = 1,621 cm-1) between 68°C and 58°C scaled by 0.1. (b) The normalized time-dependent DVE response ΔSτ = ΔS(T,τ)/ΔS(T,τ = 20 ns) plotted for three frequency channels. These channels, labeled on the equilibrium DVE spectra at T and T0 in Inset, correspond to ν⊥ (ω3 = 1,621 cm-1, open blue circles), ν∥ (ω3 = 1,677 cm-1, open green circles), and ω3 = 1,658 cm-1 (open red circles). The temperature relaxation profile determined from the transmittance change through D2O at 6 μm is shown in black. A.U., arbitrary units.
The initial rise of the difference spectrum for τ < 100 ns results mostly from the small temperature-dependent increase of transmittance in the D2O background at 6 μm. Also, these early delays could be influenced by acoustic or shock waves that result from the rapid T-jump. However, because the 100-ns DVE difference spectrum for ω3 < 1,650 cm-1 is distinctly different from the equilibrium DVE spectrum at T0, there must also be an additional fast response from the protein. Local solvation effects with a subset of solvent-exposed amide vibrations would be expected on the picosecond time scale.
The nonexponential decrease of the ν⊥ region (ω3 ≈ 1,621 cm-1) is observed on the 3-μs time scale while other spectral regions remain constant. The ν⊥ frequency is sensitive to interstrand vibrational couplings and the number of strands within the β-sheet over which the vibration is delocalized (25, 26). The presence of the major changes in this region indicates that the change results from a breakdown of interstrand interactions due to denaturing of the sheet. A stretched exponential fit of the normalized data from 20 ns to 100 μs (ΔS ∝ exp[-(τ/τ0)β]) gives τ0 = 2.6 μs and β = 0.72, implying unfolding through multiple routes. A comparison of stretched exponential and single exponential fits can be found in the supporting information.
Subsequently, the intensity of other regions decreases nonexponentially on 10-μsto ≈0.5-ms time scales. In this time window, the ν⊥ region remains unchanged. For the ω3 = 1,677 and 1,658 cm-1 relaxation traces in Fig. 2b, a fit to a sum of the stretched exponential for fast components and a single-exponential fit for slow components gives τ1 = 80 μs, β = 0.55, and τ2 = 2.8 ms and τ1 = 210 μs, β = 0.74, and τ2 = 4.0 ms, respectively. Nonexponential relaxation during the microsecond time window has previously been observed in the refolding of a cold denatured ubiquitin mutant (6).
After ≈0.5 ms, the amplitude in the ν⊥ region decreases exponentially with a time constant of ≈3 ms reaching the maximum change at 6 ms. This process is also observed at the other probing frequencies and represents a major concerted melting of the β-sheet. This drop in ν⊥ amplitude would continue; however, the T-jump reequilibrates with a 2-ms time constant. The maximum change appears after the temperature reequilibrates. At this point, the transient difference spectrum has a similar shape to the equilibrium difference spectrum ΔS∞ but only 10% of its amplitude. For times >10 ms, the temperature of the solution has reequilibrated at T0. On this time scale, the T-jump appears as an impulse, and the protein refolds with a 20-ms time constant.
Discussion
2D IR spectra and DVE spectra show distinct signatures of a two-peak structure that is characteristic of polypeptide strands in an antiparallel hydrogen-bonding registry (29). Idealized antiparallel β-sheets show two IR active bands (a+ and a-), whose symmetry leads to transition dipole moments roughly parallel and perpendicular to the strands (25, 26). Strong interstrand couplings and vibrational delocalization across strands lead to a significant red-shift of the lower-frequency a- resonance as strands are added to a growing sheet (25). However, the frequency and amplitude of the a+ transition is insensitive to the size or geometry of the sheet, remaining approximately the same for structures with two or more strands.
