Abstract
Despite the increasing availability of genome sequences from many human pathogens, the production of complete proteomes remains at a bottleneck. To address this need, a high-throughput PCR recombination cloning and expression platform has been developed that allows hundreds of genes to be batch-processed by using ordinary laboratory procedures without robotics. The method relies on high-throughput amplification of each predicted ORF by using gene specific primers, followed by in vivo homologous recombination into a T7 expression vector. The proteins are expressed in an Escherichia coli-based cell-free in vitro transcription/translation system, and the crude reactions containing expressed proteins are printed directly onto nitrocellulose microarrays without purification. The protein microarrays are useful for determining the complete antigen-specific humoral immune-response profile from vaccinated or infected humans and animals. The system was verified by cloning, expressing, and printing a vaccinia virus proteome consisting of 185 individual viral proteins. The chips were used to determine Ab profiles in serum from vaccinia virus-immunized humans, primates, and mice. Human serum has high titers of anti-E. coli Abs that require blocking to unmask vaccinia-specific responses. Naïve humans exhibit reactivity against a subset of 13 antigens that were not associated with vaccinia immunization. Naïve mice and primates lacked this background reactivity. The specific profiles between the three species differed, although a common subset of antigens was reactive after vaccinia immunization. These results verify this platform as a rapid way to comprehensively scan humoral immunity from vaccinated or infected humans and animals.
Keywords: pox virus, proteomics, vaccine, Francisella tularensis
One of the most difficult tasks in developing recombinant protein vaccines and DNA vaccines is the identification of the antigens that stimulate the most effective immune responses against the pathogen, particularly when the genome of the organism is large. The genomes of many infectious organisms have been sequenced and annotated, but to our knowledge, there is no in silico algorithm that can be used effectively to identify the target antigens or epitopes of protective T cell and Ab responses from the genomic sequence data alone. One approach to this problem of vaccine antigen identification was reported in refs. 1 and 2, in which bioinformatics were used to prioritize 570 antigens from the bacteria Neisseria meningitidis, which encodes >4,000 ORFs. A large-scale conventional cloning and expression approach led to the purification of 350 candidate antigens, which were used to immunize mice, and the antigens that produced bactericidal Abs were identified. A more ideal approach would be one that would enable a more comprehensive immunological screen of the entire proteome.
To generate a complete proteome, Liang et al. (3) developed a method for generating transcriptionally active PCR (TAP) fragments that were shown to be as active as plasmids in in vitro transfection experiments, in vivo naked DNA injections, and in vitro transcription/translation reactions (3). The TAP approach provided a basis for considering synthesis of transcriptionally active genes in high throughput by PCR, followed by in vitro protein expression to generate individual microorganism proteins on a whole-proteome scale. A recent report by Doolan et al. (4) described how whole microorganism proteomes produced in this way might be used to aid in the discovery of vaccine antigens, but this approach has not been put into practice. Liang et al. (3) reported an experiment suggesting that recombination cloning into a plasmid expression vector could be an alternative for producing transcriptionally active genes in a high-throughput manner.
Here, we describe a high-throughput approach involving a single-round PCR, followed by in vivo recombination cloning and in vitro expression, that can rapidly lead to the generation of complete microorganism proteomes. The unpurified proteins can be printed directly onto microarray chips, and the chips can be used to characterize the humoral immune-response profile from vaccinated or infected animals and humans. The approach is demonstrated here with the synthesis of the vaccinia virus proteome, and the protein microarray was used to profile the humoral immune responses from vaccinia virus-immunized mice, primates, and humans.
Experimental Procedures
PCR Amplification of Linear Acceptor Vector. Plasmid pXT7 (10 μg; 3.2 kb, KanR) was linearized with BamHI (0.1 μg/μl DNA/0.1 mg/ml BSA/0.2 units/μl BamHI; 37°C for 4 h; additional BamHI was added to 0.4 units/μl at 37°C overnight). The digest was purified by using PCR purification kit (Qiagen, Valencia, CA), quantified by fluorometry using Picogreen (Molecular Probes) according to the manufacturer's instructions, and verified by agarose gel electrophoresis (1 μg). We used 1 ng of this material to generate the linear acceptor vector in a 50-μl PCR using the following primers (0.5 μM each): 5′-CTACCCATACGATGTTCCGGATTAC and 5′-CTCGAGCATATGCTTGTCGTCGTCG. We used 0.02 units/μl TaqDNA polymerase (Fisher Scientific, buffer A)/0.1 mg/ml gelatin (Porcine, Bloom 300; Sigma, G-1890)/0.2 mM each dNTP with the following conditions: initial denaturation of 95°C for 5 min; 30 cycles of 95°C for 0.5 min, 50°C for 0.5 min, and 72°C for 3.5 min; and a final extension of 72°C for 10 min.
