Abstract
The two main recombination pathways in Escherichia coli (RecBCD and RecF) have different recombination machineries that act independently in the initiation of recombination. Three essential enzymatic activities are required for early recombinational processing of double-stranded DNA ends and breaks: a helicase, a 5′→3′ exonuclease, and loading of RecA protein onto single-stranded DNA tails. The RecBCD enzyme performs all of these activities, whereas the recombination machinery of the RecF pathway consists of RecQ (helicase), RecJ (5′→3′ exonuclease), and RecFOR (RecA-single-stranded DNA filament formation). The recombination pathway operating in recB (nuclease-deficient) mutants is a hybrid because it includes elements of both the RecBCD and RecF recombination machineries. In this study, genetic analysis of recombination in a recB (nuclease-deficient) recD double mutant was performed. We show that conjugational recombination and DNA repair after UV and gamma irradiation in this mutant are highly dependent on recJ, partially dependent on recFOR, and independent of recQ. These results suggest that the recombination pathway operating in a nuclease-deficient recB recD double mutant is also a hybrid. We propose that the helicase and RecA loading activities belong to the RecBCD recombination machinery, while the RecJ-mediated 5′→3′ exonuclease is an element of the RecF recombination machinery.
Recombination is a highly regulated biological process involved in DNA repair and the production of genetic variability. In the bacterium Escherichia coli there are two main recombination pathways: the RecBCD pathway, which is predominant in wild-type (WT) cells, and the RecF pathway, which operates in recBC sbcBC(D) mutants (19, 43). Although these pathways have different recombination machineries, they are thought to share the same sequence of events and use similar enzymatic activities for the processing of double-stranded DNA (dsDNA) ends and recombination intermediates. The major difference between the two pathways is in the initiation of recombination or presynapsis. Three key enzymatic activities required for presynapsis (helicase, 5′→3′ exonuclease, and loading of RecA protein onto prepared single-stranded DNA [ssDNA] tails) convert dsDNA ends (or dsDNA breaks) into recombinogenic filaments (24).
The RecBCD enzyme is the major component of the RecBCD recombination machinery. It is a multifunctional enzyme composed of one molecule each of the RecB, RecC, and RecD polypeptides, which are encoded by the corresponding recB, recC, and recD genes. The functional RecBCD enzyme exhibits several biochemical activities in vitro. It is a DNA helicase, a dsDNA exonuclease, a single-stranded (ss) DNA exonuclease, an ssDNA endonuclease, and an ATPase (27, 44). It also loads RecA protein onto 3′ ssDNA tails (7). The important feature of RecBCD-mediated recombination is that it is dependent on the χ sequence (5′-GCTGGTGG-3′) (17, 23, 28, 34, 37, 45, 46). The natural substrate for the RecBCD enzyme is linear dsDNA with blunt ends or with short (up to 25 nucleotides) ssDNA tails (27). The nuclease activity of the RecBCD enzyme is dependent on the concentration of free Mg2+ ions. In the presence of free Mg2+, which probably reflects the situation in vivo (27), the RecBCD enzyme binds to dsDNA ends, starts to unwind, and cuts both DNA strands until it encounters the χ site. After interaction with a χ site, the RecBCD enzyme undergoes modification. The χ-modified RecBCD enzyme retains its helicase activity, loses its 3′→5′ exonuclease activity, and enhances its 5′→3′ exonuclease activity (6). It also acquires a new activity essential for recombination, which is loading of RecA protein onto a prepared ssDNA end (7). Thus, in the RecBCD pathway all three activities essential for initiation of recombination are provided by the RecBCD enzyme itself.
The essential components of the RecF recombination machinery are RecQ (helicase), RecJ (5′→3′ exonuclease), and the RecFOR system (preparation of RecA-ssDNA filaments) (24). The mechanism by which a recombinogenic filament is prepared in the RecF pathway differs from the similar process in the RecBCD pathway. It includes replacement of SSB protein with RecA rather than direct RecA loading onto ssDNA as catalyzed by the RecBCD enzyme. RecA-ssDNA filament formation by RecFOR is not completely understood. It was shown that the RecOR complex stimulates replacement of SSB with RecA, whereas the RecFR complex prevents growth of the recombinogenic filament when it reaches a dsDNA (10, 49). However, a recent study showed that the RecFOR complex binds to the 5′ end of an ssDNA-dsDNA junction and that it loads the RecA protein from there (36). In addition to its role in the processing of dsDNA ends in a recBC sbcBC(D) mutant, the RecFOR system is essential for the recombinational repair of single-strand gaps (SSG) in WT bacteria (24, 25, 27).
