Rice D53 repression motif links two sites of TPL corepressors to mediate TPL oligomerization and nucleosome association.
Keywords: D53, TOPLESS, strigolactone, Groucho, nucleosome, corepressor
Abstract
TOPLESS are tetrameric plant corepressors of the conserved Tup1/Groucho/TLE (transducin-like enhancer of split) family. We show that they interact through their TOPLESS domains (TPDs) with two functionally important ethylene response factor–associated amphiphilic repression (EAR) motifs of the rice strigolactone signaling repressor D53: the universally conserved EAR-3 and the monocot-specific EAR-2. We present the crystal structure of the monocot-specific EAR-2 peptide in complex with the TOPLESS-related protein 2 (TPR2) TPD, in which the EAR-2 motif binds the same TPD groove as jasmonate and auxin signaling repressors but makes additional contacts with a second TPD site to mediate TPD tetramer-tetramer interaction. We validated the functional relevance of the two TPD binding sites in reporter gene assays and in transgenic rice and demonstrate that EAR-2 binding induces TPD oligomerization. Moreover, we demonstrate that the TPD directly binds nucleosomes and the tails of histones H3 and H4. Higher-order assembly of TPD complexes induced by EAR-2 binding markedly stabilizes the nucleosome-TPD interaction. These results establish a new TPD-repressor binding mode that promotes TPD oligomerization and TPD-nucleosome interaction, thus illustrating the initial assembly of a repressor-corepressor-nucleosome complex.
INTRODUCTION
Understanding the mechanistic details of how transcription factors interact with global, evolutionarily conserved coactivators and corepressors is of central importance across species. Although transcriptional repression is thought to be equally important as transcriptional activation, comparatively little is known on how transcriptional repressors interact with transcription factors and corepressor complexes and how corepressor complexes epigenetically silence gene expression. Plant hormone signaling offers a paradigm for studying transcriptional repression, because the DNA binding transcription factors, key repressors, and corepressors are known for major plant hormones such as auxin, abscisic acid, and jasmonate (1).
Strigolactones (SLs) are plant hormones that are synthesized in response to low mineral nutrients and other environmental and endogenous stimuli. They modulate many aspects of plant architecture, most notably by repressing shoot branching, a major determinant of crop yields, as well as root development, leaf senescence, flower size, and stem and hypocotyl elongation (2, 3). In addition to their function as an endogenous hormone, SLs are also exuded from roots into the soil, where they function as signaling molecules to stimulate symbiosis with mycorrhizal fungi and nitrogen-fixing bacteria to increase nutrient availability, a signal that is exploited by parasitic weeds to localize their hosts (4–6). SLs are sensed by the α/β-hydrolase DWARF14 (D14), which binds and slowly hydrolyzes SL to a form that covalently binds D14 and induces a conformational change that allows the formation of a complex between D14, the repressor DWARF 53 (D53), and SCFD3 ubiquitin ligase (7–12). Complex formation stimulates ubiquitination of D53 by SCFD3, leading to proteasomal degradation of D53, which relieves repression of D53-regulated SL-responsive genes (2, 3).
Consistent with a role as a transcriptional repressor, D53 is nuclear-localized and physically interacts with the family of TOPLESS (TPL) corepressors (9, 12). Moreover, the Arabidopsis D53 orthologs SMXL6, SMXL7, and SMXL8 repress reporter gene activity when artificially tethered to DNA, and their down-regulation induces expression of the SL-responsive transcription factor BRC1 (13, 14) and of the auxin efflux carrier PIN1 (14, 15), key factors involved in the regulation of shoot branching. Unexpectedly, for a transcriptional repressor, D53 and its homologs share secondary structure and sequence conservation throughout the protein length with members of the Clp1 double AAA domain adenosine triphosphatase (ATPase) family of protein-disaggregating and protein-remodeling machines (9, 12).
TPL and TPL-related proteins (TPRs; in rice TPR1 and TPR2) comprise a family of corepressors that interact with numerous repressors, transcription factors, and adaptors to modulate plant development and signaling (1, 16–20). They are functionally and structurally related to yeast Tup1, insect Groucho, and mammalian transducin-like enhancer of split (TLE)/Grg transcription factor–binding corepressors that function as large scaffolds to recruit repressive chromatin-modifying complexes and mediate inhibitory interactions with the Mediator complex (21, 22). All proteins of this family consist of N-terminal tetramerization domains separated by a flexible proline-rich region from one or two C-terminal seven-bladed β-propeller domains. Whereas the N-terminal tetramerization domains of yeast Tup1 (23) and human TLE (24) consist of helical coiled coil dimers of dimers, the tetramerization domain of TPL proteins, termed TPL domain (TPD), adopts a structurally unrelated helical dimer of dimers fold (25). All members of this family bind small peptide repressor motifs found in transcriptional repressors (26–36) as well as conserved chromatin-remodeling (37–39) and class 1 histone deacetylase (HDAC) complexes (40–44). In addition, yeast and animal Tup1/Groucho/TLE proteins have been shown to directly bind nucleosomes (24, 34, 43, 45–47), which has been implicated in chromatin compaction by an unknown mechanism (24, 34). In contrast, it is unknown whether plant TPL proteins bind nucleosomes.
The most widespread repressor motif in plants is the ethylene response factor–associated amphiphilic repression (EAR) motif, whose main type is characterized by the sequence LxLxL, where L represents leucine residues and the two residues flanking the first L are often acidic amino acids (48–50). The LxLxL EAR peptides of Arabidopsis thaliana IAA1 (auxin-responsive protein 1), IAA10, and NINJA (novel interactor of JAZ), repressors involved in auxin and jasmonate signaling, all bind the same conserved groove in each of the four monomers of a TPD tetramer (25), suggesting that LxLxL EAR motifs have a conserved mode of TPD binding. Here, we identify two functional EAR motifs in rice D53 EAR-3, which is found in all D53 orthologs, and EAR-2, which is only conserved in monocots. We demonstrate that the monocot-specific D53 EAR-2 motif interacts with two separate binding sites in the TPR2 TPD. We further demonstrate that the TPR2 TPD binds to nucleosomes, and the bipartite interaction of the D53 EAR-2 motif with the TPR2 TPD promotes TPD tetramer-tetramer interaction, which results in a D53-mediated stabilization of TPD corepressor-nucleosome interaction.
RESULTS
D53 binds the TPD through two different EAR motifs
Rice D53 has been shown to interact with TPL family proteins in mammalian two-hybrid (M2H) and glutathione S-transferase pulldown assays (9). D53 contains three putative EAR motifs (Fig. 1A) (9, 12). Of these, only the C-terminal motif, which we have termed EAR-3, is conserved in both monocots and dicots (fig. S1), and the corresponding motif has been shown to mediate TPL interaction in the Arabidopsis D53 orthologs SMXL6, SMXL7, and SMXL8 (13, 14). To test which of the putative motifs can mediate the interaction with the rice TPL protein TPR2, we synthesized biotinylated peptides corresponding to each of the motifs and determined their interactions with the TPD domain of TPR2 in an AlphaScreen luminescence proximity assay. To our surprise, the peptide encompassing the conserved C-terminal EAR motif did not bind the TPD, whereas the EAR-2 motif (D53 residues 794 to 808) interacted with the TPD (Fig. 1B). The core LxLxL sequence (LDLNL) of the D53 EAR-2 motif is the same as in the conserved C-terminal EAR motif in dicots, but it is not conserved in the corresponding regions of SMXL proteins in dicots (fig. S1; see also alignment of the TPDs of TPL proteins in fig. S2). The interaction depended on a set of three conserved leucine residues of the motif because a triple L796A/L799A/L801A mutation abrogated the ability of EAR-2 to interact with the TPD (Fig. 1B). Using an AlphaScreen homologous competition assay, we determined a half maximal inhibitory concentration (IC50) value of 1.4 μM using conditions where the IC50 approximates the equilibrium dissociation constant (Kd) (fig. S3A) (see Materials and Methods). The EAR-2 peptide also bound to the TPD of the other two members of the rice TPL family, TPL and TPR1 (fig. S3, B to D). Whereas the D53 EAR-3 peptide did not interact with any of the TPDs, a corresponding, one-residue longer peptide from the Arabidopsis D53 ortholog SMXL7 did bind the TPR2 TPD, although less strongly than the D53 EAR-2 peptide (fig. S3E).
Fig. 1. D53(794–808) is a functional EAR motif.
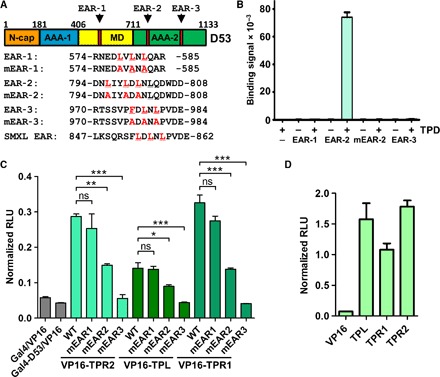
(A) Domain map of D53, with the position of three putative EAR motifs indicated. The segments with homology to the N-cap, the two AAA domains, and the middle domain (MD) of Clp-disaggregation machines are indicated. (B) AlphaScreen interaction assays between biotinylated peptides representing the three putative D53 EAR motifs and His6Sumo-tagged TPR2 TPD. (C) M2H assay between VP16 activation domain-fused TPL/TPR and Gal4 DBD-fused full-length wild-type (WT) D53 and triple EAR L→A mutant d53 [mEAR1, mEAR2, mEAR3; see mutated leucine residues in (A)]. RLU, relative luciferase units. (D) M2H assay of the interaction between the TPDs of the three TPL proteins and the 15-residue EAR-3 motif shown in (A) and (B) fused to the Gal4 DBD. ns, not significant (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001, one-way analysis of variance (ANOVA) with Tukey’s post hoc test; n = 3; error bars = SEM.
