Abstract
Salmonella enterica serovar Typhimurium LT2 harbors four temperate prophages. The lytic cycle of these phages was induced with hydrogen peroxide or mitomycin C. Microarray analysis was used to monitor the increase in phage genome copy number and the changes in RNA expression. Phage gene transcription was classified temporally, and host genes that responded to hydrogen peroxide, mitomycin C, or phage induction were also identified. A region of the serovar Typhimurium LT2 host genome encompassing hundreds of genes, flanking the Fels-1 lambdoid prophage, was amplified manyfold during lytic induction, presumably due to Fels-1 runoff replication prior to excision, a phenomenon termed escape replication. An excisionase (xis) mutant of Fels-1 also induced escape replication but did not get packaged. Gifsy-1, a lambdoid prophage that does not normally produce escape replication, did so after deletion of either its integrase or excisionase genes. Escape replication is probably widespread; large regions of host genome amplification were also observed after phage induction in serovar Typhimurium strains SL1344 and 14028s at the suspected integration site of prophage genomes.
Bacteriophages play an important role in the fitness of bacteria, in both negative and positive ways. Many phages of bacterial pathogens are known to carry virulence factors essential for successful pathogenesis of the bacterium (4). Bacteriophages are recognized as major vehicles for the lateral transfer of genes (6, 7, 10, 17, 18), and most sequenced bacterial genomes contain lysogenic phages, as well as remnants of phages. The life strategies and niches that can be occupied by bacterial strains are probably influenced by the collection of lysogenic phages associated with the strains and the flexibility and mobility of phenotypes that phages impart to the species.
In previous studies workers mapped the boundaries of four known lysogenic phages in Salmonella enterica Typhimurium LT2 by hybridizing genomic DNA from strains cured of the phages to DNA microarrays containing all protein-encoding genes in the genome (16). Here we show that induction and replication of all four lysogenic phages in LT2 can be observed by microarray analysis of the DNA content after treatment with peroxide or mitomycin C. This analysis revealed large host genome amplifications adjacent to some known and suspected phage integration sites. These amplifications are presumably a consequence of escape replication, where phage replication occurs prior to excision. This phenomenon was observed genetically decades ago (9, 27), but until now its extent had not been measured by physical methods. Deletion of integrase and excisionase genes prevents independent phage genome replication and induces escape replication in lambdoid phage (2), and we monitored this on a microarray. Gene expression was also measured by microarray analysis during the lytic cycle to reveal changes in host gene expression caused by peroxide, by mitomycin C treatment, or by the replicating phage. Changes in transcription of phage genes during the course of induction were also recorded by using microarrays, as previously shown for lytic bacteriophage T4 (15).
MATERIALS AND METHODS
Strains and growth conditions.
S. enterica serovar Typhimurium strains were grown by standard methods in Luria broth with shaking in a gyratory shaking incubator (New Brunswick, Edison, N.J.) at 220 rpm and 37°C. The strains used in this study included LT2 (Fels-1+, Fels-2+, Gifsy-2+, and Gifsy-1+), 14028s (Gifsy-2+, Gifsy-1+, and Gifsy-3+), and SL1344 (Gifsy-2+, Gifsy-1+, and SopEφ+). Knockout mutants of Fels-1, Gifsy-1, and Gifsy-2 int and xis genes were constructed, as described previously (5), with the following primers: STM0894-F (5′CGTCCAAGAGCCTCATCAGATTACCCTCTCTTCAGAAAATGTGTAGGCTGGAGCTGCTTC3′), STM0894-R (5′ACGTGGTGCCACTGCCGTGGCCCTGATGTGGTATCAGGCGCATATGAATATCCTCCTTAG3′), STM0893-F (5′CCTTTCAGTGATTCGATAACCACTTAACATCTTGTTTTATGTGTAGGCTGGAGCTGCTTC3′), STM0893-R (5′CCGGATGTCGATCTTTGACTTATTTTCTGAAGAGAGGGTACATATGAATATCCTCCTTAG3′), STM1006-F (5′CCCTCCTGACGTCCAGGAGCATTGACGAGTGTACAGCTTTGTGTAGGCTGGAGCTGCTTC3′), STM1006-R (5′TATTAGCGATGGCCCGCTGCGGGGCCACTGGAGAAAACGACATATGAATATCCTCCTTAG3′), STM1005-F (5′TGTAGCGATGCGACTGCTAACCCCTTGAATTTAAGGATTTGTGTAGGCTGGAGCTGCTTC3′), STM1005-R (5′CTGTACACTCGTCAATGCTCCTGGACGTCAGGAGGGATTACATATGAATATCCTCCTTAG3′), STM2635+2636-F (5′GAATATTAGCGATGGCCCGCTGCGGGGCCACTGGAGAGTGTAGGCTGGAGCTGCTTC3′), and STM2635+2636-R (5′AATCTTCCGTTCTGATGGACACATGCAGGGATAAATCATGCATATGAATATCCTCCTTAG3′). Knockouts were confirmed by PCR analysis with the following primers: STM0893T-F (5′CCTTTCAGTGATTCGATAACC3′), STM0893T-R (CCGGATGTCGATCTTTGACTT3′), STM0894T-F (CGTCCAAGAGCCTCATCAGAT3′), STM0894T-R (5′AGGTGGTGCCACTCCCGTGGC3′), STM1006T-F (5′CCCTCCTGACGTCCAGGAGCA3′), STM1006T-R (5′TATTAGCGATGGCCCGCTGCG3′), STM1005T-F (5′TGTAGCGATGCGACTGCTAAC3′), STM1005T-R (5′CTGTACACTCGTCAATGCTCC3′), STM2635+2636T-F (5′GAATATTAGCGATGGCCCGCT3′), and STM2635+2636T-R (5′AATCTTCCGTTCTGATGGACA3′). The mutants constructed included JF105 (LT2 STM0894 Fels-1 xis::Kan), JF106 (LT2 STM0893 Fels-1 int::Cm), JF107 (LT2 STM1006 Gifsy-2 xis::Kan), JF108 (LT2 STM1005 Gifsy-2 int::Kan), and JF109 (STM2635-2636 Gifsy-1 xis-int::Kan).
