Abstract
Mutations in genes that encode components of the phosphatidyl-inositol 3-kinase (PI3-kinase) signaling pathway are common in human cancer. The recent discovery of nonrandom somatic mutations in the PIK3CA gene of many human tumors suggests an oncogenic role for the mutated enzyme. We have determined the growth-regulatory and signaling properties of the three most frequently observed PI3-kinase mutations: E542K, E545K, and H1047R. Expressed in chicken embryo fibroblasts, all three mutants induce oncogenic transformation with high efficiency. This transforming ability is correlated with elevated catalytic activity in in vitro kinase assays. The mutant-transformed cells show constitutive phosphorylation of Akt, of p70 S6 kinase, and of the 4E-binding protein 1. Phosphorylation of S6 kinase and of 4E-binding protein 1 is regulated by the target of rapamycin (TOR) kinase and affects rates of protein synthesis. The inhibitor of TOR, rapamycin, strongly interferes with cellular transformation induced by the PI3-kinase mutants, suggesting that the TOR and its downstream targets are essential components of the transformation process. The oncogenic transforming activity makes the mutated PI3-kinase proteins promising targets for small molecule inhibitors that could be developed into effective and highly specific anticancer drugs.
Keywords: Akt, oncogenic transformation, p110α, PIK3CA
Genetic aberrations in the phosphatidylinositol 3-kinase (PI3-kinase) pathway have been detected in numerous and diverse human cancers. PIK3CA, which encodes for the catalytic subunit p110α of class IA PI3-kinase, is amplified and overexpressed in some cases of ovarian cancer (1). Mutations in the regulatory subunit p85 of PI3-kinase have been identified in ovarian and colon tumors (2). PTEN, a lipid phosphatase that counteracts the kinase activity of PI3-kinase, is frequently mutated in various tumors including those of prostate, glioblastoma, melanoma, and endometrial carcinoma (3-6). Amplification or overexpression of Akt, a downstream effector of PI3-kinase, also has been detected in ovarian, breast, and thyroid cancers (7, 8). Both Akt and PI3-kinase can function as retroviral oncoproteins, inducing rapid oncogenic transformation in vivo and in vitro. Akt is the cellular homolog of the retroviral oncogene v-akt, which was first identified in the murine lymphoma virus AKT8 (9); the catalytic subunit p110α of PI3-kinase is the cellular counterpart of the product of v-p3k, a viral oncogene found in the avian sarcoma virus 16 that induces hemangiosarcomas in chickens (10).
Recent studies have described a high frequency of mutations in the PIK3CA gene of several human cancers, including those of the colon, brain, breast, and stomach (11-13). These genetic alterations of PIK3CA consisted exclusively of the somatic missense mutations and were clustered to specific sites in the coding sequence (exons 9 and 20), clearly showing nonrandom distribution. Two of the most frequently altered residues, E542 and E545, are located within the helical domain (exon 9) of p110α. These residues are often substituted with lysine in tumors of colon and brain (12, 13). Another commonly targeted residue, H1047, resides in the kinase domain (exon 20) and is frequently substituted with an arginine in tumors of the breast, colon, and brain (11-13). When expressed in cultured cells, the H1047R mutant protein displayed higher lipid kinase activity as compared with wild-type (wt) p110α (13), suggesting a gain of function caused by the mutation.
The high incidence of these nonrandom PI3-kinase mutations detected across different types of tumors strongly suggests a functional significance in tumorigenesis. In this article, we show that PI3-kinase carrying one of the three “hot-spot” mutations (E542K, E545K, or H1047R) is able to induce transformation in cultures of chicken embryo fibroblasts (CEF). We demonstrate that the transforming activity of the mutants is correlated with increased lipid kinase activity and activation of the Akt signaling pathway and depends on the target of rapamycin (TOR) kinase.
Materials and Methods
Cell Culture and Transformation Assays. Primary cultures of CEF were prepared from White Leghorn embryos obtained from Charles River Breeding Laboratories. For focus assays, DNA was transfected into CEF by using the Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. Focus assays with infectious retroviral vectors were performed as described (14, 15). Rapamycin (Calbiochem) was added directly to the nutrient agar overlay of the infected cells. Controls received DMSO vehicle instead of rapamycin. Transfected or infected cells were fed every other day with nutrient agar for 10 or 14 days, respectively, and stained with crystal violet.