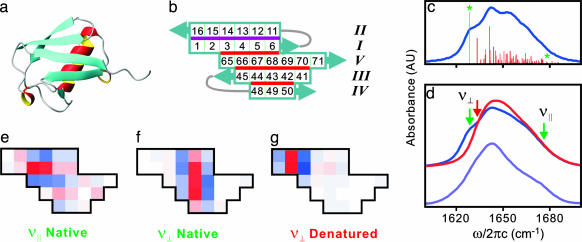
It is not immediately apparent that these features should be present in the 2D IR spectra, because the β-sheet of ubiquitin is not entirely antiparallel. We investigated the assignment by using calculations of the IR absorption spectrum of ubiquitin drawing on the crystal structure (30) and by using a local amide I Hamiltonian (21, 22). A planar projection of the β-sheet of ubiquitin (Fig. 3) shows it is composed of an N-terminal hairpin (strands I and II) and three additional antiparallel strands (III, IV, and V), with parallel registry between strand I and the C-terminal strand V. The calculated IR spectrum using the native structure has more peaks than the experimental spectrum (Fig. 3c); however, we find that by averaging over a modest variation of the amide I energies (σ = 6 cm-1), the breadth and asymmetry of the experimental line shape are reproduced (Fig. 3d). Visualization of the amide I eigenstates obtained by using the crystal structure confirms that vibrations with a+ and a- character are expected on the high- and low-frequency sides of the line, respectively. Fig. 3 e and f shows color maps of the vibrational amplitude and phase of the local amide I vibrations in the β-sheet contributing to amide I eigenstates on the high- and low-frequency sides of the absorption line. For the low-frequency (1,628 cm-1) eigenstate, the oscillators aligned between strands are in phase, but they are out of phase along the strand, the signature of a- character (26). For the high-frequency mode (1,679 cm-1), oscillators along the strands are in phase, as expected for a+ vibrations. Although these are only two eigenstates, they reflect the broader trends in the spectrum.
Fig. 3.
Modeling of amide I vibrational spectroscopy of ubiquitin. (a) Ribbon diagram of the crystal structure of ubiquitin (Protein Data Bank ID code 1UBQ) (30). (b) Two-dimensional projection of the β-sheet, with boxes numbering the peptide groups in the primary structure and indicating the numbering of strands. Amide I couplings reduced upon unfolding are indicated in red and are related to the native hydrogen-bonding contacts n(III, IV, and V). The contacts n(I and II) are shown in purple. (c) Calculated IR spectrum and stick figure of the dipole moment square for amide I eigenstates in the native structure obtained from the local amide I Hamiltonian. (d) The calculated IR absorption spectra of native structure (blue) and partially denatured structure (red), including disorder in the site energies compared with the experimental FTIR spectrum (bottom). (e–g) Color maps of the amplitude and phase of representative amide I vibrational eigenstates for the folded and denatured states. Red and blue represent opposite sign of the vibrational phase, and the depth of the color reflects the amplitude. (e and f) States from native structure with ν∥ and ν⊥ character, labeled with asterisks in c and green arrows in d.(g) State with ν⊥ character in unfolded calculation showing localization on the strand I–II hairpin. g corresponds to the transition labeled with a red arrow in d. AU, arbitrary units.
Fragmentation studies, multidimensional NMR experiments, and molecular dynamics simulation on ubiquitin have indicated that strands III, IV, and V are less conformationally stable than strands I and II and the α-helix (31–35). We therefore postulate that the 3-μs blue-shift of the ν⊥ resonances with no concerted change in ν∥ can be explained as preferential unfolding of strands III–V and a localization of the ν⊥ transition in the strand I–II hairpin. This observation can be tested by modeling changes in the amide I IR absorption spectrum upon the unfolding of strands III–V. For the calculation of the spectrum of the partially denatured state, the interstrand amide I couplings between strands I and V, V and III, and III and IV are reduced by 75%. These interactions are labeled in red in Fig. 3b. (For a direct displacement of strands, this reduction in coupling corresponds to a 60% increase in interstrand distances). Fig. 3d shows a comparison of the modeled FTIR spectra for native and denatured states with the experimental FTIR spectrum at 55°C. As the couplings are reduced, the ν⊥ region shows a significant blue-shift, whereas the ν∥ region (1,670–1,680 cm-1) remains unchanged. The localization of the ν⊥ excitation in the strand I–II hairpin in the denatured state is confirmed by visualizing the lowest-energy eigenstate in Fig. 3g.