Peroxide and mitomycin C assays.
Overnight cultures of the bacterial strains were diluted 1:50 and grown in 100 ml of Luria broth to an optical density at 600 nm of 0.35 to 0.4. DNA and RNA from untreated cells at this optical density (time zero) were extracted for comparison to the DNA and RNA from cells exposed for 5, 10, 20, 30, 40, 60, 120, and 180 min to 2 mM hydrogen peroxide or 2 μg of mitomycin C per ml (12, 20, 29). DNA was extracted from 15 ml of culture with a GenElute bacterial genomic DNA kit (Sigma, St. Louis, Mo.). RNA was prepared from 35 ml of culture by a hot phenol extraction protocol as previously described (20).
Phage isolation.
Cells grown to optical density at 600 nm of 0.35 to 0.4 were treated for 5 h with 2 μg of mitomycin C per ml. Cells were removed by centrifugation at 3,220 × g for 20 min at 4°C twice, and 30 ml of the supernatant was subjected to polyethylene glycol (PEG) precipitation (5% PEG 8000, 0.5 M NaCl) to isolate phage particles. DNA was extracted from the pellet with a GenElute bacterial genomic DNA kit. In total, 250 ng of recovered phage DNA was labeled and subsequently hybridized to the array.
Microarray construction, DNA and RNA labeling, hybridization, data acquisition, and data analysis.
Details concerning the construction of the Salmonella microarray, DNA and RNA labeling, hybridization, data acquisition, and data analysis were described previously (20). In brief, the microarray contained 4,453 whole open reading frame PCR products representing 98.9% of the S. enterica serovar Typhimurium LT2 genome and the pSLT plasmid, spotted in triplicate onto Corning Ultra-GAPS glass slides (Corning Inc., Corning, N.Y.). The median of at least three hybridization ratios per gene was recorded, and genes with signals that were less than 2 standard deviations greater than the background signal at all times were considered not detected and were removed before graphical or tabular presentation. The significance of differential expression, relative to the control, was assessed by using Cyber-T (14). Changes in gene expression were analyzed by GENESIS (25) and CAGED (22) to produce expression profiles and to perform clustering of the results. All the raw microarray data for experiments discussed below have been deposited at http://www.ncbi.nlm.nih.gov/geo under accession numbers GSE622 to GSE625, GSE806, and GSE811.
RESULTS
Phage genome amplification during the lytic cycle in S. enterica serovar Typhimurium LT2.
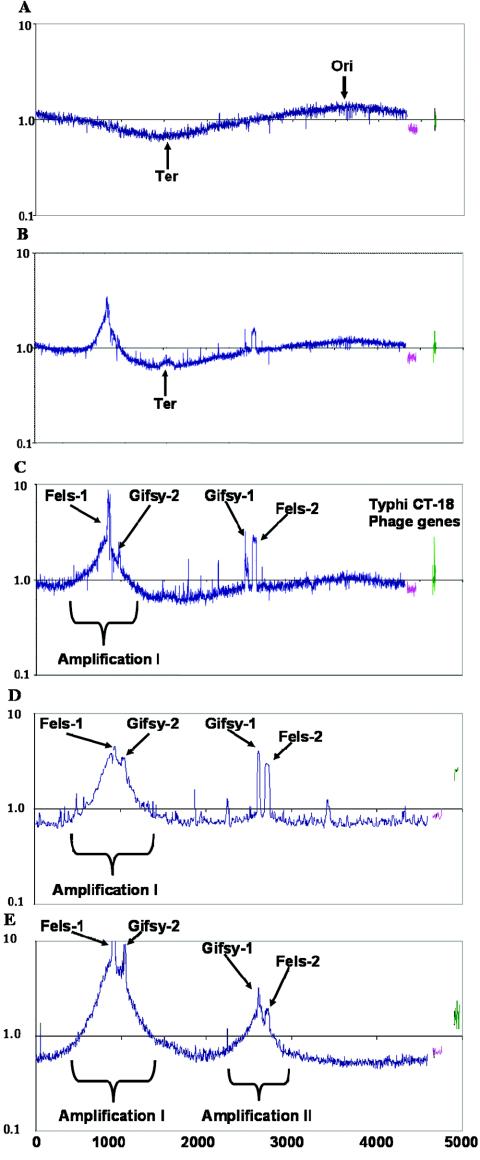
Agents that lead to DNA damage, such as hydrogen peroxide and mitomycin C, induce many lysogenic bacteriophages, often through cleavage of a phage repressor protein by activated RecA during the initiation of the SOS response (23). The lytic cycle of the four lysogenic phages present in S. enterica serovar Typhimurium strain LT2 (Fels-1, Gifsy-1, Gifsy-2, and Fels-2) was induced by hydrogen peroxide, and microarray analysis was used to measure phage and host DNA and RNA profiles. Genomic DNA was harvested from cells at 0, 60, 120, 180, and 300 min after phage induction and was compared to DNA from untreated cells. No increase in the copy number of phage genes was detected 1 h after treatment (Fig. 1A). However, at 2 and 3 h after treatment all four phage genomes were detected in higher copy numbers than the rest of the bacterial genome (Fig. 1B and C). The copy number of phage DNA within cells decreased thereafter. Fels-1 appeared to be amplified at a maximum ratio of almost ninefold relative to the genome, whereas the values for the other phages varied from two- to threefold. The observed sigmoid shape of the plotted ratios for the host genome in Fig. 1 indicates differences in growth rate between the control cells and the experimental cells, which resulted in increased gene copy number near the origin and reduced copy number near the terminus in the experimental sample (20).
