Abstract
Mutations in the MEN1 gene are associated with the multiple endocrine neoplasia syndrome type 1 (MEN1), which is characterized by parathyroid hyperplasia and tumors of the pituitary and pancreatic islets. The mechanism by which MEN1 acts as a tumor suppressor is unclear. We have recently shown that menin, the MEN1 protein product, interacts with mixed lineage leukemia (MLL) family proteins in a histone methyltransferase complex including Ash2, Rbbp5, and WDR5. Here, we show that menin directly regulates expression of the cyclin-dependent kinase inhibitors p27Kip1 and p18Ink4c. Menin activates transcription by means of a mechanism involving recruitment of MLL to the p27Kip1 and p18Ink4c promoters and coding regions. Loss of function of either MLL or menin results in down-regulation of p27Kip1 and p18Ink4c expression and deregulated cell growth. These findings suggest that regulation of cyclin-dependent kinase inhibitor transcription by cooperative interaction between menin and MLL plays a central role in menin's activity as a tumor suppressor.
Keywords: MEN1, methyltransferase, tumor suppressor
Mutations of the MEN1 gene located on chromosome 11q13 are associated with the development of a variety of endocrine neoplasms, including parathyroid hyperplasia and adenomas, pituitary adenomas, and pancreatic islet cell tumors. Tumor development is associated with deletion or mutation of the remaining MEN1 allele (1, 2). MEN1 mutations have also been reported in a variety of sporadic endocrine tumors including those commonly seen in multiple endocrine neoplasia syndrome type 1 (MEN1) as well as gastric and pulmonary carcinoid tumors (3). Men1 knockout mice have provided many insights into the role of menin in endocrine homeostasis and tumor suppression (4-7). Although Men1 knockout mice are embryonic lethal, heterozygous mice develop a variety of endocrine tumors similar to those in MEN1 patients. In this model, tumors arising from pancreatic islet cells have been most intensively studied. Heterozygous Men1 knockout mice develop progressive islet cell hyperplasia associated with loss of the other Men1 allele, which ultimately culminates in formation of insulin-producing adenomas over a 1- to 2-year period (4-7).
The mechanisms by which menin, which lacks significant homology with other proteins, functions as a tumor suppressor are unknown. Menin plays a role in regulating cellular proliferation because Men1 knockout mice show increased proliferation in neuroendocrine tissues (7), down-modulation of menin in epithelial cells stimulates proliferation (8), and menin knockout fibroblasts proliferate more rapidly than wild-type cells as assayed by tritiated thymidine incorporation (9). In addition, MEN1 cells have increased sensitivity to DNA-damaging agents. Menin interacts with proteins involved in DNA repair such as replication protein A and FANCD2, suggesting that menin plays a role in maintaining chromosomal stability (10, 11). However, recent studies of Men1 knockout mice show that pancreatic insulinomas can develop in the absence of chromosomal or microsatellite instability (12). Menin also has been reported to interact with a variety of transcription factors such as JunD and NF-κB (3, 10). Studies on these interacting proteins suggest that menin exerts its effects predominantly through inhibitory effects on transcription.
However, an alternative possibility is that menin mediates its effects through transcriptional activation of target genes. This possibility was suggested by our finding that menin exists in a histone methyltransferase complex with MLL2, a homolog of mixed lineage leukemia (MLL) (13). This complex includes other mammalian homologues of the yeast SET1 complex, including Ash2L, Rbbp5, WDR5, and hDPY30 (14, 15). Another recent report, whose findings we confirmed in this study, showed that MLL, which is fused to a variety of translocation partners in acute lymphoid and myeloid leukemias, is also associated with a similar complex (16, 17). The MLL family-menin complexes have histone methyltransferase activity specific for histone H3 lysine 4 and have transcriptional activating activity (17-19).
At the time of this study, the only reported targets regulated by menin and MLL were the clustered homeobox genes Hoxa9, c6, and c8 (13, 17, 18). Hox expression generally promotes proliferation, so it is unlikely that reduced Hox expression in menin-deficient tissues accounts for menin's role as a tumor suppressor. Given its effects on cellular proliferation, we reasoned that growth regulation by menin might be mediated through deregulation of cyclin-dependent kinase (CDK) inhibitors. CDK inhibitors of the INK4 family, which form complexes with CDK4 or CDK6 and prevent binding to D-type cyclins, or inhibitors of the CIP/KIP family, which bind to and inhibit CDK2-cyclin complexes, are attractive candidates for regulating neuroendocrine cell growth (20-22). In particular, p18Ink4c and p27Kip1 are expressed in the central nervous system and neuroendocrine tissues and play a central role in controlling cell number through cell cycle regulation (23, 24). Mutations of these CDK inhibitors in tumors are rare; however, decreased expression by means of a number of mechanisms, including methylation and haploinsufficiency, is common (25, 26). Loss of p27Kip1 expression occurs in human pituitary and parathyroid hyperplasias and adenomas, as well as pancreatic islet cell tumors, all of which are hallmarks of MEN1 (27-29). Strikingly, mice deficient for both p27Kip1 and p18Ink4c develop pituitary tumors much more rapidly than either deficiency alone, suggesting the two CDK inhibitors collaborate to suppress tumorigenesis (30).