To test which of the EAR motifs is required for TPD interaction in the context of full-length D53, we analyzed the interactions between TPD proteins and wild-type and mutant D53 by M2H assays. As shown in Fig. 1C, wild-type D53 interacted with each of the three TPD proteins, as reported (9), whereas a triple L796A/L799A/L801A mutation of the EAR-2 motif reduced and F976A/L978A/L980A mutation of the EAR-3 motif almost abolished the interaction (see expression levels in fig. S4A). This suggested that both EAR-2 and EAR-3 may be able to directly bind the TPD of rice TPL proteins but that the EAR-3 peptide used in the AlphaScreen interaction assay may be insufficient for the interaction. We therefore genetically fused two larger D53 fragments encompassing EAR-3 [58-residue D53(952–1010) and 38-residue D53(962–1000)] to the Gal4 DNA binding domain (DBD) and tested the interaction of the corresponding proteins with the TPDs in the M2H assay. As shown in fig. S5, both EAR-3 fragment–containing fusion proteins interacted robustly with all three TPDs. We then fused only the 15–amino acid EAR-3 fragment that did not interact as free peptide to the Gal4 DBD, and this protein was also able to bind the TPD (Fig. 1D). These experiments suggested that both the EAR-2 and EAR-3 motifs can interact with the TPD but that the minimal 15-residue EAR-3 fragment needs to be fused to a protein to reduce its flexibility.
Both EAR-2 and EAR-3 contribute to D53 repressor function and SL signaling
Next, we tested whether the in vitro observed EAR interactions are important for D53 repressor function. We generated expression constructs of wild-type and mutant D53 fused to the heterologous Gal4 DBD and VP16 activation domain and cotransfected them together with a Gal4-dependent luciferase reporter gene into rice protoplasts. Whereas triple L→A mutation of EAR-1 (mEAR-1; see Fig. 1A) did not change reporter gene activity, triple L→A mutations of EAR-2, EAR-3, and, most markedly, the EAR-2/EAR-3 double mutation increased reporter gene activity, indicating a severe repression defect (Fig. 2A).
Fig. 2. Mutations in both EAR-2 and EAR-3 partially compromise D53 repressor activity in protoplasts and D53-mediated SL signaling in transgenic rice.
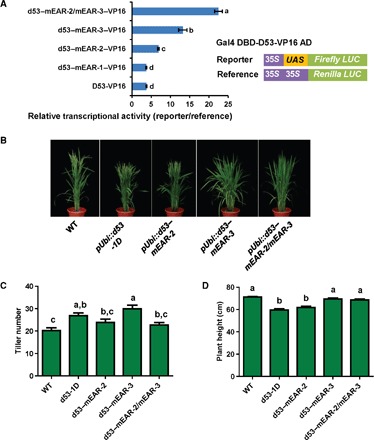
(A) D53 EAR mutant repressor activities. Gal4 DBD-D53-VP16 expression plasmids were cotransfected with a 35S promoter–Gal4 binding site (UAS)–firefly luciferase (LUC) reporter plasmid and a 35S promoter–Renilla luciferase reference plasmid into rice protoplasts; different letters above the columns indicate statistically significant differences between groups [Tukey’s honest significant difference (HSD) test, P < 0.05]. All data are presented as means ± SD. (B) Wild-type and mutant d53-overexpressing transgenic plants, tagged with 3×FLAG. (C and D) Tiller number (C) and height (D) of T2 generation transgenic plants. n = 15 plants; error bars = SEM; different letters at the top of each column indicate a significant difference at P < 0.05 determined by Tukey’s HSD test.
To further confirm that both EAR-2 and EAR-3 are functionally important in vivo, we generated transgenic rice overexpressing wild-type D53, the SL-insensitive d53 mutant allele (d53-1D) (9, 12), and d53–mEAR-2, d53–mEAR-3, and d53–mEAR-2/mEAR-3 in wild-type Nipponbare (Fig. 2, B to D) (9, 12, 51). All transgenic plants overexpressing mutant d53 genes showed a stronger tillering phenotype than did those overexpressing the wild-type D53 gene; therefore, we analyzed the effect of different EAR motifs on D53’s repression activity by using the mutant d53-1D gene. Expression of d53-1D and d53–mEAR-3 significantly increased the tiller number relative to wild type, whereas both d53–mEAR-2 and d53–mEAR-2/mEAR-3 showed a smaller increase that was not statistically significant, indicating that EAR-3 is critical for repression of tiller number (Fig. 2C). In contrast, when we determined plant height, both d53-1R and d53–mEAR-2 exhibited a dwarf phenotype, whereas d53–mEAR-3 and d53–EAR-2/mEAR-3 only exhibited smaller, statistically not significant reductions in plant height (Fig. 2D). Together, these data suggest that both EAR-2 and EAR-3 contribute to D53 repressor activity and SL responsiveness but that the two EAR motifs may have diverse functions in regulating plant height and tiller number. Deletion of the EAR-3 corresponding EAR motif in Arabidopsis also causes partial loss of SL signaling (15).
The D53 EAR-2 motif binds the interface between two TPR2 TPD tetramers
To determine the molecular details of the TPD interaction with the monocot-specific D53 EAR-2 motif, we crystallized the complex between TPR2(1–209) and the TPD-interacting EAR-2 peptide [D53(794–804)], and solved its structure at a resolution of 2.55 Å (table S1). Whereas the TPD in the complex assumed essentially the same structure as the apo TPD [Protein Data Bank (PDB) code 5C6Q] and the TPD in complexes with EAR motifs from IAA1 (5C7J), IAA10 (5C7E), and NINJA (5C6V) (25), the D53 EAR-2 peptide simultaneously bound to two different sites of the TPD: the canonical surface groove in each of the TPD monomers that is also bound by IAA1-, IAA10-, and NINJA-EAR peptides (which we refer to as “site 1,” formed by helices α5, α6, α7, α8, two short 310 helices, and a connecting loop) as well as a new binding pocket formed at the C terminus of the dimer interface near the TPD zinc finger (which we refer to as “site 2,” which is formed by the C-terminal loop and C-terminal helix α9 from one monomer and helices α1 and α9 from the other monomer; Fig. 3; see fig. S6 for the omit map of the EAR-2 peptide). The C-terminal part of EAR-2 (LDLNL) matches the consensus EAR motif and binds to site 1, the canonical EAR motif–binding groove, whereas the partially overlapping N-terminal DNLIYLDL part of the motif binds the EAR-2–specific site 2 (the C-terminal binding groove) (Fig. 3, A and B). Details of the interaction are shown in Fig. 3B and summarized in Fig. 3C.
Fig. 3. Structure of TPR2 TPD in complex with D53 EAR-2 peptide.
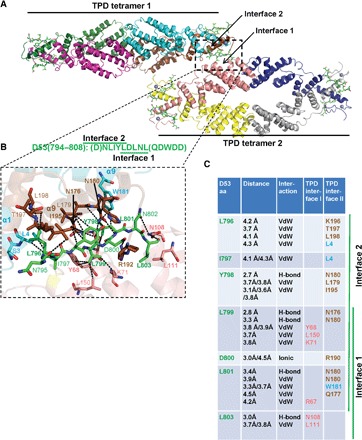
(A) Structure overview showing two TPD tetramers with an EAR-2 peptide at the TPD tetramer-tetramer interface (dashed rectangle). EAR motif peptides are shown as green stick models, the TPDs are shown as cartoon models, and the TPD Zn2+ ions are shown as gray spheres. (B) Close-up of the interface; key amino acids and bonds are shown and labeled. D794 and Q804 of the EAR-2 motif (letters in parentheses) were not resolved in the structure. (C) Main interacting residues between TPR2 TPD and the D53 EAR-2 motif peptide. Brown, TPD monomer 1; cyan, TPD monomer 2; pink, TPD monomer from interacting tetramer [same color code in (A)]; bold, key interaction residues. aa, amino acids; VdW, van der Waals bond (4.5 Å cutoff); H-bond, hydrogen bond (3.5 Å cutoff).
Mutations in either site 1 or site 2 of the TPD are insufficient to abolish the TPD–EAR-2 interaction
To test whether both binding grooves can independently form complexes with the D53 EAR motif, we first introduced mutations into the TPD that replaced key residues of the canonical EAR binding site 1 with alanine (fig. S7). Although all of these mutations abolished or strongly reduced binding to the NINJA EAR motif, which only binds to site 1 (fig. S7A) (25), they only moderately affected the interaction with the bipartite EAR-2 motif (fig. S7B). To gain further insight into binding of a site 1 mutant TPD, we determined the crystal structure of the complex between TPR2 TPD L111A/L130A and D53 EAR-2 (fig. S7, D to F). Rather than losing binding to site 1 of the TPD, the EAR motif formed new interactions with the TPD tetramer-tetramer interface by reversing its orientation relative to the wild-type TPD, which allowed the TPD to engage in less extensive alternative interactions (fig. S7E).