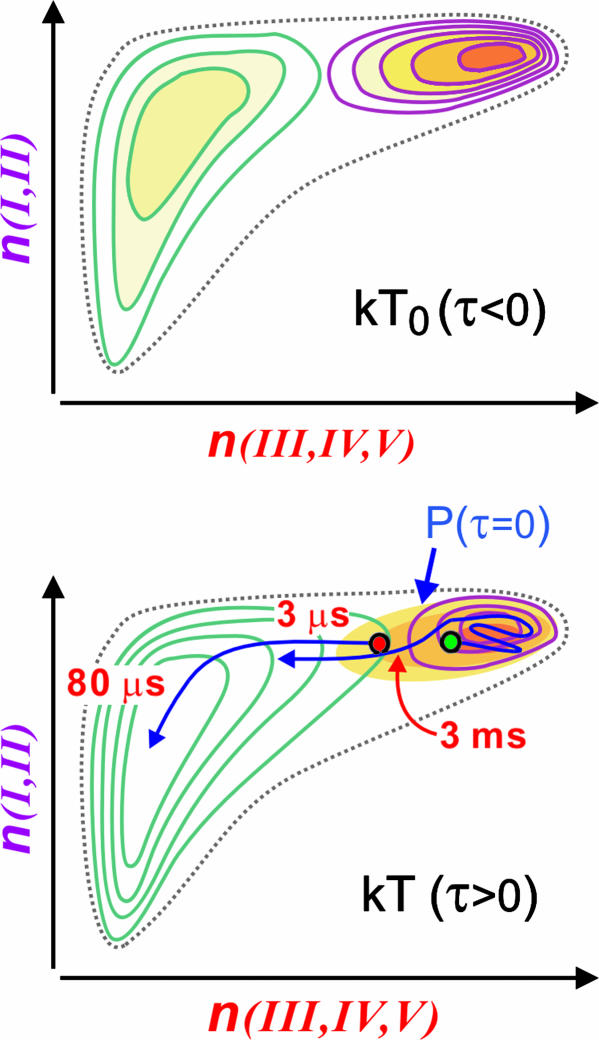
On the basis of the modeling, we propose a microscopic picture for ubiquitin unfolding in the T-jump experiments. The molecular dynamics of unfolding can be described in terms of trajectories on a microscopic free-energy landscape that is a function of numerous configurational coordinates (1–3). Because of their sensitivity to the interstrand couplings that change dramatically over the angstrom scale, the amplitude and resonance frequency of the ν∥ and ν⊥ transitions can be directly related to native interstrand contacts. We imagine that averaging the energy landscape over much of its roughness and concentrating on the native hydrogen-bonding contacts to strands III–V and the contacts in the strand I–II hairpin would lead to an energy surface at kT0 with the characteristics illustrated in Fig. 4a. The surface has a native-state energy minimum that retains most of the contacts found in the crystal structure and an extended minimum for the unfolded state that retains some residual structure. The contours indicate the preferential thermal stability of strands I and II over strands III, IV, and V. The initial temperature kT0 is already quite elevated, so a considerable fraction of the molecules are unfolded. Although this representation is just a proposal, in the future this free-energy surface can, in principle, be obtained through modeling of our 2D IR spectra.
Fig. 4.
Free-energy surfaces for ubiquitin unfolding. (a) An illustration of a reduced free-energy surface at kT0 that would be obtained by averaging over all degrees of freedom except the native hydrogen-bond contacts to strands III–V and strands I–II. (b) Same free-energy surface after T-jump at kT. The equilibrium is shifted to the unfolded side, and the barrier is shifted partially toward the native state. Shaded region shows initial population at τ = 0, with representative trajectories. A small portion of proteins with partial unfolding of the strand III–V region (red dot) unfold along a barrierless trajectory, whereas most reequilibrate in a native-like state before activated barrier crossing (green dot).
