Abstract
It is still not possible to predict whether a given molecule will have a perceived odor, or what olfactory percept it will produce. We therefore organized the crowd-sourced DREAM Olfaction Prediction Challenge. Using a large olfactory psychophysical dataset, teams developed machine learning algorithms to predict sensory attributes of molecules based on their chemoinformatic features. The resulting models accurately predicted odor intensity and pleasantness, and also successfully predicted eight among 19 rated semantic descriptors (“garlic”, “fish”, “sweet”, “fruit,” “burnt”, “spices”, “flower”, “sour”). Regularized linear models performed nearly as well as random-forest-based ones, with a predictive accuracy that closely approaches a key theoretical limit. These models help to predict the perceptual qualities of virtually any molecule with high accuracy and also reverse-engineer the smell of a molecule.
In vision and hearing, the wavelength of light and frequency of sound are highly predictive of color and tone. In contrast, it is not currently possible to predict the smell of a molecule from its chemical structure (1, 2). This stimulus-percept problem has been difficult to solve in olfaction because odors do not vary continuously in stimulus space, and the size and dimensionality of olfactory perceptual space is unknown (1, 3, 4). Some molecules with very similar chemical structures can be discriminated by humans (5, 6), and molecules with very different structures sometimes produce nearly identical percepts (2). Computational efforts developed models to relate chemical structure to odor percept (2, 7–11), but many relied on psychophysical data from a single 30-year-old study that used odorants with limited structural and perceptual diversity (12, 13).
Twenty-two teams were given a large, unpublished psychophysical dataset collected by Keller and Vosshall from 49 individuals who profiled 476 structurally and perceptually diverse molecules (14) (Fig. 1a). We supplied 4884 physicochemical features of each of the molecules smelled by the subjects, including atom types, functional groups, and topological and geometrical properties that were computed using Dragon chemoinformatic software (version 6) (Fig. 1b).
Fig. 1. DREAM Olfaction Prediction Challenge.
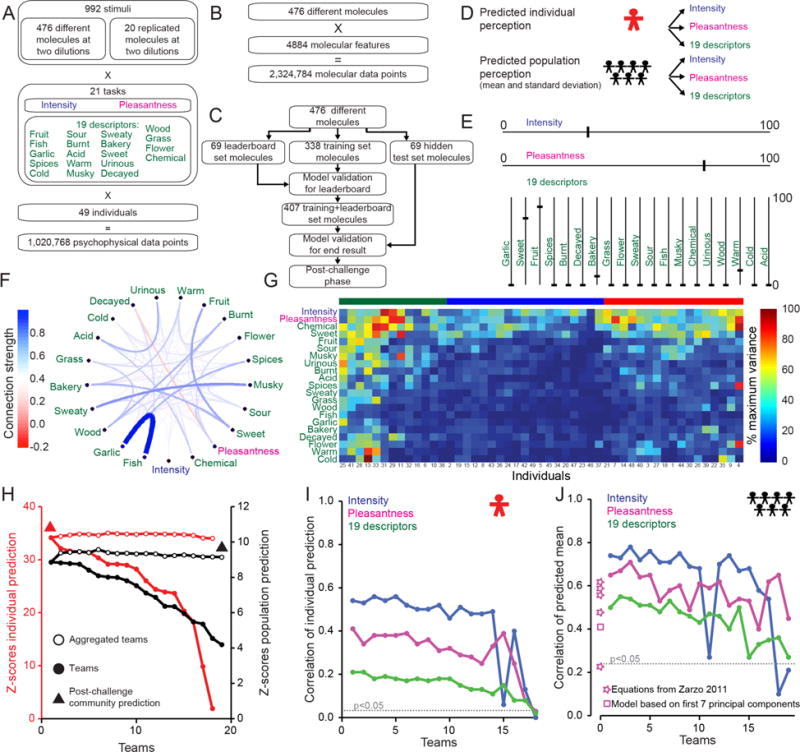
(A) Psychophysical data. (B) Chemoinformatic data. (C) DREAM challenge flowchart. (D) Individual and population challenges. (E) Hypothetical example of psychophysical profile of a stimulus. (F) Connection strength between 21 attributes for all 476 molecules. Width and color of the lines show the normalized strength of the edge. (G) Perceptual variance of 21 attributes across 49 individuals for all 476 molecules at both concentrations sorted by Euclidean distance. Three clusters are indicated by green, blue, and red bars above the matrix. (H) Model Z-scores, best performers at left. (I–J) Correlations of individual (I) or population (J) perception prediction sorted by team rank. The dotted line represents the p < 0.05 significance threshold with respect to random predictions. The performance of four equations for pleasantness prediction suggested by Zarzo (10) [from top to bottom: equations (10, 9, 11, 7, 12)] and of a linear model based on the first seven principal components inspired by Khan et al. (8) are shown.