The RecBCD and RecF pathways of recombination are completely independent in presynapsis. However, it was recently shown that in the recB1080 mutant the recombination machineries of the two pathways become interchangeable (20). The recB1080 mutation affects the nuclease center of the RecBCD enzyme, which is situated on the C-terminal portion of the RecB subunit. The consequence of this mutation is that the RecB1080CD enzyme loses its nuclease and RecA loading activities but retains its helicase activity (8, 48, 50). However, the recB1080 mutant is recombination proficient. Genetic analysis has shown that a hybrid recombination machinery operates in this mutant during initiation of recombination: the RecB1080CD enzyme (helicase) is part of the RecBCD recombination machinery, while RecJ (5′→3′exonuclease) and RecFOR (RecA-ssDNA filament formation) are parts of the RecF recombination machinery (20). In the same paper, we predicted that another hybrid pathway operates in the recB1080 recD double mutant for which we have shown that it is independent of RecFOR-mediated RecA-ssDNA filament formation (20). This result was consistent with in vitro data showing that the RecB1080C(D−) enzyme has helicase and RecA loading activities but lacks nuclease activity (4). Consequently, the hypothesis was proposed that the recombination machinery of this hybrid pathway should consist of RecB1080C(D−) (helicase and RecA loading) and RecJ (5′→3′ exonuclease) (2, 20, 32). In this report, we provide genetic evidence that supports the above hypothesis. We measured the effects of recJ and recQ mutations on recombination in a recB1080 recD background and the effects of recJ, recQ, and recFOR mutations on recombination in a recB1067 recD genetic background. The recB1067 allele is another mutation in the nuclease center of the RecB subunit. We showed that the recombination proficiency of the recB1080 recD and recB1067 recD double mutants is highly dependent on the RecJ nuclease but not on the RecQ helicase. In agreement with previous results obtained in the recB1080 recD background, RecFOR-mediated RecA-ssDNA filament formation is partially required for recombination in the recB1067 recD background.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
The bacterial strains used in this study are presented in Table 1. The P1 vir phage used for the construction of some bacterial strains was kindly provided by R. G. Lloyd, University of Nottingham, Nottingham, England. The experimental procedures used for transductions were described previously (35).
TABLE 1.
Bacterial strains used in this study
| Bacterial strain | Relevant genotype | Source or reference |
|---|---|---|
| Strains related to AB1157 | ||
| AB1157 | F−thr-1 leuB6 Δ(gpt-proA)62 hisG4 thi-1 argE3 lacY1 galK2 ara-14 xyl-5 mtl-1 tsx-33 supE44 rpsL31 kdgK51 rfbD1 mgl-51 λ−rac | 9 |
| N4634 | + recB::Tn10kan | R. G. Lloyd |
| RIK174 | + recB1080 | 21 |
| RIK144 | + recD1903::mini-Tn10 | 21 |
| RIK123 | + recB1067 | 21 |
| RIK151 | + recD1903::mini-Tn10 recB1067 | 21 |
| LMM1032 | + recJ2052::Tn10kan | D. Zahradka |
| IRB101 | + recQ1803::Tn3 | 39 |
| IRB103 | + recO1504::Tn5 | 39 |
| IIB279 | + recB1080 recQ1803::Tn3 | P1. IRB101 × RIK174 |
| IIB282 | + recB1080 recO1504::Tn5 | P1. IRB103 × RIK174 |
| IIB290 | + recB1080 recD1903::mini-Tn10 | P1. RIK144 × RIK174 |
| IIB291 | + recB1080 recD1903::mini-Tn10 recO1504::Tn5 | P1. IRB103 × IIB290 |
| IIB319 | + recB1080 recJ2052::Tn10kan | P1. LMM1032 × RIK174 |
| IIB320 | + recB1080 recD1903::mini-Tn10 recJ2052::Tn10kan | P1. LMM1032 × IIB290 |
| IIB321 | + recB1080 recD1903::mini-Tn10 recQ1803::Tn3 | P1. IRB101 × IIB290 |
| IIB340 | + recD1903::mini-Tn10 recJ2052::Tn10kan | P1. LMM1032 × RIK144 |
| IIB343 | + recB1067 recO1504::Tn5 | P1. IRB103 × RIK123 |
| IIB344 | + recB1067 recR256::Tn5 | P1. AM208 × RIK123 |
| IIB345 | + recB1067 recF400::Tn5 | P1. WA576 × RIK123 |
| IIB346 | + recB1067 recD1903::mini-Tn10 recO1504::Tn5 | P1. IRB103 × RIK151 |
| IIB347 | + recB1067 recD1903::mini-Tn10 recR256::Tn5 | P1. AM208 × RIK151 |
| IIB348 | + recB1067 recD1903::mini-Tn10 recF400::Tn5 | P1. WA576 × RIK151 |
| IIB350 | + recB1067 recJ2052::Tn10kan | P1. LMM1032 × RIK123 |
| IIB351 | + recB1067 recD1903::mini-Tn10 recJ2052::Tn10kan | P1. LMM1032 × RIK151 |
| IIB352 | + recB1067 recQ1803::Tn3 | P1. IRB101 × RIK123 |
| IIB353 | + recB1067 recD1903::mini-Tn10 recQ1803::Tn3 | P1. IRB101 × RIK151 |
| Other | ||
| KL96 | Hfr relA spoT1 thi-1 λ− | 9 |
| V2570 | Δ(recC-argA)234 hisD::kan rpsL31 λ− F− | 4 |
Media and growth conditions.