The T-jump prepares the system with a distribution of structures that is mostly centered near the native state (Fig. 4b). The shift of the barrier to the native side reflects the tendency toward a less stable native state and more stable denatured ensemble after the T-jump. For a relatively small, partially denatured subensemble (red dot in Fig. 4b), there is no significant barrier to unfolding. The T-jump places this subensemble at or near the transition state for unfolding, and we observe 1- to 500-μs nonexponential unfolding that can be described as diffusion in a multidimensional configurational space (3, 36, 37).
On the early 3-μs time scale, the rise in thermal energy in the protein and solvent allows increased configurational flexibility in the more weakly interacting and partially denatured strands III–V. Breaking of native contacts between strands reduces interstrand vibrational couplings, localizing the ν⊥ vibrational excitation on strands I and II and blue-shifting the ν⊥ transition. The fastest time scales correspond to those considered the speed limit for protein folding (38–41). In the second stage, the remaining hairpin involving the more stable strands I and II unfolds. The ν∥ transition decays only when the final interstrand interactions in hairpins are disrupted, resulting in the relaxation of theν∥ region on a 80-μs time scale. The somewhat longer time scale of relaxation in the intermediate frequency regime commonly associated with random coils and α-helices (1,658 cm-1) is more difficult to interpret but probably reflects both unfolding of antiparallel contacts and the helix and the growth of denatured amide I species. With our present methods, this central region could be influenced by interference effects arising from the projection of overlapping resonances in the nonlinear DVE response. A more detailed picture of this region can be obtained in the future by using the 2D IR experiment as a transient structural probe (42).
The majority of the population reequilibrates within the native-like minimum (green dot in Fig. 4b) and then unfolds as a 3-ms kinetic process that appears as a concerted unfolding. The time-scale separation between short-time nonexponential and long-time exponential relaxation processes does not necessarily reflect unfolding along different (parallel) coordinates. The trajectory is similar to the fast unfolding, but, because of the higher barrier, the slow process appears as the two-state kinetics seen in the equilibrium temperature-dependent experiments. This observation lends further support to the two-state model for ubiquitin folding. Previous rapid-mixing kinetic studies of ubiquitin refolding on millisecond to second time scales with NMR, fluorescence quenching, and circular dichroism have debated whether folding proceeds through an intermediate state or follows two-state kinetics (43–48). We emphasize that these long-time kinetics probe crossing of barriers ≫kT, whereas our experiments differ by revealing fast downhill evolution over a landscape with energy corrugation ≲kT.
The ability of 2D IR and DVE spectroscopy to make measurements of downhill diffusion with conformational sensitivity indicates that these experiments characterize an unfolding dynamics: a time-resolved measure of how nuclear configuration for the protein is changing. As such, it differs from a kinetics measurement that characterizes the rate of appearance or disappearance of the folded or denatured state. The structural information is not atomistic. Rather, these experiments characterize vibrations that are delocalized across extended regions of the protein, giving insight into the conformational changes to this extended region. These coordinates of mesoscopic scale are particularly appropriate for the study of large-scale conformational changes in processes such as folding or binding, where reaction coordinates are similarly collective. We also note that the conformational information is statistical in the sense that the experiment still presents an average over the influence of many degrees of freedom not directly observed in the experiment. These attributes suggest that the methods presented here open a number of avenues for making comparisons between fast folding experiments and molecular dynamics simulations.
Supplementary Material
Acknowledgments
We thank Arend Dijkstra and Jasper Knoester for discussions and for sharing a preprint on characterizing amide I vibrations of β-sheets and Minhaeng Cho for suggestions about assigning amide I site energies in β-sheets and for sharing ab initio calculations of amide I couplings found in ref. 49. This work was supported by National Science Foundation Grant CHE-0316736 and the Petroleum Research Fund of the American Chemical Society. A.T. thanks the Alfred P. Sloan Foundation and the David and Lucile Packard Foundation for their fellowship support.
Author contributions: H.S.C., M.K., and A.T. designed research; H.S.C., M.K., A.W.S., and Z.G. performed research; H.S.C. and A.T. analyzed data; and H.S.C., M.K., and A.T. wrote the paper.
Abbreviations: DVE, dispersed vibrational echo; FTIR, Fourier transform IR; T-jump, temperature jump.