Using a baseline linear model developed for the challenge and inspired by previous efforts to model perceptual responses of humans (8, 11), we divided the perceptual data into three sets. Challenge participants were provided with a training set of perceptual data from 338 molecules that they used to build models (Fig. 1c). The organizers used perceptual data from an additional 69 molecules to build a leaderboard to rank performance of participants during the competition. Towards the end of the challenge, the organizers released perceptual data from the 69 leaderboard molecules so that participants could get feedback on their model, and enable refinement with a larger training+leaderboard data set. The remaining 69 molecules were kept as a hidden test set available only to challenge organizers to evaluate the performance of the final models (Fig. 1c). Participants developed models to predict the perceived intensity, pleasantness, and usage of 19 semantic descriptors for each of the 49 individuals and for the mean and standard deviation across the population of these individuals (Fig. 1d–e).
We first examined the structure of the psychophysical data using the inverse of the covariance matrix (15) calculated across all molecules as a proxy for connection strength between each of the 21 perceptual attributes (Fig. 1f and Fig. S1). This yielded a number of strong positive interactions including those between “garlic” and “fish”, “musky” and “sweaty”, “sweet” and “bakery” and “fruit”, “acid” and “urinous”, and a negative interaction between pleasantness and “decayed” (Fig. 1f and Fig. S1a). The perception of intensity had the lowest connectivity to the other 20 attributes. To understand whether a given individual used the full rating scale or a restricted range, we examined subject-level variance across the ratings for all molecules (Fig. 1g). Applying hierarchical clustering on Euclidean distances for the variance of attribute ratings across all the molecules in the dataset, we distinguished three clusters: subjects that responded with high-variance for all 21 attributes (left cluster in green), subjects with high-variance for four attributes (intensity, pleasantness, “chemical”, “sweet”) and either low variance (middle cluster in blue) or intermediate variance (right cluster in red) for the remaining 17 attributes (Fig. 1g).
We assessed the performance of models submitted to the DREAM challenge by computing for each attribute the correlation between the predictions of the 69 hidden test molecules and the actual data. We then calculated a Z-score by subtracting the average correlations and scaling by the standard deviation of a distribution based on a randomization of the test set molecule identities. Of the 18 teams who submitted models to predict individual perception, Team GuanLab (author Y.G) was the best performer with a Z-score of 34.18 (Fig. 1h and Data File S1). Team IKW Allstars (author R.C.G.) was the best performer of 19 teams to submit models to predict population perception, with a Z-score of 8.87 (Fig. 1h and Data File S1). The aggregation of all participant models gave Z-scores of 34.02 (individual) and 9.17 (population) (Fig. 1h), and a post-challenge community phase where initial models and additional molecular features were shared across teams gave even better models with Z-scores of 36.45 (individual) and 9.92 (population) (Fig. 1h).
Predictions of the models for intensity were highly correlated with the observed data for both individuals (r = 0.56, t-test p < 10−228) and the population (r = 0.78, p < 10−9) (Fig. 1i, j). Pleasantness was also well predicted for individuals (r = 0.41, p < 10−123) and the population (r = 0.71, p <10−8) (Fig. 1i, j). The 19 semantic descriptors were more difficult to predict, but the best models performed respectably (individual: r = 0.21, p < 10−33; population: r = 0.55, p < 10−5) (Fig. 1i, j). Previously described models to predict pleasantness (8, 10) performed less well on this dataset than our best model (Fig. 1j). To our knowledge there are no existing models to predict the 19 semantic descriptors.
Random-forest (Fig. 2a and Data File S1) and regularized linear models (Fig. 2b and Data File S1) out-performed other common predictive model types for the prediction of individual and population perception (Fig. 2, Fig. S2, and Data File S1). Although the quality of the best-performing model varied greatly across attributes, it was exceptionally high in some cases (Fig. 2c), and always considerably higher than chance (dotted line in Fig. 1i), while tracking the observed perceptual values (Fig. S2 for population prediction). In contrast to most previous studies that attempted to predict olfactory perception, these results all reflect predictions of a hidden test set, avoiding the pitfall of inflated correlations due to over-fitting of the experimental data.
