Abstract
Plant growth is controlled by integration of hormonal and light-signaling pathways. BZS1 is a B-box zinc finger protein previously characterized as a negative regulator in the brassinosteroid (BR) signaling pathway and positive regulator in the light-signaling pathway. However, the mechanisms by which BZS1/BBX20 integrates light and hormonal pathways are not fully understood. Here, using a quantitative proteomic workflow, we identified several BZS1-associated proteins including light signaling components COP1 and HY5. Direct interactions of BZS1 with COP1 and HY5 were verified by yeast two-hybrid and co-immunoprecipitation assays. Overexpression of BZS1 causes a dwarf phenotype that is suppressed by the hy5 mutation, while overexpression of BZS1 fused with the SRDX transcription repression domain (BZS1-SRDX) causes a long-hypocotyl phenotype similar to hy5, indicating that BZS1’s function requires HY5. BZS1 positively regulates HY5 expression, whereas HY5 negatively regulates BZS1 protein level, forming a feedback loop that potentially contributes to signaling dynamics. In contrast to BR, strigolactone (SL) increases BZS1 level, whereas the SL responses of hypocotyl elongation, chlorophyll and HY5 accumulation are diminished in the BZS1-SRDX seedlings, indicating that BZS1 is involved in these SL responses. These results demonstrate that BZS1 interacts with HY5 and plays a central role in integrating light and multiple hormone signals for photomorphogenesis in Arabidopsis.
Keywords: light signaling, immunoprecipitation, mass spectrometry, strigolactone, BZS1/BBX20, HY5, COP1
INTRODUCTION
Plant development is particularly sensitive to light, which is both the energy source for photosynthesis and the regulatory signal (Galvão and Fankhauser, 2015). Upon germination in the dark, a seedling undergoes a developmental program named skotomorphogenesis, which is characterized by elongated hypocotyl, closed cotyledon, apical hook, and short root. Exposure to light promotes photomorphogenesis, which is characterized by short hypocotyl, open cotyledon, chloroplast development and pigment accumulation (Von Arnim and Deng, 1996; Kami et al., 2010). In addition to light, photomorphogenesis is also regulated by several hormones, including brassinosteroid (BR), auxin, gibberellin (GA) and strigolactone (SL) (de Lucas et al., 2008; Feng et al., 2008; Luo et al., 2010; Tsuchiya et al., 2010; Fan et al., 2012; Waters and Smith, 2013; Jia et al., 2014; Chaiwanon et al., 2016). The molecular mechanisms that integrate the light and hormonal signals are not fully understood.
Light signal is perceived by photoreceptors, which regulate gene expression through several classes of transcription factors (Galvão and Fankhauser, 2015). Downstream of photoreceptors, the E3 ubiquitin ligase COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) acts as a central repressor of photomorphogenesis (Deng et al., 1991; Lau and Deng, 2012). COP1 targets several transcription factors for proteasome-mediated degradation in the dark (Deng et al., 1991; Osterlund et al., 2000; Lau and Deng, 2012). Light-activated photoreceptors directly inhibit COP1’s activity, leading to the accumulation of the COP1-interacting transcription factors, such as HY5 (LONG HYPOCOTYL 5), BZS1(bzr1–1D suppressor 1), and GATA2(GATA-type transcription factor 2), which positively regulate photomorphogenesis (von Arnim and Deng, 1994; Osterlund et al., 2000; Luo et al., 2010; Lau and Deng, 2012).
Recent studies have uncovered mechanisms of signal crosstalk that integrate light signaling pathways with BR, GA, and auxin pathways (Wang et al., 2014). The transcription factors of these signaling pathways directly interact with each other in cooperative or antagonistic manners to regulate overlapping sets of target genes (Bai et al., 2012; Oh et al., 2012; Oh et al., 2014). BR has been shown to repress, through the transcription factor BZR1, the expression of positive regulators of photomorphogenesis, including the light-stabilized transcription factors GATA2 and BZS1 (Luo et al., 2010; Fan et al., 2012; Wang et al., 2012). BZS1 (also named BBX20) is a member of the B-box zinc finger protein family, which has two B-box domains at its N terminus without any known DNA binding domain (Khanna et al., 2009). It is unclear how BZS1 regulates gene expression. Recent studies have shown that SL inhibits hypocotyl elongation and promotes HY5 accumulation in Arabidopsis plants grown under light (Tsuchiya et al., 2010; Waters and Smith, 2013; Jia et al., 2014), but the molecular mechanisms through which SL signaling integrates with light and other hormone pathways remain largely unknown.