FIG. 1.
Serovar Typhimurium strains after treatment with 2 mM hydrogen peroxide or 2 μg of mitomycin C per ml. The y axis indicates the ratio of the DNA content of treated cells to the DNA content of cells before treatment on a log10 scale. The x axis indicates the S. enterica serovar Typhimurium LT2 genes in order of position on the chromosome (blue), the pSLT genes (pink), and select CT18 phage genes (green). The origin (Ori) and terminus (Ter) of replication are indicated. (A) LT2 1 h after treatment with peroxide; (B) LT2 2 h aftertreatment with peroxide; (C) LT2 3 h after treatment with peroxide; (D) LT2 Fels-1 xis mutant JF105 (STM0894::kan) 3 h after treatment with mitomycin C; (E) LT2 Gifsy-1 xis and int double mutant JF109 (STM2635-STM2636::kan) 3 h after treatment with mitomycin C.
Host genomic DNA is amplified around Fels-1 during the lytic cycle in LT2.
An area of about 800 genes surrounding Fels-1 (amplification I, STM0450 to STM1250) showed an increase in gene copy number (Fig. 1B and C). Excluding the genes encoding the Fels-1 and Gifsy-2 prophages, this area was symmetrically amplified a maximum of threefold centered at the Fels-1 genome. This phenomenon, termed escape replication, was previously observed genetically for specific genes near other lambdoid phages (9, 27). In a few lambdoid phages, DNA replication can take place prior to excision from the host genome, extending bidirectionally and terminating randomly within adjacent host genomic DNA. However, the physical extent of this amplification has never been mapped for any lambdoid phage. As only a small fraction of cells may induce phage, the amplification in these cells is presumably much greater than that shown in Fig. 1. Both Gifsy-1 and Gifsy-2 are also lambdoid phages, but they show no detectable amplification outside their respective genomes. Another small region of increased gene content (best seen in Fig. 1B) was centered at the termination region of genome replication. The cause of this deviation is not known. Mitomycin C treatment resulted in a profile very similar to that observed with hydrogen peroxide, but the onset was somewhat earlier and the DNA amplification was greater (data not shown).
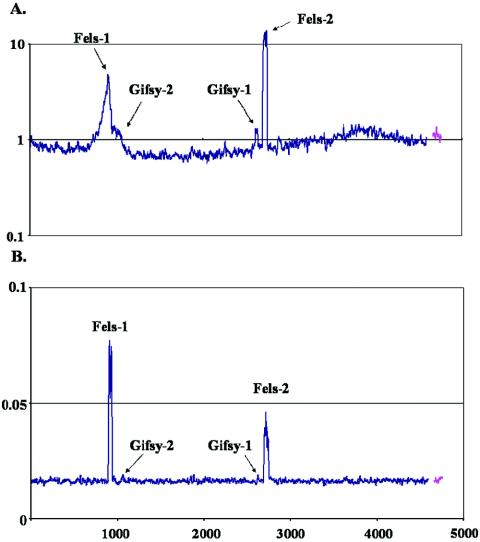
To determine whether Fels-1 was packaged and exported, growth medium from cells after 5 h of mitomycin C treatment was centrifuged to remove cells, and the supernatant was subjected to PEG precipitation to isolate phage particles. DNA was extracted from the PEG precipitate, labeled, and applied to an array. In addition, DNA was also extracted from the cells and hybridized to the array. Fels-1 and Fels-2 were detected in the supernatants, and their independent replication was still apparent in the DNA of the cells, including escape replication around Fels-1 (Fig. 2). However, the Fels-1 DNA detected in the supernatant was packaged correctly (no adjacent host genes were detected). The Gifsy phages were apparently not induced as well under these conditions, a phenomenon previously observed for mitomycin C-treated cells (6).
FIG. 2.
Fels phage particles released after mitomycin C treatment. LT2 cells growing in Luria-Bertani medium were treated with mitomycin C for 5 h. Subsequently, cells were separated from the supernatant, and phage were harvested from the supernatant fraction by PEG precipitation. DNA was isolated from both the cells and the supernatants. (A) LT2 cells 5 h after treatment with mitomycin C. The y axis indicates the ratio of the DNA content of treated cells to the DNA content of cells before treatment on a log10 scale. (B) Phage DNA harvested from LT2 supernatants 5 h after treatment with mitomycin C. The y axis indicates the contribution of every gene to the total signal of all genes represented on the microarray (expressed as percentages).
Phage and host genome amplification is not detectable in strains cured of and reinfected with Fels-1 and Fels-2.
LT2 derivatives that were cured of Fels-1 and Fels-2 by treatment with Daunorubicin and then reinfected with only Fels-1 or Fels-2 (TAQ100F1 and TAQ100F2, respectively) (1) were also tested for phage induction. In spite of having apparently normal DNA damage profiles in the Ames test (1), these cured strains did not induce any phage genomes with either peroxide or mitomycin C treatment in multiple independent experiments (data not shown). Perhaps the reintegrated phages were mutants. Alternatively, the strains cured of one or more phages by agents such as Daunorubicin may have been unable to induce any phage in numbers large enough to detect by DNA microarray analysis. Conceivably, curing may have selected for cells that were poor at sustaining phage replication; otherwise, the cells would have been lysed during curing. Alternatively, strains resistant to the action of phage-inducing agents, such as peroxide and mitomycin C, may have been selected for by Daunorubicin treatment during the curing process.