Fig. 2. Predictions of individual perception.
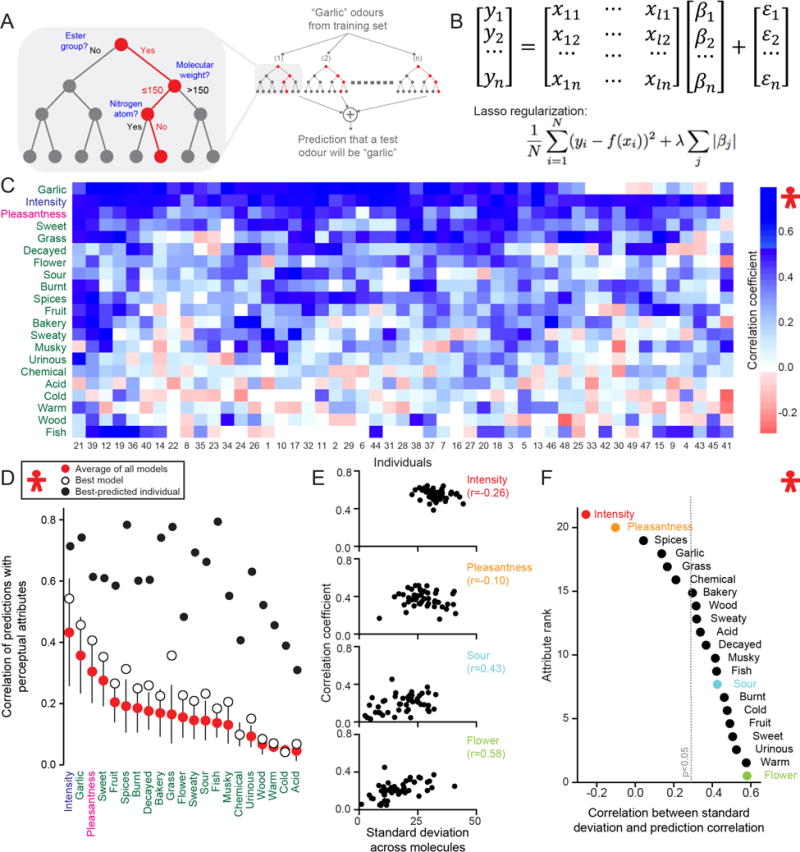
(A) Example of a random-forest algorithm that utilizes a subset of molecules from the training set to match a semantic descriptor (e.g “garlic”) to a subset of molecular features. (B) Example of a regularized linear model. For each perceptual attribute yi a linear model utilizes molecular features xij weighted by βi to predict the psychophysical data of 69 hidden test set molecules, with sparsity enforced by the magnitude of λ. (C) Correlation values of best-performer model across 69 hidden test set molecules, sorted by Euclidean distance across 21 perceptual attributes and 49 individuals. (D) Correlation values for the average of all models (red dots, mean ± s.d.), best-performing model (white dots), and best-predicted individual (black dots), sorted by the average of all models. (E) Prediction correlation of the best-performing random-forest model plotted against measured standard deviation of each subject’s perception across 69 hidden test set molecules for the four indicated attributes. Each dot represents one of 49 individuals. (F) Correlation values between prediction correlation and measured standard deviation for 21 perceptual attributes across 49 individuals, color coded as in E. The dotted line represents the p < 0.05 significance threshold obtained from shuffling individuals.
The accuracy of predictions of individual perception for the best-performing model was highly variable (Fig. 2c), but the correlation of six of the attributes was above 0.3 (Fig. 2d; white circles). The best-predicted individual showed a correlation above 0.5 for 16 of 21 attributes (Fig. 2d). We asked whether the usage of the rating scale (Fig. 1g) could be related to the predictability of each individual. Overall we observed that individuals using a narrow range of attribute ratings, measured across all molecules for a given attribute, were more difficult to predict (Fig. 2e–f, derived from the variance in Fig. 1g). The relationship between range and prediction accuracy did not hold for intensity and pleasantness (Fig. 2e–f).