Next, we mutated key site 2 interface residues to alanine. As with site 1 mutations, none of the site 2 mutations could disrupt the interaction between the mutant TPD and the D53 EAR-2 motif (fig. S8A). The small to moderate reductions in the binding signal for TPD N180A, W181A, and L198A appear to be due to general protein destabilization as well as small variations in protein amounts (fig. S8C), because these mutations decreased, to an even larger degree, the AlphaScreen signal for NINJA EAR (fig. S8B), which does not bind site 2 and has an at least 10-fold lower overall affinity for the TPD [IC50 of 16 μM (25) compared to 1.4 μM]. We were also able to crystallize the complex between D53 EAR-2 and the TPD L179A/I195A site 2 mutant and determine its structure (fig. S8, D to F). Although the resolution of this structure is relatively low (3.15 Å), in the complex, EAR-2 appears to have lost interaction with the mutated site 2 while retaining extensive interactions with site 1 (fig. S8E).
Combined mutations in both interfaces abolish TPD–EAR-2 complex formation
To further confirm modular binding to both the site 1 and site 2 interfaces, we combined both mutations to generate a TPD L111A/L130AL179A/I195A quadruple mutant protein. Whereas the site 1 (L111A/L130A) and site 2 (L179A/I195A) double mutants still retained binding to the D53 EAR-2 peptide, the quadruple mutant protein lost the ability to interact with the peptide (Fig. 4A). Moreover, when we crystallized the quadruple mutant TPD in the presence of EAR-2 peptide, the peptide was not resolved in the structure (fig. S9), indicating that the EAR motif lost the ability to form a complex with the mutant TPD even in the presence of the very high protein and peptide concentrations used for crystallization.
Fig. 4. Only mutations that affect binding of both TPD–EAR-2 interfaces disrupt the TPD–EAR-2 interaction.
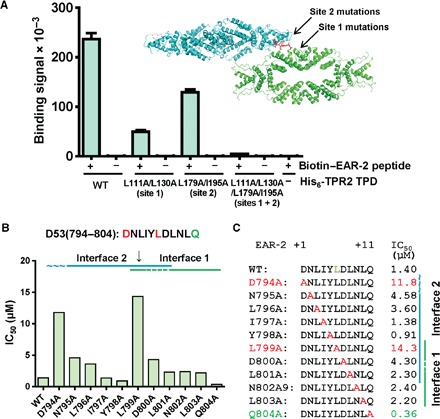
(A) TPD L111A/L130A/L179A/I195A is unable to bind the D53 EAR-2 motif. AlphaScreen interaction assay of His6Sumo-TPR2 TPD wild-type and mutant proteins with biotinylated D53 EAR-2 peptide. n = 3; error bars = SD. See fig. S4B for SDS gel with protein loading controls. (B and C) Summary of AlphaScreen homologous competition assays of D53 EAR-2 mutant peptides (see fig. S10). Concentration of untagged wild-type and D53 EAR-2 mutant peptides required to reduce 50% of the AlphaScreen biotin-D53/His6Sumo-TPD binding signal (IC50). n = 3; error bars = SD.
Finally, we performed an alanine scanning mutagenesis of the EAR-2 motif (D53 residues 704 to 804) and quantitatively determined the relative TPD affinities of each of the mutant peptides by AlphaScreen homologous competition assays (fig. S10; summarized in Fig. 4, B and C). Of the EAR-2 residues resolved in the complex crystal structure, only L799 makes substantial interactions with both the site 1 and the site 2 binding grooves of the TPD (Fig. 3C), and correspondingly, only the L799A mutant peptide was strongly compromised in its ability to compete the TPD interaction with wild-type EAR-2 peptide (IC50 increase from 1.4 to 14.3 μM; Fig. 4, B and C, and fig. S10). In addition, both terminal EAR-2 motif residues were unresolved in the complex structure containing the wild-type TPD, yet replacement of these two residues with alanine also substantially changed the strength of the TPD interaction. The N-terminal D794, although not resolved in the structure of the wild-type complex, was resolved in the TPD L111A/L130A mutant complex, in which its carboxyl group forms intrapeptide interactions with N795, L796, and Y798, suggesting that D794 may contribute to the overall conformation of the EAR-2 motif in the binding groove and therefore influence binding to both TPD sites. The C-terminal Q804 of EAR-2 is not resolved in any of the structures, but its replacement with alanine increased the strength of the interaction by more than threefold, indicating that the bulky side chain of Q804 impedes the interaction. Collectively, we have presented extensive evidence that the bipartite D53 EAR-2 motif modularly interacts with both site 1 and site 2 of the TPD and thus that either interface is sufficient for complex formation with the D53 EAR motif.
The D53 peptide mediates TPD tetramer-tetramer interaction
The ability of the EAR-2 peptide to simultaneously bind two different TPD tetramers suggests that it may mediate or stabilize the formation of oligomeric TPD assemblies. To probe for tetramer-tetramer interaction, we generated two different forms of the TPR2 TPD: one in which the TPD is biotinylated and the other in which it is His6Sumo-tagged. This allowed immobilization of the tetramers to streptavidin-coated donor beads and Ni-chelated acceptor beads, respectively, to determine their interaction by an AlphaScreen assay (Fig. 5A). The data in Fig. 5B suggest that TPD tetramers can weakly interact with each other, and this interaction is greatly enhanced by adding increasing amounts of untagged D53 EAR-2 peptide. In contrast to wild-type EAR-2, an EAR-2 peptide with a mutation in Y798, which retains high TPD binding affinity (Fig. 4, B and C) and makes key interactions with TPD site 2 but does not interact with site 1 (Fig. 3C), was unable to increase the AlphaScreen signal (Fig. 5B). This provides strong evidence that the signal increase is due to the ability of EAR-2 to simultaneously bind two separate sites on the TPD. We note that an alternative interpretation for the increased AlphaScreen signal is that EAR-2 could destabilize biotin- and His6Sumo-tagged homotetramers to promote the formation of mixed tetramers containing both biotin- and His6Sumo-tagged subunits. However, the EAR-2 peptide did not enhance the binding signal for TPD L111A/L130A (TPD site 1 mutation) (Fig. 5C) and had no effect on the stability of the TPD tetramer in a thermostability shift assay (fig. S11), consistent with the increase of the TPD-TPD AlphaScreen interaction signal being due to D53 EAR-2–mediated tetramer-tetramer interaction. We further confirmed the ability of the EAR-2 motif to induce TPD-TPD interaction by dynamic light scattering (DLS). The apo TPD forms a stable tetramer in solution with dimensions of ~160 Å × 60 Å × 40 Å (25), consistent with a tight DLS profile with a mean hydrodynamic radius of ~50 Å (100 Å diameter; Fig. 5D). Moreover, addition of NINJA EAR motif peptide or D53 EAR-2 Y798A peptide, which only binds one site of the TPD, displayed a similar TPD size distribution and average size as for the apo TPD. In marked contrast, the wild-type EAR-2 peptide caused an asymmetric broadening of the size distribution with a shift to a markedly increased hydrodynamic radius (up to a 300 Å radius; Fig. 5D and table S2), indicative of the formation of oligomeric TPD assemblies. Collectively, these data indicate that the D53 EAR-2 motif can induce or stabilize TPD tetramer-tetramer interaction and that this effect requires both TPD-EAR interfaces.
Fig. 5. D53 EAR-2 peptide induces TPR2 TPD tetramer-tetramer interaction.
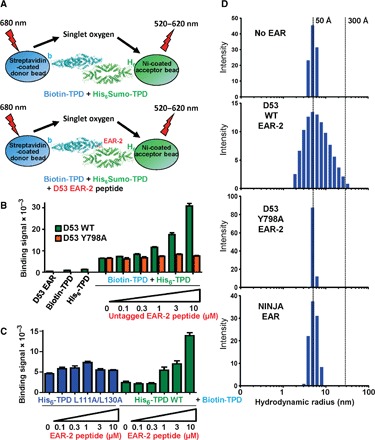
(A) Cartoon presentation of AlphaScreen oligomerization assay (see Materials and Methods for details). Biotin-tagged TPR2 TPD and His6Sumo-tagged TPR2 TPD were incubated in the absence and presence of increasing concentrations of untagged D53 EAR-2 peptide. (B) Assay with wild-type and Y798A D53 EAR-2 peptide. (C) Assay with wild-type and L111A/L130A His6Sumo-TPD. n = 3; error bars = SD. (D) DLS of TPR2 TPD in solution in the absence and presence of NINJA and D53 wild-type and Y798A EAR-2 peptides.
Because the synthesized 15–amino acid D53 EAR-3 peptide was unable to bind any of the three TPDs, we had synthesized 16–amino acid–long peptides of the corresponding motifs from the D53 orthologs SMXL6, SMXL7, and SMXL8 of Arabidopsis. These EAR peptides interacted with the TPR2 TPD in an AlphaScreen assay (shown for SMXL7 EAR in fig. S3E). However, in contrast to D53 EAR-2, they did not enhance the TPD-TPD interaction (fig. S12), even though we cannot exclude the possibility that longer D53 EAR-3 or SMXL EAR peptides might be able to do so.