Analysis of genomic amplification by phage with deleted integrase and/or excisionase genes.
Excision generally requires the phage-encoded Int and Xis proteins (2). To determine if the excisionase and integrase genes of Fels-1 were responsible for the amplification of adjacent host genes, knockout mutants with mutations in the xis and int genes in Fels-1 were constructed and analyzed. The mutation in excisionase resulted in no change to amplification I, as shown in Fig. 1D. However, replication of the phage genome independent of the host genome was not detected, suggesting that the mutation inactivated the phage. The Fels-1 integrase mutant gave similar results (data not shown). To investigate why some lambdoid phages and not others induced escape replication, knockout mutants with mutations in the xis and int genes were also constructed in Gifsy-1 and Gifsy-2. As shown in Fig. 1E, deletion of xis and int in Gifsy-1 resulted in the induction of escape replication (amplification II). The Gifsy-2 mutants were also assayed; however, if escape replication occurred, it was obscured by the Fels-1 amplification (data not shown).
Lytic cycle in other serovar Typhimurium strains.
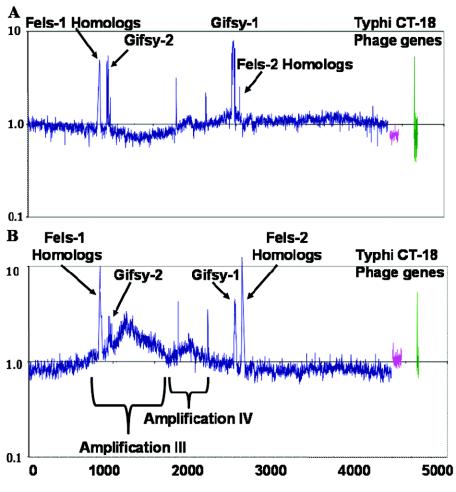
Serovar Typhimurium strains SL1344 and 14028s are known to be more virulent than strain LT2, which has a mutation in rpoS (28), and these other strains are often used in virulence assays in the laboratory (28). Both the SL1344 and 14028s genomes contain Gifsy-1 and Gifsy-2 but not Fels-1 or Fels-2 prophage (7). SL1344 and 14028s are also known to contain phages SopEφ and Gifsy-3, respectively (7). All of these prophages have been shown to encode putative virulence factors (6, 7, 10, 17, 18, 24). To determine if virulent strains also undergo phage induction or genome amplification, these strains were treated with peroxide and mitomycin C as described above. Gifsy-1 and Gifsy-2 prophages were induced in both SL1344 and 14028s 3 h after peroxide treatment (Fig. 3). Mitomycin C treatment yielded similar results (data not shown). As observed with LT2, no change in host gene copy number was observed around Gifsy-1 or Gifsy-2 in SL1344. Similarly, no escape replication occurred around the putative integration site of the unsequenced P2 phage SopEφ, which was previously mapped near centisome 60 (close to the STM2772 gene) (6, 7, 10, 17, 18). A small region of genome amplification was observed in SL1344 around pseudogene STM2003 (homologous to a phage gene).
FIG. 3.
Serovar Typhimurium strains SL1344 and 14028s after treatment with 2 mM peroxide. The y axis indicates the ratio of DNA content 3 h after treatment to the DNA content of cells before treatment on a log10 scale. The x axis indicates the S. enterica serovar Typhimurium genes in order of position on the chromosome. (A) SL1344; (B) 14028s.
Strain 14028s displayed two large areas of genome amplification during phage induction. One region was amplified approximately threefold and encompassed roughly 800 genes (region III, STM0800 to STM1600) (Fig. 3B). It was centered at icdA (STM1238), an isocitrate dehydrogenase gene homologous to a gene found in the Escherichia coli e14 prophage. Notably, icdA is located between genes encoding the virulence regulators phoPQ and the virulence factors msgA, pagD, and pagC (16). This location may be the integration site of the unsequenced lambdoid prophage, Gifsy-3, which was mapped at centisome 27-28 (7). The Gifsy-3 prophage contains the putative virulence factors pagJ and sspH1 (7). It is likely that induction of the lambdoid Gifsy-3 prophage followed by escape replication is responsible for the increased copy number of genes in this region. The other area of bacterial genome amplification after peroxide treatment in strain 14028s, region IV, included about 350 genes (STM1750 to STM2100) (Fig. 3B). This amplification seemed to be in the same location as the weak amplification observed in strain SL1344, centered at pseudogene STM2003 (Fig. 2B). Amplification IV was located near sopE2, an area previously identified as a putative phage integration site (26), and it is possible that this region was amplified by escape replication of an uncharacterized prophage in 14028s and SL1344. However, a search of SL1344 genome sequence data (http://www.sanger.ac.uk/Projects/Salmonella/) revealed no phage or other insertions at this location compared to the LT2 genome (16). Amplifications III and IV demonstrated that microarray analysis can detect changes in genome structure that may be indicative of novel prophages inserted into the genomes of unsequenced strains.
Detection of amplified phage genes by homologous gene reporters.