Bacteria were grown in a high-salt Luria broth (LB) medium composed of 10 g of Bacto Tryptone, 5 g of yeast extract, 10 g of NaCl, and water added to create a volume of 1,000 ml. Solid media for plates were supplemented with 16 g of agar per liter. M9 medium contained 0.5 g of NaCl, 1 g of NH4Cl, 3 g of KH2PO4, 7.5 g of Na2HPO4 · 2H2O, 4 g of glucose, 120 mg of MgSO4, 10 mg of CaCl2, and water added to create a volume of 1,000 ml. For minimal selective plates, M9 medium was supplemented with appropriate amino acids, 1 mg of thiamine, and 16 g of agar (33). All experiments were done with exponentially growing cells at 37°C.
Cell survival after gamma and UV irradiation.
For determination of cell survival after gamma irradiation, 0.1-ml aliquots of the appropriate dilutions of bacterial culture were plated on LB plates. Surviving cells formed visible colonies during overnight incubation at 37°C, and colonies were counted the next day. Cell survival is the ratio of the number of viable cells in a culture after administration of an appropriate dose of gamma irradiation and the number of viable cells in the culture without gamma irradiation. For gamma irradiation, a 60Co source with a dose rate of 11.4 Gy/s as measured by ferrous sulfate dosimetry was used. Bacteria were irradiated at 0°C. For UV treatment, a 30-W Philips low-pressure Hg germicidal lamp was used at a distance of 1 m. The incident dose was ∼0.25 mW/cm2, as determined with a VLX-3W UV dosimeter (Bioblock, Illkirch, France). Cell survival after UV irradiation was measured as described previously (1). Bacteria were irradiated at room temperature.
Conjugational crosses.
The procedures used for conjugational crosses were described previously (35). Hfr strain KL96 was used as the donor, and the selected marker was His+. Matings were performed in LB medium for 30 min and mixed in a 1:10 donor-to-recipient ratio with recipient and donor cells grown to an optical density at 650 nm (OD650) of 0.4. The exconjugant mixture was interrupted by vigorous agitation, serially diluted, and plated on appropriate minimal agar containing 100 μg of streptomycin per ml to counterselect donor cells. Measurements of cell viability relate to the number of CFU in the recipient cultures at an OD650 of 0.4, as determined with nonselective LB agar (41). The frequency of conjugational recombination for each experiment was corrected for the recipient's viability relative to that of the WT.
RESULTS AND DISCUSSION
Recombination in a recB1067 mutant requires RecFOR function.
In a previous paper, we have shown that the recO mutation has a strong effect in a recB1080 genetic background but has no substantial effect on recombination in a recB1080 recD background (20). This result was consistent with the biochemical evidence that inactivation (deletion) of the RecD subunit restores RecA loading activity in the RecB1080C(D−) enzyme (4). In addition to recB1080, there is another mutation in the nuclease center of RecB at position 1067, which also has a substitution of D (aspartic acid) for A (alanine) and is designated the recB1067 allele. There are no in vitro reports on the RecA loading activity of the RecB1067CD and RecB1067C(D−) enzymes. However, an earlier genetic study showed that the recB1067 mutation had the same phenotype as the recB1080 allele when UV sensitivity and λ recombination were measured (21). From these results it could be concluded that the RecB1067CD enzyme is probably inactivated for both nuclease and RecA loading activities. In accordance with the finding that the RecD subunit is an inhibitor of RecA loading (4), the RecB1067C(D−) enzyme should recover its RecA loading activity. Therefore we wanted to test genetically whether the recB1067 allele behaves similarly to the recB1080 allele in combination with the recD and recFOR mutations. We measured recombination proficiency with three in vivo assays: cell survival after UV and gamma irradiation and recombination frequency after Hfr conjugation.
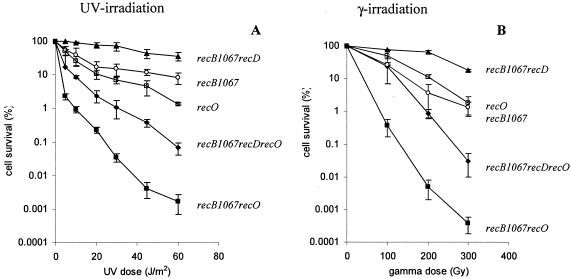
The cell survival curves after treatment with UV and gamma irradiation are presented in Fig. 1. The recB1067 recO double mutant, as well as the recB1067 recR and recB1067 recF double mutants (data not shown), was extremely sensitive to UV light and gamma irradiation, as expected if the RecB1067CD enzyme is RecA loading deficient. On the other hand, the recB1067 recD recO triple mutant, as well as the recB1067 recD recR and recB1067 recD recF triple mutants (data not shown), was more resistant, as expected if the RecB1067C(D−) enzyme is RecA loading proficient, although it was more sensitive than the recO and recB1067 single mutants and the recB1067 recD double mutant. The restoration of recombination repair ability after UV and gamma irradiation with recB1067 recD recO, as well as with the recB1080 recD recO triple mutant (data not shown), was somewhat smaller than in our previous paper (20), but the general conclusion is the same. It follows that the recB1067 allele has an effect on recombination similar to that of the recB1080 allele. These genetic data suggest that recombinational repair in the recB1067 mutant requires RecFOR-mediated RecA-ssDNA filament formation. The higher sensitivity of the recB1067 recD recO triple mutant relative to that of the recB1067 recD mutant strain can be explained by the involvement of the RecFOR system in the repair of SSG after UV treatment and by its possible role in the repair of dsDNA breaks after gamma irradiation (22, 42). An additional possibility is that the RecB1067C(D−) form of the enzyme only partially restores its RecA loading activity.