The tpl-1 mutation stabilizes an interface 1–interface 2 interaction in the absence of EAR peptides
Although loss-of-function mutations in any individual member of the TPL family have little effects due to genetic redundancy, a temperature-sensitive point mutation in Arabidopsis TPL (tpl-1; N176H) causes the marked phenotype that has given this protein family its name: At the nonpermissive temperature, shoots are converted to apical roots to generate “topless” seedlings (41, 52). We have recently shown that N176 in rice TPR2 TPD (marked in the sequence alignment in fig. S2) is surface-exposed and induces the formation of higher-order TPD oligomers, leading to severe TPD aggregation (25). We noticed that N176 is centrally located at interface 2, suggesting that N176H may cause TPD aggregation by stabilizing an inherently weak interface 1–interface 2 interaction in the absence of EAR peptides. When we model the N176H mutation into our structure of the TPD/EAR-2 complex, H176 can directly interact with Y68 of interface 1 (Fig. 6, A and B), consistent with a possible function of H176 to stabilize tetramer-tetramer interaction. Because we were unable to determine a structure of TPR2 TPD N176H due to its severe aggregation, we have used a genetic approach to experimentally address whether TPD N176H aggregation is due to an H176-mediated interaction with interface 1 residues. We individually introduced four different second-site mutations into interface 1 residues in close vicinity to N176H (N176H/R67A, N176H/Y68A, N176H/Y68R, and N176H/K71A), expressed and purified the mutant proteins, and tested their aggregation in vitro. As shown in Fig. 6C, when we centrifuged the lysate of wild-type TPD, most TPD remained as soluble protein in the supernatant, whereas >90% of TPD N176H aggregated and precipitated during centrifugation. Each of the four second-site mutations greatly suppressed the N176H aggregation phenotype, providing strong genetic support for N176H-mediating TPD aggregation by stabilizing an interface 1–interface 2 interaction (Fig. 6C). Moreover, the second-site mutations also partially suppressed the EAR-2 interaction deficit of TPD N176H (Fig. 6D).
Fig. 6. The tpl-1 (N176H) mutation can be partially rescued by mutations in interface-2.
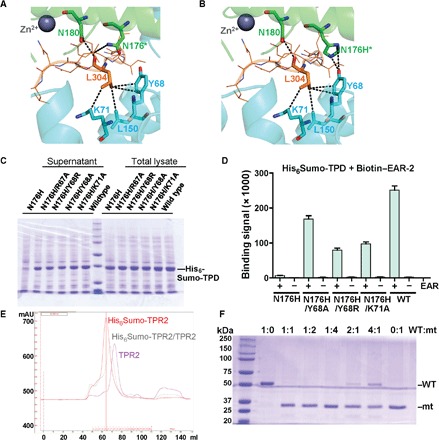
(A and B) Interface between two wild-type (N176) TPD tetramers (A) or with modeled N176H (tpl-1) mutation (B). The EAR-2 peptide is shown as orange line model, with EAR-2 L304 and its interacting residues shown in stick presentation. (C) Mutations in interface 1 residues Y68 and K71 rescue the aggregation phenotype of TPD N176H. Coomassie-stained protein gel [see (F) for size marker] of lysates of His6Sumo-TPD–expressing cells and lysate supernatant after centrifugation. (D) AlphaScreen interaction between biotin-D53 EAR-2 peptide and wild-type and mutant His6Sumo-TPD (n = 3; error bars = SD). (E) Overlay of size exclusion chromatography (SEC) profiles of TPR2 TPD, His6Sumo-TPD, and TPD/His6Sumo-TPD. mAU, milliabsorbance units. (F) TPD N176H destabilizes wild-type TPD. Different amounts of wild-type His6Sumo-TPD (WT) and untagged TPD N176H (mt) were mixed, incubated on ice for 4 hours, and centrifuged. Supernatants were analyzed by SDS–polyacrylamide gel electrophoresis (PAGE). Note that the TPD ratios are based on the amounts of pure WT and mt protein in supernatants following centrifugation (compare 1:0 and 0:1 ratios).
The dominant phenotype of the tpl-1 mutation further suggested that TPL/TPR proteins can form mixed oligomers and that mixed oligomers containing TPD N176H subunits form aggregates. As shown in Fig. 6E, when we combined separately expressed and purified His6Sumo-TPR2 TPD and untagged TPD, subunits of the different tetramers efficiently exchanged. When we further incubated wild-type His6Sumo-TPD protein with N176H mutant TPD at different ratios for 4 hours on ice and then centrifuged, wild-type TPD became largely depleted (Fig. 6F). Collectively, this suggests that the tpl-1 phenotype is due to N176H-stabilizing formation of insoluble aggregates through interface 1–interface 2 interaction, leading to TPL/TPR protein depletion in the context of cells. Additional experiments will be required to further characterize these complexes.
The TPD binds recombinant nucleosomes and histone H3 and H4 tail peptides
In addition to recruiting repressive epigenetic complexes to transcription factors, TPL homologs from yeast to humans also directly bind nucleosomes at least in part through their N-terminal tetramerization domains (24, 45–47), which has been implicated in chromatin compaction through an unknown mechanism (24, 34). Although the tetramerization domains do not share sequence or structural homology with plant TPDs, we speculated that their functions might be conserved. When we incubated biotinylated recombinant human mononucleosomes with His6Sumo-TPR2 TPD, but not with an unrelated control protein [small heterodimer partner (SHP)], we could detect a clear AlphaScreen binding signal, demonstrating that the TPD can directly bind nucleosomes (Fig. 7A). We could also detect nucleosome binding by the TPD from the related repressors TPL and TPR1 (Fig. 7A). Because many protein-nucleosome interactions are mediated through the N-terminal tails of histones, we tested TPD binding to several biotinylated histone peptides. The TPD interacted robustly with H31–20 and H41–23 and weakly with H2A1–17, H2B1–24, H3.315–34, and H411–27 (Fig. 7B). Acetylation of the histone tails is a hallmark of transcriptional activation; correspondingly, acetylation of the H4 tail at K5, K8, K12, K16, and especially K20 all decreased TPD binding (Fig. 7C), which is consistent with the preference of mammalian TLE/Gro/Grg proteins (24, 45, 46) for non- or underacetylated histones. In contrast, repressive methylations at H3 K9 and H4 K20 increased TPD binding (Fig. 7C), consistent with their role in transcription repression.
Fig. 7. TPR2 TPD binds reconstituted nucleosomes and histone tails.
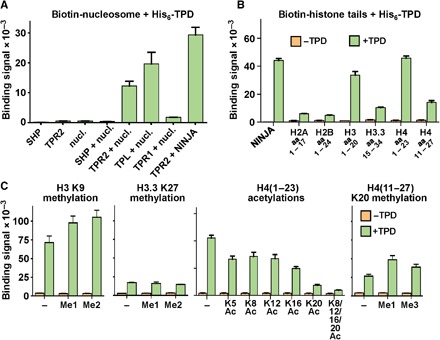
(A) AlphaScreen interaction between 100 nM biotin-tagged nucleosomes (nucl.) and 100 nM of the His6Sumo-tagged TPDs of TPL, TPR1, and TPR2. The interaction between the TPR2 TPD and the NINJA EAR motif serves as a positive control, and the interaction between the nucleosome and the unrelated nuclear receptor SHP serves as a negative control. Note that the AlphaScreen signal is dependent on proximity and therefore generates a weaker signal for biotinylated nucleosomes than for small biotinylated peptides. (B and C) AlphaScreen interaction between 100 nM His6Sumo-tagged TPR2 TPD and 100 nM biotin-tagged unmodified (B) or modified (C) histone tail peptides. n = 3; error bars = SD.
D53 EAR-2 peptide stabilizes the TPD-nucleosome interaction
Because at least four sites on a nucleosome (two H3 and two H4 tails) could independently interact with a TPD tetramer, TPD oligomerization through D53 EAR motifs might modulate the stability of TPD-nucleosome complexes through the formation of multivalent interactions. To test this hypothesis, we performed a TPD-nucleosome binding reaction in the presence of the D53 EAR-2 motif peptide. Although the TPD bound recombinant core nucleosome particles in the absence of D53 EAR-2, increasing amounts of the EAR-2 peptide markedly increased the binding signal (Fig. 8A), demonstrating that the D53 EAR motif stabilizes the TPD-nucleosome interaction. We repeated this experiment with the D53 Y798A mutant motif as well as with the NINJA EAR wild-type motif peptides, which bind only to one site of the TPD. These motifs are therefore expected to be incapable of stabilizing the TPD tetramer-tetramer formation. Both EAR motif peptides failed to enhance the TPD-nucleosome interaction, suggesting that the ability of D53 EAR-2 to stabilize TPD-nucleosome binding is due to EAR-2’s ability to promote the TPD tetramer-tetramer interaction, therefore allowing multivalent TPD-nucleosome interactions that enhance the avidity of TPD-nucleosome binding.
Fig. 8. D53 EAR-2 peptide stabilizes the binding between TPR2 TPD and recombinant nucleosomes.
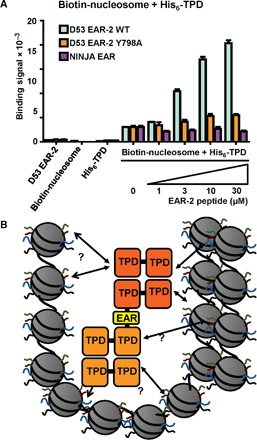
(A) AlphaScreen interaction between 100 nM biotin-tagged nucleosomes and 100 nM His6Sumo-tagged TPR2 TPD in the absence and presence of increasing concentrations of untagged D53 EAR-2 peptide. n = 3; error bars = SD. (B) Speculative model for chromatin compaction through D53 EAR-2–mediated TPD oligomerization.