In addition to genes known to be parts of lysogenic phages, several other genes were also detected at higher copy numbers in the serovar Typhimurium strains after hydrogen peroxide treatment (Fig. 1). For LT2, the PCR reporters for these genes invariably revealed significant homology with genes in the amplified phages. Therefore, cross-hybridization effects explain why these genes were detected as amplified. Generally, these genes displayed greater than 85% identity over the whole gene or 90% identity over a 100-bp window (a complete list of these reporters is provided in Table S1 in the supplemental material). Several reporters on the array detected homologous amplified genes in SL1344 that were not previously known to be present in this strain, including five Fels-1, two Fels-2, and two S. enterica serovar Typhi CT18 genes (Fig. 3A; also see Table S1 in the supplemental material). Similarly, several unknown amplified genes were observed in strain 14028s. It is likely that this hybridization was due to homologs of genes that occur in the SL1344 and/or 14028s genomes, most probably within a prophage, such as SopEφ, Gifsy-3, or an unknown prophage (7). The detection of these amplified phage genes supports the possibility that amplifications III and IV are the result of prophage insertion at these sites and that the prophages contain genes homologous to these Fels-1, Fels-2, and CT18 gene reporters.
RNA expression analysis of host and phage genomes.
RNA was harvested in the experiments whose results are presented in Fig. 1 and 3 and was analyzed by DNA microarray hybridization to determine whether host genes in the areas of the genome amplifications identified in this study had increased expression. The change in transcription compared to time zero was determined for 200 host genes in the center of each amplified region 2 and 3 h after hydrogen peroxide treatment. No increase in the average transcription in the amplified bacterial genomic regions was detected, except in region III in one strain, 14028s, where the average gene expression was 2.9-fold higher after 2 h of treatment and 3-fold higher after 3 h of treatment compared to untreated controls (Table 1). For comparison, both Gifsy-1 and Gifsy-2 displayed levels of average gene expression that were markedly elevated (more than eightfold) after 2 or 3 h of treatment. However, it is not clear if these observations were due to increased gene copy number (gene dosage) or to the SOS response caused by the compounds used to induce the phages.
TABLE 1.
Average change in gene content and expression after peroxide treatment in strain 14028s
| Genome, phage, or amplificationa | Avg changec
|
|||
|---|---|---|---|---|
| 2-hr DNA | 2-hr RNA | 3-hr DNA | 3-hr RNA | |
| Genome (not including phage) | 1.0 | 1.9 | 1.0 | 2.4 |
| Gifsy-2 (STM1005-STM1056) | 1.7 | 12.5 | 1.8 | 13.9 |
| Gifsy-1 (STM2584-STM2636) | 2.7 | 12.8 | 2.8 | 8.0 |
| Amplification I (STM0891)b | 1.2 | 1.6 | 1.2 | 2.0 |
| Amplification III (STM1238) | 2.3 | 2.9 | 2.2 | 3.0 |
| Amplification IV (STM2003) | 1.3 | 2.0 | 1.4 | 2.2 |
Amplifications were measured for 200 genes flanking the peak, excluding any prophage genes.
DNA amplification was not present in the strain.
Average value for the peroxide-treated preparation, divided by the value for the control, for the gene regions indicated.
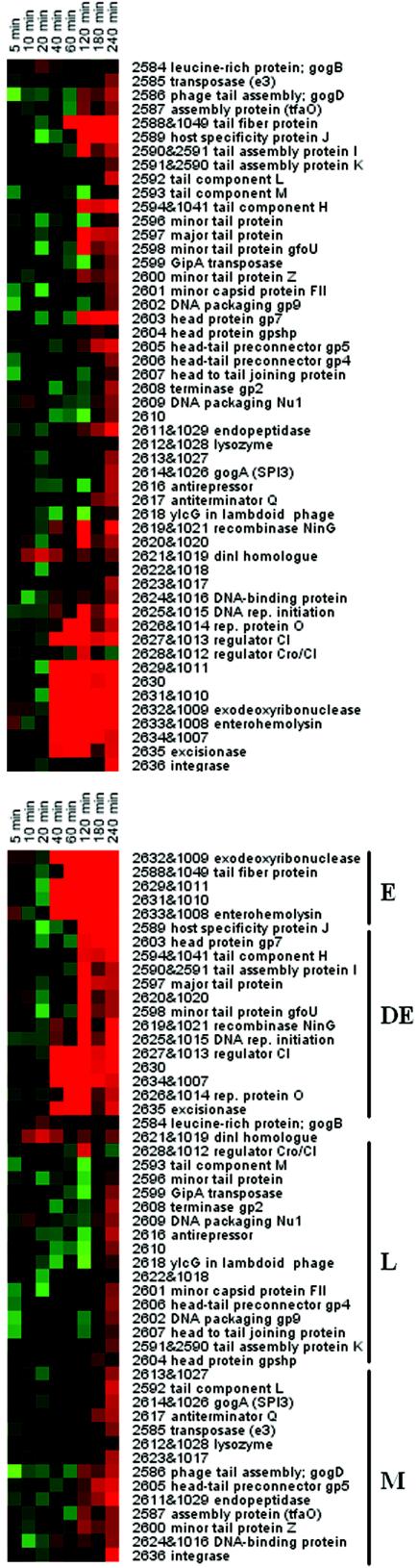
Gifsy-1, Gifsy-2, and Fels-1 are lambdoid phages (8). To analyze Gifsy-1 gene expression, strain LT2 was treated with mitomycin C as described above. Figure 4A shows that the phage genes displayed a temporal order of expression. DNA binding components, antirepressor proteins, virulence factors, and transcriptional regulators were the first proteins expressed after phage induction. DNA replication and recombinase genes were subsequently transcribed as delayed early genes. This was followed by the middle genes, including phage integration, excision, and host lysis genes. Finally, tail, head, DNA packaging, and assembly genes were expressed late in the lytic cycle. Hierarchical clustering of the Gifsy-1 genes according to their temporal transcription (Fig. 4B) confirmed the general classification into these four groups. The temporal order seen in Gifsy-1 is very similar to that observed in λ (8).