FIG. 1.
Effect of the recO mutation on DNA repair after UV (A) and gamma (B) irradiation in the WT, recB1067, and recB1067 recD genetic backgrounds. The measure for efficiency of DNA repair was cell survival after different doses of UV and gamma irradiation. The values are means of at least three independent experiments. Standard deviation bars are shown for each cell survival curve. Symbols: ○, recB1067 (strain RIK123); ▪, recB1067 recO (strain IIB343); ▴, recB1067 recD (strain RIK151); □, recO (strain IRB103); ♦, recB1067 recD recO (strain IIB346).
The relative recombination frequencies after Hfr crosses are presented in Table 2. The recB1067 mutant had moderate recombination proficiency with a relative recombination frequency of 0.14, whereas the recB1067 recO, recB1067 recR, and recB1067 recF double mutants had very weak recombination frequencies, i.e., 0.032, 0.038, and 0.038, respectively, which are comparable to the recombination proficiency of the recB null mutants. On the other hand, the recombination proficiency of the recB1067 recD recO (0.19), recB1067 recD recR (0.16), and recB1067 recD recF (0.16) triple mutants was restored to the level of the recB1067 single mutant. These data again suggest that moderate conjugational recombination proficiency in the recB1067 single mutant requires RecFOR function and that the strong recombination deficiency in the recB1067 recO, recB1067 recR, and recB1067 recF double mutants is due to a lack of essential RecA-ssDNA filament formation. Also, the higher recombination proficiency of triple mutants could be explained by the fact that the RecB1080C(D−) enzyme, and possibly the RecB1067C(D−) enzyme, has RecA loading activity (4, 20).
TABLE 2.
Recombination frequencies in Hfr-mediated conjugational crosses
| Straina | Relevant genotype | Relative viabilty | Recombination frequencyb |
|---|---|---|---|
| AB1157 | WT | 1c | 1d |
| N4634e | recB | 0.37 ± 0.075 | 0.028 ± 0.016 |
| V2570e | ΔrecBCD | 0.29 ± 0.08 | 0.005 ± 0.0029 |
| RIK123 | recB1067 | 1 ± 0.097 | 0.138 ± 0.022 |
| RIK144e | recD | 0.68 ± 0.028 | 2.64 ± 1 |
| IRB103e | recO | 0.51 ± 0.03 | 1.2 ± 0.24 |
| IRB101e | recQ | 1 ± 0.0025 | 1 ± 0.05 |
| LMM1032 | recJ | 1.4 ± 0.03 | 0.87 ± 0.083 |
| IIB343 | recB1067 recO | 0.65 ± 0.13 | 0.032 ± 0.0018 |
| IIB344 | recB1067 recR | 0.63 ± 0.12 | 0.038 ± 0.006 |
| IIB345 | recB1067 recF | 0.57 ± 0.17 | 0.038 ± 0.003 |
| IIB352 | recB1067 recQ | 0.66 ± 0.16 | 0.11 ± 0.048 |
| IIB350 | recB1067 recJ | 0.17 ± 0.05 | 0.00085 ± 0.00055 |
| IIB319 | recB1080 recJ | 0.13 ± 0.05 | 0.0015 ± 0.00033 |
| RIK151 | recB1067 recD | 1.1 ± 0.09 | 0.59 ± 0.14 |
| IIB290 | recB1080 recD | 1.1 ± 0.01 | 0.71 ± 0.11 |
| IIB346 | recB1067 recD recO | 1 ± 0.05 | 0.19 ± 0.013 |
| IIB347 | recB1067 recD recR | 1.4 ± 0.12 | 0.16 ± 0.068 |
| IIB348 | recB1067 recD recF | 1.2 ± 0.13 | 0.16 ± 0.108 |
| IIB340 | recD recJ | 1 ± 0.14 | 0.28 ± 0.138 |
| IIB351 | recB1067 recD recJ | 0.75 ± 0.007 | 0.013 ± 0.0038 |
| IIB353 | recB1067 recD recQ | 1.2 ± 0.12 | 0.39 ± 0.05 |
| IIB320 | recB1080 recD recJ | 0.75 ± 0.005 | 0.015 ± 0.002 |
| IIB321 | recB1080 recD recQ | 1 ± 0.04 | 0.39 ± 0.05 |
Mating was done with the KL96 donor; the selected marker was His+.
Values are means of at least three separate experiments with standard deviations, corrected for the viability of recipients.
WT viability of 1.0 = 2 × 108 /ml, measured at an OD650 of 0.4.