Plasmid Construction. Construction of the pBSFI vector KOZ-c-p3k encoding WT chicken PI3-kinase has been described (16). The mutant constructs containing an E542K, E545K, or H1047R substitution were generated by using the Quick Change site-directed mutagenesis kit (Stratagene) and the following primers: E542K(+), 5′-GTACGCGAGATCCT T TGTCTAAAATCACTGAGCAAGAGAAGC-3′; E542K(-), 5′-CCTTCTCTTGCTCAGTGATTTTAGACAAAGGATCTCGCGTAC-3′; E545K(+), 5′-CCTTTGTCTGAAATCACTAAGCAAGAGAAGGACTTCCTC-3′; E545K(-), 5′-GAGGAAGTCCTTCTCTTGCTTAGTGATTTCAGACAAAGG-3′; H1047R(+), 5′-GA AGCA A ATGA ATGATGCTCGCCATGGTGGCTGGACAAC-3′; and H1047R(-), 5′-GTTGTCCAGCCACCATGGCGAGCATCATTCATTTGCTTG-3′.
The mutated genes were subsequently cloned as SfiI DNA fragments into the avian retrovirus vector RCAS.Sfi. Construction of the retroviral vectors, RCAS-KOZ-c-p3k (encoding wt p110α) and RCAS-myr-Δ72-c-p3k (encoding a myristylated form of p110α with a deletion of the first 72 aa), has been described (16). In this article, the particular proteins encoded by these constructs are referred to as wt PI3-kinase and myr-PI3-kinase, respectively.
Serum Starvation and Platelet-Derived Growth Factor (PDGF) Stimulation. For serum starvation, CEF were cultured in Ham's F-10 medium (Sigma) containing 0.5% FCS and 0.1% chicken serum for 40 h. The medium was then replaced with plain F-10 medium, and the cells were further incubated for 2 h. Subsequently, the cells were stimulated with 50 ng/ml PDGF (Calbiochem) for 15 min.
Western Blotting. Western blotting was performed as described (16), with minor modifications. Briefly, cells were lysed in Passive Lysis buffer (Promega) containing a protease inhibitor mixture (Roche) (1 mM PMSF/50 mM NaF/1 mM Na3VO4). Lysates containing 20 μg of proteins were separated by SDS/PAGE and transferred to Immobilon P membranes (Millipore). After the membranes were blocked with 5% BSA/PBS/0.05% Tween-20 for 1 h at room temperature, they were incubated overnight at 4°C with primary antibodies. Anti-p110α antibody (sc-7174) was purchased from Santa Cruz Biotechnology, and anti-Akt, anti-phospho-Akt (Ser-473), anti-S6K, anti-phospho-S6K (Thr-389), anti-4E-BP1, and anti-phospho-4E-BP1 (Ser-65) antibodies were obtained from Cell Signaling Technology (Beverly, MA). Anti-p19 Gag antibody was a generous gift from Volker Vogt (Cornell University, Ithaca, NY). After rinsing with PBS-T, the blots were incubated with secondary antibodies diluted in PBS-T for 1 h at room temperature. The reactive bands were visualized by chemiluminescence (Pierce), according to the manufacturer's protocol.
PI3-Kinase Assay. In vitro PI3-kinase activity was analyzed as described (16), with minor modifications. Briefly, the immune complexes were prepared by incubating 400 μg of proteins with 5 μl of anti-p110α antibody for 2 h at 4°C, followed by 2 h of incubation with Protein A-agarose. The beads were washed three times with the lysis buffer and once with TNE (10 mM Tris, pH 7.5/100 mM NaCl/1 mM EDTA). Subsequently, the immune complexes were incubated with 50 μl of kinase reaction buffer containing 20 mM Hepes (pH 7.5), 10 mM MgCl2, 200 μg/ml phosphatidylinositol (sonicated), 60 μM ATP, and 200 μCi/ml (1 Ci = 37 GBq) [γ-32P]ATP for 5 min at room temperature. The reaction was terminated by adding 80 μl of 1 N HCl, and the phosphorylated lipids were extracted with 160 μl of chloroform/methanol (1:1). After samples were dried down, they were dissolved in chloroform and spotted onto Silica Gel 60 TLC plates (Merck). The plates were developed in a borate buffer system (17) and visualized by autoradiography.
Results
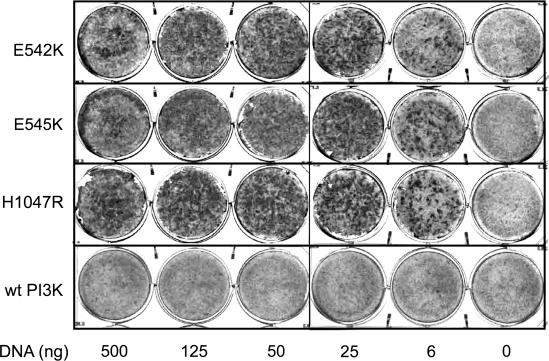
Expression of Mutant PI3-Kinase Induces Oncogenic Transformation in CEF. The three most frequent somatic missense mutations of PIK3CA observed in various cancers are E542K, E545K, and H1047R (11-13). We introduced these three mutations into the wt sequence of the chicken homolog of PIK3CA. There is strict conservation of these genes among vertebrates; 95% of avian and human protein sequences are identical, including the residues affected by mutation. The mutant PI3K genes were cloned into the retroviral expression vector RCAS and transfected into CEF. The mutant proteins induced robust cell transformation (Fig. 1). In contrast, cells transfected with the wt gene showed only few isolated foci of transformation as reported (16) (Fig. 1). These results suggest that each of the hot-spot mutations of PIK3CA is able to induce oncogenic transformation in cell culture with high efficiency.