FIG. 4.
Gene expression profile of lambdoid prophage Gifsy-1 after mitomycin C treatment. Numbers are from the STM annotation, including some homologues in Gifsy-2 (16). Names are hypothetical, based on homology with other phages. The expression ratios are the ratios compared with an untreated control and are log2 transformed. Red, increase; green, decrease; black, no change. (A) Genes listed in order of position on the serovar Typhimurium chromosome. (B) Hierarchical clustering of phage gene expression as determined by GENESIS. Putative gene expression cluster assignments are indicated. Cluster E, early; cluster DE, delayed early; cluster M, middle; cluster L, late; rep, replication.
Gifsy-2, which was also a resident in all three serovar Typhimurium strains, displayed a gene arrangement very similar to that of Gifsy-1 and a similar but delayed response to mitomycin C (see Fig. S1 in the supplemental material). As observed in Gifsy-1, Gifsy-2 DNA amplification and elevated gene expression levels appeared earlier after peroxide treatment than after mitomycin C treatment (data not shown). Fels-1 was in λ phage order, and the genes were expressed in appropriate temporal order (see Fig. S1 in the supplemental material).
Fels-2 is a member of the P2 family of phages. P2 phage genes appear to be organized like lambdoid phage genes, and the phage genome is excised prior to replication, is replicated unidirectionally, and is packaged as closed circular genomes. Peroxide treatment induced the expression of Fels-2 genes 2 h after treatment, but expression went down 3 h after treatment (data not shown). Fels-2 DNA replication was strongly induced by mitomycin C treatment and was accompanied by a sixfold increase in expression over the rest of the genome (see Fig. S2 in the supplemental material). P2 phages are not activated by RecA but may instead be activated via an SOS-independent pathway. This pathway may be more strongly induced by mitomycin C than by peroxide (13).
During the phage induction experiments, expression of over 2,000 host genes differed twofold or more compared to the expression in the time zero control. In order to define genes that are distinctively regulated in serovar Typhimurium during the phage lytic cycle, data were compared to previously published results for the close relatives serovar Typhi strain CT18 and E. coli. The analyses included studies of exposure to peroxide, superoxide, and mitomycin C, as well as the OxyR and SoxS regulons (3, 12, 19, 20, 29). Most serovar Typhimurium genes exhibited the same pattern of expression as the pattern in these bacteria; however, some genes had different transcription profiles (Table 2). The most remarkable difference was observed for ribosomal gene expression. Virtually every gene in the two major ribosomal clusters showed a twofold increase in RNA in all three serovar Typhimurium strains investigated beginning 30 min after peroxide treatment and 5 min after mitomycin C treatment. In addition, accessory proteins involved in translation and protein maturation were also induced under these conditions. Surprisingly, these genes were not induced in a close relative of serovar Typhimurium, serovar Typhi strain CT18, which showed a decrease in ribosome expression that was greater than 10-fold at 5, 10, and 30 min after peroxide treatment (12, 19, 20, 29). All of these genes were likewise down regulated or not reported for E. coli cells treated with peroxide, mitomycin C, or the superoxide generator paraquat.
TABLE 2.
Selected genes exhibiting different expression profiles in response to mitomycin C and hydrogen peroxide treatment in Salmonella and E. colia
| Gene(s) or operon(s) | LT2, mitomycin C | LT2, H2O2 | CT18, H2O2 | E. coli, mitomycin C | E. coli, H2O2c |
|---|---|---|---|---|---|
| Amino acid transport and metabolism | |||||
| gltI-L, metA-J, smvA | ↓b | ↑ | ↑ | O | |
| gcvTHRP | ↑ | O/↑ | ↓ | ↑ | |
| tyrR glpB | ↓ | ↓ | ↑ | O | |
| potA-I | O | ↑ | ↓/O | O | |
| sdaABC | ↑ | ↑ | ↓ | ↓ | |
| Carbohydrate transport and metabolism | |||||
| malGEFKSTZ, manAXYZ | ↓ | ↓ | O | ↑ | ↑ |
| mglABC | ↑ | ↓ | O | ↑ | |
| ptsGHI | ↓ | ↑ | ↓ | ↓ | |
| fba | ↑ | ↑ | ↓ | ↑ | |
| glgABPSX, nagABDET | ↓ | ↓ | ↑ | ↑ | ↑ |
| Cell envelope and surface structures | |||||
| rfbA-Z, rfaA-Z | ↓ | ↑ | O | O | |
| ompX | ↑ | ↑ | ↓ | ↑ | |
| fimA-Z, safA-D stiA-H, stdABC, stfA-G, stcA-E | O/↓ | ↑ | O/↓ | ||
| Cell motility and secretion | |||||
| fliA-Z, motAB, cheABRWYZ | ↑ | ↑ | O | O | |
| flgABCDEFGHIJKLMN | ↑ | ↓ | O | O | |
| Energy production and conservation | |||||
| glpABCDEFTQ, napABCDFH, narHIJKLPQXVWZ | ↓ | ↓ | ↑ | O/↑ | |
| atpA-I | ↑ | ↑ | ↓ | O | |
| aceEF, lpdA | ↑ | ↑ | O/↓ | O/↓ | |
| adhE, hycABCDEFGHI | ↓ | ↑ | O/↓ | O/↑ | |
| frdABCD | ↓ | ↓ | O | ↑ | |
| fdhF | ↓ | ↑ | ↑ | O | |
| Global regulatory function and transcription | |||||
| rpoN, rho | ↑ | ↑ | ↓ | O/↓ | |
| phoPQ | ↑ | ↓ | ↓ | O | |
| lrp, arcA | ↑ | ↑ | O | O | |
| lon | O | ↑ | ↓ | O | |
| lexA, | ↑ | ↑ | ↑ | O | |
| phoBR | O | ↑ | ↑ | ↑ | |
| oxyR | O | O | O | ↑ | |
| soxSR | ↓ | ↑ | ↑ | O | ↑ |
| nifU | ↑ | ↑ | O | ||
| rseABC | O | O | ↑ | O | |
| Inorganic ion transport and metabolism | |||||
| modABCEF | O/↑ | ↑ | O/↑ | O | |
| fepABCDEG, fes, fhuABCDE | ↓ | ↑ | ↑ | O/↓ | |