WT frequency of 1.0 = 39 exconjugants per 1,000 donors.
Data originally presented in reference 20.
Recombination in the recB1080 recD mutant is not dependent on RecQ helicase.
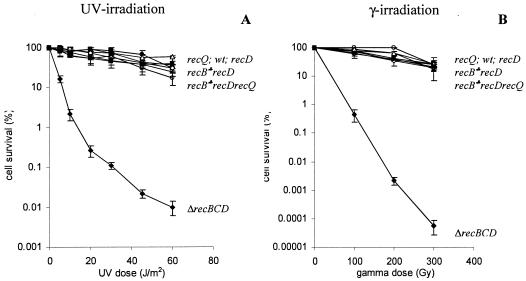
It was previously shown that recombination in the recB1080 mutant is dependent on specific RecF pathway genes (recJ, recF, recO, and recR) but not on recQ (20). The specific difference between the RecB1080CD and RecF pathways is probably due to the use of different helicases in presynapsis. The hybrid RecB1080CD pathway uses the RecB1080CD helicase, whereas the RecQ helicase participates in the RecF pathway (2, 20). Here we wanted to test whether recombination in the recB1080 recD double mutant requires RecQ. Since the RecB1080C(D−) enzyme has helicase activity in vitro (4), one would expect that recQ function is not needed for recombination in the recB1080 recD mutant. As shown in Fig. 2, the recB1080 recD recQ triple mutant was highly resistant to both UV light and gamma irradiation, similar to the recB108O recD double mutant and the recQ single mutant. On the other hand, the recB null mutant lacks all of the activities of the RecBCD enzyme (helicase, nuclease, and RecA loading), and consequently recB null mutants are recombination and repair deficient. The missing enzymatic activities cannot be replaced by the equivalent functions from the RecF pathway because of the presence of the inhibitory nucleases SbcB and SbcCD (15, 26, 29). This is not the case in WT cells since the RecBCD enzyme is able to overcome the inhibitory effect of the SbcB and SbcCD nucleases. The RecBCD enzyme has a higher affinity for dsDNA ends than do the RecQ helicase and the SbcB and SbcCD nucleases (11, 15, 18, 40). Also, it is a much more potent helicase than RecQ (18). This implies that possibly the helicase activity of the RecB1080C(D−) enzyme, together with its RecA loading activity, is essential for recombination in the recB1080 recD mutant. The same resistance to UV light and gamma irradiation was observed in the recB1067 recD recQ triple mutant (Fig. 2), suggesting a similar explanation for recombination in the recB1067 recD double mutant.
FIG. 2.
Effect of the recQ mutation on DNA repair after UV (A) and gamma (B) irradiation in the recB1067 recD and recB1080 recD genetic backgrounds. The measure for efficiency of DNA repair was cell survival after different doses of UV and gamma irradiation. The values are means of at least three independent experiments. Standard deviation bars are shown for each cell survival curve. Symbols: ○, recQ (strain IRB101); ♦, ΔrecBCD (strain V2570); ▪, recB1067 recD recQ (strain IIB353); □, recB1080 recD recQ (strain IIB321); ▴, recB1067 recD (strain RIK151); Δ, recB1080 recD (strain IIB290); ⋄, recD (strain RIK144); •, WT (strain AB1157); ♣, both recB1080 and recB1067 alleles.
Recombination proficiencies after Hfr conjugations are presented in Table 2. The relative recombination frequencies of the recB1080 recD recQ (0.39) and recB1067 recD recQ (0.39) triple mutants were similar to the relative recombination frequencies of the recB1080 recD (0.71) and recB1067 recD (0.59) double mutants and were much higher than the recombination frequencies of the recB null mutants (0.005 to 0.028). Thus, we can conclude that the RecQ helicase is not essential for conjugational recombination in the recB1080 recD and recB1067 recD mutant strains. The possible helicases used in the initiation of recombination after conjugation are RecB1080C(D−) and RecB1067C(D−), respectively.
Recombination in the recB1080 recD mutant is dependent on RecJ nuclease.