In this study, we examined whether menin regulates expression of p27Kip1 and p18Ink4c. These experiments showed that menin binds directly to the p27Kip1 and p18Ink4c loci. We confirmed that menin associates with MLL in an evolutionarily conserved complex by coimmunoprecipitation and that menin and MLL colocalize at target promoters in vivo. Interestingly, binding of MLL to transcriptional targets is highly dependent on menin, suggesting that transcriptional activation by menin involves MLL recruitment. Point mutations that occur in MEN1 patients were found to inhibit menin binding to target promoters. Finally, we found that expression of p27Kip1 is decreased in tumors from MEN1 patients compared with normal neuroendocrine tissues. In aggregate, our data suggest that cooperative interaction and transcriptional activation of p27Kip1 and possibly p18Ink4c loci by menin and MLL are important mechanisms for menin-mediated tumor suppression.
Materials and Methods
Cell Lines. Mll knockout and wild-type fibroblasts have been described (18). Menin knockout and wild-type fibroblasts were established in the laboratories of X.H. as described (11). For comparisons of growth and CDK inhibitor expression, particular care was given to seeding cells at comparable densities.
Antibodies. Anti-Ash2 (Bethyl Laboratories, Montgomery, TX), anti-RbBP5 (Bethyl laboratories; catalog no. A300-109A), anti-Flag (M2, Sigma) antibodies were obtained commercially. Anti-MLL N-terminal polyclonal antibody for Western blot and for immunohistochemistry were from Bethyl Laboratories (catalog nos. A300-087A and A300-086A, respectively). Anti-MLL C-terminal antibody was obtained from S. Korsmeyer (Dana-Farber Cancer Institute, Harvard Medical School, Boston). Anti-menin was from Bethyl Laboratories (catalog no. A300-105A). Anti-p27 rabbit polyclonal antibody for immunohistochemistry is from Santa Cruz Biotechnology (C-19; catalog no. sc-528). Anti-p27 antibody for Western blots is from BD Biosciences (no. 610241). Anti-vinculin and β-actin antibodies are from Sigma (catalog nos. V9131 and A5316, respectively). Horseradish peroxidase (HRP)-conjugated sheep anti-rabbit Ig(H+L) and HRP-conjugated sheep anti-mouse antibodies (Roche Diagnostics) were used as secondary reagents for Western blotting.
Immunoaffinity Purification of MLL1-Containing Complex. A human WDR5 cDNA was subcloned into the pIRES-neo mammalian expression vector (Clontech/BD Biosciences), adding the FLAG epitope to the WDR5 N-terminal. The FLAG-WDR5 construct was transfected into HeLa cells, and stable transformants were selected by using 0.5 mg/ml G418. G418-resistant clones were analyzed for expression of Flag-WDR5, and the highest expressing clone was expanded. Nuclear extracts were obtained from 20L f-WDR5 cells by a modified Dignam procedure (31) and dialyzed against BC100 buffer [20 mM Tris, pH 7.8/0.2 mM EDTA/20% glycerol/1 mM DTT/0.2 mM PMSF/proteinase inhibitor mixture (Roche Diagnostics) at 1× strength] before loading onto a phosphocellulose column P11. Bound proteins were eluted sequentially with BC100, BC300, BC500, and BC850. An aliquot of the BC500 fraction (≈2 mg) was incubated with anti-FLAG M2 agarose (Sigma). The bound proteins were washed three times with buffer BC500 and eluted with BC100 containing 0.25 mg/ml Flag peptide (Sigma). The elution fraction was analyzed by immunoblotting.
Luciferase Assays. Sequences from -1100 to -27 of the human p27Kip1 promoter were amplified by PCR, confirmed by sequencing, and cloned into the KpnI and HindIII sites of the luciferase reporter pTAL-Luc (Clontech/BD Biosciences). The p18Ink4c promoter-luciferase reporter, which contains sequences from -922 to -1 in the human p18Ink4c promoter was a gift of C. Labrie (Université Laval, Quebec). For luciferase assays, 500 ng of expression vector (pSPORT-menin, pSPORT-L22R, or A242V menin mutants or pcDNAIII control) was transfected with 500 ng of luciferase reporter and 250 ng of RL-CMV (Promega) into Men1-/- cells in six-well plates by using FuGENE 6 (Roche Diagnostics). Cells were harvested 48 h later and Dual Luciferase assays (Promega) were performed as described (27).
Immunohistochemical Staining. Paraffin-embedded normal and tumor tissues were obtained with institutional review board (IRB) approval from the files of the Mayo Clinic. Additional normal parathyroid and pancreas tissues were obtained with IRB approval from the Cooperative Human Tissue Network. Antigen retrieval was performed on sections to be stained for menin and MLL by boiling in 10 mM sodium citrate (pH 6.0) for 10 min. Tissues were stained for p27Kip1 by using antigen retrieval as described (17).