Immunoprecipitation (IP) of protein complexes followed by mass spectrometry analysis (IP-MS) is a powerful method for identifying interacting partners and posttranslational modifications of a protein of interest (Ni et al., 2013; Wang et al., 2013; Ni et al., 2014). In particular, research in animal systems has shown that combining stable isotope labeling with IP-MS can quantitatively distinguish specific interacting proteins from non-specific background proteins (Blagoev et al., 2003; Trinkle-Mulcahy et al., 2008; Hubner et al., 2010). Stable isotope labeling in Arabidopsis (SILIA) has been established as an effective method of quantitative mass spectrometry (Li, 2012; Yang et al., 2013); however, combination of SILIA with IP-MS (SILIA-IP-MS) has yet to be established.
To further characterize the molecular function of BZS1, we performed SILIA-IP-MS analysis of the BZS1 protein complex, and identified several BZS1-accociated proteins. Among those are COP1, HY5, and BZS1’s homologs STH2/BBX21 and STO/BBX24. We further showed that BZS1 directly interacts with HY5, and positively regulates HY5 RNA and protein levels. Genetic analysis indicated that HY5 is required for BZS1 to inhibit hypocotyl elongation and promote anthocyanin accumulation. In addition, BZS1 is positively regulated by SL at both transcriptional and translational levels. Plants overexpressing a dominant-negative form of BZS1 (fusion with the SRDX transcription repressor domain) show an elongated-hypocotyl phenotype and reduced sensitivity to SL, similar to the hy5 mutant. Our results demonstrated that BZS1 acts through HY5 to promote photomorphogenesis and is a crosstalk junction of light, BR and SL signals. This study further advances our understanding of the complex network that integrates multiple hormonal and environmental signals.
RESULTS
Identifying BZS1 protein complex by a quantitative proteomics workflow
In order to elucidate the biochemical mechanism of BZS1 function, we performed a SILIA-IP-MS analysis of the BZS1 protein complex. We transformed Arabidopsis with a construct that overexpresses a BZS1 protein fused with the yellow fluorescence protein (YFP) at the C-terminus driven by the constitutive 35S promoter (35S::BZS1-YFP, named BZS1-YFP). A transgenic line that showed mild dwarf and dark-green-leaf phenotypes, resembling the bzs1-D mutant (Fan et al., 2012), was selected for the analysis. Pair-wised comparison was designed to seperately compare BZS1-YFP and 35S::YFP (named YFP) transgenic plants with non-transgenic wild type, to determine proteins associated with BZS1-YFP and YFP alone, respectively. The isotope-labeling was reversed in replicate experiments (IP1 and IP2) to minimize false positives (Fig. S1).
To obtain complete 15nitrogen (15N) labeling of young seedlings, we first grew BZS1-YFP, YFP and wild-type plants hydroponically in medium containing 15N, and obtained stable isotope-labeled seeds (Fig. S2A). These 15N-labeled seeds and regular 14N seeds were grown again on corresponding 15N or 14N medium to obtain 5-day-old seedlings for further analysis (Fig. S2B). For each pair of isotope-labeled sample and control, equal amount of tissues was mixed, and the protein extract was used for immunoprecipitation using the GFP-trap beads. The immunoprecipitated proteins were separated in SDS-PAGE, gel bands were in-gel digested, and the tryptic peptides were analyzed by mass spectrometry (Figs. S1 and S2C).
Mass spectrometry analyses of the two BZS1-YFP immunoprecipitation experiments identified 514 and 383 proteins, respectively, with 279 proteins identified in both repeats (Table S1A). A smaller number of proteins were identified in the YFP experiments (312 in IP1 and 329 in IP2, with 254 identified in both IPs) (Table S1B). Quantitation of isotope ratios showed median ratios of 1.16 and 1.23 for the two BZS1-YFP experiments, and 1.0 and 0.92 for the two YFP control experiments. The protein ratios of the YFP control datasets had standard deviation (SD) of 0.23 and 0.57 (after removing obvious isotope bias and one potential YFP-interacting protein). Using 2× median as cutoff, 16 proteins were enriched in BZS1-YFP compared to wild-type control in the two repeat experiments. The YFP and wild type comparison identified 2 proteins that were enriched over 2× median, presumably due to association with YFP or false discovery, suggesting a false discovery rate <0.8% (2 of 254 quantified). The 15 proteins enriched by BZS1-YFP( not including BZS1 itself) were not enriched by YFP alone, and thus were considered BZS1-associated proteins (Table S2). Among the BZS1-associated proteins are COP1 and HY5, two key regulators of the light signaling pathways, as well as BZS1/BBX20’s homologs STH2/BBX21 and STO/BBX24 (Tables 1 and S1C).