PCR Amplification of ORF Insert. We used 1–10 ng of genomic DNA as template in a 50-μl PCR. The following primers were used (0.5 μM each): 5′-CATATCGACGACGACGACAAGCATATGCTCGAG (20-mer ORF specific at the 5′ end) and 5′-ATCTTAAGCGTAATCCGGAACATCGTATGGGTA (20-mer ORF specific at the 3′ end). We used 0.02 units/μl TaqDNA polymerase (buffer A, Fisher Scientific)/0.1 mg/ml gelatin (Bloom 300, Porcine; G-1890, Sigma)/0.2 mM each dNTP. Conditions were as follows: initial denaturation of 95°C for 5 min; 30 cycles of 20 sec at 95°C, 30 sec at 50°C, and 60 sec/kb at 72°C (1–3 min on average based on ORF size); and a final extension of 72°C for 10 min. PCR products that were more difficult to produce were reamplified by using a 30-sec annealing time at 45°C and 40°C, instead of 0.5 min at 50°C. The PCR product was visualized by agarose gel electrophoresis (3 μl). For quantification, the product was purified (PCR purification kit, Qiagen) and quantified by fluorometry.
In Vivo Recombination Cloning Method. Competent cells were prepared in our laboratory by growing DH5α cells at 18°C in 500 ml of SOB (super optimal broth) medium (2% tryptone/0.5% yeast extract/10 mM NaCl/2.5 mM KCl/20 mM MgSO4) to an OD of 0.5–0.7. The cells were washed and suspended in 10 ml of prechilled PCKMS buffer (10 mM Pipes/15 mM CaCl2/250 mM KCl/55 mM MnCl2/5% sucrose, pH 6.7) on ice, and 735 μl of DMSO was added dropwise with constant swirling. The competent cells were frozen on dry ice–ethanol in 100-μl aliquots and stored at -80°C. Each transformation consisted of the following: 10 μl of competent DH5α and 10 μl of DNA mixture (40 ng of PCR-generated linear vector/10 ng of PCR-generated ORF fragment; molar ratio, 1:1; vector, 1-kb ORF fragment). For transformation, the purification of PCR product was unnecessary. The mixture was incubated on ice for 45 min, heat shocked at 42°C for 1 min, and chilled on ice for 1 min; mixed with 250 μl of SOC (super optimal catabolizer) medium (2% tryptone/0.55% yeast extract/10 mM NaCl/10 mM KCl/10 mM MgCl2/10 mM MgSO4/20 mM glucose); incubated at 37°C for 1 h; diluted into 3 ml of LB medium supplemented with 50 μg of kanamycin per ml (LB Kan 50); and incubated with shaking overnight. The plasmid was isolated and purified from this culture, without colony selection.
Immunoblots and Microarrays. Plasmid templates used for in vitro transcription/translation were prepared by using QIAprep Spin Miniprep kits (Qiagen), including the “optional” step, which contains protein denaturants to deplete RNase activity. In vitro transcription/translation reactions (RTS 100 Escherichia coli HY kits; Roche) were set up in 0.2-ml PCR 12-well strip tubes and incubated for 5 h at 30°C, according to the manufacturer's instructions. For immunodot blotting, 0.3 μl of whole rapid-translation system reactions were spotted manually onto nitrocellulose and allowed to air dry before blocking in 5% nonfat milk powder in TBS containing 0.05% Tween 20. Blots were probed with vaccinia immune globulin (VIG) (Cangene, Winnipeg, MB, Canada) diluted to 1:1,000 in blocking buffer with or without 10% E. coli lysate. The following three different batches of VIG were used (lot nos.; quantities are given in parentheses): 1730204 (56 mg/ml), 1730208 (53 mg/ml), and 1730302 (56 mg/ml). Bound human Abs were detected by incubation in alkaline phosphatase-conjugated goat anti-human IgA/IgG/IgM (H+L) secondary Ab (Jackson ImmunoResearch) and visualized with nitroblue tetrazolium (nitro-BT) developer. Routinely, dot blots were also stained with both anti-poly-His mAb (clone, His-1; H-1029, Sigma) and with rat anti-hemagglutinin (HA) mAb (clone, 3F10; 1 867 423, Roche), followed by alkaline phosphatase-conjugated goat anti-mouse IgG (H+L) (BioRad) or goat anti-rat IgG (H+L) (Jackson ImmunoResearch) secondary Abs, respectively, to confirm the presence of recombinant protein. For microarrays, 10 μl of 0.125% Tween 20 was mixed with 15 μl of rapid-translation system reaction (to give a final concentration of 0.05% Tween 20), and 15-μl volumes were transferred to 384-well plates. The plates were centrifuged at 1,600 × g to pellet any precipitate, and supernatant was printed without further purification onto nitrocellulose-coated