Results
Menin Is Associated with an MLL-Containing Histone Methyltransferase Complex in Nonhematopoietic Cells. The association of menin with histone methyltransferase complexes has important implications for its ability to act as a transcriptional activator. However, some controversy has arisen regarding the composition of menin and MLL complexes. We initially reported that menin was associated with an evolutionary complex including MLL2 (13). Subsequent inspection of mass spectroscopy spectra revealed that MLL peptides were also present in the menin immunoprecipitation, and, consistent with this finding, the recently reported biochemical purification of MLL also revealed menin (17). However, other published complexes of either trx or MLL-associated proteins lacked menin (19, 32). We wished to confirm whether menin and MLL were associated in nonhematopoietic cells. These experiments showed that menin and the previously reported homologs of yeast Set1 complex proteins coimmunoprecipitate with MLL-containing complexes from HeLa cells expressing FLAG-tagged WDR5. Proteins coimmunoprecipitating with WDR5 included the proteolytic cleavage products of MLL, MLLN and MLLC, hASH2, RBBP5, and hDPY30 (Fig. 1 A and B). Additional experiments showed that the menin-MLL interaction is mediated by sequences within the N-terminal 260 aa of MLL. Reciprocal immunoprecipitations with antibodies directed against MLL and menin immunoprecipitated the other, confirming that MLL and menin interact at endogenous levels (data not shown). Histone methyltransferase activity specific for lysine 4 could be precipitated by anti-menin antibodies in MLL knockout cells (data not shown). This and previously published work (13, 17) suggest that MLL and MLL2 are associated with similar but distinct complexes.
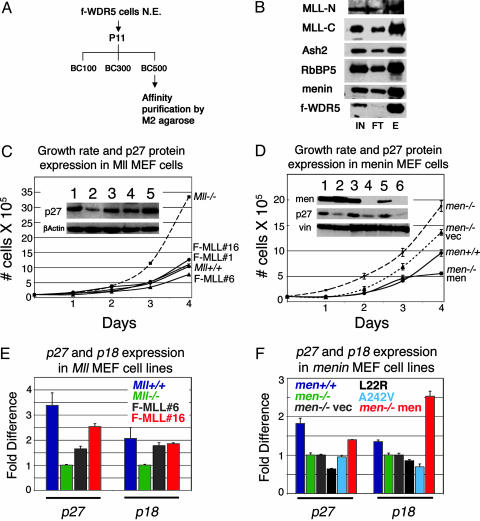
Fig. 1.
MLL and menin copurify in a complex that regulates expression of CDK inhibitors. (A) Strategy for immunoprecipitating MLL complex from HeLa cells expressing FLAG-tagged WDR-5. N.E., nuclear extract. (B) Western blots showing that menin and mammalian homologues of yeast SET1 complexes copurify with MLL. IN, input; FT, flow-through; E, eluate. (C) Mll-/- cells have a faster growth rate than Mll+/+ cells or three independent Mll-/- cells expressing FLAG-tagged MLL (F-MLL#1, F-MLL#6, and F-MLL#16). Growth curves were generated by plating cells at 1 × 105 in T75 flasks and counting cells at the same time point in successive 24-h periods by using Trypan blue exclusion. (Inset) Higher levels of p27Kip1 protein in Mll+/+ (lane 1), F-MLL#1 (lane 3), F-MLL#6 (lane 4), and F-MLL#16 (lane 5) compared with Mll-/- (lane 2) cell lines. (D) Menin-null cells (Men1-/-) and null cells with an empty expression vector (Men1-/- vec) have a faster growth rate than menin wild-type (Men1+/+) cells and null cells expressing exogenous menin (Men1-/- menin). (Inset) Higher p27Kip1 levels in slower Mll+/+ (lane 1), Men1+/+ (lane 3), and Men1-/- menin (5) compared with faster growing cells that are Mll-/- (lane 2), Men1-/- (lane 4), or Men1-/- cells with an empty expression vector (lane 6). Western blot for menin shows lack of expression in Men1-/- (lane 4) or Men1-/- cells with an empty expression vector (lane 6) and restoration of menin expression in Men1-/- menin cells (lane 5). (E) Real-Time PCR quantification using TaqMan probes shows higher expression of p27Kip1 and p18Ink4c transcripts in Mll+/+ (dark blue), F-MLL#6 (gray), and F-MLL#16 (red) cell lines compared with Mll-/- (green) cells. Gapdh and β-actin were both used as internal reference standards. TaqMan probe and primer sequences are available on request. (F) Quantitative RT-PCR shows higher expression of p27Kip1 and p18Ink4c transcripts in menin wild-type (Men1+/+, dark blue) or null cells with menin reexpression (Men1-/- men, red) compared with menin-null cells (Men1-/-, green) or menin-null cells harboring an empty expression vector (Men1-/- vec, gray). Cell lines that express patient-derived menin point mutants (L22R, black; A242V, light blue) show defective restoration of p27Kip1 and p18Ink4c expression (Fig. 2F Inset shows Western blot for menin expression in these cell lines).
Menin- and MLL-Deficient Cells Show Increased Cell Growth and Decreased p27Kip1 and p18Ink4c Expression. Despite global deficiencies in Hox gene expression, whose expression is generally associated with proliferation, both Men1 and Mll knockout fibroblasts grow faster than wild-type cells (Fig. 1 C and D). Conversely, reexpression of menin or MLL in Men1- or Mll-null fibroblasts, respectively, slows growth rates to levels comparable with those of wild-type cells (Fig. 1 C and D).