Fig. 1.
Cellular transformation induced by mutant PI3-kinase (PI3K). CEF were transfected with RCAS vectors encoding PI3-kinase mutants or wt as indicated by using various amounts of DNA. The cultures were overlaid with nutrient agar and fixed and stained with crystal violet on day 10.
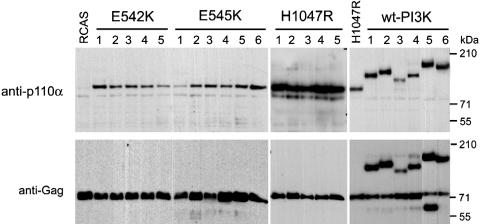
Oncogenic Transformation by Mutant PI3-Kinase Does Not Require Fusion to Viral Gag Sequences. Previous studies have shown that the catalytic subunit of wt cellular PI3-kinase does not transform cells (16). However, expression of PI3-kinase by the replication-competent retroviral vector RCAS can generate fusion proteins in which PI3-kinase is joined at the N terminus to viral Gag sequences. Such Gag fusions of PI3-kinase induce oncogenic transformation because the membrane address supplied by the Gag domain makes the cellular PI3-kinase constitutively active and independent of the regulatory subunit p85. Because the RCAS vector was also used to express mutant PI3-kinase, it was important to rule out fusion to Gag as an explanation for the transforming activity of the mutant proteins. Therefore, individual foci of PI3-kinase-transformed cells were isolated and grown into mass cultures containing ≈4 × 106 transformed cells. Protein extracts of these cultures, containing the progeny of a single viral particle, were then analyzed by Western blotting using anti-Gag and anti-PI3-kinase antibodies. No evidence of Gag fusions was seen in cultures derived from 15 individual foci induced by mutant PI3-kinase (Fig. 2). The Western blots showed only the separately expressed 76-kDa Gag protein, derived from the vector, as well as the p110α proteins that were of the expected size in each of the foci. As controls, we tested several of the rare transformed cell foci induced by wt PI3-kinase and found Gag-p110α fusions in each of these (Fig. 2). These results demonstrate the intrinsic transforming potential of mutant PI3-kinase, independent of fusion to viral Gag sequences.
Fig. 2.
Characterization of single transformed cell foci. Individual foci of transformed CEF transfected with either mutant or wt PI3-kinase (PI3K) constructs were isolated and grown until they reached confluency in 100-mm plates. After cell lysis, 20 μg of the lysates were separated on a 3-8% gradient SDS/polyacrylamide gel, and the transferred blot was probed with antibodies as indicated.
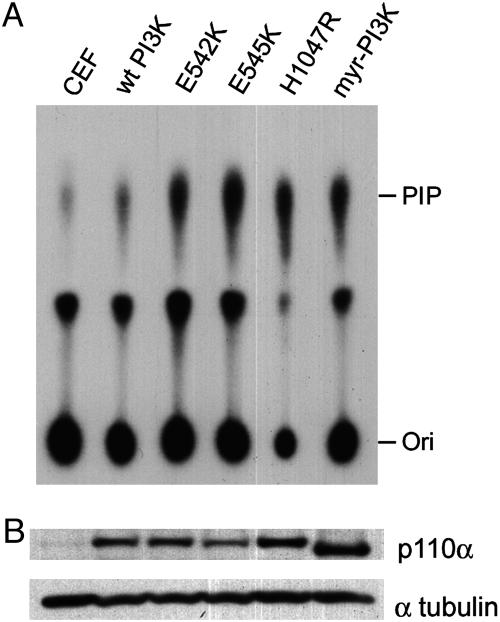
Mutant PI3-Kinase Proteins Display Enhanced Lipid Kinase Activity. PI3-kinase is a lipid kinase that catalyzes the phosphorylation of inositol phospholipids at the D3 position of the inositol ring. To determine whether this enzymatic activity is enhanced in the mutant proteins, in vitro kinase assays were performed. The levels of D3-phosphorylated inositol phospholipid, the product of PI3-kinase, were higher in CEF transfected with the mutant PI3-kinase expression than in CEF transfected with WT PI3-kinase (Fig. 3A). Mutant enzyme activities were similar to those of myristylated PI3-kinase, which has enhanced lipid kinase activity because of its constitutive membrane localization (16). These results demonstrate that mutations in the helical domain (E542K and E545K), as well as in the kinase domain (H1047R), can increase the enzymatic activity of p110α. The correlation between elevated lipid kinase and transforming activities strongly implicates gain of enzymatic function mediated by cancer-specific hot-spot mutations of PIK3CA as a dominant growth-promoting signal.
