Abstract
INTRODUCTION
Cells synthesize glucose if deprived of it, and destroy gluconeogenic enzymes upon return to glucose-replete conditions. Gluconeogenesis (de novo synthesis of glucose) is, in effect, a reversal of glycolysis, in which glucose is converted to pyruvate. Some enzymatic steps are shared between gluconeogenesis and glycolysis, but other steps are confined to one of the two pathways. In the yeast Saccharomyces cerevisiae, the gluconeogenesis-specific enzymes are fructose-1,6-bisphosphatase (Fbp1), isocitrate lyase (Icl1), malate dehydrogenase (Mdh2), and phosphoenolpyruvate carboxykinase (Pck1).
We found that Gid4, a subunit of the oligomeric GID ubiquitin ligase, is the recognition component of a proteolytic pathway termed the Pro/N-end rule pathway, which conditionally destroys gluconeogenic enzymes. The N-end rule pathway is a set of proteolytic systems whose unifying feature is their ability to recognize and polyubiquitylate proteins containing N-terminal degradation signals called N-degrons, thereby causing the degradation of these proteins by the proteasome. In eukaryotes, the previously known branches of this system are the Arg/N-end rule pathway and the Ac/N-end rule pathway. The Arg/N-end rule pathway targets specific unacetylated N-terminal residues of cellular proteins, including Asn, Gln, Glu, Asp, Arg, Lys,His, Leu, Phe, Tyr, Trp, Ile, and Met (if Met is followed by a bulky hydrophobic residue). The pathway’s other branch, called the Ac/N-end rule pathway, targets proteins for degradation by recognizing their Nα-terminally acetylated (Ntacetylated) residues. About 90% of human proteins are cotranslationally and irreversibly Nt-acetylated. Many, possiblymost, Nt-acetylated proteins bear N-degrons of the Ac/N-end rule pathway.
RATIONALE
We wished to identify the recognition component of themultisubunit GID ubiquitin ligase and also to determine whether GID, which was known to mediate the conditional degradation of gluconeogenic enzymes, might recognize them through their N-terminal Pro residues, and also through Pro at position 2.
RESULTS
The successful strategy involved a version of two-hybrid assay for in vivo protein interactions. Themain discovery identified Gid4, a subunit of the GID ubiquitin ligase, as the recognition component (termed Pro/N-recognin) of the Pro/N-end rule pathway. Gid4 was shown to target the gluconeogenic enzymes Fbp1, Icl1, and Mdh2 (and possibly other yeast proteins as well) through the binding to their N-terminal Pro residues in the presence of cognate adjacent sequence motifs. Pck1, the fourth gluconeogenic enzyme, contains Pro at position 2. Gid4 was also required for the degradation of Pck1 through the ability ofGid4 to target the Pro residue of Pck1 at position 2. The properties of Gid4 discovered so far indicate that its substrate binding groove can recognize either the N-terminal Pro residue or Pro at position 2 in the presence of cognate adjacent sequence motifs. The recognition flexibility of Gid4 suggests that the true diversity of Gid4 substrates is only beginning to be determined. Subunits of the S. cerevisiae GID ubiquitin ligase have counterparts in animals and plants. Thus, the discovery that yeast Gid4 is the Pro/N-recognin will facilitate the understanding of the Pro/N-end rule pathway in other eukaryotes as well.
CONCLUSION
With our identification of the Gid4-mediated Pro/N-end rule pathway that specifically recognizes the N-terminal Pro residue, all 20 amino acids of the genetic code have now been shown to act, in specific sequence contexts, as destabilizing N-terminal residues. Thus, most proteins in a cell may be conditionally short-lived N-end rule substrates, either as full-length proteins or as protease generated natural protein fragments.
Graphical Abstract
The Pro/N-end rule pathway and the Gid4 Pro/N-recognin. (Top left) Degradation of the P-Mdh2 (Pro-Mdh2) gluconeogenic enzyme in yeast cells under glucose-replete conditions. (Top right) Two-hybrid protein binding assays with Gid4 vis-à-vis the wild-type P-Fbp1 gluconeogenic enzyme (row 1), its S-Fbp1 mutant (row 2), and analogous pairs of the P-Icl1/S-Icl1 (rows 3 and 4) and P-Pyk2/S-Pyk2 (rows 5 and 6) proteins. Note the binding of Gid4 to P-Fbp1 and P-Icl1. (Bottom) The Pro/N-end rule pathway and the Gid4 Pro/N-recognin.TheGid4 subunit of the multisubunit GID ubiquitin ligase binds to theN-terminal Pro (P) residue of a cellular protein substrate if it also contains a cognate adjoining sequencemotif. The targeted substrate is polyubiquitylated and processively destroyed by the 26S proteasome. The same pathway can also recognize substrates in which Pro is at position 2.
When glucose is low or absent, cells synthesize it through gluconeogenesis, an adenosine triphosphate (ATP)–consuming process. Gluconeogenesis is, in effect, a reversal of glycolysis, in which glucose is converted to pyruvate, with production of ATP and reduced nicotinamide adenine dinucleotide (NADH) (fig. S1). Some enzymatic steps are shared between gluconeogenesis and glycolysis, but other steps are confined to one of the two pathways (1, 2). In the yeast S. cerevisiae, the main gluconeogenesis-specific enzymes are fructose-1,6-bisphosphatase (Fbp1), isocitrate lyase (Icl1), malate dehydrogenase (Mdh2), and phosphoenolpyruvate carboxykinase (Pck1) (fig. S1) (3–12). Mammalian counterparts of these enzymes are regulated by insulin and other hormones, and become essential for viability during fasting. Aberrant control of gluconeogenic enzymes can promote cancer, diabetes, and other human diseases (13, 14).
When S. cerevisiae grows on ethanol or acetate, the gluconeogenic enzymes are expressed and long-lived. Transition to a medium containing glucose inhibits the synthesis of these enzymes and induces their degradation (fig. S2, A and B). Screens for mutants unable to destroy Fbp1 identified subunits of the ubiquitin (Ub) ligase termed GID (glucose-induced degradation) (3, 6–8). The GID Ub ligase of S. cerevisiae contains eight distinct subunits and is conserved among eukaryotes, including animals and plants (5, 6, 15–17).
A 1998 study by Wolf and colleagues suggested that the GID/proteasome-dependent degradation of Fbp1, Icl1, and Mdh2—three of the four yeast gluconeogenic enzymes—may be mediated by their common property of bearing the N-terminal proline (Pro) residue (4). We decided to investigate this suggestion and found that Gid4 (also known as Vid24) is a substrate-targeting subunit of the GID Ub ligase.
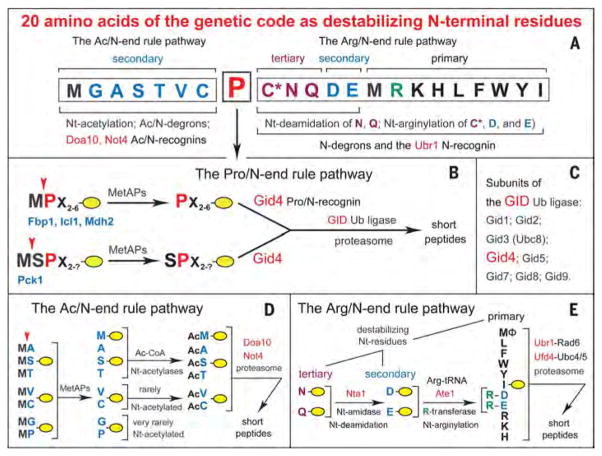
The N-end rule pathway is a set of proteolytic systems whose unifying feature is their ability to recognize and polyubiquitylate proteins containing N-terminal degradation signals called N-degrons, thereby causing the degradation of these proteins by the proteasome (18–27). The main determinant of an N-degron is a destabilizing N-terminal residue of a protein. Recognition components of the N-end rule pathway, called N-recognins, are E3 Ub ligases that can target N-degrons (Fig. 1).