DISCUSSION
D53 has two TPD-binding EAR motifs: A conserved C-terminal motif and a central, novel bipartite EAR
The D53 protein is a central component of the SL signaling pathway and is required for repression of SL target genes. Its protein level is directly regulated by SL through the formation of SL-dependent D14-D3-D53 receptor complexes, in which D53 is ubiquitinated and targeted for proteasomal degradation in a hormone-dependent manner (7, 9, 12). Although direct interaction of a D53/SMXL protein with an SL-responsive transcription factor, such as BRC1, has not been shown, repression has been shown to require EAR-dependent interaction between D53/SMXL and TPL/TPR (9, 13–15). Here, we have found that, in addition to the C-terminal EAR-3 motif that is conserved in all D53 orthologs, monocot D53 has a novel, bipartite EAR motif, EAR-2. EAR-2 consists of the C-terminal LxLxL motif that binds the canonical TPD binding site 1 and an N-terminal DNLIYLDL motif that binds the previously unrecognized TPD binding site 2. We have further shown that both motifs contribute to repression and SL signaling, but EAR-2 may be more important for tiller number and EAR-3 may be more important for plant height. Deletion or triple L→A mutation of the conserved C-terminal EAR motif of Arabidopsis SMXL7 also led to only partial loss of SL signaling. This includes a very small reduction in plant height that was statistically significant only for the deletion, but not the L→A mutation (15), very similar to the phenotype of the L→A mutation of the corresponding EAR-3 in rice. It is therefore possible that SMXL7 and other dicot D53 orthologs have, in addition to the identified C-terminal EAR motif, an unidentified nonhomologous EAR motif that is functionally analogous to EAR-2.
The TPD of TPL family proteins directly binds nucleosomes and the tails of histones H3 and H4
TPRs from yeast to human have been shown to bind to nucleosomes, and this interaction is mediated at least in part through interaction of their N-terminal tetramerization domains with the tails of histones H3 and H4 (24, 34, 43, 45–47). Although the TPD does not share sequence homology with the tetramerization domains of Tup1 and TLE/Groucho, our finding that the TPD of all three rice TPL/TPR proteins directly binds nucleosomes and has the same histone H3 and H4 tail specificity extends the functional conservation among this family of proteins. Moreover, TPL interacts with the Mediator complex (53) and recruits class 1 HDACs, which promiscuously deacetylate acetyllysines of histone tails (54, 55). The TPD shares with its yeast and animal counterparts a higher binding affinity for non- or underacetylated histone tails, suggesting that deacetylation by TPL-recruited HDACs may promote a more stable formation of nucleosome/TPL/HDAC complexes through positively reinforcing interactions, which may contribute to the propagation of repressive chromatin structures. In addition to nucleosome binding, the N termini of all family members form extended helical dimers of dimers and interact with repression motif peptides, whereas the C termini form seven-bladed β-propeller domains that may independently interact with nucleosomes and other classes of repression motif peptides, as shown for Tup1 and TLE (21, 56). All family members recruit chromatin-remodeling and class I HDAC complexes, which are required for their corepressor function.
D53 EAR motif–induced TPD oligomerization allows the formation of highly multivalent TPD interactions
Structural, mutational, and biochemical data provide conclusive evidence that the bipartite D53 EAR-2 motif mediates TPD tetramer-tetramer interactions through simultaneous interactions with both binding sites. Nucleosome binding becomes markedly stabilized by EAR-2 motif peptide, which depends on the ability of the EAR motif to mediate TPD tetramer-tetramer interactions. Because nucleosomes are likely to make multiple TPD interactions through their four TPD-binding histone tails, we propose the formation of multivalent interactions with oligomeric TPDs as a likely mechanism (see model in Fig. 8B). In addition, induced TPD oligomerization may also stabilize the interaction between the TPD and other multivalent complexes.
D53 may regulate formation of TPD-nucleosome complexes and higher-order chromatin structures
The ability of the D53 EAR-2 motif to stabilize TPD-nucleosome complex formation suggests that D53, which is rapidly degraded in response to SLs, may have a regulatory role in enhancing TPL-mediated formation of repressive chromatin structures. This may occur by more efficiently bringing chromatin-remodeling and histone-modifying complexes to their substrate or by directly affecting higher-order nucleosome structure. Notably, the functional TPL homologs Tup1 and TLE have been implicated in nucleosome repositioning and chromatin compaction (24, 34, 57–61). Chromatin compaction is a hallmark of transcriptionally repressed, silenced chromatin, and full-length TLE as well as its N-terminal tetramerization domain have been shown to directly compact nucleosomes in a reconstituted system (34). The extensive functional similarity between TPL, TLE, and Tup1 suggests that TPL could also function in chromatin compaction. Known chromatin compaction factors such as HP1, PRC2, Sir2/3/4, and TLE are all proposed to condense chromatin by often weak auto-oligomerization, which allows multivalent interactions with noncontiguous regions of chromatin. If TPL does compact chromatin, then D53-induced TPL self-association could present a mechanism for regulation of chromatin compaction other than by site-specific recruitment.
What is the function of the putative D53 protein remodeling activity?
D53 has similarity to double AAA proteins, which form hexamers consisting of two stacked hexameric rings with a total of 12 AAA ATPase domains. Consistently, insect cell–purified recombinant rice D53 is hexameric, and D53 hexamers appear to interact with TPD tetramers (fig. S13). The similarity is closest to Hsp104/ClpB-type double AAA chaperones, which also contain N-terminal (N) domains implicated in substrate binding and middle (M) domains that are bound by aggregate-associated Hsp70 to activate Hsp104 (62). We used I-TASSER (63–65) to generate a structural homology model of D53 with high confidence in the overall fold, which places both EAR-2 and EAR-3 on the surface of the lower ring (fig. S14).
AAA proteins are molecular machines that are functionally characterized by their adenosine triphosphate (ATP)–dependent protein disaggregation and protein complex assembly and disassembly activities, in which substrates are separated from aggregates or complexes by threading through a central pore (66, 67). Protein threading is mediated in part by transient substrate binding by conserved tyrosine residues that stick into the pore. ATP hydrolysis leads to conformational changes in the tyrosine-containing pore loops to generate pulling forces (62, 66). Many AAA proteins use DNA/protein complexes as substrate, and there is precedence for double AAA proteins with functions in transcriptional repression. The best-known examples are mammalian reptin and pontin (and their yeast homologs Rvb1 and Rvb2), AAA proteins that together can form a heterododecameric double AAA ring but lack N and M domains (68, 69). They interact directly or indirectly with transcription factors and are integral components of the INO80 and SWR chromatin-remodeling complexes and the NuA4 histone acetyltransferase complex (68, 70–73) and have been linked to chromatin decondensation at the end of mitosis (74). Similarly, the N and M domain–containing AAA domain of the yeast Sir3 component of the Sir2/3/4 chromatin-condensing corepressor complex is required for gene silencing in vivo (75), directly binds chromatin (75), and interacts with histones H3 and H4 in vitro (76).
Although we have shown that the monomeric D53 EAR-2 motif peptide is sufficient to induce tetramer-tetramer interaction and stabilize TPD-nucleosome complex formation, the likely hexameric full-length D53 oligomer would contain six EAR-2 and six EAR-3 motifs that would be available for binding of multiple nucleosome-interacting TPDs, adding additional layers of multivalent interactions. In addition, given the role of other known transcriptional AAA proteins in chromatin remodeling and condensation, it is tempting to speculate that the physical vicinity in a D53-TPL-chromatin complex could allow a putative D53 remodeling activity to contribute to nucleosome repositioning and/or higher-order chromatin reorganization. We note that the conserved hydrophobic tyrosine in the putative pore loop is replaced in D53 by a positively charged lysine (GKTG instead GYVG), which appears to be more suitable to thread negatively charged DNA rather than unfolded polypeptide chains (see also model in fig. S13). In an alternative, nonmutually exclusive model, D53 remodeling activity might be required to rearrange the constitutive SCFD3-D53 complex (9) to trigger D53 ubiquitination in the presence of D14 and SL. Notably, the SL-insensitive d53-1D mutant differs from wild type by precise deletion of the pore loop. This mutation does not affect the SL-dependent interaction of D53 with D14 but abolishes D53 ubiquitination (9, 12), thereby linking a putative D53 threading activity to SCFD3 function.
In summary, our crystal structure of the TPD in complex with the D53 EAR-2 motif peptide revealed a novel, bipartite TPD binding mode that allows the EAR-2 motif to mediate TPD oligomerization and TPD-nucleosome interaction. Although much work remains to be done to understand its biological significance, this observation, together with our findings that the TPD binds nucleosomes and that EAR-2 enhances this interaction, links SL-dependent D53 protein levels to stabilization of TPD-nucleosome complexes and formation of repressive chromatin structures.
MATERIALS AND METHODS
DNA constructs and reagents
The TPL/TPR expression clones were described previously (25). For the M2H assay, the full-length D53 open reading frame was cloned into Gal4 plasmid pM (Clontech). Cloning of full-length rice TPL, TPR1, and TPR2 open reading frames as fusions with an N-terminal VP16 activation domain was described previously (25). Site-directed mutagenesis was performed using the QuikChange method (Stratagene). All expression constructs and mutations were confirmed by DNA sequencing. Nonhistone peptides were synthesized by Peptide 2.0 Inc. Histone peptides were synthesized by the High-Throughput Peptide Synthesis and Array Core Facility at the University of North Carolina at Chapel Hill. Biotinylated recombinant human nucleosomes were produced by EpiCypher Inc.