Table 1.
Light signaling components identified as BZS1-associated proteins
| Gene ID | PSM in-IP1 | PSM-in IP2 | IP1 ratio (BZS1-YFP/Col) | IP2ratio (BZS1-YFP/Col) | Description | Reference |
|---|---|---|---|---|---|---|
| AT1G75540 | 10 | 12 | 1024 | 1024 | B-box protein, STH2/BBX21 | Datta et al., 2007 |
| AT1G06040 | 2 | 2 | 5.88 | 11.12 | B-box protein, STO/BBX24 | Indorf, Cordero et al. 2007 |
| AT5G11260 | 4 | 1 | 6.25 | 1024 | bZIP transcription factor, HY5 | Oyama et al., 1997 |
| AT2G32950 | 1 | 2 | 5.56 | 3.83 | COP1 | Deng et al., 1991 |
BZS1-associated proteins known to be involved in light signaling are listed. PSM is the number of peptide-spectrum matches. The protein ratio is derived from peptide ratios using kernel density estimation as described by Liu et al. (2014). All the quantified peptides of STH2, STO, HY5 and COP1 show a ratio over 2× median of each IP, as listed in Table S1C.
BZS1 interacts with COP1, HY5 and STH2
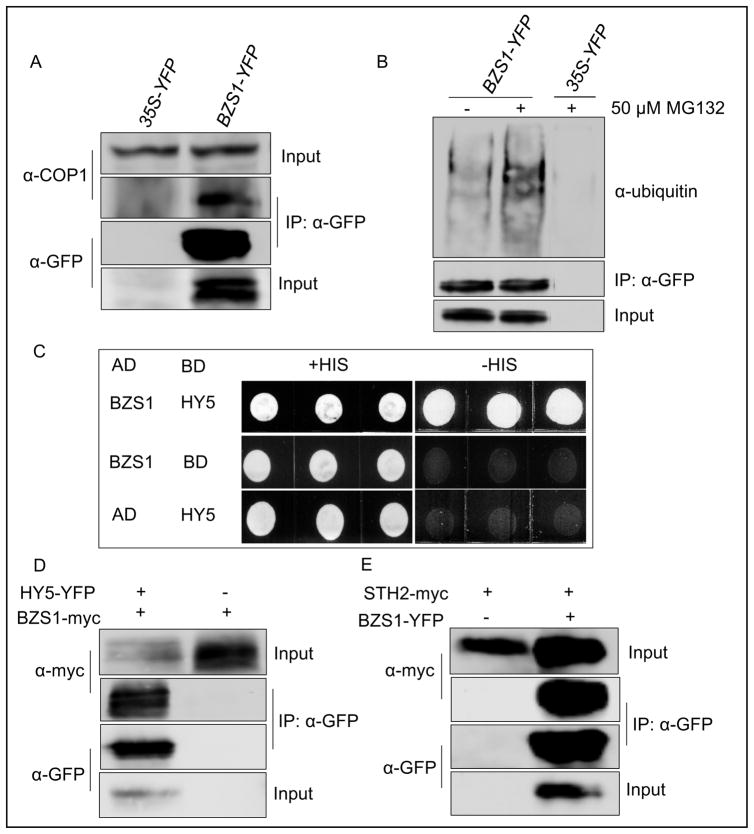
To verify the interaction between BZS1 and COP1 in vivo, we performed immunoprecipitation of BZS1-YFP from the BZS1-YFP transgenic Arabidopsis seedlings using anti-GFP antibody, and probed the immunoblot with anti-COP1 antibody. The results showed that COP1 co-immunoprecipitates with BZS1-YFP (Fig. 1A), confirming that BZS1 interacts with COP1 in plants. Consistent with BZS1’s interaction with the COP1 E3 ubiquitin ligase, the immunoprecipitated BZS1-YFP can be detected by anti-ubiquitin antibody, and the level of ubiquitination was increased by treatment with proteasome inhibitor MG132 (Fig. 1B).
Fig. 1. BZS1 interacts with COP1, HY5 and STH2/BBX21.