Fig. 1. Twenty amino acids of the genetic code as destabilizing N-terminal residues.
(A) N-terminal residues are indicated by single-letter abbreviations. Twenty DNA-encoded amino acids are arranged to delineate three branches of the S. cerevisiae N-end rule pathway, including its proline-specific branch, indicated by the vertical arrow. N-terminal Met is cited twice, because it can be recognized by the Ac/N-end rule pathway (as Nt-acetylated Met) and by the Arg/N-end rule pathway (as the unacetylated N-terminal Met). N-terminal Cys is also cited twice, because it can be recognized by the Ac/N-end rule pathway (as Nt-acetylated Cys) and by the Arg/N-end rule pathway [as an oxidized, Nt-arginylatable N-terminal Cys, denoted as Cys* and formed in multicellular eukaryotes but apparently not in S. cerevisiae (22), at least in the absence of stress]. (B) The Pro/N-end rule pathway and Gid4.We have identified this subunit of the GID Ub ligase as the Pro/N-recognin of the Pro/N-end rule pathway, whose functions include the conditional degradation of the gluconeogenic enzymes Fbp1, Icl1, Mdh2, and Pck1. A yellow oval denotes the remainder of a protein substrate. (C) Subunits of the S. cerevisiae GID Ub ligase, including Gid4. (D) The Ac/N-end rule pathway. (E) The Arg/N-end rule pathway. Amino acid abbreviations: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T,Thr; V, Val; W,Trp; Y,Tyr.
Regulated degradation of proteins and their natural fragments by the N-end rule pathway mediates a remarkably broad range of biological processes, including the sensing of heme, O2, NO, and short peptides; the control of subunit stoichiometries in oligomeric proteins; the elimination of misfolded and otherwise abnormal proteins; the degradation of proteins after their translocation to the cytosol from membrane-enclosed compartments; regulation of apoptosis and repression of neurodegeneration; regulation of DNA repair, transcription, replication, and chromosome cohesion/segregation; regulation of G proteins, cytoskeletal proteins, autophagy, peptide import, meiosis, immunity, circadian rhythms, fat metabolism, cell migration, cardiovascular development, spermatogenesis, and neurogenesis; the functioning of adult organs, including the brain, muscle, testis, and pancreas; and the regulation of leaf and shoot development, senescence, NO/O2 sensing, and many other processes in plants (22–29).
The eukaryotic N-end rule pathway has been known to comprise two branches. One branch, the Arg/N-end rule pathway, targets unacetylated N-terminal residues (18, 30–36). N-terminal Arg, Lys, His, Leu, Phe, Tyr, Trp, Ile, and Met (if Met is followed by a bulky hydrophobic residue) are directly recognized by the N-recognin Ubr1 in S. cerevisiae and by several N-recognins in multicellular eukaryotes (22, 30). In contrast, N-terminal Asn, Gln, Glu, and Asp (as well as Cys, under some conditions) are destabilizing because of their preliminary modifications, including Nα-terminal deamidation (Nt-deamidation) and Nα-terminalarginylation (Nt-arginylation) (Fig. 1E) (21, 22, 25, 37).
The pathway’s other branch, called the Ac/N-end rule pathway, targets proteins for degradation by recognizing their Nt-acetylated residues (Fig. 1D) (19, 20, 30, 38–40). N-degrons and Ub ligases of the Ac/N-end rule pathway are called Ac/N-degrons and Ac/N-recognins, respectively. At least 60% of S. cerevisiae proteins and about 90% of human proteins are cotranslationally and irreversibly Nt-acetylated by ribosome-associated Nt-acetylases (39). Many, possibly most, Nt-acetylated proteins bear Ac/N-degrons, which are regulated through their steric shielding in cognate protein complexes (20, 38).
Degradation of gluconeogenic enzymes requires N-terminal proline
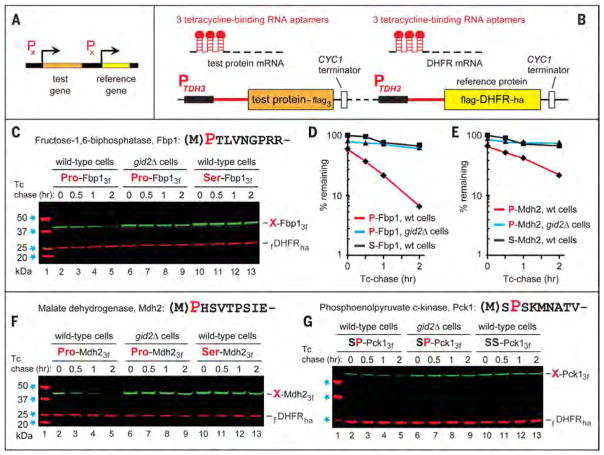
We began by designing a version of the promoter reference technique (PRT) (Fig. 2, A and B). In this method, one measures, during a chase, the ratio of a test protein to a long-lived reference protein, the mouse dihydrofolate reductase (DHFR). Epitope-tagged reference and test proteins were coexpressed from two identical PTDH3 promoters containing additional DNA elements developed by others (41). Once transcribed into an mRNA, these elements form 5′-RNA aptamers that can bind to tetracycline, thereby repressing translation in cis (Fig. 2B). This PRT design improves degradation assays by providing a reference and by allowing chases that involve a gene-specific repression of translation. The latter option avoids a global shutoff of translation by inhibitors such as cycloheximide. This advantage is particularly important if an essential component of a relevant proteolytic pathway is itself short-lived.
Fig. 2. The promoter reference technique and the degradation of gluconeogenic enzymes.
(A and B) The promoter reference technique (PRT). (C) Lane 1, kDa markers. Tetracycline-based chases were performed at 30°C during transition from ethanol to glucose media with wild-type (lanes 2 to 5 and 10 to 13) or gid2Δ S. cerevisiae (lanes 6 to 9) expressing either wild-type P-Fbp13f (Pro-Fbp1) or S-Fbp13f (Ser-Fbp1). At the indicated times of a chase, proteins in cell extracts were fractionated by SDS–polyacrylamide gel electrophoresis followed by immuno-blotting with antibody to Flag. (D) Quantification of data in (C). All chases in this study were performed at least twice and yielded results that differed by less than 10%. (E) Quantification of data in (F). (F) Same as (C) but with P-Mdh23f and S-Mdh23f. (G) Same as (C) but with SP-Pck13f (Ser-Pro-Pck1) and SS-Pck13f (Ser-Ser-Pck1). The N-terminal sequences of Fbp1, Mdh2, and Pck1 are shown at the upper right of (C), (F), and (G), respectively. The bands of test proteins and the fDHFRha reference protein are indicated.