Protein expression and purification
For crystallization, the His6Sumo-tagged wild-type and mutant TPR2 TPD proteins were expressed in Escherichia coli BL21 (DE3) cells and purified as described (25). Briefly, cleared lysates were passed through a Ni-chelating Sepharose column, followed by proteolytic cleavage of the tag using Ulp1 Sumo protease, removal of the tag by repassing through a Ni-chelating high-performance Sepharose column, and SEC. For AlphaScreen assays, His6Sumo-TPD was purified from 50-ml cultures without tag cleavage to allow binding to Ni-chelated acceptor beads, as described (25). To prepare biotinylated TPD for binding to streptavidin-coated donor beads, we followed the methods described previously (77).
Crystallization
Purified rice TPR2 TPD protein was concentrated to about 10 mg/ml. The TPR2 TPD protein was mixed with D53 EAR-2 peptide at a molar ratio of 1:2 before setting up crystallization trials. Initial crystallization conditions were examined using commercially available Hampton Research screening kits. Crystal optimization trays were set up manually using the sitting-drop method at 20°C. Rice TPR2 TPD plus EAR-2 peptide crystals were grown in a well solution of 25% (w/v) polyethylene glycol (PEG) 3350, 0.2 M ammonium sulfate, and 0.1 M bis-tris (pH 6.5). Crystals were rod-shaped with a size of 100 to 200 μm and diffracted x-rays to about 2.6 Å at the Life Sciences Collaborative Access Team (LS-CAT) of the Advanced Photon Source (APS) synchrotron. The site 1–mutated TPD (L111A + L130A)/EAR-2 peptide crystals were grown in a well solution containing 20% (w/v) PEG 3350 and 0.2 M potassium phosphate monobasic (pH 4.8). The site 1–mutated TPD (L179A + I195A)/EAR-2 peptide crystals were grown in a well solution containing 20% (w/v) PEG 3350 and 0.2 M lithium sulfate monohydrate (pH 6). The site 1 and site 2 quadruple mutant TPD (L111A + L130A + L179A + I195A) crystals were grown in a well solution of 15% (w/v) PEG 4000, 0.2 M magnesium chloride, and tris-HCl (pH 8.5).
Data collection and structure determination
All crystals were transferred to the well solution with 22% ethylene glycol as cryoprotectant before flash-freezing in liquid nitrogen. Data collections were performed at the sector 21-ID LS-CAT beamlines of the APS synchrotron. The rice TPR2 TPD/EAR-2 peptide, TPR2 TPD(L111A + L130A)/EAR-2 peptide, TPR2 TPD(L179A + I195A)/EAR-2 peptide, and TPD(L111A + L130A + L179A + I195A) complex structures were determined by molecular replacement using the apo TPR2 TPD structure (PDB code 5C6Q) (25) as searching model. The initial model generated by the PHENIX AutoBuild program (78) was refined using several cycles of REFMAC and PHENIX refine programs (79). The D53 peptide model was built using Coot (80) based on the electron density map. All structure figures were prepared using PyMol (81).
Homology modeling
The homology model was generated by the I-TASSER server (63–65) with a C-score of −1.35. We have also generated a homology model with Phyre2 (82–84), with >90% confidence for all nonloop regions (79% of the sequence). Both models agreed well with each other with the same domain organization and comparable placement of the pore loop and the EAR and Walker motifs.
M2H assays
To construct the Gal4 DBD-D53 plasmid, full-length, codon-optimized D53 coding sequence was synthesized by GeneWiz with flanking restriction sites. The restriction fragment was cloned into the Gal4 plasmid pM (Clontech). The full-length TPL/TPR1/TPR2-VP16 activation domain constructs were described previously (25). Gal4 fusion constructs (25 ng) were cotransfected with VP16 fusion constructs (25 ng), together with 100 ng of pG5-Luc reporter and 5 ng of phRG-TK/Renilla (Promega) control into AD293 cells using FuGENE 6 (Promega) according to the manufacturer’s instructions. Cells were harvested 24 hours after transfection and lysed in 1× passive lysis buffer (Promega). Luciferase/Renilla activities were measured with the Dual Luciferase Kit (Promega), and data were plotted as relative activities (Luciferase activity:Renilla activity).
AlphaScreen assay
Luminescence proximity AlphaScreen (PerkinElmer) assays were performed using a hexahistidine detection kit. His6Sumo-tagged wild-type and mutant TPD proteins (50 nM) and biotinylated peptides (50 nM) were incubated with streptavidin-coated donor beads (5 μg/ml) and Ni-chelated acceptor beads (5 μg/ml) in 50 mM Mops (pH 7.4), 100 mM NaCl, and bovine serum albumin (0.1 mg/ml) for 1.5 hours at room temperature. Donor beads contain a photosensitizer, which can convert ambient oxygen into short-lived singlet oxygen upon light activation at 680 nm. When the acceptor beads are brought close enough to the donor beads by interaction between His6-tagged proteins and biotinylated proteins or peptides, singlet oxygen can diffuse from the donor to the acceptor beads and transfer energy to the thioxene derivatives of the acceptor beads, resulting in light emission at 520 to 620 nm. For the nucleosome interaction assay, we used 100 nM His6Sumo-tagged TPR2 TPD and 100 nM biotinylated nucleosomes, and for the interaction between His6-tagged TPD and biotinylated TPD, we used 400 nM of each protein. Note that the biotinylated proteins are much larger than biotinylated peptides and therefore yield much weaker proximity signals. For the competition assays, untagged peptides at increasing concentrations were added in the presence of a constant concentration (50 nM) of tagged protein and peptide. The IC50 values were obtained from curve fitting using the competitive inhibitor model in GraphPad Prism. All competitions have fulfilled conditions where IC50 ~ Kd, that is, [ligand plus its homologous competitor]total > [receptor]total at IC50 and [competitor]eq ~ [competitor]total at IC50 and Kd > 10 × [ligand]total.
Transcriptional activity assay in rice protoplasts
To generate the Gal4 DBD-D53-VP16 AD construct, the full-length D53 open reading frame was cloned into the plasmid containing the Gal4 DBD-VP16 activation domain (85). Site-directed mutagenesis was performed using the QuikChange method (Stratagene). The plasmids containing Gal4 DBD-D53mEAR1-VP16 AD/Gal4 DBD-D53mEAR2-VP16 AD/Gal4 DBD-D53mEAR3-VP16 AD/Gal4 DBD-D53mEAR2mEAR3-VP16 AD, 35SLUC, and pRTL were transformed into rice protoplasts by PEG-mediated transformation method (86), whereas plasmids containing Gal4 DBD-D53-VP16 AD, 35SLUC, and pRTL were used as a negative control. After incubation in the dark for 14 hours, the luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions, and data were plotted as relative activities (Luciferase activity:Renilla activity).
Plant materials and growth conditions
To generate the Ubi::d53-3×FLAG plasmid series, the full-length d53 open reading frame and its various motif mutation forms were amplified, and the polymerase chain reaction (PCR) products were then cloned into the binary vector pTCK303 (87) using the In-Fusion Advantage PCR Cloning Kit (catalog no. PT4065, Clontech) and sequenced. The pUbi::d53-1D-3×FLAG, pUbi::d53-mEAR2-3×FLAG, pUbi::d53-mEAR3-3×FLAG, and pUbi::d53-mEAR2/mEAR3-3×FLAG plasmids were introduced into rice (Oryza sativa L. subspecies japonica) wild-type Nipponbare. All transgenic plants were generated using Agrobacterium-mediated transformation of rice calli, as described previously (88). Rice plants were cultivated in Hainan (18°20′N, 109°38′E), China.
Dynamic light scattering
Freshly SEC-purified TPR2 TPD was incubated with or without 100 μM EAR motif peptides from D53 (wild type), NINJA, and D53 (Y798A) for 1 hour on ice. DLS data of the TPD preparations were collected using the microCUVETTE (Wyatt Technology) on a DynaPro NanoStar (Wyatt Technology) instrument. Each sample was equilibrated at 25°C before measurements and then analyzed in 10 replicates. Software 7.1.7 was used to calculate intensity-based particle size and number distributions.
Thermostability shift assay
For thermostability shift assays, TPR2 TPD (200 μg/ml, 8 μM) was incubated for 1 hour with or without 100 μM D53 EAR-2 peptide at room temperature in 0.1% SYPRO Orange dye, 20 mM tris-HCl (pH 8), and 200 mM NaCl. Samples were heated in a StepOnePlus Real-Time thermocycler (Life Technologies) from 20° to 85°C.
TPD N176H aggregation assay
The His6Sumo-tagged TPR2 TPD(N176H) protein was expressed in E. coli BL21 (DE3) cells and purified as described (25). The His6Sumo tag was cleaved by Ulp1 Sumo protease and removed by chromatography through a Ni-chelating high-performance Sepharose column, followed by SEC. Purified His6Sumo-tagged wild-type and TPR2 TPD(N176H) proteins were mixed at indicated ratios, incubated on ice for 1 hour, and centrifuged at 16,100g for 10 min. Supernatants were collected and separated by SDS-PAGE.