Fig. 3.
In vitro PI3-kinase activity of mutant proteins. (A) PI3-kinase (PI3K) activity was measured with the immunoprecipitated lysates of CEF that were transfected with WT, mutant (E542K, E545K, H1047R), or myristylated p110α expression construct. PIP, phosphoinositol phosphate; Ori, origin. (B) Expression of different p110α proteins. Twenty micrograms of the indicated cell lysates used for the measurement of the PI3-kinase activity were separated on a 3-8% gradient SDS/polyacrylamide gel, and the transferred blot was probed with anti-p110α antibody. Myristylated p110α expression construct contains a deletion of the first 72 aa, as noted in Materials and Methods.
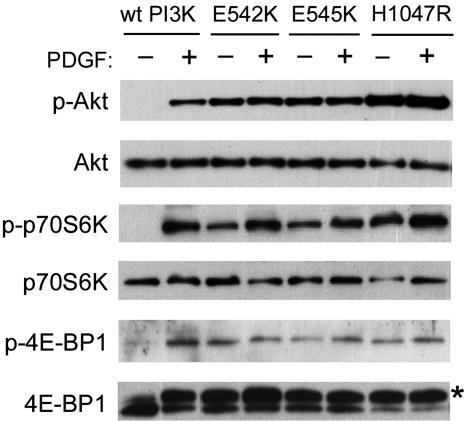
Mutant PI3-Kinase Induces Constitutive Activation of Akt Signaling in CEF. One of the downstream targets of PI3-kinase is Akt, a serine-threonine protein kinase that regulates multiple biological processes including survival, proliferation, cell cycle, and protein synthesis (18, 19). Akt binds to the product of PI3-kinase (D3-phosphorylated inositol phospholipid) with its pleckstrin homology domain and subsequently becomes activated by phosphorylation through the actions of PI3-kinase-dependent kinases, PDK1 and PDK2. To test for the enhanced Akt activity in the cells expressing the mutant PI3-kinase proteins, the levels of Akt phosphorylation were determined with phosphospecific Akt antibody. In serum-starved CEF expressing wt PI3-kinase from the RCAS vector, phosphorylation of Akt required treatment of the cells with PDGF, an activator of PI3-kinase signaling (Fig. 4). In contrast, serum-starved CEF transfected with mutant PI3-kinase showed constitutive phosphorylation of Akt. The level of this phosphorylation was even higher than the level found in the PDGF-treated CEF transfected with wt PI3-kinase (Fig. 4). A control Western blot with the phosphorylation-independent antibody against Akt showed similar Akt expression levels in wt and mutant-transfected cells. PDGF stimulation had no effect on the overall cellular Akt content (Fig. 4).
Fig. 4.
Constitutive activation of Akt signaling by mutant p110α. CEF that were transfected with wt or the mutant p110α were serum starved for 42 h and subsequently stimulated with PDGF for 15 min. Cells were then lysed, and the proteins were separated on a 4-20% gradient SDS/polyacrylamide gel. The transferred blot was probed with antibodies directed against total proteins or antibodies recognizing p-S473 Akt, p-T389 S6K, or p-S65 4E-BP1. Anti-4E-BP1 antibody detects both phosphorylated (*) and nonphosphorylated forms of the protein.
The oncogenic transforming activity of Akt correlates with the phosphorylation of the downstream targets, p70 S6 kinase (S6K) and 4E-binding protein-1 (4E-BP1) (20). The phosphorylation-mediated activation of S6K and the inhibition of 4E-BP1 results in the stimulation of mRNA translation (reviewed in refs. 21 and 22). Immunoblots with phosphospecific antibodies against S6K and 4E-BP1 revealed constitutive phosphorylation of S6K and 4E-BP1 in CEF expressing mutant PI3-kinase proteins under conditions of serum starvation (Fig. 4). In cells transfected with wt PI3-kinase, the S6K and 4E-BP1 phosphorylation levels were either undetectable or extremely low. There was no difference between WT and mutant PI3-kinase-expressing cells in the overall levels of S6K and 4E-BP1. These results suggest that the transforming activity of mutant PI3-kinase proteins may be mediated through the constitutive activation of Akt and alterations in translational controls after the phosphorylation of S6K and 4E-BP1.