We next compared the results of predicting individual and population perception. The seven best predicted attributes overall (intensity, “garlic”, pleasantness, “sweet”, “fruit”, “spices”, “burnt”) were the same for both individuals and the population (Fig. 2d and Fig. 3a except “fish”). Similarly, the seven attributes that were the most difficult to predict (“acid”, “cold”, “warm”, “wood”, “urinous”, “chemical”, “musky”) were the same for both individual and population predictions (Fig. 2d and Fig. 3a), and except for a low correlation for “warm”, these attributes are anti-correlated or un-correlated to the “familiarity” attribute (14). This suggests some bias in the predictability of more familiar attributes, perhaps due to a better match to a well-defined reference molecule (14), and that in this categorization individual perceptions are similar across the population. For the population predictions, the first ten attributes have a correlation above 0.5 (Fig. 3a). The connectivity structure in Fig. 1f follows the model’s performance for the population (Fig. 3a). “Garlic”/“fish” (p < 10−4), “sweet”/“fruit” (p < 10−3) and “musky”/“sweaty” (p < 10−3) are pairs with strong connectivity that were also similarly difficult to predict.
Fig. 3. Predictions of population perception.
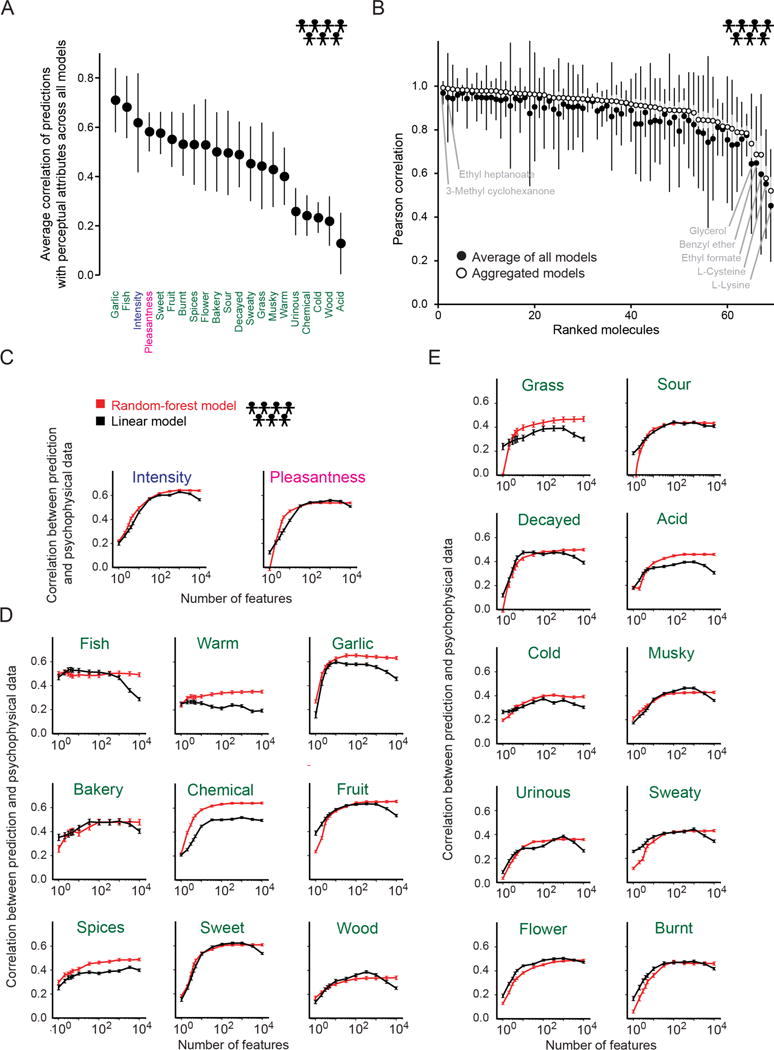
(A), Average of correlation of population predictions. Error bars indicate standard deviations calculated across models. (B) Ranked prediction correlation for 69 hidden test set molecules produced by aggregated models (open black circles, standard deviation indicated with grey bars) and the average of all models (solid black dots, standard deviation indicated with black bars). (C–E) Prediction correlation with increasing number of Dragon features using random-forest (red) or linear (black) models. Attributes are ordered from top to bottom and left to right by the number of features required to obtain 80% of the maximum prediction correlation using the random-forest model. Plotted are intensity and pleasantness (C), and attributes that required six or fewer (D) or more than six features (E). The combined training+leaderboard set of 407 molecules was randomly partitioned 250 times to obtain error bars for both types of models.