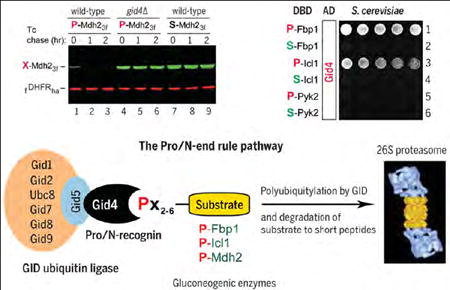
After 18 hours in ethanol medium at 30°C, S. cerevisiae were shifted to a glucose medium while the synthesis of the 348-residue, C-terminally Flag-tagged P-Fbp13f (bearing N-terminal Pro) and the coexpressed 187-residue DHFR reference was repressed by tetracycline, initiating a chase (Fig. 2, C and D). Under these conditions, P-Fbp13f [its N-terminal Met is cotranslationally removed by Met-aminopeptidases (42)] was short-lived in wild-type cells (t1/2 < 30 min) but was stable in gid1Δ, gid2Δ, gid3Δ, and gid4Δ mutants that lacked specific subunits of the GID Ub ligase (Fig. 1C, Fig. 2, C and D, and fig. S3, A and C). In contrast, the S-Fbp13f mutant, bearing N-terminal Ser instead of Pro, was long-lived even in wild-type cells (Fig. 2, C and D). Degradation assays with another gluconeogenic enzyme, the 377-residue cytosolic P-Mdh23f and its S-Mdh23f mutant (bearing N-terminal Ser), produced similar results (Fig. 2, E and F).
Gid4 as a recognition subunit of GID
Given these findings, we wished to identify an N-recognin of the GID Ub ligase. The successful strategy used a version of a two-hybrid assay in which the positions of two-hybrid–specific protein moieties, called the activation domain (AD) and DNA binding domain (DBD), were such that at least one member of a fusion pair contained the original, unobstructed N terminus (43–45) (fig. S2C). Controls included immunoblotting to verify expression of two-hybrid fusions (fig. S2D) and “exchanges” of DBD and AD domains between two interacting proteins. Other controls verified that binding-positive two-hybrid fusions did not stay positive in assays with just one of two fusions. The results summarized below passed all of these and additional controls.
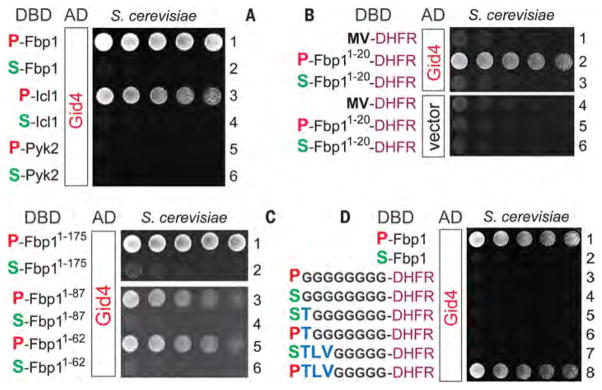
We discovered, using two-hybrid assays, that S. cerevisiae Gid4 binds to wild-type P-Fbp1 but does not bind to the S-Fbp1, A-Fbp1, T-Fbp1, C-Fbp1, V-Fbp1, and G-Fbp1 mutants (bearing, respectively, N-terminal Ser, Ala, Thr, Cys, Val, and Gly), in agreement with degradation assays that examined P-Fbp1 versus S-Fbp1 (Fig. 2, C and D, Fig. 3A, and figs. S4 and S5). These and other two-hybrid results did not depend on whether a yeast strain involved was wild-type in regard to GID or lacked the GID subunits Gid2, Gid5, and Gid9.
In contrast to Gid4, all other “canonical” GID subunits, specifically Gid1, Gid2, Gid5, Gid7, Gid8, and Gid9 (Fig. 1C), did not bind to P-Fbp1 (figs. S4, B and C, and S6, B and C).
The remarkable specificity of Gid4 for N-terminal Pro of P-Fbp1 (Fig. 3, A and D, and fig. S4) recurred with the binding of Gid4 to P-Icl1, another gluconeogenic enzyme. Specifically, the S-Icl1 mutant did not bind to Gid4, in contrast to wild-type P-Icl1 (Fig. 3A and fig. S5B).
C-terminal truncations of P-Fbp1, from its full length of 348 residues to 175, 87, and 62 residues, respectively, retained the binding of Gid4 to the resulting P-Fbp11-175, P-Fbp11-87, and P-Fbp11-62 proteins, and also retained the specificity for N-terminal Pro (Fig. 3C).
P-Fbp11-20-DHFR-DBD, a two-hybrid fusion containing the first 20 N-terminal residues of P-Fbp1, the 21-kDa DHFR moiety, and the DBD domain, exhibited binding to Gid4, and the interaction remained exquisitely specific for N-terminal Pro of P-Fbp11-20-DHFR-DBD (Fig. 3B). The analogous two-hybrid fusion containing only eight N-terminal residues of P-Fbp1 also bound to Gid4 and also retained the specificity for N-terminal Pro (fig. S5B).
Similar to the results with P-Fbp11-20-DHFR-DBD (Fig. 3B), Gid4 also bound to the analogous two-hybrid fusions that contained 20 N-terminal residues of the gluconeogenic enzymes P-Icl1 and P-Mdh2. The binding remained specific for N-terminal Pro (fig. S4D).
Fig. 3. Gid4 recognizes N-terminal Pro and at least four adjoining residues.
In this and other figures describing two-hybrid results, the Pro residues (N-terminal in this figure) are in red, mutant residues (Ser in this figure) are in green, and wild-type residues (other than Pro) are in blue. Expression of His3, the reporter of two-hybrid assays (44) in otherwise His−cells, was a function of the binding affinity between test proteins. Histidine-lacking plates were incubated for 2 days at 30°C to detect the growth of His+ cells. (A) Rows 1 and 2: Gid4 binds to P-Fbp1 but not to S-Fbp1. Rows 3 and 4: Gid4 binds to P-Icl1 but not to S-Icl1. Rows 5 and 6: Gid4 binds neither to P-Pyk2 nor to S-Pyk2. (B) Row 1: Gid4 does not bind to Met-Val-DHFR-DBD (MV-DHFR, a negative control). Rows 2 and 3: Gid4 binds to P-Fbp11-20-DHFR-DBD, a two-hybrid fusion containing the first 20 N-terminal residues of P-Fbp1, but does not bind to the otherwise identical S-Fbp11-20-DHFR-DBD fusion. Rows 4 to 6: Same as rows 1 to 3, but with vector alone instead of Gid4 (negative controls). (C) Rows 1 and 2: The N-terminally truncated P-Fbp11-175, but not the otherwise identical S-Fbp11-175, binds to Gid4. Rows 3 and 4: Same as rows 1 and 2, but with P-Fbp11-87 and S-Fbp11-87, respectively. Rows 5 and 6: Same as rows 1 and 2, but with P-Fbp11-62 and S-Fbp11-62, respectively. (D) Rows 1 and 2: Same as (A), rows 1 and 2, but independent two-hybrid assays. Rows 3 and 4: X-G8-DHFR-DBD (X = P or S) (see text). Rows 5 and 6: XT-G7-DHFR-DBD (X = P or S). Rows 7 and 8: XTLV-G5-DHFR-DBD (X = P or S). Note the binding of Gid4 to PTLV-G5-DHFR-DBD.