FAST glass slides (Schleicher & Schuell) by using an OmniGrid 100 microarray printer (Genomic Solutions, Ann Arbor, MI). Proteome chips were probed with VIG, serum from vaccinia infected macaques (a gift from Jay Hooper, U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, MD) or mouse serum pooled from five BALB/c mice on day 19 after infection with 5 × 105 plaque-forming units of vaccinia strain WR (administered i.p.). For all staining, slides were first blocked for 30 min in protein array-blocking buffer (Schleicher & Schuell) before incubation in primary Ab for 2 h. Abs were visualized with Cy3-conjugated secondary Abs (Jackson ImmunoResearch) and scanned in a ScanArray 4000 laser confocal scanner (GSI Lumonics, Billerica, MA). Fluoresence intensities were quantified by using quantarray software (GSI Lumonics). VIG has high titers of anti-E. coli Abs that mask any antigen-specific responses when using whole rapid-translation system reactions on dot blots and arrays. This masking was overcome by the addition of E. coli lysate to the VIG. E. coli lysate was produced from a 1l stationary-phase culture of E. coli (DH5α) resuspended in 25 ml of TBS/Tween 20 and sonicated with a probe of 2 cm in diameter. We stored 1-ml aliquots at -80°C. Mouse sera lack endogenous anti-E. coli reactivity and, therefore, do not require pretreatment with E. coli lysate to reduce background.
Results
An approach has been developed that allows ≥384 genes derived from genomic template from a microorganism to be expressed and printed onto microarray chips very rapidly (in as little as 3 days for most genes), and the chips can be used to scan the humoral immune response from vaccinated or infected animals and humans. The overall approach involves PCR, recombination cloning, in vitro transcription/translation, and microarray printing.
In Vivo Recombination Cloning in E. coli. A linear T7 vector encoding an N-terminal His tag and a C-terminal HA tag was generated by extensive restriction digestion, followed by PCR. ORFs from vaccinia virus genomic DNA were amplified by using gene-specific primers containing 33 nucleotide extensions complementary to the ends of the linear T7 vector. Mixtures of PCR-amplified ORFs and linear T7 vector were introduced into chemically competent E. coli, resulting in antibiotic-resistant cells containing plasmid with the PCR fragment directionally inserted in-frame. This high-efficiency recombination cloning method enables hundreds of PCR fragments amplified from genomic template to be cloned rapidly.
Linear Vector. The plasmid used to generate the linear recombination vector is shown in Fig. 5, which is published as supporting information on the PNAS web site. This vector contains a T7 promoter, a prokaryotic ribosome-binding sequence (Shine-Delgarno), followed by ATG start codon, a 10× His sequence, a spacer sequence in front of the first codon of the ORF to be cloned, a BamHI site, and a T7 terminator. The vector was double-digested at the BamHI site to eliminate residual circular vector, because incompletely digested vector creates background colonies that lack insert. This linearized vector was amplified further by PCR to dilute out any residual circular vector and to generate inventory of the linear recombination vector.
PCR of ORFs from Genomic Template. Each ORF was amplified from genomic template by using gene-specific primers (Fig. 5). The 5′ oligonucleotide contained 53 nucleotides, of which 33 nucleotides constitute the 5′ extension and 20 nucleotides make up the gene-specific sequence. The 3′ custom oligonucleotide also contains 53 nucleotides, of which 33 nucleotides comprise the 3′ extension and 20 nucleotides are specific to the gene of interest. A gel showing a set of PCR products amplified from vaccinia is shown in Fig. 1A. For genes shorter than 1,000 bp, the success rate for getting the predicted PCR product was >99%. For these short genes, failures could be recovered by ordering new primers, indicating that most of the failures were caused by errors in olignucleotide synthesis. Longer genes (>2,000 bp) can be produced under different PCR conditions favoring amplification of longer products.
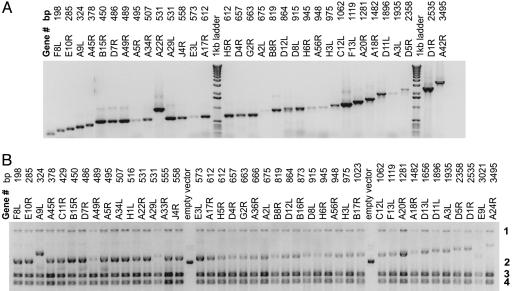
Fig. 1.