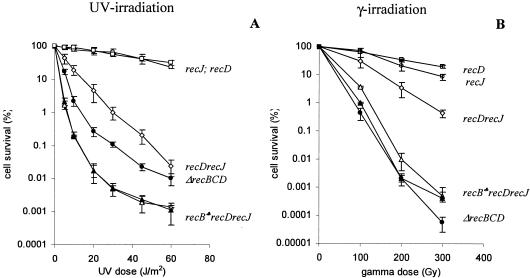
The genetic analysis described so far suggests that two essential activities (helicase and RecA loading) in initiation of recombination in the recB1080 recD and recB1067 recD double mutants are parts of the RecBCD recombination machinery. Since these double mutants produce nuclease-deficient forms of the RecBCD enzyme and are recombination proficient, it was expected that RecJ 5′→3′ exonuclease is required for efficient recombination. Consistent with this, it was shown that recB1067 recD recJ mutants are extremely sensitive to UV irradiation (21). Here we wanted to test whether the RecJ nuclease is also necessary for double-strand break repair after gamma irradiation and for conjugational recombination in the recB1080 recD and recB1067 recD backgrounds. We compared the recombination proficiencies of recB1080 recD recJ and recB1067 recD recJ triple mutants with those of recB1080 recD and recB1067 recD double mutants. Figure 3 shows the cell survival curves after treatment with UV light and gamma irradiation. The recB1080 recD recJ and recB1067 recD recJ triple mutants were extremely sensitive to both UV light and gamma irradiation, while the recB1080 recD and recB1067 recD double mutants, as well as the recJ single mutant, were resistant. This result strongly suggests that recombination in the recB1080 recD and recB1067 recD double mutants requires RecJ-mediated 5′→3′ exonuclease and that the recombination pathway operating in these strains is hybrid since it uses elements from both the RecBCD and RecF recombination machineries. This is further supported by data from the Hfr-mediated conjugational recombination presented in Table 2. The triple mutants recB1080 recD recJ (0.015) and recB1067 recD recJ (0.013) had extremely low recombination frequencies down to the level of recB null mutants (0.005 to 0.028), whereas the recB1080 recD (0.71) and recB1067 recD (0.59) double mutants and the recJ (0.87) single mutant were highly recombination proficient.
FIG. 3.
Effect of the recJ mutation on DNA repair after UV (A) and gamma (B) irradiation in the recB1080 recD, recB1067 recD, and recD genetic backgrounds. The measure for efficiency of DNA repair was cell survival after different doses of UV and gamma irradiation. The values are means of at least three independent experiments. Standard deviation bars are shown for each cell survival curve. Symbols: ○, recJ (strain LMM1032); □, recD (strain RIK144); •, ΔrecBCD (strain V2570); ⋄, recD recJ (strain IIB340); ▴, recB1067 recD recJ (strain IIB351); ▵, recB1080 recD recJ (strain IIB320); ♣, both recB1080 and recB1067 alleles.
Weak requirement of RecJ nuclease for recombination in the recD mutant.
We also compared the recombination of the recB1080 recD and recB1067 recD double mutants with that of the recD single mutant, which is traditionally considered to be nuclease deficient (3, 12, 44). In addition to deficiency in DNA degradation, recD mutants show normal recombination, DNA repair, and cell viability, which are abolished in recB null mutants (3, 12, 27). This is in agreement with the biochemical properties of the RecBC(D−) enzyme, which has helicase activity (weaker than that of the WT enzyme) (25) and constitutive (χ-independent) RecA loading activity (14). Consequently, recombination operating in the recD mutant is χ independent and strand exchange can occur close to dsDNA ends or dsDNA breaks (12, 44). The RecD subunit is important for expression of nuclease and RecA loading activities of the RecBCD enzyme, for which its proper interaction with the RecB and RecC subunits is crucial (4, 5, 50, 51). Also, the RecD subunit contains a fast helicase domain (16, 47). Since the RecBC(D−) enzyme is nuclease deficient, recombination in the recD mutant is partially dependent on RecJ nuclease (31, 32). In fact, this recombination pathway was the first described hybrid pathway. However, there is some controversy about the importance of the RecJ nuclease for recombination in a recD mutant. First, it was shown that the recJ mutation has a substantial effect on cell survival after UV treatment (31) and on conjugational recombination (32). In contrast, another study has shown that the effect of the recJ mutation on conjugational recombination in recD mutants is marginal (30). We tested again the effect of the recJ mutation on recombinational repair after UV treatment and gamma irradiation and on conjugational recombination in a recD mutant. The recD recJ double mutant was substantially more sensitive than the recD and recJ single mutants after UV irradiation, although this strain was more resistant than the recB1080 recD recJ and recB1067 recD recJ mutant strains (Fig. 3A). This effect of the recJ mutation on recombinational repair in the recD background after UV treatment was similar to that reported earlier (31). However, the cell survival of the recD recJ double mutant after gamma irradiation was only slightly lower than the cell survival of the recD and recJ single mutants (Fig. 3B). The effect on conjugational recombination was also marginal (Table 2) and similar to the data of Lloyd and Buckman (30). The relative frequency of conjugational recombination in the recD recJ double mutant was close to 0.3, whereas in the recB1080 recD recJ and recB1067 recD recJ triple mutants the frequencies were 0.015 and 0.013, respectively. These results suggest that the RecJ nuclease in recD mutants is more important for recombinational repair of SSG rather than processing of dsDNA ends and breaks. Consistent with this is the higher sensitivity after UV treatment of the recB1080 recD recJ and recB1067 recD recJ triple mutants relative to that of the ΔrecB null mutant (Fig. 3A). The reason for this higher sensitivity of triple mutants is that in the triple mutants both components of recombinational repair (dsDNA break repair and SSG repair) are deficient whereas in the recB null mutant only the repair of dsDNA breaks is deficient. In contrast, the triple mutants were slightly more resistant than the recB null mutant after gamma irradiation (Fig. 3B) since this repair is predominately RecBCD-mediated double-strand break repair. The simplest explanation for the weak requirement of the RecJ nuclease in double-strand break repair and conjugational recombination could be that the RecBC(D−) enzyme still retains some residual nuclease activity (13, 21, 38, 51), which can contribute to a substantial amount of recombinational events. When the nuclease activity of the RecBCD enzyme is completely abolished because of mutations in the nuclease center (as in the recB1080 recD and recB1067 recD double mutants, as well as in the recB1080 and recB1067 single mutants), recombination is highly dependent on the RecJ nuclease. An additional possibility is that the recB (nuclease-deficient) mutants are also partially deficient in RecA loading.