Readout of a two-hybrid assay involves activation of a transcriptional promoter through a promoter-proximal interaction between test protein moieties (43). To verify the specificity of interaction between Gid4 and P-Fbp1 (Fig. 3 and fig. S4) through a technique distinct from two-hybrid, we used the split-Ub assay. In this method, test proteins are expressed as fusions, respectively, to a C-terminal half of Ub (Cub) and to its mutant N-terminal half (Nub) (46, 47). Interactions between test protein moieties would reconstitute a quasi-native Ub moiety from Nub and Cub. The resulting in vivo cleavage of a Cub-containing fusion by deubiquitylases immediately downstream from the reconstituted Ub moiety serves as a readout of this technique (fig. S7A). Our findings with split-Ub assays were in agreement with those by two-hybrid assays, in that Gid4 interacted with a split-Ub–based P-Fbp1 but did not interact with the otherwise identical S-Fbp1 mutant (fig. S7, B and C). Two-hybrid and split-Ub assays do not distinguish a direct interaction between test protein moieties from their interaction through another, third protein. Although unlikely (for example, because GID subunits other than Gid4 did not bind to P-Fbp1 in two-hybrid assays; see item 2 above), the possibility of a “third-party” protein being involved in the observed binding of Gid4 to P-Fbp1 is not precluded.
Gid4 recognizes N-terminal Pro and five adjoining residues
To define N-terminal Pro–containing motifs that can be specifically recognized by Gid4, we began with two-hybrid fusions comprising N-terminal Pro (versus N-terminal Ser) followed by eight Gly residues, X-G8-DHFR-DBD (X = P or S); Gly is the smallest amino acid. These fusions did not bind to Gid4 (Fig. 3D). Gid4 also did not bind to PT-G7-DHFR-DBD, containing the first two residues of wild-type P-Fbp1 (Fig. 3D). However, PTLV-G5-DHFR-DBD, containing the first four residues of P-Fbp1 (PTLV-Fbp1), interacted with Gid4 and required N-terminal Pro for the binding (Fig. 3D and fig. S5A).
How tolerant is the interaction between Gid4 and the N-terminal region of PTLV-Fbp1 to changes of residues near Pro? This question remains to be addressed through a combinatorial-scale two-hybrid screen, but we carried out its useful approximation (Fig. 4 and figs. S8 to S10). First, we included positions 5 and 6 as well (PTLVNG-Fbp1). Second, we mutated wild-type residues of PTLVNG-Fbp1 at each of its positions (1 to 6) to 19 other residues while keeping residues at other positions unchanged. The results identified specific alterations that were “allowed” (i.e., were compatible with robust binding of a fusion to Gid4), “suboptimal” (a decreased binding), or “disallowed.” The latter residues abrogated the binding of Gid4 to a fusion, despite its N-terminal Pro residue (Fig. 4 and figs. S8 to S10).
Fig. 4. Specificity of the Gid4 Pro/N-recognin.
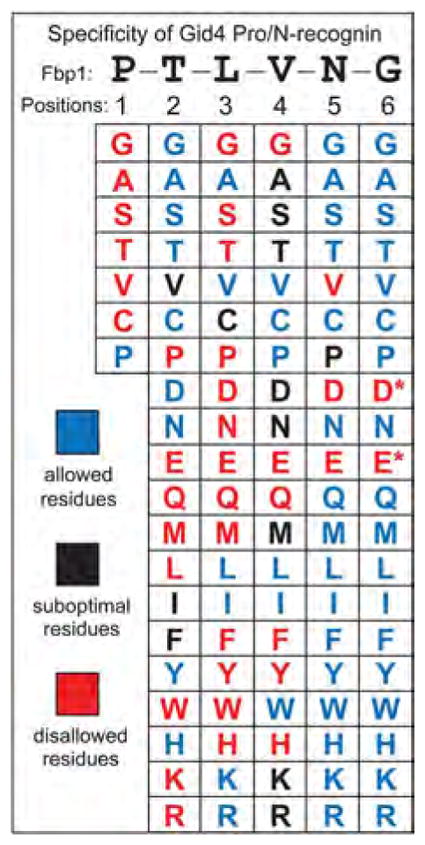
Individual residues of PTLVNG, the N-terminal sequence of wild-type P-Fbp1, were mutated to other residues while keeping residues at other positions unchanged.Two-hybrid assays were carried out to examine the binding of Gid4 to the P-X-L-V-G5-DHFR-DBD, P-T-X-V-G5-DHFR-DBD, P-T-L-X-G5-DHFR-DBD, P-T-L-V-X-G5-DHFR-DBD, and P-T-L-V-N-X-G5-DHFR-DBD fusions (X = 20 different residues) (>100 two-hybrid assays total; figs. S8 to S10). The residues are cited in this summary of the binding data as those that were “allowed (blue) (i.e., were compatible with robust binding of a fusion to Gid4), “suboptimal (black), or “disallowed (red). The latter residues abrogated the binding of Gid4 to a fusion, despite its N-terminal Pro. A residue at position 1 was varied less extensively for the reason described in the text. The Asp (D) and Glu (E) residues at position 6 are marked with asterisks to indicate that these residues were “suboptimal-like at this position, rather than abrogating the binding to Gid4. See figs. S8 to S10 for the two-hybrid data that underlie this summary.
N-terminal Pro was the sole allowed residue at position 1 (Fig. 3A and figs. S4A and S5A). Position 1 was varied less extensively than positions 2 to 6 (Fig. 4) because the initially present N-terminal Met would be (cotranslationally) cleaved off by Met-aminopeptidases, but only if the second residue, to be made N-terminal by the cleavage, was not larger than Val (42).
Gly, Ala, Ser, Cys, Asp, Asn, Tyr, His, and Thr were the allowed residues at position 2 (Thr is the position-2 residue of wild-type PTLVNG-Fbp1), whereas Pro, Glu, Gln, Met, Leu, Trp, Lys, and Arg were disallowed (Fig. 4 and figs. S8 and S9). Position 3 was the most restrictive one: Ala, Val, Ile, Lys, Arg, and Leu were the only allowed residues (Leu is the position-3 residue of wild-type PTLVNG-Fbp1) (Fig. 4). Position 4 was less restrictive, and position 5 still less, with the disallowed residues being only Val, Asp, and Glu (Fig. 4 and figs. S8 and S9). Position 6, the one remotest from N-terminal Pro, still partly disallowed Asp and Glu (Fig. 4 and fig. S10), which suggests that the substrate-binding groove of Gid4 is long enough to interact with up to six N-terminal residues of a substrate that bears N-terminal Pro.
The first four residues of PTLV-Fbp1, in the PTLV-G5-DHFR-DBD fusion, sufficed to confer the Pro-specific interaction with Gid4 (Fig. 3D, Fig. 4, and figs. S5A and S6A). We asked the same question about the first four residues of PHSV-Mdh2 and PIPV-Icl1, two other gluconeogenic enzymes. PHSV-G5-DHFR-DBD did bind to Gid4 and required N-terminal Pro for the binding (fig. S6A). However, PIPV-G5-DHFR-DBD did not bind to Gid4, despite the presence of N-terminal Pro (fig. S6A). But a longer N-terminal segment of PIPV-Icl1, its first 20 residues, did suffice for the Pro-specific binding to Gid4 (fig. S4D). We discuss the resulting understanding below after describing yet another feature of Gid4: its ability to recognize Pro at position 2.