PCR amplification and recombination cloning of the vaccinia virus proteome. (A) A series of genes of increasing size from vaccinia were amplified according to the procedures described in the methods section. (B) The PCR products shown in A were transformed into competent E. coli. After transformation and overnight growth, the cells were lysed in phenol-chloroform and the total nucleic acid was isolated and run the agarose gel. Band 1 shows genomic DNA, and band 2 is supercoiled empty vector. Plasmids with insert migrate more slowly depending on the size of the insert. Bands 3 and 4 are 23S and 16S rRNAs. Routinely, purified DNA Miniprep DNA was prepared from the overnight culture without colony selection. The resulting plasmids are sequence verified and used directly in in vitro transcription/translation reactions.
Transformation and Overnight Culture. Transformation of the DH5α competent cells was accomplished with a mixture of PCR fragment and linear vector in a molar ratio of 1:1, with 50 ng of total DNA used in the transformation. The competent cells were transformed, grown overnight, and observed for turbidity due to bacterial growth. Under these conditions, cloning efficiency was >90%, but if the cells were plated for colony selection on the day of transformation, the observed success rate was lower. The rate of successful transformation progressively declined as the total DNA used for transformation was reduced to 25 and 10 ng (data not shown). The process was streamlined by omitting colony selection before using the QIAprep Spin Miniprep kit. Fig. 1B shows a “cracking gel” (phenol-lysed bacteria showing total nucleic acid) from overnight cultures by using the PCR fragments shown above. The top band on these gels (band 1) is genomic DNA, the bottom two bands (bands 3 and 4) are 23S and 16S rRNA, and the central band (band 2) is the plasmid formed by recombination with linear vector and PCR fragment. Empty vector is included on this gel for reference. Of the 42 plasmids shown in Fig. 1, only 1 (E9L) lacks insert of the appropriate size. To calibrate the efficiency of the overall system, a test set of genes from the bacterium Francisella tularensis were amplified, cloned, and expressed. The data show (Table 1, which is published as supporting information on the PNAS web site) that, of 1,933 genes attempted, 96% were successfully amplified and 93% were successfully cloned.
In Vitro Transcription and Translation. The proteins encoded on recombinant pXT7 plasmids, with or without prior cloning by colony selection, were expressed in an E. coli-based cell-free in vitro transcription/translation system that was supplemented with T7 RNA polymerase. The in vitro transcription/translation reactions (≤25 μl) were incubated for 5 h at 30°C, and the unpurified reactions were resolved on SDS/PAGE gels. The gels were routinely checked for expression by immunodot blots or Western blots probed with anti-His tag Ab. The Western blots (Fig. 6, which is published as supporting information on the PNAS web site) show expression of the His-tagged products of the predicted molecular weights.
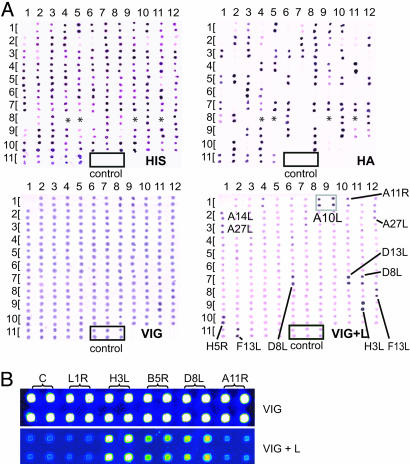
Nondenatured proteins from the cell-free reactions could also be detected on immunodot blots (Fig. 2A). Membranes were probed with either anti-His tag Ab, anti-HA tag Ab, VIG, or VIG plus 10% E. coli lysate. The anti-His and HA tag Abs show no cross-reactivity with other proteins in the in vitro reactions and, therefore, were used routinely for monitoring the expression of large numbers of reactions. Of 112 different expressed proteins, only 4 were negative for both the His and HA tags (marked with an asterisk). To evaluate the overall efficiency of expression without prior colony picking, 390 F. tularensis genes that were prepared from overnight cultures were expressed in vitro. The reactions were spotted onto nitrocellulose and probed with either anti-His or anti-HA Ab. The results (Table 1) show that only 7% were HA and His negative. It is apparent that VIG has high titers of anti-E. coli Abs that mask any reactivity to vaccinia proteins. However, the addition of E. coli lysate to VIG reduces this background to a level such that the detection of specific vaccinia proteins by VIG is possible.