References
- 1.Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. (1997) Annu. Rev. Phys. Chem. 48, 545-600. [DOI] [PubMed] [Google Scholar]
- 2.Dobson, C. M., Sali, A. & Karplus, M. (1998) Angew. Chem. Int. Ed. 37, 868-893. [DOI] [PubMed] [Google Scholar]
- 3.Dill, K. A. (1999) Protein Sci. 8, 1166-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snow, C. D., Nguyen, H., Pande, V. S. & Gruebele, M. (2002) Nature 420, 102-106. [DOI] [PubMed] [Google Scholar]
- 5.Snow, C. D., Qiu, L., Du, D., Gai, F., Hagen, S. J. & Pande, V. S. (2004) Proc. Natl. Acad. Sci. USA 101, 4077-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabelko, J., Ervin, J. & Gruebele, M. (1999) Proc. Natl. Acad. Sci. USA 96, 6031-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leeson, D. T., Gai, F., Rodriguez, H. M., Gregoret, L. M. & Dyer, R. B. (2000) Proc. Natl. Acad. Sci. USA 97, 2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, W. Y. & Gruebele, M. (2004) Biophys. J. 87, 596-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, C.-Y., Getahun, Z., Zhu, Y., Klemke, J. W., DeGrado, W. F. & Gai, F. (2002) Proc. Natl. Acad. Sci. USA 99, 2788-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, W. A., Muñoz, V., Hagen, S. J., Jas, G. S., Lapidus, L. J., Henry, E. R. & Hofrichter, J. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 327-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruebele, M., Sabelko, J., Ballew, R. & Ervin, J. (1998) Acc. Chem. Res. 31, 699-707. [Google Scholar]
- 12.Goldbeck, R. A., Thomas, Y. G., Chen, E., Esquerra, R. M. & Kliger, D. S. (1999) Proc. Natl. Acad. Sci. USA 96, 2782-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto, K., Mizutani, Y. & Kitagawa, T. (2000) Biophys. J. 79, 485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyer, R. B., Gai, F., Woodruff, W. H., Gilmanshin, R. & Callender, R. H. (1998) Acc. Chem. Res. 31, 709-716. [Google Scholar]
- 15.Callender, R. H., Dyer, R. B., Gilmanshin, R. & Woodruff, W. H. (1998) Annu. Rev. Phys. Chem. 49, 173-202. [DOI] [PubMed] [Google Scholar]
- 16.Phillips, C. M., Mizutani, Y. & Hochstrasser, R. M. (1995) Proc. Natl. Acad. Sci. USA 92, 7292-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmanshin, R., Williams, S., Callender, R. H., Woodruff, W. H. & Dyer, R. B. (1997) Proc. Natl. Acad. Sci. USA 94, 3709-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams, S., Causgrove, T. P., Gilmanshin, R., Fang, K. S., Callender, R. H. & Woodruff, W. H. (1996) Biochemistry 35, 691-697. [DOI] [PubMed] [Google Scholar]
- 19.Werner, J. H., Dyer, R. B., Fesinmeyer, R. M. & Andersen, N. H. (2002) J. Phys. Chem. B 106, 487-494. [Google Scholar]
- 20.Bredenbeck, J., Helbing, J., Sieg, A., Schrader, T., Zinth, W., Renner, C., Behrendth, R., Moroder, L., Wachtveitl, J. & Hamm, P. (2003) Proc. Natl. Acad. Sci. USA 100, 6452-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krimm, S. & Bandekar, J. (1986) Adv. Protein Chem. 38, 181-364. [DOI] [PubMed] [Google Scholar]
- 22.Hamm, P., Lim, M. & Hochstrasser, R. M. (1998) J. Phys. Chem. B 102, 6123-6138. [Google Scholar]