Pck1 is targeted for degradation via its proline at position 2
The S. cerevisiae SP-Pck1 (SPSK-Pck1) is the fourth gluconeogenic enzyme (fig. S1). The N terminus–proximal Pro of SP-Pck1 is at position 2, in contrast to P-Fbp1, P-Icl1, and P-Mdh2. The degradation of SP-Pck1 was mediated by the GID Ub ligase, and the Pro2 residue of SP-Pck1 was required for its targeting by GID. Specifically, SP-Pck1 was stabilized in either gid2Δ or gid4Δ mutants, whereas SS-Pck1, which contained Ser2 (instead of wild-type Pro2), was a stable protein even in wild-type cells (Fig. 2G, fig. S3, B, D, and E, and fig. S11).
Although Gid4 was required for the degradation of SPSK-Pck1, we found that Gid4 did not bind to wild-type SPSK-Pck1 in two-hybrid assays, in contrast to the binding of Gid4 to P-Fbp1, P-Icl1, and P-Mdh2 (figs. S4A and S12A). To increase the likelihood of detecting binding of Gid4 to SPSK-Pck1, we performed two-hybrid assays that encompassed both N-terminal and C-terminal fusions of the AD domain to Gid4. However, no interaction of SPSK-Pck1 with Gid4 was observed (figs. S4A and S12A). We also examined the other GID subunits (Gid1, Gid2, Gid5, Gid7, Gid8, and Gid9) for their binding to SPSK-Pck1, with negative results as well (fig. S6, B and C).
PTLV-Fbp1, PIPV-Icl1, and PHSV-Mdh2, which interacted with Gid4 (see above), shared not only N-terminal Pro but also Val4. We therefore asked whether replacing Lys4 of wild-type SPSK-Pck1 with Val4 might suffice for detecting an interaction between Gid4 and the resulting (mutant) SPSV-Pck1. Remarkably, SPSV-Pck11-20-DHFR-DBD, bearing the first 20 residues of wild-type Pck1 as well as the Lys → Val mutation at position 4, interacted with Gid4 in two-hybrid assays, whereas the “wild-type” fusion, SPSK-Pck11-20-DHFR-DBD, did not (fig. S13, A to C). Given this result, we also constructed the same mutation in the 549-residue SPSK-Pck1. The full-length SPSV-Pck1 mutant (with Val4 instead of Lys4) clearly interacted with Gid4 in two-hybrid assays, in contrast to wild-type SPSK-Pck1, but the apparent affinity was lower than, for example, with PTLV-Fbp1 (fig. S13C).
In contrast to the first 20 residues of SPSV-Pck11-20-DHFR-DBD (with Val4 instead of wild-type Lys4), its first four residues, in the SPSV-G5-DHFR-DBD fusion, did not suffice for the binding to Gid4 (fig. S13C versus fig. S12, B and C). Additional two-hybrid assays showed that SPSVMNA, the first eight residues (including the transiently present N-terminal Met residue) of the mutant Pck1 sequence (containing Val4 instead of wild-type Lys4), were sufficient for conferring the binding to Gid4 (fig. S12B). Further analyses showed that Pro at position 2 was required, in the context of SPSVMNA-G2-DHFR-DBD, for the binding of SPSVMNA to Gid4, because the replacement of Pro2 with Ser2 abrogated the binding (figs. S12B and S13B). In addition, the change of wild-type Ser1 of SPSVMNA to Ala1, Thr1, or Pro1 also abolished the interaction of resulting fusions with Gid4 (figs. S12B and S13B).
Analogy between substrate-binding properties of Gid4 and MHC proteins
At least six N-terminal residues of a substrate were “sensed” by Gid4 (Fig. 4). This span of the binding groove of Gid4 and its “flexibility” in recognizing either N-terminal Pro (e.g., in P-Fbp1, P-Icl1, and P-Mdh2) or Pro at position 2 (e.g., in the N-terminal sequence SPSVMNA) are reminiscent of the binding grooves of major histo-compatibility complex (MHC) proteins, which interact with short peptides as a part of antigen presentation in vertebrates. The peptide-binding groove of MHC I is closed at both ends (it accommodates peptides up to ~9 residues long), whereas the binding groove of MHC II is open at both ends and can accommodate longer peptides (48, 49). In the language of this analogy, the binding groove of Gid4 is open at least at its “C-terminal” end, as Gid4 can bind to N-terminal sequences of protein-sized polypeptides (Figs. 2 and 3).
Is the binding groove of Gid4 open or closed at its “N-terminal” end? To address this question, we extended the Gid4-binding N-terminal sequence SPSVMNA by one residue (Gly, Ala, Ser, or Thr). Tellingly, none of the resulting XSPSVMNA-G2-DHFR-DBD fusions (X = Gly, Ala, Ser, Thr) interacted with Gid4, in contrast to SPSVMNA-G2-DHFR-DBD (fig. S12, B and C). Thus, the binding groove of Gid4 is likely to be closed at its “N-terminal” end, similarly to the groove of MHC I.
Yet another analogy between the binding grooves of MHC proteins and Gid4 is the pronounced nonuniformity in the recognition of individual residues of a bound peptide by an MHC groove. The side moieties of strongly bound (“anchor”) residues of an MHC-bound peptide are buried in pockets within the MHC binding groove. In contrast, side moieties of other MHC-bound residues of the same peptide make minor or no contacts with the groove, thereby allowing a variety of residues to be accommodated at such positions (48, 49). If this MHC-based understanding is applied to the setting of Gid4, then a Pro residue at positions 1 or 2 of a Gid4 substrate can be viewed as an “anchor” residue. Specifically, that Pro would be bound by the same (or nearly the same) region of the binding groove of Gid4, irrespective of whether the Pro residue is at position 1 or position 2 of a bound substrate.
A prediction of this model is that the pattern of “allowed” versus “disallowed” residues downstream of Pro (see, e.g., Fig. 4) would differ depending on whether the Pro (anchor) residue is at position 1 or at position 2. That would be so because the substrate-binding groove of Gid4 would “count” downstream residues starting from the bound anchor Pro residue. To address this model, we took advantage of the fact that the Leu residue, in the Gid4-binding N-terminal sequence PTLV of PTLV-Fbp1, is an allowed residue at position 3 of PTLV. In contrast, Leu is disallowed (abrogates the binding) if it is present at position 2, in the mutant sequence PLLV (Fig. 4 and figs. S8 and S9).
Given this logic, we compared the binding of PSLVMNA-G2-DHFR-DBD and SPLVMNA-G2-DHFR-DBD to Gid4. In the first fusion, Pro is at position 1 (followed by Ser2 and Leu3), whereas in the second fusion, Pro is at position 2. Although Leu is at the “absolute” position 3 in both fusions, Leu happens to be at position 2 (a disallowed position in Fig. 4) relative to the Pro-2 residue (a putative anchor residue) of SPLVMNA-G2-DHFR-DBD. Remarkably, PSLVMNA-G2-DHFR-DBD bound to Gid4, whereas SPLVMNA-G2-DHFR-DBD did not (fig. S12C). These results, predicted by the anchor-residue model, provide further support for the analogy between the binding groove of Gid4 and the grooves of MHC (48, 49).