A: Co-immunoprecipitation of BZS1 with COP1. BZS1-YFP was immunoprecipitated with anti-GFP polyclonal antibody and the immunoblot was probed with anti-COP1 antibody or anti-GFP monoclonal antibody. B: BZS1 was ubiquitinated. Three-day-old dark-grown BZS1-YFP seedlings were treated with mock or 50 μM MG132 for 24 h before immunoprecipitation. The immunoblot was probed with either anti-GFP monoclonal antibody or anti-ubiquitin antibody. C: Interaction of BZS1 with HY5 in yeast two-hybrid assay. D and E: Co-immunoprecipitation of BZS1 with HY5 (D) or STH2 (E). Vectors containing the indicated constructs were co-transformed into Nicotiana benthamiana. HY5-YFP (D) or BZS1-YFP (E) was immunoprecipitated with anti-GFP polyclonal antibody and the immunoblot was probed with anti-myc or anti-GFP monoclonal antibody.
We further confirmed the direct interaction of BZS1 and HY5 by yeast two-hybrid assays (Fig. 1C). Further, when transiently co-expressed in Nicotiana benthamiana, the BZS1-myc protein was co-immunoprecipitated by the HY5-YFP protein (Fig. 1D), confirming their interaction in plant cells. Similarly, the STH2-myc protein was co-immunoprecipitated by BZS1-YFP (Fig. 1E). These results confirmed the SILIA-IP-MS results that BZS1 interacts with COP1, HY5, and STH2/BBX21.
The function of BZS1 depends on HY5
To determine the functional relationship between BZS1 and HY5, we first compared previously published transcriptomic data from BZS1-overexpression plants with chromatin immunoprecipitation-microarray data of HY5 direct target genes (Lee et al., 2007; Fan et al., 2012). The result showed that 56.3% of BZS1-activated genes are HY5 targets while only 13% of BZS1-repressed genes are HY5 targets (Fig. 2A). Such significant overlap between BZS1-activated and HY5-bound genes suggests that BZS1 interacts with HY5 to activate gene expression.
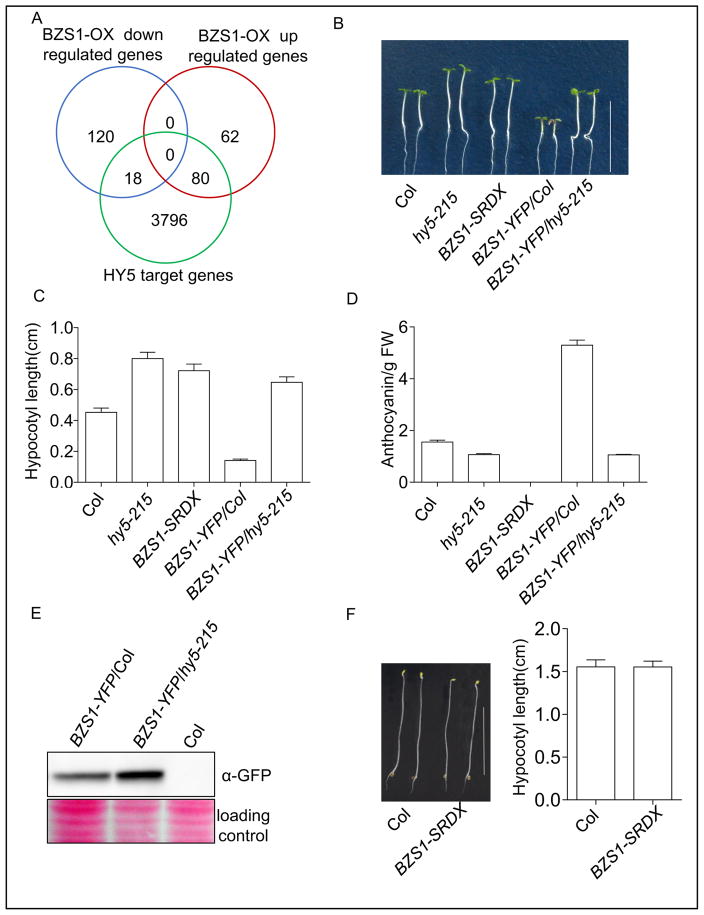
Fig. 2. The function of BZS1 is dependent on HY5.