Fig. 2.
The unpurified proteins expressed in vitro can be used for serology on immunodot blots or microarrays. (A) Immunodot blots of 112 different vaccinia proteins (encoded by 124 plasmids cloned by colony selection) expressed in vitro. The control reaction lacked plasmid template; empty vector controls generate a positive signal because of the expression of a small 10× His-positive fragment (data not shown). Volumes of 0.3 μl of each reaction were spotted directly onto nitrocellulose in duplicate and probed with either anti-His tag Ab (Upper Left), anti-HA tag Ab (Upper Right) VIG (Lower Left), or VIG plus 10% E. coli lysate (VIG+L; Lower Right). The identity of each pair of spots is given in Table 2, which is published as supporting information on the PNAS web site. Proteins marked with an asterisk in Upper were negative when probed with both anti-His and anti-HA Abs. E. coli lysate unmasks vaccinia proteins recognized by VIG; dots considered positive by visual estimation are indicated. (B) A pilot microarray constituting five vaccinia proteins expressed from DNA with colony selection and probed with VIG, preabsorbed with or without E. coli lysate. Interspot distance, 0.3 mm.
Microarrays and Serological Screening. E. coli lysate treatment of serum was also effective to reduce E. coli background reactivity on microarrays. A pilot microarray consisting of vaccinia proteins probed with VIG, with and without E. coli lysate, is shown in Fig. 2B. The effect of high titers of anti-E. coli Abs, as detected in dot blots, is also obvious on microarrays. This high background that is present also in the control preparations masks specific reactivity to vaccinia proteins. Addition of 10% E. coli lysate to VIG before probing the microarray reduced the E. coli background, revealing the specific reactivity.
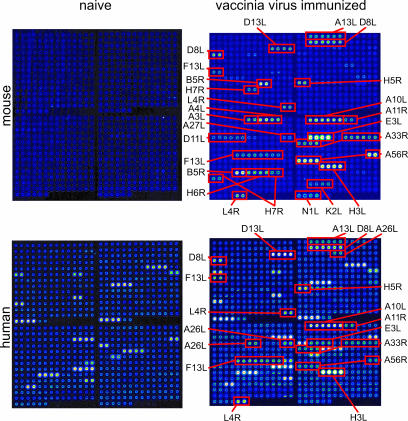
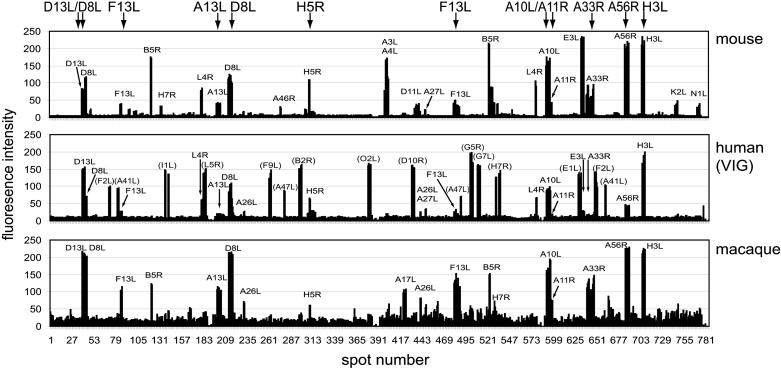
Fig. 3 shows results from an array consisting of 376 reactions comprising 194 different ORFs, which we estimate to represent >95% of the complete vaccinia virus proteome. This array was screened with human VIG or sera from mice before and after vaccination with vaccinia virus. Naïve nonimmunized mice lack reactivity against all of the proteins on the array, but sera from vaccinia virus-immunized mice react with a subset of the antigens on the array. Unlike naïve mice, nonimmunized humans react with a subset of 13 antigens on the array, which are thought to represent nonspecific cross-reactions by Abs to other environmental antigens. After immunization with vaccinia virus, another subset of reactive antigens develops. Quantification of these data, plus vaccinia-immune macaque sera, is shown in Fig. 4. VIG recognizes 14 vaccinia-specific proteins. Similar profiles are also seen in sera from macaque and mice. Although there are species-specific responses (for example, A3L or A4L in mice only), there are many proteins recognized in common by humans and either animal model and 11 proteins recognized by all three species (Table 4, which is published as supporting information on the PNAS web site). These antigens would be priority candidates for the preclinical testing of a vaccine for use in humans. Overall, responses to viral structural proteins dominate the response, with more than one-half of these antigens being envelope proteins. The proteins that were seropositive included those with and without transmembrane domains or signal peptides, and isoelectric point ranges from 4 to 10. Moreover, several of these proteins have been reported to produce humoral responses in animals and humans, whereas others have not. Thus, the methodology described here identifies immunologically reactive antigens, not all of which would be identified by conventional predictive approaches. Data obtained with the arrays are in agreement with immunoblots that we have reported (5). Notably, in vaccinated humans, we see a rapid and strong increase in reactivity against a subset of dominant antigens (H3L, D13L, and A10L) after boosting many years after primary immunization.