Supplementary Material
Acknowledgments
We thank S. Grant and M. Martin for administrative support. Funding: This work was supported by the Outstanding Young Scientist Foundation of CAS (Y.J.), the Youth Innovation Promotion Association of CAS (Y.J.), the Van Andel Research Institute (to S.B.R., H.E.X., and K.M.), Ministry of Science and Technology (China) (grants 2012ZX09301001, 2012CB910403, 2013CB910600, XDB08020303, and 2013ZX09507001), NSF (grant 91217311 to H.E.X.), and NIH [grants DK071662 (to H.E.X.), GM102545 and GM104212 (to K.M.), and CA181343 (to S.B.R.)]. We thank the staff members of the LS-CAT of the APS for assistance in data collection at the beamlines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). The use of APS was supported by the Office of Science of the U.S. Department of Energy, under contract no. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Author contributions: H.E.X., Y.J., and K.M. conceived the project. J.K., Y.J., S.B.R., Y.W., H.E.X., J.L., and K.M. designed the experiments. H.M., J.D., J.K., Y.H., X.G., T.-H.X., H.Y., and J.S.B. performed and/or interpreted the experiments. H.E.X. and K.M. wrote the manuscript with support from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All structure data were deposited in the PDB under accession codes 5J9K (TPD wild type), 5JA5 (TPD L111A/L130A), 5JHP (TPD L179A/I195A), and 5JGC (TPD L111A/L130A/L179A/I195A). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Correspondence and requests for materials should be addressed to K.M. (Karsten.Melcher@vai.org) or J.L. (jyli@genetics.ac.cn).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1601217/DC1
table S1. X-ray data collection and refinement statistics for TPR2-TPD + D53 EAR-2 peptide complex structures.
table S2. D53 EAR-2 peptide increases the average hydrodynamic radius of the TPR2 TPD.
fig. S1. Sequence alignment of the EAR motifs of D53 homologs from monocots and dicots.
fig. S2. Sequence alignment of the TPD from monocots and dicots.
fig. S3. Interaction of D53 EAR-2 peptide with the TPDs of TPL, TPR1, and TPR2.
fig. S4. Expression and protein loading controls.
fig. S5. M2H interaction between EAR-3–containing D53 fragments and TPL proteins.
fig. S6. Fo-Fc omit map of the bound EAR peptide contoured at 2σ in the binding pocket of the TPR2 TPD.
fig. S7. Mutations in TPR2 TPD site 1 do not abolish binding of the D53 EAR motif.
fig. S8. D53 EAR-2 peptide binds site 1 in TPR2 TPD site 2 mutation L179A/L195A.
fig. S9. TPR2 TPD L111A/L130A/L179A/I195A fails to bind the D53 EAR-2 motif.
fig. S10. Relative binding strength of wild-type and mutant D53 EAR-2 motif peptides.
fig. S11. The D53 EAR-2 peptide does not change the stability of the TPR2 TPD.
fig. S12. The EAR peptides of SMXL6, SMXL7, and SMXL8 do not induce TPR2 TPD tetramer-tetramer interaction.
fig. S13. Analysis of GFP-D53 and GFP-D53/TPD complexes by fluorescence-detection SEC.
fig. S14. D53 homology model.
REFERENCES AND NOTES
- 1.Krogan N. T., Long J. A., Why so repressed? Turning off transcription during plant growth and development. Curr. Opin. Plant Biol. 12, 628–636 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett T., Leyser O., Strigolactone signalling: Standing on the shoulders of DWARFs. Curr. Opin. Plant Biol. 22, 7–13 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Smith S. M., Li J., Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 21, 23–29 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Al-Babili S., Bouwmeester H. J., Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186 (2015). [DOI] [PubMed] [Google Scholar]
- 5.de Saint Germain A., Bonhomme S., Boyer F.-D., Rameau C., Novel insights into strigolactone distribution and signalling. Curr. Opin. Plant Biol. 16, 583–589 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Waldie T., McCulloch H., Leyser O., Strigolactones and the control of plant development: Lessons from shoot branching. Plant J. 79, 607–622 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Yao R., Ming Z., Yan L., Li S., Wang F., Ma S., Yu C., Yang M., Chen L., Li Y., Yan C., Miao D., Sun Z., Yan J., Sun Y., Wang L., Chu J., Fan S., He W., Deng H., Nan F., Li J., Rao Z., Lou Z., Xie D., DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Hamiaux C., Drummond R. S. M., Janssen B. J., Ledger S. E., Cooney J. M., Newcomb R. D., Snowden K. C., DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., Meng X., Liu G., Yu H., Yuan Y., Yi W., Zhao L., Ma H., He Y., Wu Z., Melcher K., Qian Q., Xu H. E., Wang Y., Li J., DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L.- H., Zhou X. E., Wu Z. S., Yi W., Xu Y., Li S., Xu T.-H., Liu Y., Chen R.-Z., Kovach A., Kang Y., Hou L., He Y., Xie C., Song W., Zhong D., Xu Y., Wang Y., Li J., Zhang C., Melcher K., Xu H. E., Crystal structures of two phytohormone signal-transducing α/β hydrolases: Karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L.-H., Zhou X. E., Yi W., Wu Z., Liu Y., Kang Y., Hou L., de Waal P. W., Li S., Jiang Y., Scaffidi A., Flematti G. R., Smith S. M., Lam V. Q., Griffin P. R., Wang Y., Li J., Melcher K., Xu H. E., Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., Wu F., Mao H., Dong W., Gan L., Ma W., Gao H., Chen J., Yang C., Wang D., Tan J., Zhang X., Guo X., Wang J., Jiang L., Liu X., Chen W., Chu J., Yan C., Ueno K., Ito S., Asami T., Cheng Z., Lei C., Zhai H., Wu C., Wang H., Zheng N., Wan J., D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S. M., Li J., Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J. P., Abbas A., Leyser O., Nelson D. C., SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Y., Ward S., Li P., Bennett T., Leyser O., SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28, 1581–1601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Causier B., Ashworth M., Guo W., Davies B., The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., Karmarkar V., Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13, 137–144 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Lee J. E., Golz J. F., Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal. Behav. 7, 86–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szemenyei H., Hannon M., Long J. A., TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Smith Z. R., Long J. A., Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464, 423–426 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal M., Kumar P., Mathew S. J., The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 67, 472–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turki-Judeh W., Courey A. J., Groucho: A corepressor with instructive roles in development. Curr. Top. Dev. Biol. 98, 65–96 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Matsumura H., Kusaka N., Nakamura T., Tanaka N., Sagegami K., Uegaki K., Inoue T., Mukai Y., Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications. J. Biol. Chem. 287, 26528–26538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chodaparambil J. V., Pate K. T., Hepler M. R. D., Tsai B. P., Muthurajan U. M., Luger K., Waterman M. L., Weis W. I., Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 33, 719–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke J., Ma H., Gu X., Thelen A., Brunzelle J., Li J., Xu H. E., Melcher K., Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 1, e1500107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brantjes H., Roose J., van De Wetering M., Clevers H., All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29, 1410–1419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein R. E., Jimenez G., Cook O., Gur D., Paroush Z., Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development 126, 3747–3755 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Jiménez G., Paroush Z., Ish-Horowicz D., Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 11, 3072–3082 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komachi K., Redd M. J., Johnson A. D., The WD repeats of Tup1 interact with the homeo domain protein α2. Genes Dev. 8, 2857–2867 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Muhr J., Andersson E., Persson M., Jessell T. M., Ericson J., Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104, 861–873 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Orian A., Delrow J. J., Rosales Nieves A. E., Abed M., Metzger D., Paroush Z., Eisenman R. N., Parkhurst S. M., A Myc-Groucho complex integrates EGF and Notch signaling to regulate neural development. Proc. Natl. Acad. Sci. U.S.A. 104, 15771–15776 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paroush Z., Finley R. L. Jr., Kidd T., Wainwright S. M., Ingham P. W., Brent R., Ish-Horowicz D., Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79, 805–815 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O., Clevers H., The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395, 608–612 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Sekiya T., Zaret K. S., Repression by Groucho/TLE/Grg proteins: Genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell 28, 291–303 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzamarias D., Struhl K., Functional dissection of the yeast Cyc8–Tup1 transcriptional co-repressor complex. Nature 369, 758–761 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Zhang H., Levine M., Ashe H. L., Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev. 15, 261–266 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogas J., Kaufmann S., Henderson J., Somerville C., PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 13839–13844 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Rider S. D. Jr., Henderson J. T., Fountain M., Chuang K., Kandachar V., Simons A., Edenberg H. J., Romero-Severson J., Muir W. M., Ogas J., The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283, 22637–22648 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z., Xu F., Zhang Y., Cheng Y. T., Wiermer M., Li X., Zhang Y., Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. U.S.A. 107, 13960–13965 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krogan N. T., Hogan K., Long J. A., APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long J. A., Ohno C., Smith Z. R., Meyerowitz E. M., TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Chen G., Fernandez J., Mische S., Courey A. J., A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 13, 2218–2230 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davie J. K., Trumbly R. J., Dent S. Y. R., Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22, 693–703 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J., Suka N., Carlson M., Grunstein M., TUP1 utilizes histone H3/H2B–specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7, 117–126 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Flores-Saaib R. D., Courey A. J., Analysis of Groucho–histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression. Nucleic Acids Res. 28, 4189–4196 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palaparti A., Baratz A., Stifani S., The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 272, 26604–26610 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Edmondson D. G., Smith M. M., Roth S. Y., Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10, 1247–1259 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Kagale S., Links M. G., Rozwadowski K., Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152, 1109–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kagale S., Rozwadowski K., Small yet effective: The ethylene responsive element binding factor-associated amphiphilic repression (EAR) motif. Plant Signal. Behav. 5, 691–694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kagale S., Rozwadowski K., EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwata N., Satoh H., Omura T., Linkage studies in rice. Linkage groups of 6 genes newly described. Jpn. J. Breed. 27 (suppl. 1), 250–251 (1977). [Google Scholar]