The effects of N terminus–proximal residues of a protein on its binding to Gid4 are likely to be mutually dependent (cooperative or anticooperative) rather than strictly position-specific. For example, a particularly favorable residue, among the allowed ones (Fig. 4 and figs. S8 to S10), may partly compensate for a binding-interfering effect of an adjacent suboptimal residue, or even of a disallowed residue. This model would account for the observed specific binding of PIPV-Icl1 and PHSV-Mdh2 to Gid4 (fig. S4D), despite the presence of N terminus–proximal residues, in both of these Gid4 substrates, which would be either disallowed or suboptimal in the N-terminal sequence context of PTLV-Fbp1, the other Gid4 substrate (Fig. 4).
The reason for the apparent absence of binding of Gid4 to wild-type SPSK-Pck1, in contrast to the binding of Gid4 to the position-4 mutant SPSV-Pck1, is unclear (fig. S13, A to C). A parsimonious possibility is that SPSK-Pck1 does bind to Gid4 [thereby accounting for the Gid4/Gid2-dependent degradation of SPSK-Pck1 (Fig. 2G and figs. S3B and S11)], but with a binding affinity that is below the detection threshold of two-hybrid assays. One prediction of this (to be verified) explanation is that the rate of degradation of the mutant SPSV-Pck1 would be higher than that of wild-type SPSK-Pck1 (Fig. 2G and figs. S3B and S11).
Are there nongluconeogenic substrates of Gid4?
Although most subunits of the GID Ub ligase (Fig. 1C) are expressed constitutively, previous work suggested that the Gid4 subunit was absent from cells growing on either ethanol or glucose, and that Gid4 was transiently up-regulated, in part through becoming longer-lived, during transitions from a nonfermentable carbon source (such as ethanol) to glucose (6, 7). However, when we ectopically expressed P-Fbp13f (versus the S-Fbp13f mutant) and P-Mdh23f (versus the S-Mdh23f mutant) in cells growing on glucose, we found that these gluconeogenic enzymes were rapidly destroyed in wild-type cells, that their degradation required N-terminal Pro, and that this degradation was abolished in gid4Δ cells (Fig. 5 and fig. S14, A and B). Because the expression of P-Fbp1, P-Icl1, P-Mdh2, and SP-Pck1 is normally repressed by glucose at the level of transcription (5, 50), the continued activity of the Pro/N-end rule pathway under glucose-replete conditions (Fig. 5 and fig. S14, A and B) suggested that nongluconeogenic substrates of this pathway exist as well.
Fig. 5. Degradation of P-Mdh2 in cells under glucose-replete conditions.
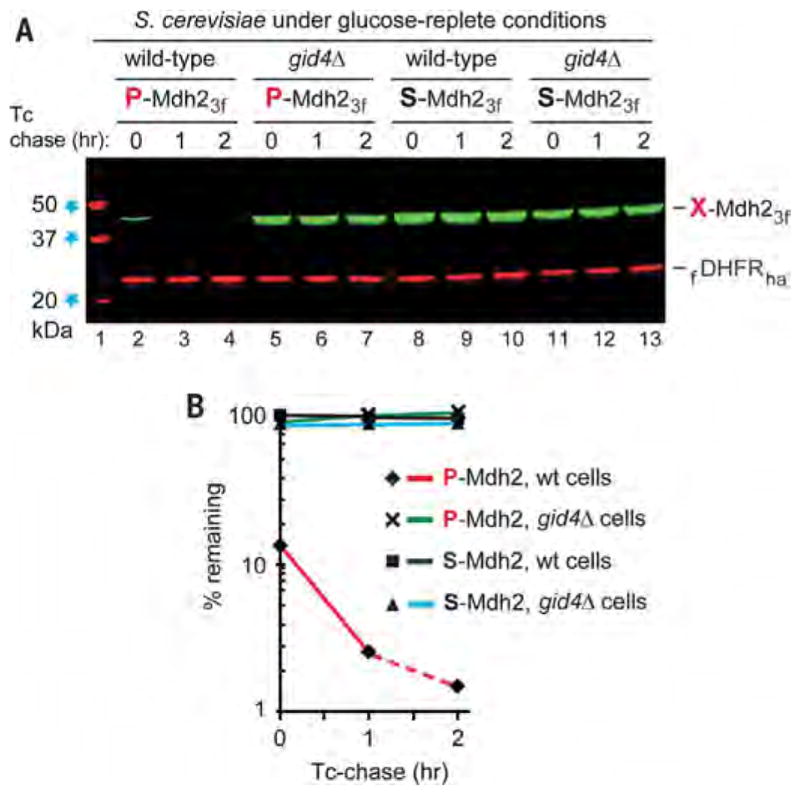
(A) Tetracycline-based, reference-based chases were performed as described in the legend to Fig. 2, at 30°C with indicated S. cerevisiae strains expressing either P-Mdh23f or S-Mdh23f, but with cells that did not undergo glucose deprivation. Lane 1: kDa markers. Lanes 2 to 4: Wild-type cells expressing P-Mdh23f. Lanes 5 to 7: Same as lanes 2 to 4 but with gid4Δ cells. Lanes 8 to 10: Same as lanes 2 to 4 but with S-Mdh23f. Lanes 11 to 13: Same as lanes 8 to 10 but with gid4Δ cells. (B) Quantification of data in (A). All chases in this study were performed at least twice and yielded results that differed by less than 10%. Note the time-zero, before-chase degradation of more than 80% of P-Mdh23f in wild-type cells (lanes 2 to 4).The bands of X-Mdh23f and fDHFRha are indicated.
S. cerevisiae DNA encodes ~300 proteins that would be expected to bear N-terminal Pro after the cotranslational removal of N-terminal Met (figs. S15 to S17). We asked whether some of these diverse proteins (which are largely unrelated to gluconeogenesis) might interact with Gid4 in two-hybrid assays, be targeted for degradation by the GID Ub ligase, or both. The results of this continuing investigation are summarized below.
The first 20 residues of some nongluconeogenic N-terminal Pro–bearing yeast proteins (figs. S15 to S17) could confer a specific interaction with Gid4. A positive example is the PFVK-Oye21-20-DHFR-DBD (fig. S4D). A negative example is PESR-Pyk21-20-DHFR-DBD, which did not bind to Gid4, despite its N-terminal Pro (fig. S4D).
If the first 20 residues of an N-terminal Pro–bearing protein could confer its binding to Gid4, this did not necessarily imply the binding of the full-length protein. For example, PFVK-Oye21-20-DHFR-DBD bound to Gid4, but the corresponding full-length PFVK-Oye2 did not (fig. S4D). [Gid4 can bind to both entities, as shown by the example of the full-length gluconeogenic enzyme PTLV-Fbp1 and its PTLV-Fbp11-20-DHFR-DBD derivative (Fig. 3, A, B, and D).] Thus, features of a full-length N-terminal Pro–bearing protein, such as the extent of steric exposure (and/or flexibility) of its N-terminal region, can be either permissive or non-permissive in regard to its interaction with Gid4.