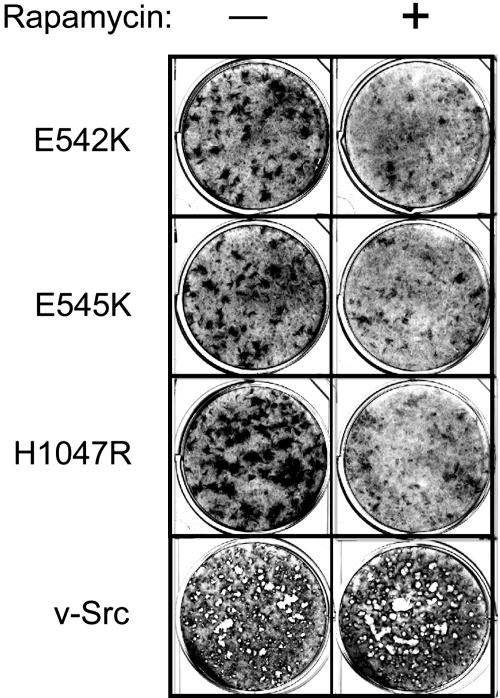
Rapamycin Inhibits Transformation Induced by Mutant PI3-Kinase Proteins. The protein kinase termed TOR is a major signaling component downstream of Akt (20, 21). TOR functions as a regulator of translation and mediates the phosphorylation of S6K and of 4E-BP1. Rapamycin, a specific and effective inhibitor of TOR, blocks oncogenic transformation induced by constitutively active PI3-kinase or Akt (20). To determine whether the mutant PI3-kinase-induced oncogenicity involves the TOR-dependent process, the inhibitory effect of rapamycin on the transforming ability of the mutant PI3-kinase was explored. As shown in Fig. 5, rapamycin also interferes with oncogenic transformation induced by the cancer-derived mutant PI3-kinase proteins, although some residual transforming activity remained at the low drug concentrations (5 ng/ml). The same concentration of rapamycin had no effect on the transforming ability of the Src oncoprotein (Fig. 5). Sensitivity to inhibition by rapamycin and constitutive activation of S6K and inactivation of 4E-BP1 implicate translational controls in the oncogenic activities of mutant PI3-kinase proteins.
Fig. 5.
Rapamycin inhibits transformation induced by mutant p110α. CEF were inoculated (100 μl) with 100-fold dilutions of retroviral vectors expressing the indicated mutant p110α or with the PR-A strain of Rous sarcoma virus expressing the v-Src oncoprotein. The cells were overlaid with nutrient agar supplemented with 5 ng/ml rapamycin or solvent only. After 14 days, the cultures were fixed and stained.
Discussion
Extensive sequencing analysis of PI3-kinase genes in human cancers has revealed mutations in the PIK3CA gene, encoding the catalytic subunit p110α of PI3-kinase. A functional significance of these mutations is suggested by four observations: (i) the mutations are nonrandomly distributed, with a high percentage found in exon 9, encoding residues of the helical domain, and in exon 20, encoding the kinase domain (11-13); (ii) most of the mutations are nonsynonymous; (iii) the mutations affect preferentially residues that are highly conserved in evolution and therefore are likely of critical functional importance; and (iv) one of the mutations has been shown to cause a gain in enzymatic activity (13).
Our studies offer strong evidence for the oncogenic potential of the mutant proteins. We introduced mutations identified in three hot spots of the human PIK3CA gene into the corresponding residues of the chicken PI3-kinase gene encoding the p110α protein. The avian p110α shows 95% identity with its human counterpart, and the three mutated residues are conserved in the two species. Expressed in CEF, these mutated p110α proteins induce oncogenic transformation (Fig. 1). Previous work has shown that wt p110α can acquire transforming activity if it is provided with a constitutive membrane address, either in the form of a myristylation or farnesylation signal or by fusion to viral Gag sequences (16). Clonal analysis of transformed cell foci induced by the mutant p110α proteins has ruled out such secondary mechanisms of activation for the mutants because they induce oncogenic transformation without any additional alterations to their structure (Fig. 2).
This intrinsic oncogenic activity of the mutant p110α proteins is also reflected in their enzymatic properties. All three mutants display enhanced lipid kinase activity as compared with the wt protein (Fig. 3). These results are consistent with previous work showing increased enzymatic activity of the human p110α protein with the H1047R mutation that is identical to one of the three mutations tested here (13). Mutations in the helical domain of p110α (E542K and E545K) also confer enhanced enzymatic activity. Structural data that could explain this effect of mutations in the helical domain are not available, but it is conceivable that the difference in charge created by the glutamate-to-lysine substitution has a conformational effect that alters enzyme activity. The strict conservation of the affected residues in the avian and human sequences suggests that the corresponding mutations in the helical domain of the human p110α protein will also cause a gain of function. The correlation between transforming potential of mutated p110α and enhanced enzymatic activity implicates abnormally active PI3-kinase in human cancer as a growth-promoting protein that can convey selective advantage to the cell.