- 23.Torii, H. & Tasumi, M. (1992) J. Chem. Phys. 95, 3379-3387. [Google Scholar]
- 24.Khalil, M., Demirdöven, N. & Tokmakoff, A. (2003) J. Phys. Chem. A 107, 5258-5279. [Google Scholar]
- 25.Cheatum, C. M., Tokmakoff, A. & Knoester, J. (2004) J. Chem. Phys. 120, 8201-8215. [DOI] [PubMed] [Google Scholar]
- 26.Demirdöven, N., Cheatum, C. M., Chung, H. S., Khalil, M., Knoester, J. & Tokmakoff, A. (2004) J. Am. Chem. Soc. 126, 7981-7990. [DOI] [PubMed] [Google Scholar]
- 27.Chung, H. S., Khalil, M. & Tokmakoff, A. (2004) J. Phys. Chem. B 108, 15332-15342. [Google Scholar]
- 28.Byler, D. M. & Susi, H. (1986) Biopolymers 25, 469-487. [DOI] [PubMed] [Google Scholar]
- 29.Miyazawa, T. (1960) J. Chem. Phys. 32, 1647-1652. [Google Scholar]
- 30.Vijay-Kumar, S., Bugg, C. E. & Cook, W. J. (1987) J. Mol. Biol. 194, 531-544. [DOI] [PubMed] [Google Scholar]
- 31.Jourdan, M. & Searle, M. S. (2000) Biochemistry 39, 12355-12364. [DOI] [PubMed] [Google Scholar]
- 32.Harding, M. M., Williams, D. H. & Woolfson, D. N. (1991) Biochemistry 30, 3120-3128. [DOI] [PubMed] [Google Scholar]
- 33.Cox, J. P. L., Evans, P. A., Packman, L. C., Williams, D. H. & Woolfson, D. N. (1993) J. Mol. Biol. 234, 483-492. [DOI] [PubMed] [Google Scholar]
- 34.Stockman, B. J., Euvrard, A. & Scahill, T. A. (1993) J. Biomol. NMR 3, 285-296. [DOI] [PubMed] [Google Scholar]
- 35.Alonso, D. O. V. & Daggett, V. (1995) J. Mol. Biol. 247, 501-520. [DOI] [PubMed] [Google Scholar]
- 36.Socci, N. D., Onuchic, J. N. & Wolynes, P. G. (1998) Proteins 32, 136-158. [PubMed] [Google Scholar]
- 37.Gruebele, M. (1999) Annu. Rev. Phys. Chem. 50, 485-516. [DOI] [PubMed] [Google Scholar]
- 38.Hagen, S. J., Hofrichter, J., Szabo, A. & Eaton, W. A. (1996) Proc. Natl. Acad. Sci. USA 93, 11615-11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubelka, J., Hofrichter, J. & Eaton, W. A. (2004) Curr. Opin. Struct. Biol. 14, 76-88. [DOI] [PubMed] [Google Scholar]
- 40.Yang, W. Y. & Gruebele, M. (2003) Nature 423, 193-197. [DOI] [PubMed] [Google Scholar]
- 41.Wang, T., Zhu, Y. & Gai, F. (2004) J. Phys. Chem. B 108, 3694-3697. [Google Scholar]
- 42.Bredenbeck, J., Helbing, J., Behrendt, R., Renner, C., Moroder, L., Wachtveitl, J. & Hamm, P. (2003) J. Phys. Chem. B 107, 8654-8660. [Google Scholar]
- 43.Briggs, M. S. & Roder, H. (1992) Proc. Natl. Acad. Sci. USA 89, 2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khorasanizadeh, S., Peters, I. D. & Roder, H. (1996) Nat. Struct. Biol. 3, 193-205. [DOI] [PubMed] [Google Scholar]
- 45.Krantz, B. A. & Sosnick, T. R. (2000) Biochemistry 39, 11696-11701. [DOI] [PubMed] [Google Scholar]
- 46.Gladwin, S. T. & Evans, P. A. (1996) Folding Des. 1, 407-417. [DOI] [PubMed] [Google Scholar]
- 47.Qin, Z., Ervin, J., Larios, E., Gruebele, M. & Kihara, H. (2002) J. Phys. Chem. B 106, 13040-13046. [Google Scholar]
- 48.Larios, E., Li, J. S., Schulten, K., Kihara, H. & Gruebele, M. (2004) J. Mol. Biol. 340, 115-125. [DOI] [PubMed] [Google Scholar]
- 49.Ham, S. & Cho, M. (2003) J. Chem. Phys. 118, 6915-6922. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.