Fig. 3.
Protein microarray analysis of Abs generated during vaccinia infection. Shown are scans of a vaccinia virus proteome microarrays, probed with normal mouse serum (Upper Left) serum pooled from five vaccinia-immunized mice on day 19 after infection (Upper Right), vaccinia-naive human serum (Lower Left) or human VIG (Lower Right). Vaccinia-specific proteins recognized are indicated. Unannotated proteins in Lower are recognized by vaccinia-naïve human sera and, therefore, are considered nonspecific “background.” The identity of each pair of spots is given in Table 3, which is published as supporting information on the PNAS web site. All of the arrays shown were probed with serum preabsorbed with E. coli lysate. For mouse serum, which lacks anti-E. coli reactivity, there was no difference in the signal intensities against vaccinia antigens with or without lysate treatment (data not shown). No signals were seen by using secondary Ab alone.
Fig. 4.
Signal intensities of the microarrays shown in Fig. 3 probed with pooled mouse sera and VIG. Proteins annotated in parenthesis in Middle are the nonspecific proteins also seen by naïve human sera. Also shown are data from a Dryvax-immunized macaque on day 10 after monkeypox challenge (animal CH39 in ref. 14). Spots: 1–196, top left subarray; 197-392, top right, 393–588, bottom left, 589–784, bottom right. Proteins recognized by all three species are listed along the top. Relative intensities of a particular protein spot are proportional to the titer of the specific Abs in each serum.
Discussion
The PCR cloning method described relies on the principle that E. coli cells are able to recombine homologous sequences with high fidelity and speed (6). Several articles have appeared that suggest the usefulness of in vivo recombination in E. coli for PCR cloning (3, 6–9). Most of these approaches rely on the use of electroporation of certain strains of E. coli that are RecA-negative and RecE- and RecT-positive. Hence, the method has been referred to as ET recombination cloning. These reported methods are <90% efficient and require >40 bp of sequence homology between each end of the PCR fragment and the corresponding homology regions of circular or linearized vector. For the method reported here, linear DNA fragments were generated by PCR to include sequences on the 5′ and 3′ ends that are homologous with end sequences on a linearized vector. When the PCR fragments and the linear vector are mixed and transformed into competent E. coli, endogenous bacterial recombinase activity is able to join the two DNA fragments resulting in a circular plasmid. This method allows large numbers of PCR fragments to be cloned into bacterial or mammalian expression vectors in a high-throughput manner.
The method also uses chemically competent DH5α E. coli cells and relatively short 33-bp homologous sequences for recombination. Parrish et al. (10) reported an in vivo recombination approach for cloning PCR products by using an enzyme-restricted linear vector and ordinary chemically competent E. coli cells. The method, which used a 24-bp homologous sequence, showed 75% cloning efficiency but required colony picking to identify recombinant clones. The method reported here gives >90% cloning efficiency, which eliminates the necessity of the colony-picking step. Using the clonal mixture without colony selection streamlines the high-throughput cloning procedure and reduces the impact of PCR errors in the amplified ORFs. Several additional steps also contribute to robust performance. First, precautions were taken to eliminate residual circular plasmid from the linear vector. This removal is accomplished by double digestion of the circular vector template followed by PCR amplification to produce the linear vector. Second, a relatively high concentration of DNA was used during transformation. We found 50 ng of total DNA at a 1:1 molar ratio of PCR fragment/linear vector to be optimal. Last, the transformed competent cells were allowed to grow overnight before DNA isolation. This step allows time for a relatively low number of transformed cells to expand.
Theoretically, cell-free protein-expression systems offer many advantages over in vivo approaches, such as a simple and straightforward protocol and small reaction volumes applicable to high-throughput applications, the ability to be used with both circular and linear DNA templates, and the ability to express toxic proteins. In our experience, >90% of the target proteins were expressed at a level of 5–20 μg/ml (data not shown). Of the protein present in these reactions, 99% is from E. coli lysate and only 1% is from the target protein. Nevertheless, these crude reactions can be spotted directly onto nitrocellulose microarrays without purification or concentration at levels sufficient to capture detectable antigen-specific Abs from the serum of vaccinia-immunized humans and animals.