We analyzed the quality of model predictions for specific molecules in the population (Fig. 3b). The correlation between predicted and observed attributes exceeded 0.9 (t-test p < 10−4) for 44 of 69 hidden test set molecules when we used aggregated models, and 28 of 69 when we averaged all models (Data File S1). The quality of predictions varied across molecules, but for every molecule the aggregated models exhibited higher correlations (Fig. 3b). The two best-predicted molecules were 3-methyl cyclohexanone followed by ethyl heptanoate. Conversely, the five molecules that were most difficult to predict were L-lysine and L-cysteine, followed by ethyl formate, benzyl ether, and glycerol (Fig. 3b and Fig. S3).
To better understand how the models successfully predicted the different perceptual attributes, we first asked how many molecular features were needed to predict a given population attribute. While some attributes required hundreds of features to be optimally predicted (Fig. 3c–e), both the random-forest and linear models achieved prediction quality of at least 80% of that optimum with far fewer features. By that measure, the algorithm to predict intensity was the most complex, requiring fifteen molecular features to reach the 80% threshold (Fig. 3c). “Fish” was the simplest, requiring only one (Fig. 3d). Although Dragon features are highly correlated, these results are remarkable because even those attributes needing the most molecular features to predict required only a small fraction of the thousands of chemoinformatic features.
We asked what features are most important for predicting a given attribute (Fig. S4, Fig. S5, Fig. S6, and Data File S1). The Dragon software calculates a large number of molecular features, but is not exhaustive. In a post-challenge phase (Fig. 1h, triangles), four of the original teams attempted to improve their model predictions by using additional features. These included Morgan (16) and NSPDK (17), which encode features through the presence or absence of particular substructures in the molecule; experimentally derived partition coefficients from EPI Suite (18); and the common names of the molecules. We used cross-validation on the whole dataset to compare the performance of the same models using different subsets of Dragon and these additional molecular features. Only Dragon features combined with Morgan features yielded decisively better results than Dragon features alone both for random-forest (Fig. 4a) and linear (Fig. 4b) models. We then examined how the random-forest model weighted each feature (Data File S1 for a similar analysis using the linear model). As observed previously, intensity was negatively correlated with molecular size, but was positively correlated with the presence of polar groups, such as phenol, enol, and carboxyl features (Fig. S6a) (1, 7). Predictions of intensity relied primarily on Dragon features.
Fig. 4. Quality of predictions.
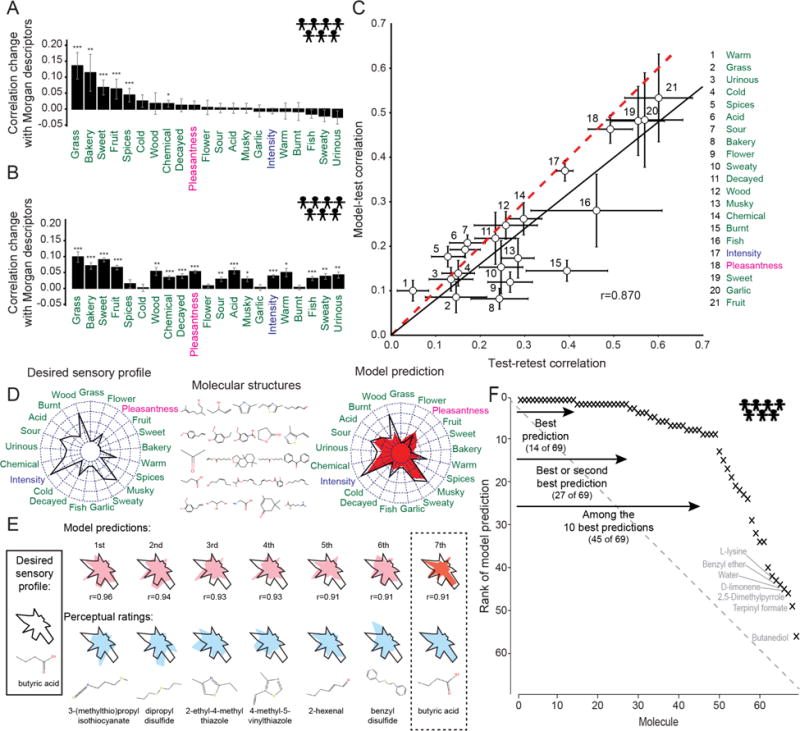
(A–B) Community phase predictions for random-forest (A) and linear (B) models using both Morgan and Dragon features for population prediction. The training set was randomly partitioned 250 times to obtain error bars *p < 0.05, **p < 0.01, ***p < 0.001 corrected for multiple comparisons (FDR). (C) Comparison between correlation coefficients for model predictions and for test-retest for individual perceptual attributes using the aggregated predictions from linear and random-forest models. Error bars reflect standard error obtained from jackknife resampling of the retested molecules. Linear regression of the model-test correlation coefficients against the test-retest correlation coefficients yields a slope of 0.80 ± 0.02 and a correlation of r = 0.870 (black line) compared to a theoretically optimal model (perfect prediction given intra-individual variability, dashed red line). Only the model-test correlation coefficient for “burnt” (15) was statistically distinguishable from the corresponding test-retest coefficient (p < 0.05 with FDR correction). (D) Schematic