If the first 20 residues of a natural N-terminal Pro–bearing protein did not bind to Gid4, it meant (in cases examined so far) that the full-length protein also did not bind to Gid4. An example is the absence of binding by both PESR-Pyk21-20-DHFR-DBD and the full-length PESR-Pyk2 (Fig. 3A and fig. S4). Another example of a full-length protein that contained N-terminal Pro but did not bind to Gid4 is PTLY-Ald2 (fig. S4D).
Degradation assays with nongluconeogenic, N-terminal Pro–bearing natural yeast proteins (specifically, proteins other than P-Fbp1, P-Icl1, and P-Mdh2) have not yet identified a substrate of the Gid4-dependent Pro/N-end rule pathway. The N-terminal Pro–bearing proteins Ald2, Hri1, Naa10, Nvj3, Pex21, Pyk2, Rpl12a, Snx3, Swc3, and Yhr020w have been examined by degradation assays (figs. S15 to S17). We show one example of these degradation assays with PVSE-Yhr020w, the yeast proline-tRNA synthetase, a long-lived protein despite its N-terminal Pro (fig. S14C). These ongoing analyses are far from complete, as the examined N-terminal Pro–bearing yeast proteins constitute less than 4% of the full set (figs. S15 to S17). In sum, while N-terminal Pro–bearing physiological substrates of the S. cerevisiae Pro/N-end rule pathway other than P-Fbp1, P-Icl1, and P-Mdh2 have not yet been pinpointed, they are likely to exist and would be expected to be identified through the approaches described above.
Gid4 and the Pro/N-end rule pathway
We have identified Gid4, a subunit of the S. cerevisiae GID Ub ligase, as the recognition component of the GID-mediated proteolytic system termed the Pro/N-end rule pathway (Fig. 1B). One function of this system is a rapid down-regulation of gluconeogenesis, through the destruction of gluconeogenic enzymes, upon a return of cells to glucose-replete conditions. Gid4, the Pro/N-recognin of the Pro/N-end rule pathway, is shown here to target the gluconeogenic enzymes P-Fbp1, P-Icl1, and P-Mdh2 (and possibly other proteins as well) through the binding to their N-terminal Pro residues and adjacent sequence motifs (Figs. 1, 3, and 4).
Gid4 was also required for the GID-mediated degradation of SP-Pck1, the fourth gluconeogenic enzyme (Fig. 2G and figs. S3B and S11). Degradation of SP-Pck1 involved the recognition of its internal (position-2) Pro residue (Fig. 2G and fig. S11). However, in two-hybrid assays, Gid4 did not bind to either full-length SP-Pck1 or its N-terminal fragment (figs. S4A and S12A). Nonetheless, Gid4 could also be shown to recognize Pro at position 2, provided that the sequence of SP-Pck1 proximal to Pro2 was altered by one residue (figs. S12 and S13). A possible explanation of this result is mentioned above.
The properties of Gid4 discovered so far indicate that its substrate-binding groove, likely to be analogous to peptide-binding grooves of antigen-presenting MHC proteins (see above), can recognize either the N-terminal Pro residue or Pro at position 2 in the presence of cognate adjacent sequence motifs. The S. cerevisiae genome encodes ~300 proteins that are expected to bear N-terminal Pro (figs. S15 to S17). Nongluconeogenic substrates of the Pro/N-end rule pathway that bear N-terminal Pro remain to be identified. The recognition flexibility of the substrate-binding groove of Gid4 (discussed above; see also Fig. 4 and figs. S12 and S13) suggests that the true diversity of Gid4 substrates is only beginning to be determined.
A notable aspect of GID-mediated processes is the dichotomy between the GID/proteasome-mediated degradation of gluconeogenic enzymes (Fig. 1B) and the “alternative” degradation of the same enzymes through an autophagy-related pathway called VID (vacuole import and degradation) (4–7, 10, 11). If S. cerevisiae is grown on a nonfermentable carbon source such as ethanol for less than a day before returning cells to glucose (i.e., the regimen of the present study), the involvement of VID is negligible (4–7). However, a much longer growth on a nonfermentable carbon source results (after return to glucose) in the VID-mediated degradation of gluconeogenic enzymes (8). Remarkably, both proteolytic processes require the Gid4 Pro/N-recognin of the present work as well as the other subunits of the GID Ub ligase (5, 8). It is unclear why two distinct mechanisms have evolved to conditionally destroy specific proteins bearing Pro/N-degrons. The design of a circuit for transitions between the proteasome-based and the VID-based protein degradation is also unknown. How the ability of Gid4 to recognize Pro/N-degrons (Fig. 1B) is used by VID (as distinguished from the proteasome) remains to be understood as well.
Subunits of the S. cerevisiae GID Ub ligase (Fig. 1, B and C) have sequelogous [similar in sequence (51)] counterparts in animals and plants (15–17). Metazoan sequelogs of GID subunits form a complex that acts as a Ub ligase (15, 17). Thus, the discovery that yeast Gid4 is the Pro/N-recognin (Figs. 1B and 3) will facilitate the understanding of the Pro/N-end rule pathway in other eukaryotes as well.
The Arg/N-end rule pathway (Fig. 1E) does not recognize N-terminal Pro (22). In addition, the N-terminal Pro residue is not Nt-acetylated, in contrast to other N-terminal residues (39, 52). Consequently, N-terminal Pro cannot confer recognition by the Ac/N-end rule pathway or by the Arg/N-end rule pathway on a protein bearing this residue (Fig. 1, D and E). Nonetheless, owing to the identification of the GID-mediated proteolytic system as the Pro/N-end rule pathway, all 20 amino acids of the genetic code have now been shown to act, in specific sequence contexts, as destabilizing N-terminal residues (Fig. 1A). Thus, most proteins in a cell may be conditionally short-lived N-end rule substrates, either as full-length proteins or as protease-generated natural protein fragments.
Materials and methods
Yeast strains, media, and genetic techniques
Standard techniques were used for transformation with DNA and for construction of specific strains (table S1). S. cerevisiae media included YPD, SD, SE, and SC (see supplementary materials and methods). The alternative carbon sources, in either liquid or plate media, were 2% ethanol or 2% glucose.
Construction of plasmids
Polymerase chain reaction (PCR) and the Gateway cloning method (Invitrogen) were used for constructions of plasmids that are cited in table S2. The oligonucleotide primers are cited in table S3. The Escherichia coli strains DH5α, SUREII (Stratagene), and STBL2 (Invitrogen) (table S1) were used for cloning and maintaining plasmids. See supplementary materials and methods for details.
Tetracycline-chase assays and immunoblotting
Most S. cerevisiae protein degradation assays of this study used a version of the PRT (Fig. 2, A and B). These reference-based, PRT-based degradation assays, which used tetracycline-mediated chases, are described in supplementary materials and methods. Immunoblotting was carried out using standard techniques. The results were quantified using the Odyssey 9120 scanner (Li-Cor).
Two-hybrid and split-ubiquitin binding assays
Two initial plasmids for two-hybrid assays were pGADCg and pGBKCg (table S2). These assays were carried out largely as described (43–45). Split-Ub assays (46, 47) for mapping interactions between Gid4 and (M)P-Fbp1 versus (M)S-Fbp1 involved cotransformations of S. cerevisiae JD52 (table S1) with pCSJ418 (expressing NUb-Gid4-Flag) and the plasmids pCSJ473 or pCSJ474 (table S2), which expressed, respectively, (M)P-Fbp1 or (M)S-Fbp1 linked to the CUb-R-Ura3 moiety. See supplementary materials and methods for details of both assays.
