Abstract
Transcription of the dinB gene, encoding DNA polymerase IV, is induced by the inhibition of cell wall synthesis at different levels. Using the β-lactam antibiotic ceftazidime, a PBP3 inhibitor, as a model, we have shown that this induction is independent of the LexA/RecA regulatory system. Induction of dinB transcription mediated by ceftazidime produces an increase in the reversion of a +1 Lac frameshift mutation.
The Escherichia coli SOS system comprises at least 30 different genes, some of them encoding proteins involved in DNA repair pathways (7). When activated by single-strand DNA (which can be created by multiple events, such as DNA damage, stalled replication, and conjugation, etc.), the coprotease activity of the RecA protein promotes the self-cleavage of the LexA protein (the SOS repressor) and, consequently, induces the SOS response (3). Among the known SOS genes are polB, dinB, and umuDC, encoding the specialized DNA polymerases II, IV, and V (Pol II, Pol IV, and Pol V), respectively (8). These enzymes are able to bypass DNA lesions that block chain elongation by the replicative DNA polymerase. When operating on nonsubstrate templates or copy-noncognate lesions, these enzymes exhibit reduced fidelity and, consequently, produce mutations in the newly synthesized DNA strand (8, 19). Thus, as a consequence of DNA damage or a cessation of DNA replication, the mutation rate of bacteria is increased (17). Finally, the importance of these enzymes for the long-term survival of E. coli has been demonstrated (23).
The activity of Pol IV is involved in adaptive mutation (21). Overproduction of Pol IV produces a mutator phenotype (10). Thus, its synthesis has to be regulated in normal (nonstressed) cells to keep its mutagenic activity under control. It has recently been described that, in addition to DNA damage or DNA replication, other types of stress, including stationary phase, can trigger the transcription of dinB (11). This SOS-independent regulation indicates that induction of Pol IV is part of a more general stress-regulated response. This result has suggested to some researchers that DNA polymerase IV may be part of a mechanism to produce mutants when the population is under extreme conditions (4, 11, 13).
There are a large number of situations that can produce stress in bacteria. Some of them, such as the presence of antibiotics, are specifically used to produce different types of vital stress in bacteria and to eliminate them from humans. Thus, it is tempting to test whether different antibiotics, as stress producers, can potentially induce dinB transcription and as a result increase the mutation frequency.
It was previously shown that some antibiotics used in clinical practice, such as quinolones, are good inducers of the SOS system and, consequently, increase the mutation frequency (16). This increase in mutation frequency was shown to be due to the activity of Pol V (24). Also, streptomycin, an aminoglycoside antibiotic, is known to promote mistranslation and induce a recA- and umuDC-independent mutator phenotype (18).
In this work, we have investigated the capacity of different antibiotic families to induce the transcription of the dinB gene and to increase the rate at which mutations are produced. Our results show that some antibiotics, known to act as cell wall synthesis inhibitors at different steps but without having any effect on DNA (damage or replication) or translation are able to induce dinB transcription. Using the β-lactam antibiotic ceftazidime (CAZ), which is a widely used PBP3 inhibitor, as a model, we have demonstrated that this induction occurs independently of the LexA/RecA regulators. In addition, we have shown that CAZ can increase, although slightly, the reversion frequency of a +1 Lac frameshift mutation in a dinB-dependent way.
Antibiotics that induce dinB transcription.