An important downstream target of PI3-kinase is the serinethreonine kinase Akt (18, 19). Binding of Akt to the product of PI3-kinase, D3-phosphorylated inositol phospholipid, occurs at cellular membranes and facilitates the activation of Akt by two kinases: PDK1, phosphorylating threonine 308 (23, 24), and DNA-dependent protein kinase, formerly called PDK2, targeting serine 473 of Akt (25). Increased PI3-kinase activity commonly induces elevated levels of phosphorylated Akt (16, 26, 27). The mutant PI3-kinase proteins meet this expectation; in cells expressing these proteins, Akt is constitutively phosphorylated, both in the presence and absence of growth factor (Fig. 4). In contrast, in cells expressing wt p110α, Akt is activated only in the presence of PDGF. Akt affects numerous cellular processes related to growth and survival (reviewed in refs. 19, 28, and 29). It plays an important role in the resistance of cancer cells to drug-induced apoptosis (30, 31) and mediates changes in protein synthesis that are essential for maintaining the oncogenic cellular phenotype (20). The effect of Akt on protein synthesis is reflected in constitutive phosphorylation of S6K and 4E-BP1 (20). S6K is activated by phosphorylation; it selectively affects the translation of specific mRNAs by unknown mechanisms (32-34). The 4E-BP1 is a negative regulator of translation; its inactivation by phosphorylation increases the supply of the eukaryotic initiation factor 4E (35-38). Numerous mRNAs that encode growth-promoting proteins depend on amply available 4E for efficient translation (39, 40). Cells expressing the mutant p110α proteins show constitutive phosphorylation of both S6K and 4E-BP1 (Fig. 4), suggesting the involvement of aberrant translational control in mutant PI3-kinase-induced oncogenic transformation.
The phosphorylation of S6K and 4E-BP1 requires the activity of the protein kinase TOR (reviewed in refs. 21 and 22). Rapamycin, an inhibitor of TOR, interferes with this phosphorylation. Rapamycin also inhibits the mutant-induced cellular transformation (Fig. 5), suggesting that this transformation depends on TOR and its downstream targets that control protein synthesis. High sensitivity to the rapamycin derivative CCI-779 has previously been detected in cells deficient in PTEN, the phosphatase antagonist of PI3-kinase, and in tumors of PTEN heterozygous mice (41, 42). In the absence or reduction of PTEN, the activity of the PI3-kinase-Akt signaling pathway is elevated. The growth regulatory defect in these cells therefore resembles that in cells transformed by the PI3-kinase gain-of-function mutants.
A kinase with a cancer-specific somatic mutation is an ideal target for a small molecule inhibitor that could be developed into an effective anticancer drug. Specificity of such inhibitor for the mutant enzyme would minimize undesirable side effects that could arise from interference with the functions of the wt protein. Recent clinical experience with the cancer drugs Gleevec (Imatinib), directed against the Bcr-Abl oncoprotein (43, 44), and Iressa (Gefitinib), effective against mutants of the EGF receptor (45, 46), suggest that synthesizing and optimizing such inhibitors is an achievable goal.
Acknowledgments
We thank Dr. Masahiro Aoki for technical advice on the PI3-kinase assay and Hershey Filoteo for assistance in cell culture. This work was supported by grants from the National Cancer Institute. This work is manuscript no. 17013-MEM of The Scripps Research Institute.
Author contributions: P.K.V. designed research; S.K. and A.G.B. performed research; S.K., A.G.B., and P.K.V. analyzed data; and S.K., A.G.B., and P.K.V. wrote the paper.
Abbreviations: CEF, chicken embryo fibroblasts; PI3-kinase, phosphatidylinositol 3-kinase; TOR, target of rapamycin; PDGF, platelet-derived growth factor; S6K, p70 S6 kinase; 4E-BP, eukaryotic translational initiation factor 4E-binding protein; wt, wild type.