- 52.Long J. A., Woody S., Poethig S., Meyerowitz E. M., Barton M. K., Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129, 2797–2806 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Ito J., Fukaki H., Onoda M., Li L., Li C., Tasaka M., Furutani M., Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. U.S.A. 113, 6562–6567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robyr D., Suka Y., Xenarios I., Kurdistani S. K., Wang A., Suka N., Grunstein M., Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109, 437–446 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Suka N., Suka Y., Carmen A. A., Wu J., Grunstein M., Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8, 473–479 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Buscarlet M., Stifani S., The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 17, 353–361 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Fleming A. B., Pennings S., Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 20, 5219–5231 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kastaniotis A. J., Mennella T. A., Konrad C., Torres A. M. R., Zitomer R. S., Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol. 20, 7088–7098 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B., Reese J. C., Ssn6-Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J. Biol. Chem. 276, 33788–33797 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Lin Y.-S., Carey M. F., Ptashne M., Green M. R., GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell 54, 659–664 (1988). [DOI] [PubMed] [Google Scholar]
- 61.Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T., Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J. 10, 3033–3041 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mogk A., Kummer E., Bukau B., Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y., The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanson P. I., Whiteheart S. W., AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell. Biol. 6, 519–529 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V., AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43 (1999). [PubMed] [Google Scholar]
- 68.Jha S., Gupta A., Dar A., Dutta A., RVBs are required for assembling a functional TIP60 complex. Mol. Cell. Biol. 33, 1164–1174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matias P. M., Baek S. H., Bandeiras T. M., Dutta A., Houry W. A., Llorca O., Rosenbaum J., The AAA+ proteins Pontin and Reptin enter adult age: From understanding their basic biology to the identification of selective inhibitors. Front. Mol. Biosci. 2, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jha S., Shibata E., Dutta A., Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 28, 2690–2700 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jónsson Z. O., Dhar S. K., Narlikar G. J., Auty R., Wagle N., Pellman D., Pratt R. E., Kingston R., Dutta A., Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem. 276, 16279–16288 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Jónsson Z. O., Jha S., Wohlschlegel J. A., Dutta A., Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 16, 465–477 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Mouse Genome Sequencing Consortium, Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Antonarakis S. E., Attwood J., Baertsch R., Bailey J., Barlow K., Beck S., Berry E., Birren B., Bloom T., Bork P., Botcherby M., Bray N., Brent M. R., Brown D. G., Brown S. D., Bult C., Burton J., Butler J., Campbell R. D., Carninci P., Cawley S., Chiaromonte F., Chinwalla A. T., Church D. M., Clamp M., Clee C., Collins F. S., Cook L. L., Copley R. R., Coulson A., Couronne O., Cuff J., Curwen V., Cutts T., Daly M., David R., Davies J., Delehaunty K. D., Deri J., Dermitzakis E. T., Dewey C., Dickens N. J., Diekhans M., Dodge S., Dubchak I., Dunn D. M., Eddy S. R., Elnitski L., Emes R. D., Eswara P., Eyras E., Felsenfeld A., Fewell G. A., Flicek P., Foley K., Frankel W. N., Fulton L. A., Fulton R. S., Furey T. S., Gage D., Gibbs R. A., Glusman G., Gnerre S., Goldman N., Goodstadt L., Grafham D., Graves T. A., Green E. D., Gregory S., Guigó R., Guyer M., Hardison R. C., Haussler D., Hayashizaki Y., Hillier L. W., Hinrichs A., Hlavina W., Holzer T., Hsu F., Hua A., Hubbard T., Hunt A., Jackson I., Jaffe D. B., Johnson L. S., Jones M., Jones T. A., Joy A., Kamal M., Karlsson E. K., Karolchik D., Kasprzyk A., Kawai J., Keibler E., Kells C., Kent W. J., Kirby A., Kolbe D. L., Korf I., Kucherlapati R. S., Kulbokas E. J., Kulp D., Landers T., Leger J. P., Leonard S., Letunic I., Levine R., Li J., Li M., Lloyd C., Lucas S., Ma B., Maglott D. R., Mardis E. R., Matthews L., Mauceli E., Mayer J. H., McCarthy M., McCombie W. R., McLaren S., McLay K., McPherson J. D., Meldrim J., Meredith B., Mesirov J. P., Miller W., Miner T. L., Mongin E., Montgomery K. T., Morgan M., Mott R., Mullikin J. C., Muzny D. M., Nash W. E., Nelson J. O., Nhan M. N., Nicol R., Ning Z., Nusbaum C., O’Connor M. J., Okazaki Y., Oliver K., Overton-Larty E., Pachter L., Parra G., Pepin K. H., Peterson J., Pevzner P., Plumb R., Pohl C. S., Poliakov A., Ponce T. C., Ponting C. P., Potter S., Quail M., Reymond A., Roe B. A., Roskin K. M., Rubin E. M., Rust A. G., Santos R., Sapojnikov V., Schultz B., Schultz J., Schwartz M. S., Schwartz S., Scott C., Seaman S., Searle S., Sharpe T., Sheridan A., Shownkeen R., Sims S., Singer J. B., Slater G., Smit A., Smith D. R., Spencer B., Stabenau A., Stange-Thomann N., Sugnet C., Suyama M., Tesler G., Thompson J., Torrents D., Trevaskis E., Tromp J., Ucla C., Ureta-Vidal A., Vinson J. P., Von Niederhausern A. C., Wade C. M., Wall M., Weber R. J., Weiss R. B., Wendl M. C., West A. P., Wetterstrand K., Wheeler R., Whelan S., Wierzbowski J., Willey D., Williams S., Wilson R. K., Winter E., Worley K. C., Wyman D., Yang S., Yang S. P., Zdobnov E. M., Zody M. C., Lander E. S., Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Magalska A., Schellhaus A. K., Moreno-Andrés D., Zanini F., Schooley A., Sachdev R., Schwarz H., Madlung J., Antonin W., RuvB-like ATPases function in chromatin decondensation at the end of mitosis. Dev. Cell 31, 305–318 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Ehrentraut S., Hassler M., Oppikofer M., Kueng S., Weber J. M., Mueller J. W., Gasser S. M., Ladurner A. G., Ehrenhofer-Murray A. E., Structural basis for the role of the Sir3 AAA+ domain in silencing: Interaction with Sir4 and unmethylated histone H3K79. Genes Dev. 25, 1835–1846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M., Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell 80, 583–592 (1995). [DOI] [PubMed] [Google Scholar]
- 77.Zhang F., Yao J., Ke J., Zhang L., Lam V. Q., Xin X.-F., Zhou X. E., Chen J., Brunzelle J., Griffin P. R., Zhou M., Xu H. E., Melcher K., He S. Y., Structural basis of JAZ-mediated repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., Hung L.-W., Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 81.W. DeLano, The PyMol Molecular Graphics System (DeLano Scientific, 2002). [Google Scholar]
- 82.Bennett-Lovsey R. M., Herbert A. D., Sternberg M. J. E., Kelley L. A., Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70, 611–625 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kelley L. A., Sternberg M. J. E., Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Lu Z., Yu H., Xiong G., Wang J., Jiao Y., Liu G., Jing Y., Meng X., Hu X., Qian Q., Fu X., Wang Y., Li J., Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bart R., Chern M., Park C.-J., Bartley L., Ronald P. C., A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2, 13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Z., Chen C., Xu Y., Jiang R., Han Y., Xu Z., Chong K., A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22, 409–417 (2004). [Google Scholar]
- 88.Hiei Y., Ohta S., Komari T., Kumashiro T., Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1601217/DC1
table S1. X-ray data collection and refinement statistics for TPR2-TPD + D53 EAR-2 peptide complex structures.
table S2. D53 EAR-2 peptide increases the average hydrodynamic radius of the TPR2 TPD.
fig. S1. Sequence alignment of the EAR motifs of D53 homologs from monocots and dicots.
fig. S2. Sequence alignment of the TPD from monocots and dicots.
fig. S3. Interaction of D53 EAR-2 peptide with the TPDs of TPL, TPR1, and TPR2.
fig. S4. Expression and protein loading controls.
fig. S5. M2H interaction between EAR-3–containing D53 fragments and TPL proteins.
fig. S6. Fo-Fc omit map of the bound EAR peptide contoured at 2σ in the binding pocket of the TPR2 TPD.
fig. S7. Mutations in TPR2 TPD site 1 do not abolish binding of the D53 EAR motif.
fig. S8. D53 EAR-2 peptide binds site 1 in TPR2 TPD site 2 mutation L179A/L195A.
fig. S9. TPR2 TPD L111A/L130A/L179A/I195A fails to bind the D53 EAR-2 motif.
fig. S10. Relative binding strength of wild-type and mutant D53 EAR-2 motif peptides.
fig. S11. The D53 EAR-2 peptide does not change the stability of the TPR2 TPD.
fig. S12. The EAR peptides of SMXL6, SMXL7, and SMXL8 do not induce TPR2 TPD tetramer-tetramer interaction.
fig. S13. Analysis of GFP-D53 and GFP-D53/TPD complexes by fluorescence-detection SEC.
fig. S14. D53 homology model.