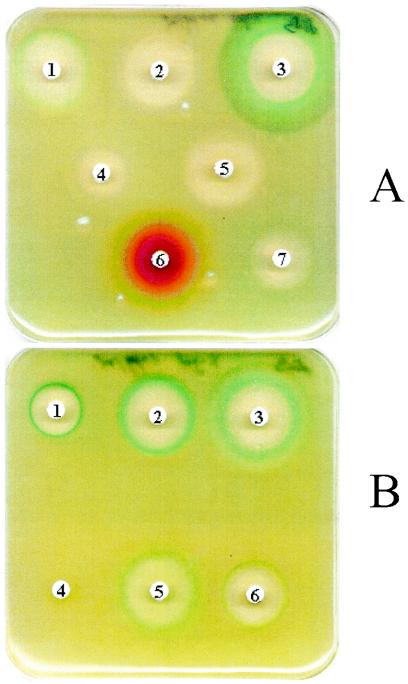
Using a disk plate assay, we tested the capacity of different antibiotic families to induce dinB transcription. Aliquots of 100 μl from overnight cultures of strain GW1030 [dinB1::MudI (Ap lac)], harboring a dinB::lacZ operon fusion (9), were resuspended in soft Luria-Bertani (LB) agar, mixed, and plated onto LB plates containing 50 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml. Disks containing different antibiotics were put onto the seeded bacteria, and the plates were incubated for 24 h at 30°C. We found that, as expected, quinolones, such as ciprofloxacin (Fig. 1A) and nalidixic acid (NAL) and norfloxacin (results not shown), produced a strong blue band in the border of the inhibition halo, reflecting their SOS induction capacity. In contrast, other structurally unrelated antibiotics, such as tetracycline, amikacin, rifampin, erythromycin, and chloramphenicol, did not show any apparent induction (Fig. 1A). However, different β-lactam antibiotics, such as ceftazidime (Fig. 1A), aztreonam, and imipenem (Fig. 1B), also induced the transcription of the dinB::lacZ operon fusion, as deduced from the blue band in the border of the inhibition halo.
FIG. 1.
Disk plate assays showing the effects of different antibiotics on the transcription of the dinB::lacZ operon fusion. (A) Antibiotics belonging to different families (the charge of each disk [in micrograms] is in parentheses) were as follows: on disk 1, ceftazidime (30); on disk 2, chloramphenicol (50); on disk 3, ciprofloxacin (5); on disk 4, erythromycin (30); on disk 5, tetracycline (50); on disk 6, rifampin (100); and on disk 7, amikacin (30). (B) Antibiotics affecting the cell wall synthesis at different steps (the charge of each disk [in micrograms] is in parentheses) were as follows: on disk 1, amoxicillin-clavulanate (30); on disk 2, imipenem (10); on disk 3, fosfomycin (50); on disk 4, no antibiotic; on disk 5, aztreonam (30); and on disk 6, d-cycloserine (100). All disks were purchased from Oxoid Ltd., except those containing rifampin and d-cycloserine, which were prepared in our laboratory.
The results obtained with β-lactam antibiotics suggested that cell wall damage may affect dinB transcription. Consequently, we tested the effects of several antibiotics known to affect different steps in cell wall physiology. Results shown in Fig. 1B indicate that the inhibition of cell wall biosynthesis (via fosfomycin or d-cycloserin), elongation by inhibition of PBP2 (via imipenem), or septation by inhibition of PBP3 (via aztreonam) induces dinB transcription. These results indicate that, independently of the level at which it is produced, cell wall synthesis inhibition leads to the induction of dinB transcription.
CAZ-mediated dinB induction is independent of LexA and RecA.