| katE | O | ↑ | O | ↑ | |
| katG | ↓ | ↓ | ↑ | ↑ | ↑ |
| ydiE | ↓ | ↑ | ↑ | O | |
| narK, nirCD, nrfDA | ↓ | ↓ | O | O | |
| Lipid metabolism | |||||
| accABCD, fadABDEFHILZ | ↑ | ↑ | ↓ | ↓ | |
| SPI1 | |||||
| invA-J, sipA-D, prgHIJK | ↑ | ↓ | ↓ | ||
| spaO-S | ↑ | O | O | ||
| sitA-D | O | ↑ | ↑ | ||
| SPI2 | |||||
| ssaA-V, ssrAB, sseA-G | O/↓ | ↑ | ↑ | ||
| Translation ribosome structure and biogenesis | |||||
| rplA-Y, rpmA-G, rpsA-V, infABC, tufAB | ↑ | ↑ | ↓ | O | O |
DISCUSSION
In this study we measured gene content and gene expression in Salmonella during the induction of its resident prophages. One of the unexpected results of this study was the frequent detection of escape replication (9, 27) and the overall magnitude of amplifications caused by this phenomenon. Generally, once the lytic cycle begins in a lambdoid phage, the phage DNA is excised from the host genome as a closed circular structure. Initially, replication takes place in theta mode (i.e., bidirectionally). Linear copies are then created by the rolling circle method of replication, which creates a long concatemer consisting of many phage genomes. These genomes are later cleaved at the cos site during packaging as a linear genome into nascent phage heads (8). Escape replication, in which bidirectional runoff replication presumably takes place prior to excision, has been observed only in some lambdoid phages (9, 27). As we were looking at the bulk property of the population, we did not know if this replication was occurring in a subset of cells, whereas other cells contained phage that excised normally. However, in the cells in which escape replication took place we found that the physical extent of this amplification could be striking. The escape replication caused by Fels-1 in strain LT2 encompassed more than 15% of the entire host genome and resulted in manyfold amplification (Fig. 1C). As this amplification presumably occurred in only the subset of cells with lytic phages, the actual increase in the copy number of genes in this area within a cell must have been substantial. We also found that escape replication could be induced in other lambdoid phages by deletion of either genes encoding the integration or excision machinery (Gifsy-1 in Fig. 1E).
The observation of genome amplification III and amplification IV in the unsequenced strain 14028S (Fig. 3) implies that temperate prophages may be integrated into the genomes at these locations. Gifsy-3 is an inducible lysogenic phage that was previously mapped close to the location of amplification III and is therefore a strong candidate for insertion at that site (7). Detection of an amplified gene homologous to the sspH2 reporter on the microarray provided circumstantial evidence that Gifsy-3, containing the homologous sspH1 gene, is amplified under these conditions. Taken together, this evidence suggests that Gifsy-3 is induced by DNA damage and is most likely responsible for amplification III. Due to the symmetry of genome amplification IV and the presence of phage genes in this area in LT2, it is possible that a new lambdoid uncharacterized phage, or another element with a bidirectional replication origin, is responsible for amplification IV.
Phage gene expression analysis confirmed the results of previous studies and assignment of phage genes to early, delayed-early, middle, and late groups. One notable observation was that the virulence factors encoded by the phages were on the opposite coding DNA strand with respect to the phage structural genes. These factors included nanH and sodCIII of Fels-1, gtgA, gtgB, and sodCI of Gifsy-2, and gogB of Gifsy-1 (6, 7, 10, 11, 17, 18, 21). Consistent with this orientation, these genes did not seem to be transcribed at the same time as adjacent genes, nor did they fit into one specific regulatory cluster (data not shown). This may have been because these genes were controlled by regulators present within the host, which may not have been activated under the conditions tested.
The three serovar Typhimurium strains had some intriguing RNA responses to both peroxide and mitomycin C that were not seen in serovar Typhi strain CT18 or E. coli K-12 under similar conditions (12, 19, 20, 29). Among these unique responses was the induction of operons encoding genes involved in protein synthesis, carbohydrate transport, aerobic-anaerobic metabolism, and cell surface structures (Table 2). The regulation of these pathways in serovar Typhimurium but not in serovar Typhi strain CT18 or E. coli K-12 may have been due to any of a number of unique aspects of serovar Typhimurium. However, one commonality between serovar Typhi strain CT18 and the E. coli strains used in previous studies is the rather unusual lack of inducible prophages in the genomes. Thus, it likely that induction of the resident phage and the concomitant production of phage particles were responsible for the overexpression of protein synthesis genes seen in LT2. Further investigation by comparing isogenic strains that differ only in the presence of inducible phage is required to confirm this possibility. Certainly, experiments with any bacteria that could induce lysogenic phage have to take into account the likely dramatic effects of phage induction when the results are interpreted.