for reverse-engineering a desired sensory profile from molecular features. The model was presented with the experimental sensory profile of a molecule (spider plot, left) and tasked with searching through 69 hidden test set molecules (middle) to find the best match (right, model prediction in red). Spider plots represent perceptual data for all 21 attributes, with the lowest rating at the center and highest at the outside of the circle. (E) Example where the model selected a molecule with a sensory profile 7th closest to the target, butyric acid. (F) Population prediction quality for the 69 molecules in the hidden test set when all 19 models are aggregated. The overall area under the curve (AUC) for the prediction is 0.83, compared to 0.5 for a random model (grey dotted line) and 1.0 for a perfect model.
There is already anecdotal evidence that some chemical features are associated with a sensory attribute. For example, sulfurous molecules are known to smell “garlic” or “burnt”, but no quantitative model exists to confirm this. Our model confirms that the presence of sulfur in the Dragon descriptors used by the model correlated positively with both “burnt” (r = 0.661 p < 10−62 Fig. S4a) and “garlic” (r = 0.413 p < 10−22 Data File S1). Pleasantness was predicted most accurately using a mix of both Dragon and Morgan/NSPDK features. For example, pleasantness correlated with both molecular size (r = .160 p < 10−3) (9), and similarity to paclitaxel (r = 0.184 p < 10−4) and citronellyl phenylacetate (r = 0.178 p < 10−4)(Fig. S6b). “Bakery” predictions were driven by similarity to the molecule vanillin (r = 0.45 p < 10−24)(Fig. S4b). Morgan features improved prediction in part by enabling a model to template-match target molecules against reference molecules for which the training set contains perceptual data. Thus, structural similarity to vanillin or ethyl vanillin predicts “bakery” without recourse to structural features.
Twenty of the molecules in the training set were rated twice (“test” and “retest”) by each individual, providing an estimate of within-individual variability for the same stimulus. This within-individual variability places an upper limit on the expected accuracy of the optimal predictive model. We calculated the test-retest correlation across individuals and molecules for each perceptual attribute. This value of the observed correlation provides an upper limit to any model, because no model prediction should produce a better correlation than data from an independent trial with an identical stimulus and individual. To examine the performance of our model compared to the theoretically best model, we calculated a correlation coefficient between the prediction of a top-performing random-forest model and the test data. All attributes except “burnt” were statistically indistinguishable from the test-retest correlation coefficients evaluated at the individual-level (Fig. 4c). The slope for the best linear fit of the test-retest and model-test correlation coefficients was 0.80 ± 0.02, with a slope of 1 expected for optimal performance (Fig. 4c). Similar results were obtained using model-retest correlation. Thus, given this dataset, performance of the model is close to that of the theoretically optimal model.
We evaluated the specificity of the predictions of the aggregated model by calculating how frequently the predicted sensory profile had a better correlation with the actual sensory profile of the target molecule than it did with the sensory profiles of any of the other 68 molecules in the hidden test set (Fig. 4d–e). For 14 of 69 molecules, the highest correlation coincided with the actual sensory profile (p < 10−11). For an additional 20% it was second highest and 65% of the molecules ranked in the top ten predictions (Fig. 4f and Data File S1; AUC = 0.83). The specificity of the aggregated model shows that its predictions could be used to reverse-engineer a desired sensory profile using a combination of molecular features to synthesize a designed molecule.