Concluding remarks.
In this study we have shown that recombinational processing of dsDNA ends and breaks in the nuclease-deficient recB recD double mutants uses elements of both the RecBCD and RecF recombination machineries. We propose that the helicase and RecA loading activities are provided by the RecB1080C(D−) [or RecB1067C(D−)] enzyme, while the 5′→3′ exonuclease activity is a function of the RecJ nuclease. These concerted activities can produce a recombinogenic filament that leads to DNA strand exchange and finally to DNA repair and/or production of new genotypic variants.
Acknowledgments
We are grateful to R. S. Myers (University of Miami School of Medicine) for sending us bacterial strains (RIK123 and RIK151) and Mary Sopta (Ruđer Bošković Institute) for correcting our English.
This work was supported by the Croatian Ministry of Science (grant 0098070).
REFERENCES
- 1.Al-Deib, A. A., A. A. Mahdi, and R. G. Lloyd. 1996. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J. Bacteriol. 178:6782-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amundsen, S. K., and G. R. Smith. 2003. Interchangeable parts of the Escherichia coli recombination machinery. Cell 112:741-744. [DOI] [PubMed] [Google Scholar]
- 3.Amundsen, S. K., A. F. Taylor, A. M. Chaudhury, and G. R. Smith. 1986. recD: the gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 83:5558-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amundsen, S. K., A. F. Taylor, and G. R. Smith. 2000. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination, and DNA repair. Proc. Natl. Acad. Sci. USA 97:7399-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amundsen, S. K., A. F. Taylor, and G. R. Smith. 2002. A domain of RecC required for assembly of the regulatory RecD subunit into the Escherichia coli RecBCD holoenzyme. Genetics 161:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson, D. G., and S. C. Kowalczykowski. 1997. The recombinational hot spot χ is a regulatory element that switches the polarity of DNA degradation by RecBCD enzyme. Genes Dev. 11:571-581. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocation RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90:77-86. [DOI] [PubMed] [Google Scholar]
- 8.Anderson, D. G., J. J. Churchill, and S. C. Kowalczykowski. 1999. A single mutation, recBD1080A, eliminates RecA protein loading but not Chi recognition by RecBCD enzyme. J. Biol. Chem. 274:27139-27144. [DOI] [PubMed] [Google Scholar]
- 9.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 10.Bork, J. M., M. M. Cox, and R. B. Inman. 2001. The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J. 20:7313-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brody, R. S., K. G. Doherty, and P. D. Zimmerman. 1986. Processivity and kinetics of the reaction of exonuclease I from Escherichia coli with polydeoxyribonucleotides. J. Biol. Chem. 261:7136-7143. [PubMed] [Google Scholar]
- 12.Chaudhury, A. M., and G. R. Smith. 1984. A new class of Escherichia coli recBC mutants: implications for the role of RecBCD enzyme in homologous recombination. Proc. Natl. Acad. Sci. USA 81:7850-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H. W., D. E. Randle, M. Gabbidon, and D. A. Julin. 1998. Functions of the ATP hydrolysis subunits (RecB and RecD) in the nuclease reactions catalyzed by the RecBCD enzyme from Escherichia coli. J. Mol. Biol. 278:89-104. [DOI] [PubMed] [Google Scholar]
- 14.Churchill, J. J., D. G. Anderson, and S. C. Kowalczykowski. 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independent of χ, resulting in constitutive recombination activation. Genes Dev. 13:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connelly, J. C., E. S. de Leau, E. A. Okely, and D. R. Leach. 1997. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J. Biol. Chem. 272:19819-19826. [DOI] [PubMed] [Google Scholar]
- 16.Dillingham, M. S., M. Spies, and S. C. Kowalczykowski. 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 423:893-897. [DOI] [PubMed] [Google Scholar]
- 17.Dixon, D. A., and S. C. Kowalczykowski. 1993. The recombination hotspot, Chi, is a regulatory sequence that acts by attenuating the nuclease activity of the Escherichia coli RecBCD enzyme. Cell 73:87-96. [DOI] [PubMed] [Google Scholar]
- 18.Harmon, F. G., and S. C. Kowalczykowski. 2001. Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J. Biol. Chem. 276:232-243. [DOI] [PubMed] [Google Scholar]
- 19.Horii, Z.-I., and A. J. Clark. 1973. Genetic analysis of the RecF pathway of genetic recombination in Escherichia coli K-12: isolation and characterization of mutants. J. Mol. Biol. 80:327-344. [DOI] [PubMed] [Google Scholar]
- 20.Ivančić-Baće, I., P. Peharec, S. Moslavac, N. Škrobot, E. Salaj-Šmic, and K. Brčić-Kostić. 2003. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics 163:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jockovich, M. E., and R. S. Myers. 2001. Nuclease activity is essential for RecBCD recombination in Escherichia coli. Mol. Microbiol. 41:949-962. [DOI] [PubMed] [Google Scholar]