We therefore examined whether expression of p18Ink4c and p27Kip1 is altered in Mll and Men1 knockout fibroblast lines. Western blot analysis showed that both p18Ink4c and p27Kip were expressed at reduced levels in Mll knockout cells (Fig. 1C and data not shown) and that p27Kip1 was expressed at reduced levels in menin knockout cells (Fig. 1D). Quantitative RT-PCR showed decreased expression of both p27Kip1 and p18Ink4c in Mll-null cells (Fig. 1E, blue versus green bars), which could be restored by MLL expression (Fig. 1E, red and gray bars). Expression of p27Kip1 and p18Ink4c was similarly reduced in menin null cells (Fig. 1F, blue versus green bars). Stable expression of menin (red bars), but not the patient derived menin point mutants L22R (black bars) or A242V (light blue bars), restored expression of both CDKs. The magnitude of these changes is likely to be significant because haploinsufficiency of either p27Kip1 or p18Ink4c results in either spontaneous or chemically induced tumorigenesis (25, 26).
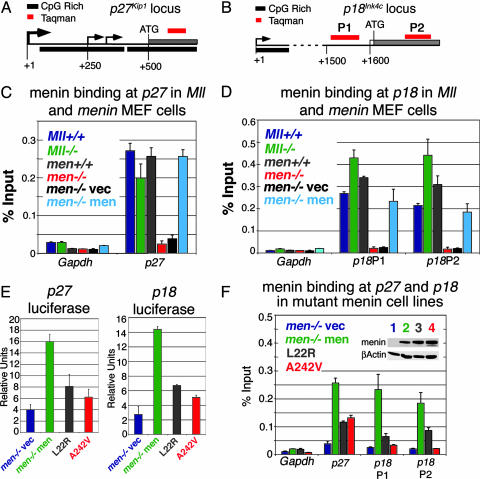
Menin and MLL Bind Directly to the p27Kip1 and p18Ink4c Loci. Chromatin immunoprecipitation (ChIP) with anti-menin antibodies was then done to determine whether menin directly regulated p27Kip1 and p18Ink4c. These experiments showed menin bound directly to the coding regions of both genes (Fig. 2 C and D). Signal was abolished in Men1 knockout cells (red bars) and is restored by menin reexpression (light blue bars) confirming the specificity of the antibody (Fig. 2 C and D). No binding of menin was detected to Gapdh in either cell type, indicating that menin does not bind to all transcriptionally active loci. Only minimal differences in menin binding to the p27Kip1 and p18Ink4c loci were detected between Mll+/+ and Mll-/- cells (Fig. 2 C and D, blue versus green bars), indicating that MLL is not required for menin binding. Luciferase reporter assays showed that menin reexpression in Men1-null cells activated transcription from both p27Kip1 and p18Ink4c promoters approximately 4- to 5-fold (Fig. 2E, green versus blue bars). Importantly, the menin mutants L22R and A242V were impaired in their ability to activate both promoters (gray and red bars). ChIP studies suggested that this deficiency was the result of defective recruitment. Men1-null cells were established stably expressing comparable levels of either wild type or the L22R or A242V menin mutants (Fig. 2F Inset). ChIP with menin antibodies showed that the amount of the L22R or A242V menin mutants associated with the p27Kip1 and p18Ink4c loci was greatly decreased compared with wild-type menin (Fig. 2F, gray and red bars).
Fig. 2.
Point mutations in menin interfere with binding and transcriptional regulation of CDK inhibitor loci. (A and B) Schematic of the p27Kip1 (A) and p18Ink4c (B) loci. Large arrow shows a major transcription start site at p27Kip1 conserved in both mice and humans. Smaller arrows show minor transcription start sites. Black bars are CpG rich regions; gray bars are exons. Red bar represents TaqMan primer/probe set. The initial ATG is indicated. (C and D) ChIP with an anti-menin antibody quantified with Real-Time PCR. TaqMan primer and probe sequences are available on request. (C) Menin binds to the p27 coding region in cells that are Mll+/+ (dark blue), Mll-/- (green), Men1+/+ (gray), or Men1-/- with reexpressed menin (light blue), but not in Men1-null cells or at the Gapdh locus in any cell type (Men-/-, red; Men1-/- with an empty vector, black). (D) Menin binds to the p18Ink4c coding region in cells that are Mll+/+ (dark blue), Mll-/- (green), Men1+/+ (gray), or Men1-/- with reexpressed menin (light blue). No binding is seen in Men1-null cells or at the Gapdh locus in any cell type (Men1-/-, red; Men1-/- with empty vector, black). (E) Luciferase assays on Men1-/- cells cotransfected with menin expression vector and p27Kip1 or p18Ink4c luciferase reporter shows menin activates transcription from the p27Kip1 or p18Ink4c promoters (green versus blue bars). The L22R (gray) and A242V (red) mutants show impaired ability to activate transcription. (F) Binding of the menin mutants L22R (gray) and A242V (red) are reduced at the p27Kip1 and p18Ink4c loci compared with wild-type menin (Men1-/- with reexpressed menin, green). (Inset) Western blot shows the L22R (gray lane 3) and A242V (red lane 4) mutants are expressed at similar levels to wild-type menin protein (green lane 2). Binding and menin expression in Men1-/- cells with an empty vector are shown as controls.