References
- 1.Shayesteh, L., Lu, Y., Kuo, W. L., Baldocchi, R., Godfrey, T., Collins, C., Pinkel, D., Powell, B., Mills, G. B. & Gray, J. W. (1999) Nat. Genet. 21, 99-102. [DOI] [PubMed] [Google Scholar]
- 2.Philp, A. J., Campbell, I. G., Leet, C., Vincan, E., Rockman, S. P., Whitehead, R. H., Thomas, R. J. & Phillips, W. A. (2001) Cancer Res. 61, 7426-7429. [PubMed] [Google Scholar]
- 3.Cantley, L. C. & Neel, B. G. (1999) Proc. Natl. Acad. Sci. USA 96, 4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang, S. I., Puc, J., Li, J., Bruce, J. N., Cairns, P., Sidransky, D. & Parsons, R. (1997) Cancer Res. 57, 4183-4186. [PubMed] [Google Scholar]
- 5.Yokoyama, Y., Wan, X., Shinohara, A., Takahashi, S., Takahashi, Y., Niwa, K. & Tamaya, T. (2000) Int. J. Mol. Med. 6, 47-50. [DOI] [PubMed] [Google Scholar]
- 6.Celebi, J. T., Shendrik, I., Silvers, D. N. & Peacocke, M. (2000) J. Med. Genet. 37, 653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringel, M. D., Hayre, N., Saito, J., Saunier, B., Schuppert, F., Burch, H., Bernet, V., Burman, K. D., Kohn, L. D. & Saji, M. (2001) Cancer Res. 61, 6105-6111. [PubMed] [Google Scholar]
- 8.Bellacosa, A., de Feo, D., Godwin, A. K., Bell, D. W., Cheng, J. Q., Altomare, D. A., Wan, M., Dubeau, L., Scambia, G. & Masciullo, V. (1995) Int. J. Cancer 64, 280-285. [DOI] [PubMed] [Google Scholar]
- 9.Bellacosa, A., Testa, J. R., Staal, S. P. & Tsichlis, P. N. (1991) Science 254, 274-277. [DOI] [PubMed] [Google Scholar]
- 10.Chang, H. W., Aoki, M., Fruman, D., Auger, K. R., Bellacosa, A., Tsichlis, P. N., Cantley, L. C., Roberts, T. M. & Vogt, P. K. (1997) Science 276, 1848-1850. [DOI] [PubMed] [Google Scholar]
- 11.Bachman, K. E., Argani, P., Samuels, Y., Silliman, N., Ptak, J., Szabo, S., Konishi, H., Karakas, B., Blair, B. G., Lin, C., et al. (2004) Cancer Biol. Ther. 3, e49-e52. [DOI] [PubMed] [Google Scholar]
- 12.Broderick, D. K., Di, C., Parrett, T. J., Samuels, Y. R., Cummins, J. M., McLendon, R. E., Fults, D. W., Velculescu, V. E., Bigner, D. D. & Yan, H. (2004) Cancer Res. 64, 5048-5050. [DOI] [PubMed] [Google Scholar]
- 13.Samuels, Y., Wang, Z., Bardelli, A., Silliman, N., Ptak, J., Szabo, S., Yan, H., Gazdar, A., Powell, S. M., Riggins, G. J., et al. (2004) Science 304, 554. [DOI] [PubMed] [Google Scholar]
- 14.Bos, T. J., Monteclaro, F. S., Mitsunobu, F., Ball, A. R., Jr., Chang, C. H., Nishimura, T. & Vogt, P. K. (1990) Genes Dev. 4, 1677-1687. [DOI] [PubMed] [Google Scholar]
- 15.Duff, R. G. & Vogt, P. K. (1969) Virology 39, 18-30. [DOI] [PubMed] [Google Scholar]
- 16.Aoki, M., Schetter, C., Himly, M., Batista, O., Chang, H. W. & Vogt, P. K. (2000) J. Biol. Chem. 275, 6267-6275. [DOI] [PubMed] [Google Scholar]
- 17.Walsh, J. P., Caldwell, K. K. & Majerus, P. W. (1991) Proc. Natl. Acad. Sci. USA 88, 9184-9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheid, M. P. & Woodgett, J. R. (2001) Nat. Rev. Mol. Cell. Biol. 2, 760-768. [DOI] [PubMed] [Google Scholar]
- 19.Vivanco, I. & Sawyers, C. L. (2002) Nat. Rev. Cancer 2, 489-501. [DOI] [PubMed] [Google Scholar]
- 20.Aoki, M., Blazek, E. & Vogt, P. K. (2001) Proc. Natl. Acad. Sci. USA 98, 136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris, T. E. & Lawrence, J. C., Jr. (2003) Sci. STKE 2003, re15. [DOI] [PubMed] [Google Scholar]
- 22.Hay, N. & Sonenberg, N. (2004) Genes Dev. 18, 1926-1945. [DOI] [PubMed] [Google Scholar]
- 23.Alessi, D. R., James, S. R., Downes, C. P., Holmes, A. B., Gaffney, P. R., Reese, C. B. & Cohen, P. (1997) Curr. Biol. 7, 261-269. [DOI] [PubMed] [Google Scholar]