- 22.Kidane, D., H. Sanchez, J. C. Alonso, and P. L. Graumann. 2004. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol. Microbiol. 52:1627-1639. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, I., H. Murialdo, J. M. Crasemann, M. M. Stahl, and F. W. Stahl. 1982. Orientation of cohesive end site cos determines the active orientation of χ sequence in stimulating recA, recBC-mediated recombination in phage λ lytic infections. Proc. Natl. Acad. Sci. USA 79:5981-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 25.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kushner, S. R., T. Nagaishi, A. Templin, and A. J. Clark. 1971. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl. Acad. Sci. USA 68:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam, S. T., M. M. Stahl, K. D. McMilin, and F. W. Stahl. 1974. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 77:425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd, R. G., and C. Buckman. 1991. Overlapping functions of recD, recJ and recN provide evidence of three epistatic groups of genes in Escherichia coli recombination and DNA repair. Biochimie 73:313-320. [DOI] [PubMed] [Google Scholar]
- 31.Lloyd, R. G., M. C. Porton, and C. Buckman. 1988. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol. Gen. Genet. 212:317-324. [DOI] [PubMed] [Google Scholar]
- 32.Lovett, S. T., C. Luisi-De Luca, and R. D. Kolodner. 1988. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics 120:37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maršić, N., S. Roje, I. Stojiljković, E. Salaj-Šmic, and Ž. Trgovčević. 1993. In vivo studies on the interaction of RecBCD enzyme and λ Gam protein. J. Bacteriol. 175:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMilin, K. D., M. M. Stahl, and F. W. Stahl. 1974. Rec-mediated recombinational hot spot activity in bacteriophage lambda. I. Hot spot activity associated with spi − deletions and bio substitutions. Genetics 77:409-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337-1347. [DOI] [PubMed] [Google Scholar]
- 37.Myers, R. S., and F. W. Stahl. 1994. Chi and the RecBCD enzyme of Escherichia coli. Annu. Rev. Genet. 28:49-70. [DOI] [PubMed] [Google Scholar]
- 38.Palas, K. M., and S. R. Kushner. 1990. Biochemical and physical characterization of exonuclease V from Escherichia coli. Comparison of the catalytic activities of the RecBC and RecBCD enzymes. J. Biol. Chem. 265:3447-3454. [PubMed] [Google Scholar]
- 39.Paškvan, I., E. Salaj-Šmic, I. Ivančić-Baće, K. Zahradka, Ž. Trgovčević, and K. Brčić-Kostić. 2001. The genetic dependence of RecBCD-Gam mediated double strand end repair in Escherichia coli. FEMS Microbiol. Lett. 205:299-303. [DOI] [PubMed] [Google Scholar]
- 40.Roman, L. J., and S. C. Kowalczykowski. 1989. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry 28:2863-2873. [DOI] [PubMed] [Google Scholar]
- 41.Ryder, L., M. C. Whitby, and R. G. Lloyd. 1994. Mutation of recF, recJ, recO, recQ, or recR improves Hfr recombination in resolvase-deficient ruv recG strains of Escherichia coli. J. Bacteriol. 176:1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sargentini, N. J., and K. C. Smith. 1986. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, uvrD, and umuC genes in the repair of X-ray-induced DNA double-strand breaks in Escherichia coli. Radiat. Res. 107:58-72. [PubMed] [Google Scholar]
- 43.Smith, G. R. 1989. Homologous recombination in E. coli: multiple pathways for multiple reasons. Cell 58:807-809. [DOI] [PubMed] [Google Scholar]
- 44.Smith, G. R. 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35:243-274. [DOI] [PubMed] [Google Scholar]
- 45.Spies, M., P. R. Bianco, M. S. Dillingham, N. Handa, R. J. Baskin, and S. C. Kowalczykowski. 2003. A molecular throttle: the recombination hotspot chi controls DNA translocatin by the RecBCD helicase. Cell 114:647-654. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, A. F., and G. R. Smith. 1992. RecBCD enzyme is altered upon cutting DNA at a chi recombination hotspot. Proc. Natl. Acad. Sci. USA 89:5226-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, A. F., and G. R. Smith. 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423:889-893. [DOI] [PubMed] [Google Scholar]
- 48.Wang, J., R. Chen, and D. A. Julin. 2000. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem. 275:507-513. [DOI] [PubMed] [Google Scholar]
- 49.Webb, B. L., M. M. Cox, and R. B. Inman. 1997. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91:347-356. [DOI] [PubMed] [Google Scholar]
- 50.Yu, M., J. Souaya, and D. A. Julin. 1998. The 30-kDa C-terminal domain of the RecBCD protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 95:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, M., J. Souaya, and D. A. Julin. 1998. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 283:797-808. [DOI] [PubMed] [Google Scholar]