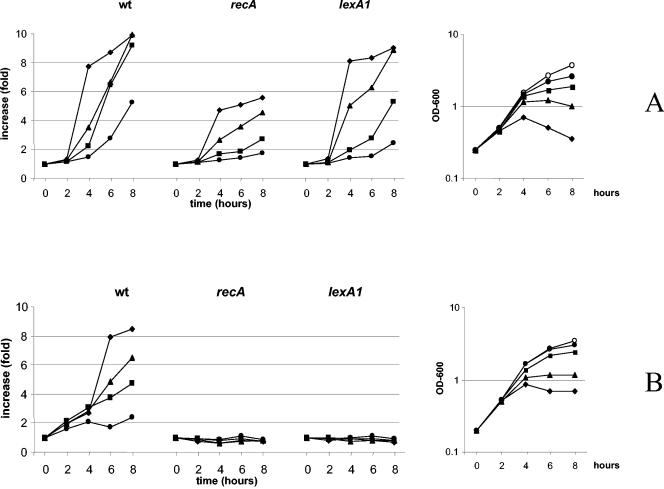
To understand better the molecular mechanism involved in dinB induction, we examined dinB expression using the β-lactam antibiotic CAZ as a model. As a part of the SOS regulon, dinB transcription is regulated by LexA and RecA. Thus, we tested the induction of the dinB::lacZ fusion in recA-defective and lexA1 Ind− backgrounds. Liquid cultures of strain GW1030 and its derivatives GW1030-recA Δ(srlR-recA)306::Tn10 and GW1030-lexA1 (lexA1 malE::Tn5), constructed by P1 transduction (14), were exposed to many different CAZ concentrations around the MIC, which, under our experimental conditions (i.e., LB agar, a large amount of inoculum, and 30°C), was determined to be 1.0 μg/ml. The β-galactosidase levels were determined after different exposure times (14). Figure 2A shows that the dinB transcription induction mediated by ceftazidime is time and concentration dependent in the wild-type context. Surprisingly, CAZ was able to induce the transcription of the dinB::lacZ fusion when SOS induction is blocked (i.e., in ΔrecA or lexA1 backgrounds), although the relative values of induction in the ΔrecA context are lower than in both the wild-type and lexA1 backgrounds, indicating that the effect may be not fully recA independent. The same experiments were performed in parallel using nalidixic acid as the inducer. In this case (Fig. 2B), dinB::lacZ transcription was induced in strain GW1030 but not in its ΔrecA or lexA1 derivatives.
FIG. 2.
Effects of the Δ(srlR-recA)306::Tn10 and lexA1 genetic backgrounds on the ceftazidime-mediated induction of transcription of the dinB::lacZ fusion. The curves on the left of each panel indicate the transcription levels (n-fold) between treated and nontreated cultures (calculated by assigning a value of 1 to the transcription level of the nontreated culture at each time point). Concentrations of CAZ (A) and NAL (B) expressed in micrograms per milliliter were as follows: 1 (♦), 2 (▴), 4 (▪), and 8 (•). The mean values of results from three independent experiments are displayed. The curves on the right of each panel represent the growth curves without and with CAZ or NAL for strain GW1030 (symbols are as described for the transcription levels; ○ indicates the values obtained without CAZ). Curves for recA and lexA1 derivatives are not presented because they are similar to those for strain GW1030. OD-600, optical density at 600 nm; wt, wild type.
CAZ produces an increase in the frequency of Lac+ revertants of strain SMR4562 that is mediated by DNA polymerase IV.
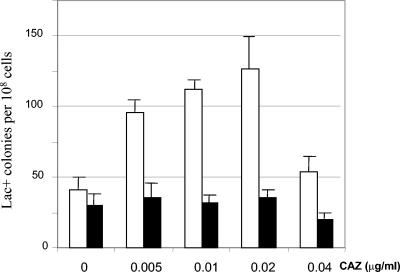
Because overproduction of DNA polymerase IV causes a mutator phenotype (10, 22), it was possible that the CAZ-mediated increase in dinB transcription could also cause an increase in the mutation frequency. As CAZ produces filamentous multinucleated cells when it inhibits its PBP3 target, the classical method for calculating the mutation frequency may lead to an overestimation of this frequency. To avoid this potential problem, we modified the classical experiment described by Cairns and Foster (5). About 108 SMR4562 cells carrying the allele lac +1 frameshift in the F128 episome {Δ(lac-proAB) thi ara [F′ proAB lacI33ΩlacZ]} (12) or SMR5830 cells (dinB10 [F′ dinB10 proAB+ lacI33ΩlacZ]) (13) from overnight LB agar stationary-phase cultures were resuspended in 4 ml of soft minimal agar and plated onto minimal M9 plates containing lactose as the only carbon source. The plates were incubated for 2 h at 37°C, and then soft agar containing appropriate quantities of CAZ was added to the plates. By this procedure, we ensured that every Lac+ colony was derived from a single plated cell, independently of the number of nucleoids present per filamented cell after the CAZ challenge. The numbers of Lac+ colonies and of viable cells were determined before and 1 and 2 days after the CAZ treatment for both treated and untreated cells. The viable counts (obtained by taking plugs from the lactose plates) were similar for control plates and