Supplementary Material
Acknowledgments
We thank P. Bjorkman (Caltech) for her comments on the manuscript, and A. Melnykov and other members of the Varshavsky laboratory for discussions and helpful suggestions. Supported by NIH grants DK039520 and GM031530 (A.V.).
Footnotes
REFERENCES AND NOTES
- 1.Fraenkel DG. Yeast Intermediary Metabolism. Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- 2.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman M, Chiang HL. Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics. 1996;143:1555–1566. doi: 10.1093/genetics/143.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hämmerle M, et al. Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome-catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J Biol Chem. 1998;273:25000–25005. doi: 10.1074/jbc.273.39.25000. [DOI] [PubMed] [Google Scholar]
- 5.Menssen R, et al. Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J Biol Chem. 2012;287:25602–25614. doi: 10.1074/jbc.M112.363762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santt O, et al. The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell. 2008;19:3323–3333. doi: 10.1091/mbc.E08-03-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regelmann J, et al. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: A genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell. 2003;14:1652–1663. doi: 10.1091/mbc.E02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alibhoy AA, Chiang HL. Vacuole import and degradation pathway: Insights into a specialized autophagy pathway. World J Biol Chem. 2011;2:239–245. doi: 10.4331/wjbc.v2.i11.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giardina BJ, Chiang HL. Fructose-1,6-bisphosphatase, malate dehydrogenase, isocitrate lyase, phosphoenolpyruvate carboxykinase, glyceraldehyde-3-phosphate dehydrogenase, and cyclophilin A are secreted in Saccharomyces cerevisiae grown in low glucose. Commun Integr Biol. 2013;6:e27216. doi: 10.4161/cib.27216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alibhoy AA, Giardina BJ, Dunton DD, Chiang HL. Vid30 is required for the association of Vid vesicles and actin patches in the vacuole import and degradation pathway. Autophagy. 2012;8:29–46. doi: 10.4161/auto.8.1.18104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CR, Wolfe AB, Cui D, Chiang HL. The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J Biol Chem. 2008;283:26116–26127. doi: 10.1074/jbc.M709922200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minard KI, McAlister-Henn L. Glucose-induced degradation of the MDH2 isozyme of malate dehydrogenase in yeast. J Biol Chem. 1992;267:17458–17464. [PubMed] [Google Scholar]
- 13.Li B, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balsa-Martinez E, Puigserver P. Cancer cells hijack gluconeogenic enzymes to fuel cell growth. Mol Cell. 2015;60:509–511. doi: 10.1016/j.molcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi N, et al. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8a and ARMC8b are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 16.Francis O, Han F, Adams JC. Molecular phylogeny of a RING E3 ubiquitin ligase, conserved in eukaryotic cells and dominated by homologous components, the muskelin/RanBPM/CTLH complex. PLOS ONE. 2013;8:e75217. doi: 10.1371/journal.pone.0075217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfirrmann T, et al. RMND5 from Xenopus laevis is an E3 ubiquitin-ligase and functions in early embryonic forebrain development. PLOS ONE. 2015;10:e0120342. doi: 10.1371/journal.pone.0120342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 19.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MK, Oh SJ, Lee BG, Song HK. Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway. Proc Natl Acad Sci USA. 2016;113:12438–12443. doi: 10.1073/pnas.1612620113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014;24:603–611. doi: 10.1016/j.tcb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Dougan DA, Micevski D, Truscott KN. The N-end rule pathway: From recognition by N-recognins, to destruction by AAA+ proteases. Biochim Biophys Acta. 2012;1823:83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Eldeeb M, Fahlman R. The N-end rule: The beginning determines the end. Protein Pept Lett. 2016;23:343–348. doi: 10.2174/0929866523666160108115809. [DOI] [PubMed] [Google Scholar]
- 29.Graciet E, Wellmer F. The plant N-end rule pathway: Structure and functions. Trends Plant Sci. 2010;15:447–453. doi: 10.1016/j.tplants.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Kim HK, et al. The N-terminal methionine of cellular proteins as a degradation signal. Cell. 2014;156:158–169. doi: 10.1016/j.cell.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piatkov KI, Brower CS, Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci USA. 2012;109:E1839–E1847. doi: 10.1073/pnas.1207786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piatkov KI, Oh J-H, Liu Y, Varshavsky A. Calpain-generated natural protein fragments as short-lived substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2014;111:E817–E826. doi: 10.1073/pnas.1401639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brower CS, Piatkov KI, Varshavsky A. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell. 2013;50:161–171. doi: 10.1016/j.molcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cha-Molstad H, et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. 2015;17:917–929. doi: 10.1038/ncb3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash S, Inobe T, Hatch AJ, Matouschek A. Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol. 2009;5:29–36. doi: 10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadas B, Piatkov KI, Brower CS, Varshavsky A. Analyzing N-terminal arginylation through the use of peptide arrays and degradation assays. J Biol Chem. 2016;291:20976–20992. doi: 10.1074/jbc.M116.747956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SE, et al. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015;347:1249–1252. doi: 10.1126/science.aaa3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aksnes H, Drazic A, Marie M, Arnesen T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem Sci. 2016;41:746–760. doi: 10.1016/j.tibs.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Dörfel MJ, Lyon GJ. The biological functions of Naa10—From amino-terminal acetylation to human disease. Gene. 2015;567:103–131. doi: 10.1016/j.gene.2015.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kötter P, Weigand JE, Meyer B, Entian K-D, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: Substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49:5588–5599. doi: 10.1021/bi1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vidal M, Fields S. The yeast two-hybrid assay: Still finding connections after 25 years. Nat Methods. 2014;11:1203–1206. doi: 10.1038/nmeth.3182. [DOI] [PubMed] [Google Scholar]
- 44.Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763–2788. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stellberger T, et al. Improving the yeast two-hybrid system with permutated fusions proteins: The Varicella Zoster Virus interactome. Proteome Sci. 2010;8:8. doi: 10.1186/1477-5956-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dünkler A, Müller J, Johnsson N. Detecting protein-protein interactions with the split-ubiquitin sensor. Methods Mol Biol. 2012;786:115–130. doi: 10.1007/978-1-61779-292-2_7. [DOI] [PubMed] [Google Scholar]
- 48.Batalia MA, Collins EJ. Peptide binding by class I and class II MHC molecules. Biopolymers. 1997;43:281–302. doi: 10.1002/(SICI)1097-0282(1997)43:4<281::AID-BIP3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 49.Yaneva R, Schneeweiss C, Zacharias M, Springer S. Peptide binding to MHC class I and II proteins: New avenues from new methods. Mol Immunol. 2010;47:649–657. doi: 10.1016/j.molimm.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Hung G-C, Brown CR, Wolfe AB, Liu J, Chiang H-L. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 51.Varshavsky A. ‘Spalog’ and ‘sequelog’: Neutral terms for spatial and sequence similarity. Curr Biol. 2004;14:R181–R183. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Goetze S, et al. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLOS Biol. 2009;7:e1000236. doi: 10.1371/journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.