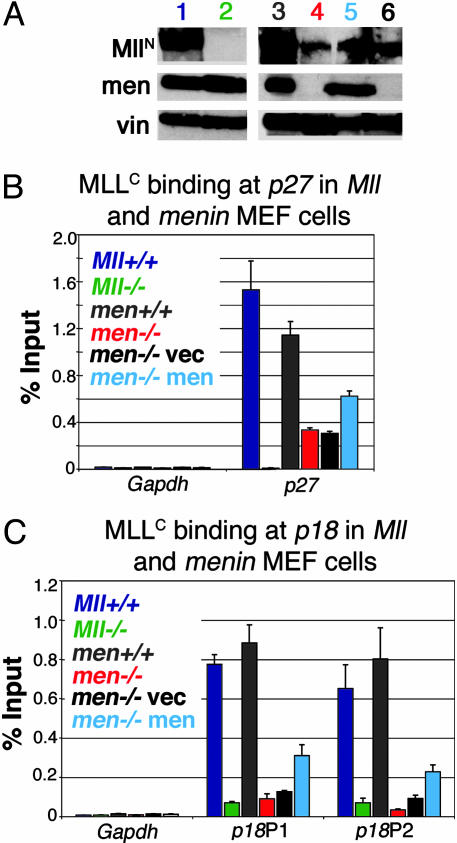
MLL Association with p27Kip1 and p18Ink4c Loci Is Menin-Dependent. MLL increases transcription and interacts with menin, suggesting that transcriptional activation by menin involves MLL recruitment to target promoters. To investigate this possibility, we performed ChIP to localize Mll binding in Mll and Men1 wild-type and knockout cells. Surprisingly, lower MLL protein levels were seen in Men1-null cell compared with wild-type cells (Fig. 3A). ChIP showed that MLL directly binds to the p27Kip1 and p18Ink4c loci (Fig. 3 B and C) and that, in the absence of menin, a drastic reduction is seen in MLL binding (Fig. 3 B and C, red versus gray bars). Menin reexpression had minimal effects on Mll protein levels (Fig. 3A) but resulted in increased binding at the p27Kip1 and p18Ink4c loci (Fig. 3 B and C, light blue bars). Overall, the data suggest that transcriptional regulation by menin involves increasing MLL protein association with target loci.
Fig. 3.
MLL binding to CDK inhibitor loci is reduced in the absence of menin. (A) Western blots for Mll and menin expression in cell lines used for ChIP. Although Men-/- cells (lane 4, red) express lower levels of Mll than Men1 wild-type cells (lane 3, gray), Men-/- cells with empty expression vector (lane 6, black) or menin expression vector (lane 5, light blue) show comparable levels of Mll expression. men, menin; vin, vinculin loading control. For comparison, Mll expression is also shown in Mll+/+ (lane 1, dark blue) and Mll-/- cells (lane 2, green). (B) ChIP with anti-MLLC antibody quantitated with Real-Time PCR shows that Mll binding to the p27Kip1 coding region is much higher in cells expressing menin. (C) Mll binding to the p18Ink4c coding region is also much higher in cells expressing menin. Binding to both p27Kip1 and p18Ink4c is high in Mll+/+ (dark blue), Men1+/+ (gray), or Men1-/- with menin reexpression (light blue) compared with Men1-null cells (Men1-/-, red), or Men1-/- with an empty expression vector (black). Negligible binding is detected in Mll-/- cells or at the Gapdh locus in any cell type.
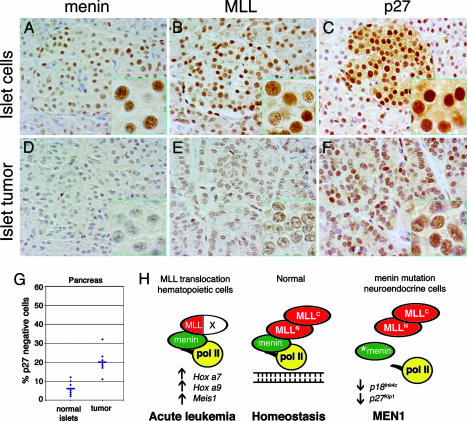
MLL Is Coexpressed with Menin in Endocrine Tissues. We then examined normal tissues and tumors from MEN1 patients for menin, MLL, and CDK inhibitor expression. Pancreas, pancreatic adenomas, and normal and hyperplastic parathyroids were obtained from the files of the Mayo Clinic with additional normal tissues provided by the Cooperative Human Tissue Network. Immunohistochemical staining showed that both menin and MLL are expressed at high levels in neuroendocrine tissues, including pancreatic islets and parathyroid (Fig. 5, which is published as supporting information on the PNAS web site). Reduced expression of menin was seen in most islet cell tumors compared with normal islets in the same sections (Fig. 4 A versus D). Reduced expression of MLL was also seen in the MEN1 tumors (Fig. 4 B versus E). These results, along with the finding of less Mll expression in Men-/- cells (Fig. 3A), suggest that MLL expression could be either directly or indirectly regulated by menin. However, ChIP experiments to date have not shown menin binding to Mll coding regions.
Fig. 4.
MEN1 tumors show decreased p27Kip1 expression compared with normal endocrine tissues. (A-F) Immunoperoxidase staining for menin, MLL, and p27Kip1 in pancreatic tissues from a representative MEN1 patient. (A) Immunoperoxidase staining shows that menin is expressed in most morphologically normal islet cells. Expression is almost totally abolished in adenoma from the same tissue section (D). (B) MLL is expressed in essentially all normal islet cells. Expression is markedly decreased in adenoma from the same tissue section (E). (C) Immunoperoxidase staining for p27Kip1 shows high-level expression in 97% of islet cells (500 nuclei count). Expression is decreased both in number of cells (82%; 500 nuclei count) and intensity of staining in adenoma from the same tissue section(F). (G) Plot of percentage of p27Kip1-negative cells in normal islets and islet tumors based on 500 nuclei counts (n = 7). The blue horizontal bar represents average percent negative cells. (H) Model for role of MLL in normal homeostasis through regulation of Hox genes and CDK inhibitors (see text).