Finally, to ensure that the performance of our model would extend to new subjects, we trained it on random subsets of 25 subjects from the DREAM dataset and consistently predicted the attribute ratings of the mean across the population of the 24 left out subjects (Fig. S7A). To test our model across new subjects and new molecules, we took advantage of a large unpublished dataset comprising 403 volunteers who rated the intensity and pleasantness of 47 molecules, of which only 32 overlapped with the stimuli used in the original study (Data File S1). Using a random forest model trained on the original 49 DREAM challenge subjects and all the molecules, we are able to show that the model robustly predicts the average perception of all of these molecules across the population (Fig. S7B).
The DREAM Olfaction Prediction Challenge has yielded models that generated high-quality personalized perceptual predictions. This work significantly expands on previous modelling efforts (2, 3, 7–11) because it predicts not only pleasantness and intensity, but also 8 out of 19 semantic descriptors of odor quality. The predictive models enable the reverseengineering of a desired perceptual profile to identify suitable molecules from vast databases of chemical structures and closely approach the theoretical limits of accuracy when accounting for within-individual variability. Although highly significant, there is still much room for improving in particular the individual predictions. While the current models can only be used to predict the 21 attributes, the same approach could be applied to a psychophysical dataset that measured any desired sensory attribute (e.g. “rose”, “sandalwood”, or “citrus”). How can the highly predictive models presented here be further improved? Recognizing the inherent limits of using semantic descriptors for odors (12–14), alternative perceptual data such as ratings of stimulus similarity will be important (11).
What do our results imply about how the brain encodes an olfactory percept? We speculate that for each molecular feature there must be some quantitative mapping, possibly one to many, between the magnitude of that feature and the spatiotemporal pattern and activation magnitude of the associated olfactory receptor. If features rarely or never interact to produce perception, as suggested by the strong relative performance of linear models in this challenge, then these feature-specific patterns must sum linearly at the perceptual stage. Peripheral events in the olfactory sensory epithelium, including receptor binding and sensory neuron firing rates might have non-linearities, but the numerical representation of perceptual magnitude must be linear in these patterns. It is possible that stronger non-linearity will be discovered when odor mixtures or the temporal dynamics of odor perception are investigated. Many questions regarding human olfaction remain that may be successfully addressed by applying this method to future datasets that include more specific descriptors; more molecules that represent different olfactory percepts than those studied here; and subjects of different genetic, cultural, and geographic backgrounds.
Results of the DREAM Olfaction Prediction Challenge may accelerate efforts to understand basic mechanisms of ligand-receptor interactions, and to test predictive models of olfactory coding in both humans and animal models. Finally, these models have the potential to streamline the production and evaluation of new molecules by the flavor and fragrance industry.
Supplementary Material
One Sentence Summary.
Results of a crowdsourcing competition show that it is possible to accurately predict and reverse-engineer perceptual attribute values for individuals smelling pure molecules.
Acknowledgments
This research was supported in part by grants from the National Institutes of Health (R01DC013339 to JDM, R01MH106674, R01 EB021711 to R.C.G., UL1RR024143 to Rockefeller University), the Russian Science Foundation (#14-24-00155 to M.D.K.), the Slovenian Research Agency (P2-0209 to B.Z.), the Research Fund KU Leuven (C.V.), and IWT-SBO Nemoa (L.S.). A.K. was supported by a Branco Weiss Science in Society Fellowship. G.S is employee of IBM Research. G.T is partly funded by the Hungarian Academy of Sciences. L.B.V. is an investigator of the Howard Hughes Medical Institute. P.C.B. has the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario and a Terry Fox Research Institute New Investigator Award and a CIHR New Investigator Award. C.V. acknowledges the Research Fund KU Leuven. K.R. is supported by a Grant from the Council of Scientific and Industrial Research and ERC PoC ‘SNIPER’.
Footnotes
Author Contributions: All authors together interpreted the results, approved the design of the figures and the text, which were prepared by A.K., R.C.G., J.D.M., L.B.V., and P.M. LBV is a member of the scientific advisory board of International Flavors & Fragrances, Inc. (IFF) and receives compensation for these activities. IFF was one of the corporate sponsors of the DREAM Olfaction Prediction Challenge. Y.I. is employed by Ajinomoto Co., Inc. J.D.M. is a member of the scientific advisory board of Aromyx and receives compensation and stock for these activities.
Weblinks for data and models are provided below. On the web pages, individual predictions are known as “Subchallenge 1,” and population prediction as “Subchallenge 2.”