Because the in vitro expression system that we used contains E. coli lysate, our serological screening method depends on the competitive blocking of anti-E. coli Abs that are present in human sera that otherwise overwhelm any specific antigen detection. For mouse sera, which lack endogenous E. coli reactivity, there is no difference in the vaccinia-specific reactivity profile with or without E. coli lysate treatment, indicating that lysate pretreatment does not deplete vaccinia-specific reactivity (data not shown). However, we are also generated microarrays and profiling immune responses against prokaryotic organisms such as F. tularensis and M. tuberculosis that might exhibit more cross-reactivity to E. coli proteins. Nickel-coated slides could be used to selectively capture the His-tagged proteins and eliminate E. coli proteins as an alternative way to reduce E. coli background reactivity. Eukaryotic expression systems, which avoid E. coli, also can be used if posttranslational modification of the antigen is desirable. We are also aware that HA-tagged proteins could generate background reactivity from individuals previously exposed to influenza. However, VIG, which is pooled from multiple donors, shows no evidence of such background, and vaccinia-specific signals have been detected in nearly all of the ≥20 individual human sera that we have screened so far.
One of the most difficult tasks in developing a recombinant protein subunit vaccine or DNA vaccine is the identification of the antigens that will stimulate the most effective immune response against the pathogen, particularly when the genome of the organism is large (11). A comprehensive way to accomplish this task would be to obtain each of the structural, metabolic, and regulatory antigens of the pathogen and test their protective immunity individually or as mixtures in the vaccine (12). Although this approach may work for small viruses encoding several antigens, it is not practical for large viruses like smallpox, or bacteria, which encode hundreds or thousands of antigens. Our high-throughput platform for serological screening allows screening of the proteins without the risk of losing them through purification. We have found that sera reveals reactivity skewed toward vaccinia envelope proteins. Despite the simplicity of this method, its nonrandom detection of antigens gives us confidence that this method provides biologically meaningful data. Although here we have used secondary Abs to detect IgA/IgM/IgG, the detection of specific Ig isotypes is readily achieved with the appropriate isotype-specific secondary Abs (13).
The method in its current format is designed to detect protein antigens only. Clearly, other surface antigens that contain lipid or polysaccharide are not detected. However, this format is not likely to be a serious limitation if the goal is to identify protein antigens that can be expressed in E. coli and formulated for use as subunit vaccines or as antigens in diagnostic tests. The significance of the vaccinia proteins detected by VIG and sera from human vaccinees has implications for rational vaccine design and diagnostics. However, general observations can be made about the properties of the antigens detected. First, the presence of hydrophobic transmembrane domains of the vaccinia envelope proteins did not cause major insolubility problems that prevented them from being printed successfully onto the arrays. Although we are aware that the small sample of proteins presented here may not be typical of transmembrane proteins in general, at the time of writing this article, we have expressed >190 proteins from vaccinia and >1,700 proteins from F. tularensis. Of these proteins, >90% were detected by anti-His and/or anti-HA Abs on the dot blots. Second, because the proteins are printed directly from the in vitro transcription/translation reactions, conformations may be preserved that would otherwise be lost in Western blots. Third, we believe that there is a wider applicability of this technique for the identification of strongly immunogenic proteins. For example, some of the vaccinia proteins detected by VIG or immune sera are not found on the virion surface and, therefore, cannot be the targets of neutralizing Abs. In this study, we used a proteomic approach to identify the targets recognized by vaccinia neutralizing sera. These data lead us to a different set of candidate antigens than those investigated for subunit vaccines that were chosen by predictive means. In particular, we found several instances of Abs to nonenvelope proteins that would not have been identified by any predictive algorithm. These antigens, which are probably released from infected cells, are unlikely to elicit neutralizing Abs. Whether or not their immunogenicity contributes to protection in other ways remains to be determined. However, we are interested in the possibility that these Abs are acting as markers for recognition of these antigens by T cells.
In summary, we have developed a simple and robust high-throughput proteome-synthesis process that is useful for determining antigen-specific Ab profiles from vaccinated or infected animals and humans. More widespread application of this general approach could contribute to basic infectious disease research and more rapid discovery of antigens for vaccines and diagnostics applications.