MEN1 Tumors Show Decreased p27Kip1 Expression Compared with Normal Endocrine Tissues. We focused on assessing p27Kip1 expression because minimal nuclear staining for p18Ink4c was seen in human neuroendocrine tissues. Nonneoplastic islets from seven MEN1 patients showed strong expression of p27Kip1 (Fig. 4C). Variable but reduced intensity of expression was seen in all islet cell tumors in the same sections, which showed an increase in the proportion of nonstaining cells (20.00% versus 5.43%, paired t test, P = 0.01) (Fig. 4 F and G). Similar results were seen in parathyroids from normal patients (n = 5) compared with hyperplastic parathyroids from MEN1 patients (Fig. 6, which is published as supporting information on the PNAS web site); however, the levels were heterogeneous and of lower statistical significance (34.4% versus 20.00%, unpaired t test, P = 0.11, n = 5).
Discussion
Although the majority of attention has focused on menin as a transcriptional repressor, our work suggests that menin functions as a tumor suppressor through transcriptional activation of CDK inhibitors. Menin has been shown to bind nonspecifically to DNA; however, the mechanisms responsible for its recruitment to specific chromosomal sites are not known. One possibility is that menin binds with higher affinity to yet-to-be-identified DNA sequences. In addition, menin interacts with the serine 5 phosphorylated C-terminal domain of RNA polymerase II (13) as well as a number of transcription factors, including activator protein-1 (AP-1) family members JunD and c-Jun, NF-κB, and mothers against decapentaplegic (SMAD) family members (10). These findings raise the possibility that menin is targeted to transcriptionally active promoters through interactions with promoter-bound RNA polymerase II and/or transcription factors. Although other activities may be affected, our data suggest that tumor-associated menin mutations decrease menin recruitment to target promoters.
Menin interacts with MLL as part of an evolutionarily conserved histone methylase complex, possibly explaining the ability of menin to activate transcription. Furthermore, both menin and MLL colocalize at target genes in vivo. Menin binding to transcriptional targets is apparently not dependent on MLL. However, menin dramatically increases the amount of MLL bound at the p27Kip1 and p18Ink4c loci, suggesting that it either directly or indirectly promotes MLL recruitment to these targets. Once recruited, MLL could enhance transcription by a number of mechanisms. One possibility is that transcription is enhanced by histone H3 lysine 4 methylation mediated by the SET domain (18, 19). However, to date, we have not detected a difference in histone methylation at either the Hox, p27Kip1, or p18Ink4c loci in Men+/+ vs. Men-/- cells. One alternative possibility that the MLL menin complex methylates non-histone components at target promoters such as general transcription factors. In addition, MLL interacts with CREB-binding protein (CBP) (33) as well as the SWI/SNF chromatin-remodeling complex (34, 35), and either or both of these may also contribute to transcriptional activation at target promoters.
These findings extend the realm of menin- and MLL-regulated genes beyond Hox genes to kinase inhibitors involved in cell cycle regulation. This dual role is highly reminiscent of Bmi1, a mammalian Polycomb group protein that antagonizes MLL function and negatively regulates Hox gene expression. The abilities of Bmi to promote neural and hematopoietic stem cell renewal, as well as to cooperate with Myc in oncogenesis, are both linked to its negative regulation of another CDK, p16Ink4a (36).
The N-terminal MLL sequences required for interaction with menin are retained in leukemogenic MLL fusion proteins (17), raising the question of whether MLL fusion proteins up-regulate CDK inhibitors in hematopoietic cells and, if so, how this regulatory pathway is bypassed to allow for leukemogenesis. One possible explanation is that the relative importance of the targets depends on the cell type, which affects what targets are expressed. In early hematopoietic progenitor cells, where A cluster Hox genes are expressed at high levels (37) but p27Kip1 is either not expressed or expressed at low levels (38), MLL fusion proteins predominantly up-regulate Hox gene expression, resulting in leukemia (16). In keeping with this finding, we have found that activation of a tamoxifen-inducible form of MLL-ENL only transiently upregulates p27Kip1 but persistently up-regulates the leukemogenic targets Hoxa9 and Meis1 (T.A.M. and J.L.H., unpublished results). In pancreatic islet cells, these leukemogenic target genes are minimally expressed (39), but p27Kip1 is abundant, suggesting that the predominant effect of alterations in the menin-MLL axis is decreased CDK expression resulting in deregulated cell growth (Fig. 4H).
Finally, this work raises the surprising possibility that loss-of-function MLL plays a role in neoplasia. Deletions of the 11q23.3 locus spanning the MLL gene are extremely common in a variety of tumors, including pituitary and follicular adenomas, neuroblastoma, and malignant melanoma, and, in some cases, of mantle cell lymphoma and acute lymphoblastic leukemia (40-42). MLL does not meet criteria for a “classic” tumor suppressor gene because Mll knockout mice are embryonic lethal and Mll heterozygous mice do not show a higher rate of tumors. Nonetheless, MLL suppresses growth of a variety of cell types and interacts with at least two tumor suppressors, menin and INI1 (34), which is mutated in rhabdoid and other primitive neuroectodermal tumors, warranting additional studies to assess whether MLL is a “tumor susceptibility and resistance gene” (43).