Model details and code from the best-performing team for individual prediction (Team GuanLab; authors Y.G. and B. P):
https://www.synapse.org/#!Synapse:syn3354800/wiki/ (see files folder for code)
Model details and code for the best-performing team for population prediction (Team IKW Allstars; author R.C.G.):
https://www.synapse.org/#!Synapse:syn3822692/wiki/231036 (check ipython notebooks)
DREAM Olfaction challenge description, participants, leaderboards and datasets:
https://www.synapse.org/#!Synapse:syn2811262/wiki/78368
Model descriptions and predictions:
https://www.synapse.org/#!Synapse:syn2811262/wiki/78388
Code and details to reproduce analysis for scoring and to reproduce all the analysis for the Figures:
References and Notes
- 1.Boelens H. Structure-activity relationships in chemoreception by human olfaction. Trends Pharmacol Sci. 1983;4:421–426. [Google Scholar]
- 2.Sell C. Structure-odor relationships: On the unpredictability of odor. Angew Chem Int Edit. 2006;45:6254–6261. doi: 10.1002/anie.200600782. [DOI] [PubMed] [Google Scholar]
- 3.Koulakov AA, Kolterman BE, Enikolopov AG, Rinberg D. In search of the structure of human olfactory space. Frontiers in systems neuroscience. 2011;5:65. doi: 10.3389/fnsys.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro JB, Ramanathan A, Chennubhotla CS. Categorical dimensions of human odor descriptor space revealed by non-negative matrix factorization. PLoS ONE. 2013;8:e73289. doi: 10.1371/journal.pone.0073289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laska M, Teubner P. Olfactory discrimination ability for homologous series of aliphatic alcohols and aldehydes. Chem Senses. 1999;24:263–270. doi: 10.1093/chemse/24.3.263. [DOI] [PubMed] [Google Scholar]
- 6.Boesveldt S, Olsson MJ, Lundstrom JN. Carbon chain length and the stimulus problem in olfaction. Behavioural brain research. 2010;215:110–113. doi: 10.1016/j.bbr.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Edwards PA, Jurs PC. Correlation of odor intensities with structural properties of odorants. Chem Senses. 1989;14:281–291. [Google Scholar]
- 8.Khan RM, Luk CH, Flinker A, Aggarwal A, Lapid H, Haddad R, Sobel N. Predicting odor pleasantness from odorant structure: Pleasantness as a reflection of the physical world. J Neurosci. 2007;27:10015–10023. doi: 10.1523/JNEUROSCI.1158-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kermen F, Chakirian A, Sezille C, Joussain P, Le Goff G, Ziessel A, Chastrette M, Mandairon N, Didier A, Rouby C, Bensafi M. Molecular complexity determines the number of olfactory notes and the pleasantness of smells. Sci Rep. 2011;1:206. doi: 10.1038/srep00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarzo M. Hedonic judgments of chemical compounds are correlated with molecular size. Sensors. 2011;11:3667–3686. doi: 10.3390/s110403667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snitz K, Yablonka A, Weiss T, Frumin I, Khan RM, Sobel N. Predicting odor perceptual similarity from odor structure. PloS Comp Biol. 2013;9:e1003184. doi: 10.1371/journal.pcbi.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dravnieks A. Odor quality: semantically generated multidimensional profiles are stable. Science. 1982;218:799–801. doi: 10.1126/science.7134974. [DOI] [PubMed] [Google Scholar]
- 13.Dravnieks A. Atlas of odor character profiles. ASTM; Philadelphia: 1985. [Google Scholar]
- 14.Keller A, Vosshall LB. Olfactory perception of chemically diverse molecules. BMC Neurosci. 2016;17:55. doi: 10.1186/s12868-016-0287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prill R, Vogel R, Cecchi G, Altan-Bonnet G, Stolovitzky G. Noise-driven causal inference in biomolecular networks. PLoS ONE. 2015;10:e0125777. doi: 10.1371/journal.pone.0125777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 17.Costa F, De Grave K. Fast neighborhood subgraph pairwise distance kernel. Proceedings of the 26th International Conference on Machine Learning. 2010:255–262. [Google Scholar]
- 18.EPA. (United States Environmental Protection Agency, Washington, DC, USA, 2014), pp. Estimation Programs Interface Suite for Microsoft Windows, v 4.11. (Episuite).
- 19.Varshney KR, Varshney LR. Olfactory signal processing. Digit Signal Process. 2016;48:84–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