for plates containing 0.005, 0.01, or 0.02 μg of CAZ/ml. Plates with 0.04 μg of CAZ/ml showed small decreases in viable counts, and those with higher concentrations of CAZ showed clear decreases in the number of viable cells at concentrations above 0.08 μg/ml. This result is consistent with the MIC of CAZ in minimal M9 medium for strains SMR4562 and SMR5830 (0.1 to 0.2 μg/ml). Ten independent experiments were carried out with strain SMR4562 and its dinB10 derivative. Mutant frequencies were calculated as the mean number of Lac+ colonies per 108 plated cells for CAZ-treated and nontreated SMR4562 and its dinB10 derivative 3 days after CAZ addition. Figure 3 shows that CAZ concentrations of 0.005, 0.01, and 0.02 μg/ml produced small increases in the rate of Lac reversion in strain SMR4562, with a maximum increase at 0.02 μg/ml. As stated above, at concentrations above 0.04 μg of CAZ/ml, cell viability is reduced and, even if the increase in dinB transcription is proportional to the CAZ concentration, the number of Lac+ colonies is below the limit of detection under our experimental conditions. Contrary to what happens with strain SMR4562, CAZ addition does not increase the frequency of Lac reversion in the strain lacking dinB, SMR5830 dinB10, indicating that this increase is dinB dependent.
FIG. 3.
Number of Lac+ revertants after CAZ treatment. Bars represent the mean numbers of Lac+ colonies per 108 plated cells of CAZ-treated and nontreated SMR4562 (open bars) and its dinB10 derivative (filled bars) after 3 days of incubation. T bars represent standard deviations.
There is evidence of interactions between the SOS response and other stress response pathways. In E. coli, induction of some SOS genes may occur as a function of growth phase (1, 6) or in resting bacteria in structured habitats (20). The stationary-phase sigma factor RpoS positively controls transcription of the dinB gene in E. coli (11). Thus, one can speculate that inhibition of cell wall synthesis by CAZ may cause an RpoS-dependent dinB response. To test this hypothesis, we studied the induction of the dinB::lacZ fusion in an rpoS-defective derivative of GW1030 (constructed by P1 transduction). Induction of dinB transcription occurred also in the rpoS-deficient background (data not shown). Thus, the induction of dinB transcription produced by inhibition of cell wall synthesis seems to be independent of RpoS.
It was previously shown that chromosome replication stop induces a septation arrest through induction of transcription of the SOS gene sulA (15). However, to our knowledge, this is the first time that induction of a DNA polymerase of the SOS system, due to a stop in cell wall synthesis, is described. Experiments are being performed in our laboratory to decipher the molecular basis of the induction of dinB transcription by cell wall synthesis inhibition.
Whether this increase in mutation frequency produced by β-lactam treatment, mediated by DNA polymerase IV, is important for the acquisition of resistance mutations in nature remains to be demonstrated. However, the results presented here are of particular concern. The paradoxical possibility that antibiotic treatments increase the rate of antibiotic resistance acquisition exists (2).
ADDENDUM IN PROOF
A recent article by Miller et al. (C. Miller, L. E. Thomsen, C. Gaggero, R. Mosseri, H. Ingmer, and S. N. Cohen, Science, 305:1629-1631, 2004) provided evidence that PBP-3 inhibition resulted in the induction of the SOS response in E. coli. Although part of the results presented here are similar, our study gives different results in some respects. (i) Miller et al. described that the induction of SOS mediated by β-lactams required RecA and LexA. Here we demonstrate that the induction of dinB is independent of RecA and LexA. (ii) Whereas Miller et al. reported that the SOS induction was exclusively produced by inhibition of cell septation (PBP3 inhibition), we demonstrate that dinB transcription is induced by inhibition of cell wall biosynthesis at different steps.