Supplementary Material
Acknowledgments
We thank S. Korsmeyer for anti MLL-C antibodies and Mll+/+ and Mll-/- cells and Dr. C. Labrie for the p18Ink4c luciferase plasmid. J.L.H. and X.H. were supported by grants from the National Institutes of Health and by a Specialized Center of Research (SCOR) grant from the Leukemia and Lymphoma Society of America. Some tissues were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute.
Author contributions: X.H., R.G.R., M.M., and J.L.H. designed research; T.A.M., C.M.H., R.L., Z.Y., O.R.-R., Y.D., R.W.S., C.K., and D.G. performed research; V.A.L., X.H., R.G.R., and M.M. contributed new reagents/analytic tools; M.M. and J.L.H. analyzed data; and J.L.H. wrote the paper.
Abbreviations: MEN1, multiple endocrine neoplasia syndrome type 1; MLL, mixed lineage leukemia; CDK, cyclin-dependent kinase; ChIP, chromatin immunoprecipitation.
References
- 1.Chandrasekharappa, S. C., Guru, S. C., Manickam, P., Olufemi, S. E., Collins, F. S., Emmert-Buck, M. R., Debelenko, L. V., Zhuang, Z., Lubensky, I. A., Liotta, L. A., et al. (1997) Science 276, 404-407. [DOI] [PubMed] [Google Scholar]
- 2.Lemmens, I., Van de Ven, W. J., Kas, K., Zhang, C. X., Giraud, S., Wautot, V., Buisson, N., De Witte, K., Salandre, J., Lenoir, G., et al. (1997) Hum. Mol. Genet. 6, 1177-1183. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal, S. K., Lee Burns, A., Sukhodolets, K. E., Kennedy, P. A., Obungu, V. H., Hickman, A. B., Mullendore, M. E., Whitten, I., Skarulis, M. C., Simonds, W. F., et al. (2004) Ann. N. Y. Acad. Sci. 1014, 189-198. [DOI] [PubMed] [Google Scholar]
- 4.Crabtree, J. S., Scacheri, P. C., Ward, J. M., Garrett-Beal, L., Emmert-Buck, M. R., Edgemon, K. A., Lorang, D., Libutti, S. K., Chandrasekharappa, S. C., Marx, S. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolino, P., Tong, W. M., Herrera, P. L., Casse, H., Zhang, C. X. & Wang, Z. Q. (2003) Cancer Res. 63, 4836-4841. [PubMed] [Google Scholar]
- 6.Biondi, C., Gartside, M., Tonks, I., Paterson, C., Hayward, N. K. & Kay, G. F. (2002) Genesis 32, 150-151. [DOI] [PubMed] [Google Scholar]
- 7.Crabtree, J. S., Scacheri, P. C., Ward, J. M., McNally, S. R., Swain, G. P., Montagna, C., Hager, J. H., Hanahan, D., Edlund, H., Magnuson, M. A., et al. (2003) Mol. Cell. Biol. 23, 6075-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratineau, C., Bernard, C., Poncet, G., Blanc, M., Josso, C., Fontaniere, S., Calender, A., Chayvialle, J. A., Zhang, C. X. & Roche, C. (2004) J. Biol. Chem. 2, 24477-24484. [DOI] [PubMed] [Google Scholar]
- 9.Schnepp, R. W., Hou, Z., Wang, H., Petersen, C., Silva, A., Masai, H. & Hua, X. (2004) Cancer Res. 64, 6791-6796. [DOI] [PubMed] [Google Scholar]
- 10.Poisson, A., Zablewska, B. & Gaudray, P. (2003) Cancer Lett. 189, 1-10. [DOI] [PubMed] [Google Scholar]
- 11.Jin, S., Mao, H., Schnepp, R. W., Sykes, S. M., Silva, A. C., D'Andrea, A. D. & Hua, X. (2003) Cancer Res. 63, 4204-4210. [PubMed] [Google Scholar]
- 12.Scacheri, P. C., Kennedy, A. L., Chin, K., Miller, M. T., Hodgson, J. G., Gray, J. W., Marx, S. J., Spiegel, A. M. & Collins, F. S. (2004) Cancer Res. 64, 7039-7044. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, C. M., Rozenblatt-Rosen, O., Milne, T. A., Copeland, T. D., Levine, S. S., Lee, J. C., Hayes, D. N., Shanmugam, K. S., Bhattacharjee, A., Biondi, C. A., et al. (2004) Mol. Cell 13, 587-597. [DOI] [PubMed] [Google Scholar]
- 14.Miller, T., Krogan, N. J., Dover, J., Erdjument-Bromage, H., Tempst, P., Johnston, M., Greenblatt, J. F. & Shilatifard, A. (2001) Proc. Natl. Acad. Sci. USA 98, 12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy, P. L., Griesenbeck, J., Kornberg, R. D. & Cleary, M. L. (2002) Proc. Natl. Acad. Sci. USA 99, 90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess, J. L. (2004) Trends Mol. Med. 10, 500-507. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama, A., Wang, Z., Wysocka, J., Sanyal, M., Aufiero, D. J., Kitabayashi, I., Herr, W. & Cleary, M. L. (2004) Mol. Cell. Biol. 24, 5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne, T. A., Briggs, S. D., Brock, H. W., Martin, M. E., Gibbs, D., Allis, C. D. & Hess, J. L. (2002) Mol. Cell 10, 1107-1117. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura, T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C. M. & Canaani, E. (2002) Mol. Cell 10, 1119-1128. [DOI] [PubMed] [Google Scholar]