Supplementary Material
Acknowledgments
We thank Rick Titball and his colleagues (The Defence Science and Technology Laboratory, Porton Down, U.K.) for the annotated genome sequence of F. tularensis genomic DNA from the Schu4 strain; Paul Gershon (University of California, Irvine) for vaccinia virus genomic DNA; Jay Hooper (U.S. Army Medical Research Institute of Infectious Diseases) for the gift of sera from vaccinia-immunized macaques; and Denis Heck (Microarray Facility, University of California, Irvine) for chip printing. We also thank Denise Doolan (Naval Medical Research Center) and Ian Gibbons for helpful discussions. This work was supported by National Institute of Allergy and Infectious Diseases Grants U01AI056464 and 1U01AI061363-01. The bioinformatics and primer design in this work was supported by National Institutes of Health Biomedical Informatics Training Program Grant 5T15LM007743 and National Science Foundation Grant MRI EIA-0321390 (to P.B. and the Institute for Genomics and Bioinformatics).
Author contributions: D.H.D., X.L., P.B., L.P.V., and P.L.F. designed research; D.H.D., X.L., J.E.H., S.H., Y.M., K.M.R., T.T.N., M.K.-D., and S.C. performed research; A.R. and P.B. contributed new reagents/analytic tools; D.H.D., X.L., J.E.H., S.C., and P.L.F. analyzed data; and D.H.D., X.L., and P.L.F. wrote the paper.
Abbreviations: HA, hemagglutinin; VIG, vaccinia immune globulin.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY871843–AY872187).
References
- 1.Pizza, M., Scarlato, V., Masignani, V., Giuliani, M. M., Arico, B., Comanducci, M., Jennings, G. T., Baldi, L., Bartolini, E., Capecchi, B., et al. (2000) Science 287, 1816-1820. [DOI] [PubMed] [Google Scholar]
- 2.Tettelin, H., Saunders, N. J., Heidelberg, J., Jeffries, A. C., Nelson, K. E., Eisen, J. A., Ketchum, K. A., Hood, D. W., Peden, J. F., Dodson, R. J., et al. (2000) Science 287, 1809-1815. [DOI] [PubMed] [Google Scholar]
- 3.Liang, X., Teng, A., Braun, D. M., Felgner, J., Wang, Y., Baker, S. I., Chen, S., Zelphati, O. & Felgner, P. L. (2002) J. Biol. Chem. 277, 3593-3598. [DOI] [PubMed] [Google Scholar]
- 4.Doolan, D. L., Aguiar, J. C., Weiss, W. R., Sette, A., Felgner, P. L., Regis, D. P., Quinones-Casas, P., Yates, J. R., III, Blair, P. L., Richie, T. L., et al. (2003) J. Exp. Biol. 206, 3789-3802. [DOI] [PubMed] [Google Scholar]
- 5.Crotty, S., Felgner, P., Davies, H., Glidewell, J., Villarreal, L. & Ahmed, R. (2003) J. Immunol. 171, 4969-4973. [DOI] [PubMed] [Google Scholar]
- 6.Oliner, J. D., Kinzler, K. W. & Vogelstein, B. (1993) Nucleic Acids Res. 21, 5192-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shashikant, C. S., Carr, J. L., Bhargava, J., Bentley, K. L. & Ruddle, F. H. (1998) Gene 223, 9-20. [DOI] [PubMed] [Google Scholar]
- 8.Zhang, Y., Buchholz, F., Muyrers, J. P. & Stewart, A. F. (1998) Nat. Genet. 20, 123-128. [DOI] [PubMed] [Google Scholar]
- 9.Zhang, Y., Muyrers, J. P., Testa, G. & Stewart, A. F. (2000) Nat. Biotechnol. 18, 1314-1317. [DOI] [PubMed] [Google Scholar]
- 10.Parrish, J. R., Limjindaporn, T., Hines, J. A., Liu, J., Liu, G. & Finley, R. L., Jr. (2004) J. Proteome Res. 3, 582-586. [DOI] [PubMed] [Google Scholar]
- 11.Felgner, P. L. & Liang, X. (1999) Nat. Biotechnol. 17, 329-330. [DOI] [PubMed] [Google Scholar]
- 12.Johnston, S. A. & Barry, M. A. (1997) Vaccine 15, 808-809. [DOI] [PubMed] [Google Scholar]
- 13.Bacarese-Hamilton, T., Mezzasoma, L., Ardizzoni, A., Bistoni, F. & Crisanti, A. (2004) J. Appl. Microbiol. 96, 10-17. [DOI] [PubMed] [Google Scholar]
- 14.Hooper, J. W., Thompson, E., Wilhelmsen, C., Zimmerman, M., Ichou, M. A., Steffen, S. E., Schmaljohn, C. S., Schmaljohn, A. L. & Jahrling, P. B. (2004) J. Virol. 78, 4433-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.