Lambdoid phages are clearly capable of clean excision and replication without detectable host genome amplification (as seen for wild-type Gifsy-1 and Gifsy-2). Thus, when a wild-type phage does cause escape replication, this is presumably a mechanistic choice that occurs because it provides a fitness advantage to the phage and perhaps even to the host. One possible reason to use this replication strategy is to increase the amount of particular host proteins encoded by the amplified region. However, an increase in host gene expression in the amplified regions was not detected in most of our experiments. Alternatively, genome amplification provides multicopy targets for both homologous and illegitimate recombination and could enhance the frequency of packaging of host DNA or increase the production of chimeric phages carrying new combinations of virulence factors. Any of these consequences could conceivably confer an evolutionary advantage to the phage and even to the host strain. Interestingly, for example, when Fels-1 amplifies adjacent host genes, it also amplifies Gifsy-2, and this could make the exchange of genes between these phages more likely.
In summary, DNA microarrays proved to be an effective way to measure temporal changes in phage and host RNA and genomic DNA during lytic growth of temperate bacteriophages. Escape replication may be quite common in S. enterica serovar Typhimurium and can involve very large portions of the host genome. Other lambdoid phages that do not normally cause escape replication do so after mutation of genes for integration and excision. Escape replication of unknown phages could be observed by virtue of amplification of the host genome adjacent to the presumptive site of phage integration. Occasionally, a gene from an uncharacterized phage cross-hybridized with a gene on the array, allowing induction of this additional phage to be monitored.
Supplementary Material
Acknowledgments
We thank Ken Sanderson (University of Calgary, Calgary, Alberta, Canada) and Gaelle Rondeau (Sidney Kimmel Cancer Center) for critical reading of the manuscript. We also thank the anonymous reviewers for their helpful suggestions. We are grateful for the policy of the Sanger Centre to make sequences available before completion.
This research was funded by NIH grants R01AI34829 (to M.M.) and R21 AI057733 and by the generosity of Sidney Kimmel.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Affolter, M., C. Parent-Vaugeois, and A. Anderson. 1983. Curing and induction of the Fels 1 and Fels 2 prophages in the Ames mutagen tester strains of Salmonella typhimurium. Mutat. Res. 110:243-262. [DOI] [PubMed] [Google Scholar]
- 2.Ball, C. A., and R. C. Johnson. 1991. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J. Bacteriol. 173:4032-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, N. R., R. M. Wong, and M. McClelland. 2000. Analysis of the SOS response in Salmonella enterica serovar Typhimurium using RNA fingerprinting by arbitrarily primed PCR. J. Bacteriol. 182:3490-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. [DOI] [PubMed] [Google Scholar]
- 7.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 8.Friedman, D. I. 1992. Interaction between bacteriophage lambda and its Escherichia coli host. Curr. Opin. Genet. Dev. 2:727-738. [DOI] [PubMed] [Google Scholar]
- 9.Fukasawa, T., K. Hirai, T. Segawa, and K. Obonai. 1978. Regional replication of the bacterial chromosome induced by derepression of prophage lambda. IV. Escape synthesis of gal operon in phage 82. Mol. Gen. Genet. 167:83-93. [DOI] [PubMed] [Google Scholar]
- 10.Hardt, W. D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, T. D., N. Figueroa-Bossi, M. Wang, S. Uzzau, L. Bossi, and J. M. Slauch. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khil, P. P., and R. D. Camerini-Otero. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89-105. [DOI] [PubMed] [Google Scholar]
- 13.Liu, T., S. K. Renberg, and E. Haggard-Ljungquist. 1997. Derepression of prophage P2 by satellite phage P4: cloning of the P4 epsilon gene and identification of its product. J. Virol. 71:4502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 15.Luke, K., A. Radek, X. Liu, J. Campbell, M. Uzan, R. Haselkorn, and Y. Kogan. 2002. Microarray analysis of gene expression during bacteriophage T4 infection. Virology 299:182-191. [DOI] [PubMed] [Google Scholar]
- 16.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 17.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirold, S., W. Rabsch, H. Tschape, and W. D. Hardt. 2001. Transfer of the Salmonella type III effector sopE between unrelated phage families. J. Mol. Biol. 312:7-16. [DOI] [PubMed] [Google Scholar]
- 19.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porwollik, S., J. Frye, L. D. Florea, F. Blackmer, and M. McClelland. 2003. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 31:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabsch, W., S. Mirold, W. D. Hardt, and H. Tschape. 2002. The dual role of wild phages for horizontal gene transfer among Salmonella strains. Berl. Muench. Tieraerztl. Wochenschr. 115:355-359. [PubMed] [Google Scholar]
- 22.Ramoni, M. F., P. Sebastiani, and I. S. Kohane. 2002. Cluster analysis of gene expression dynamics. Proc. Natl. Acad. Sci. USA 99:9121-9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts, J. W., C. W. Roberts, and N. L. Craig. 1978. Escherichia coli recA gene product inactivates phage lambda repressor. Proc. Natl. Acad. Sci. USA 75:4714-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207-208. [DOI] [PubMed] [Google Scholar]
- 26.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 27.Willard, M., and H. Echols. 1968. Role of bacteriophage DNA replication in lambda-dg escape synthesis. J. Mol. Biol. 32:37-46. [DOI] [PubMed] [Google Scholar]
- 28.Wilmes-Riesenberg, M. R., J. W. Foster, and R. Curtiss. 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect. Immun. 65:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.