Our results suggest the existence of an additional SOS-independent pathway for the induction of dinB mediated by the inhibition of the cell wall synthesis.
Acknowledgments
We thank I. Matic and J. A. Ayala for helpful comments; S. Rosenberg and G. Walker for kindly providing strains SMR4562 and SMR5830 and strain GW1030, respectively; F. Baquero for kindly hosting the first steps of this work in his laboratory; and D. Hochberg for critically reading the manuscript.
This work was supported in part by grant 01/0020-02 from the Ministerio de Sanidad y Consumo (FIS) and grant BMC2001-0012 from the Ministerio de Ciencia y Tecnología. J.-M.G.-G. is the recipient of an astrobiology training fellowship from INTA. M.-R.B. was supported in part by grant QLK2-CT-2001-873 from the European Committee.
REFERENCES
- 1.Baquero, M. R., M. Bouzon, J. Varea, and F. Moreno. 1995. sbmC, a stationary-phase induced SOS Escherichia coli gene, whose product protects cells from the DNA replication inhibitor microcin B17. Mol. Microbiol. 18:301-311. [DOI] [PubMed] [Google Scholar]
- 2.Blázquez, J. 2003. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. 37:1201-1209. [DOI] [PubMed] [Google Scholar]
- 3.Bonner, C. A., S. Hays, K. McEntee, and M. F. Goodman. 1990. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:7663-7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brégeon, D., I. Matic, M. Radman, and F. Taddei. 1999. Inefficient mismatch repair: genetic defects and down regulation. J. Genet. 78:21-28. [Google Scholar]
- 5.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dri, A. M., and P. L. Moreau. 1993. Phosphate starvation and low temperature as well as ultraviolet irradiation transcriptionally induce the Escherichia coli LexA-controlled gene sfiA. Mol. Microbiol. 8:697-706. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez de Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Wodgate. 2000. Identification of additional genes belonging to the lexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg, E. C., R. Wagner, and M. Radman. 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296:1627-1630. [DOI] [PubMed] [Google Scholar]
- 9.Kenyon, C. J., and G. C. Walker. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. USA 77:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, S. R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding DNA Pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 166:207-215. [DOI] [PubMed] [Google Scholar]
- 11.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenzie, G. J., D. B. Magner, P. L. Lee, and S. M. Rosenberg. 2003. The dinB operon and spontaneous mutation in Escherichia coli. J. Bacteriol. 185:3972-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips, I., E. Culebras, F. Moreno, and F. Baquero. 1987. Induction of the SOS response by new 4-quinolones. J. Antimicrob. Chemother. 20:631-638. [DOI] [PubMed] [Google Scholar]
- 17.Radman, M., F. Taddei, and I. Matic. 2000. Evolution-driving genes. Res. Microbiol. 151:91-95. [DOI] [PubMed] [Google Scholar]
- 18.Ren, L., M. S. Rahman, and M. Z. Humayun. 1999. Escherichia coli cells exposed to streptomycin display a mutator phenotype. J. Bacteriol. 181:1043-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 20.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tompkins, J. D., J. L. Nelson, J. C. Hazel, S. L. Leugers, J. D. Stumpf, and P. L. Foster. 2003. Error-prone polymerase, DNA polymerase IV, is responsible for transient hypermutation during adaptive mutation in Escherichia coli. J. Bacteriol. 185:3469-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner, J., and T. Nohmi. 2000. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 182:4587-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. USA 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ysern, P., B. Clerch, M. Castaño, I. Gilbert, J. Barbé, and M. Llagostera. 1990. Induction of SOS genes in Escherichia coli and mutagenesis in Salmonella typhimurium by fluoroquinolones. Mutagenesis 5:63-66. [DOI] [PubMed] [Google Scholar]