- 20.Polyak, K., Lee, M. H., Erdjument-Bromage, H., Koff, A., Roberts, J. M., Tempst, P. & Massague, J. (1994) Cell 78, 59-66. [DOI] [PubMed] [Google Scholar]
- 21.Guan, K. L., Jenkins, C. W., Li, Y., Nichols, M. A., Wu, X., O'Keefe, C. L., Matera, A. G. & Xiong, Y. (1994) Genes Dev. 8, 2939-2952. [DOI] [PubMed] [Google Scholar]
- 22.Sherr, C. J. & Roberts, J. M. (1995) Genes Dev. 9, 1149-1163. [DOI] [PubMed] [Google Scholar]
- 23.Zindy, F., Nilsson, L. M., Nguyen, L., Meunier, C., Smeyne, R. J., Rehg, J. E., Eberhart, C., Sherr, C. J. & Roussel, M. F. (2003) Cancer Res. 63, 5420-5427. [PubMed] [Google Scholar]
- 24.Zindy, F., Cunningham, J. J., Sherr, C. J., Jogal, S., Smeyne, R. J. & Roussel, M. F. (1999) Proc. Natl. Acad. Sci. USA 96, 13462-13467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai, F., Pei, X. H., Godfrey, V. L. & Xiong, Y. (2003) Mol. Cell. Biol. 23, 1269-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fero, M. L., Randel, E., Gurley, K. E., Roberts, J. M. & Kemp, C. J. (1998) Nature 396, 177-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd, R. V., Jin, L., Qian, X. & Kulig, E. (1997) Am. J. Pathol. 150, 401-407. [PMC free article] [PubMed] [Google Scholar]
- 28.Lidhar, K., Korbonits, M., Jordan, S., Khalimova, Z., Kaltsas, G., Lu, X., Clayton, R. N., Jenkins, P. J., Monson, J. P., Besser, G. M., et al. (1999) J. Clin. Endocrinol. Metab. 84, 3823-3830. [DOI] [PubMed] [Google Scholar]
- 29.Buchwald, P. C., Akerstrom, G. & Westin, G. (2004) Clin. Endocrinol. (Oxford) 60, 389-393. [DOI] [PubMed] [Google Scholar]
- 30.Franklin, D. S., Godfrey, V. L., Lee, H., Kovalev, G. I., Schoonhoven, R., Chen-Kiang, S., Su, L. & Xiong, Y. (1998) Genes Dev. 12, 2899-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petruk, S., Sedkov, Y., Smith, S., Tillib, S., Kraevski, V., Nakamura, T., Canaani, E., Croce, C. M. & Mazo, A. (2001) Science 294, 1331-1334. [DOI] [PubMed] [Google Scholar]
- 33.Ernst, P., Wang, J., Huang, M., Goodman, R. H. & Korsmeyer, S. J. (2001) Mol. Cell. Biol. 21, 2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, C. W. & Orkin, S. H. (2004) Nat. Rev. Cancer 4, 133-142. [DOI] [PubMed] [Google Scholar]
- 35.Rozenblatt-Rosen, O., Rozovskaia, T., Burakov, D., Sedkov, Y., Tillib, S., Blechman, J., Nakamura, T., Croce, C. M., Mazo, A. & Canaani, E. (1998) Proc. Natl. Acad. Sci. USA 95, 4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, I. K., Morrison, S. J. & Clarke, M. F. (2004) J. Clin. Invest. 113, 175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pineault, N., Helgason, C. D., Lawrence, H. J. & Humphries, R. K. (2002) Exp. Hematol. 30, 49-57. [DOI] [PubMed] [Google Scholar]
- 38.Furukawa, Y., Kikuchi, J., Nakamura, M., Iwase, S., Yamada, H. & Matsuda, M. (2000) Br. J. Haematol. 110, 663-673. [DOI] [PubMed] [Google Scholar]
- 39.Mizusawa, N., Hasegawa, T., Ohigashi, I., Tanaka-Kosugi, C., Harada, N., Itakura, M. & Yoshimoto, K. (2004) Gene 331, 53-63. [DOI] [PubMed] [Google Scholar]
- 40.Monni, O. & Knuutila, S. (2001) Leuk. Lymphoma 40, 259-266. [DOI] [PubMed] [Google Scholar]
- 41.Guo, C., White, P. S., Weiss, M. J., Hogarty, M. D., Thompson, P. M., Stram, D. O., Gerbing, R., Matthay, K. K., Seeger, R. C., Brodeur, G. M., et al. (1999) Oncogene 18, 4948-4957. [DOI] [PubMed] [Google Scholar]
- 42.Herbst, R. A., Gutzmer, R., Matiaske, F., Mommert, S., Casper, U., Kapp, A. & Weiss, J. (1999) Int. J. Cancer 80, 205-209. [DOI] [PubMed] [Google Scholar]
- 43.Sherr, C. J. (2004) Cell 116, 235-246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.