- 24.Williams, M. R., Arthur, J. S., Balendran, A., van der Kaay, J., Poli, V., Cohen, P. & Alessi, D. R. (2000) Curr. Biol. 10, 439-448. [DOI] [PubMed] [Google Scholar]
- 25.Feng, J., Park, J., Cron, P., Hess, D. & Hemmings, B. A. (2004) J. Biol. Chem. 279, 41189-41196. [DOI] [PubMed] [Google Scholar]
- 26.Liu, A. X., Testa, J. R., Hamilton, T. C., Jove, R., Nicosia, S. V. & Cheng, J. Q. (1998) Cancer Res. 58, 2973-2977. [PubMed] [Google Scholar]
- 27.Franke, T. F., Yang, S. I., Chan, T. O., Datta, K., Kazlauskas, A., Morrison, D. K., Kaplan, D. R. & Tsichlis, P. N. (1995) Cell 81, 727-736. [DOI] [PubMed] [Google Scholar]
- 28.Downward, J. (2004) Semin. Cell. Dev. Biol. 15, 177-182. [DOI] [PubMed] [Google Scholar]
- 29.Fresno Vara, J. A., Casado, E., de Castro, J., Cejas, P., Belda-Iniesta, C. & Gonzalez-Baron, M. (2004) Cancer Treat. Rev. 30, 193-204. [DOI] [PubMed] [Google Scholar]
- 30.Bacus, S. S., Altomare, D. A., Lyass, L., Chin, D. M., Farrell, M. P., Gurova, K., Gudkov, A. & Testa, J. R. (2002) Oncogene 21, 3532-3540. [DOI] [PubMed] [Google Scholar]
- 31.Knuefermann, C., Lu, Y., Liu, B., Jin, W., Liang, K., Wu, L., Schmidt, M., Mills, G. B., Mendelsohn, J. & Fan, Z. (2003) Oncogene 22, 3205-3212. [DOI] [PubMed] [Google Scholar]
- 32.Dufner, A. & Thomas, G. (1999) Exp. Cell Res. 253, 100-109. [DOI] [PubMed] [Google Scholar]
- 33.Jefferies, H. B., Fumagalli, S., Dennis, P. B., Reinhard, C., Pearson, R. B. & Thomas, G. (1997) EMBO J. 16, 3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwab, M. S., Kim, S. H., Terada, N., Edfjall, C., Kozma, S. C., Thomas, G. & Maller, J. L. (1999) Mol. Cell. Biol. 19, 2485-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunn, G. J., Hudson, C. C., Sekulic, A., Williams, J. M., Hosoi, H., Houghton, P. J., Lawrence, J. C., Jr., & Abraham, R. T. (1997) Science 277, 99-101. [DOI] [PubMed] [Google Scholar]
- 36.Gingras, A. C., Gygi, S. P., Raught, B., Polakiewicz, R. D., Abraham, R. T., Hoekstra, M. F., Aebersold, R. & Sonenberg, N. (1999) Genes Dev. 13, 1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gingras, A. C., Kennedy, S. G., O'Leary, M. A., Sonenberg, N. & Hay, N. (1998) Genes Dev. 12, 502-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burnett, P. E., Barrow, R. K., Cohen, N. A., Snyder, S. H. & Sabatini, D. M. (1998) Proc. Natl. Acad. Sci. USA 95, 1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gingras, A. C., Raught, B. & Sonenberg, N. (1999) Annu. Rev. Biochem. 68, 913-963. [DOI] [PubMed] [Google Scholar]
- 40.Sonenberg, N. & Gingras, A. C. (1998) Curr. Opin. Cell Biol. 10, 268-275. [DOI] [PubMed] [Google Scholar]
- 41.Neshat, M. S., Mellinghoff, I. K., Tran, C., Stiles, B., Thomas, G., Petersen, R., Frost, P., Gibbons, J. J., Wu, H. & Sawyers, C. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Podsypanina, K., Lee, R. T., Politis, C., Hennessy, I., Crane, A., Puc, J., Neshat, M., Wang, H., Yang, L., Gibbons, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Druker, B. J., Sawyers, C. L., Kantarjian, H., Resta, D. J., Reese, S. F., Ford, J. M., Capdeville, R. & Talpaz, M. (2001) N. Engl. J. Med. 344, 1038-1042. [DOI] [PubMed] [Google Scholar]
- 44.O'Brien, S. G., Guilhot, F., Larson, R. A., Gathmann, I., Baccarani, M., Cervantes, F., Cornelissen, J. J., Fischer, T., Hochhaus, A., Hughes, T., et al. (2003) N. Engl. J. Med. 348, 994-1004. [DOI] [PubMed] [Google Scholar]
- 45.Pao, W., Miller, V., Zakowski, M., Doherty, J., Politi, K., Sarkaria, I., Singh, B., Heelan, R., Rusch, V., Fulton, L., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 13306-13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natale, R. B. (2004) Semin. Oncol. 31, 23-30. [DOI] [PubMed] [Google Scholar]