A: Overlaps of BZS1-regulated genes (Fan et al., 2012) with HY5 target genes (Lee et al., 2007). B: Phenotypes of wild-type (Col), hy5-215, BZS1-SRDX, BZS1-YFP/Col, and BZS1-YFP/hy5-215 seedlings grown under constant red light (20 μmol m–2s–1) for 4 days. Scale bar, 1 cm. C: Hypocotyl length measurement of seedlings described in (B). Error bars represent SD (n = 30). Significant differences are marked as asterisks, P < 0.001. D: Anthocyanin content of the indicated seedlings grown under continuous white light (100 μmol m–2s–1) for 5 days. Error bars represent SD (n = 30). Significant differences are marked as asterisks, P < 0.001. FW, fresh weight. E: Western blot analysis of BZS1 protein accumulation in the hy5 mutant. Total proteins were extracted from 4-day-old seedlings grown under continuous white light (100 μmol m–2s–1). F: Hypocotyl length of BZS1-SRDX seedlings grown in the dark for 4 days. Error bars represent SD (n = 30). Scale bar, 1 cm.
Fusing a transcription repressor domain, such as the SRDX domain, to a transcription activator has been shown to have a dominant negative effect (Hiratsu et al., 2003). Overexpression of the BZS1-SRDX fusion sequence driven by 35S promotor in Arabidopsis caused a long-hypocotyl phenotype and reduced anthocyanin accumulation (Figs. 2B–D and S3), which were similar to the phenotypes of loss-of-function mutant hy5-215 but opposite to the phenotypes caused by BZS1 overexpression, further supporting that BZS1 functions as a transcription activator together with HY5. The BZS1-SRDX plants grown in the dark did not show any obvious phenotype (Fig. 2F), consistent with HY5 and BZS1 being degraded in the dark.
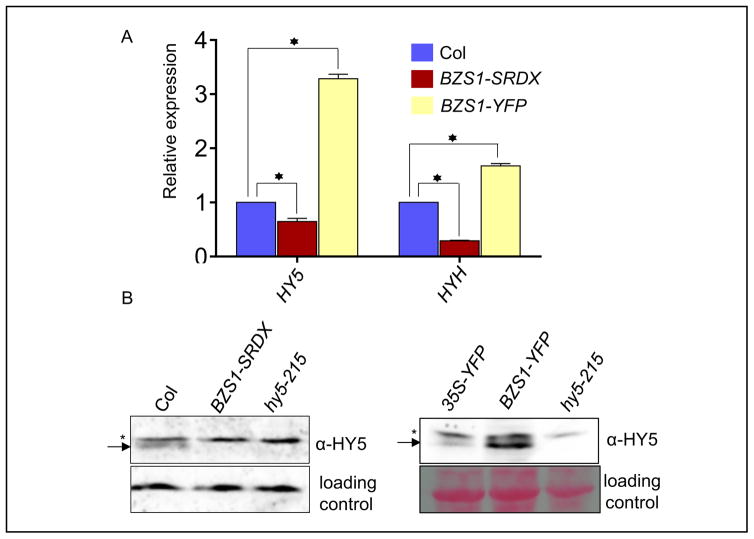
To further investigate whether BZS1 function requires HY5, we crossed BZS1-YFP with hy5-215. The BZS1-YFP/hy5-215 plants showed similar phenotypes of long hypocotyls and low anthocyanin accumulation as hy5-215 (Fig. 2B–D), demonstrating that BZS1 activity requires HY5. Interestingly, the BZS1-YFP protein accumulates at a higher level in the hy5-215 mutant than in wild-type background (Fig. 2E), suggesting that HY5 negatively regulates BZS1 accumulation while required for BZS1 function. On the other hand, the RNA levels of HY5 and HYH are higher in BZS1-YFP line but lower in BZS1-SRDX seedlings as compared with those in wild type (Fig. 3A). Immunoblot analysis also confirmed that the HY5 protein level was increased in the BZS1-YFP line and reduced in the BZS1-SRDX line (Fig. 3B). These results indicated that BZS1 and HY5 proteins not only interact directly, but also influence each other’s protein abundance.
Fig. 3. BZS1 positively regulates HY5.
A: Quantitative RT-PCR analysis of HY5 and HYH expression levels in wild-type (Col), BZS1-SRDX and BZS1-YPF plants. Seedlings were grown under constant white light for five days. The actin2 gene was used as an internal reference. Significant differences from wild type are marked as asterisks, P < 0.01. B: Western blot analysis of HY5 protein accumulation in BZS1-SRDX and BZS1-YFP plants. The arrows indicate HY5 protein and the asterisks mark a non-specific band. Seedlings were grown under continuous white light for 4 days and total proteins were immunoblotted with anti-HY5 antibody. The loading controls were a non-specific band on the blot (left) and the Ponceau S staining of total proteins (right).
BZS1 is a positive regulator of strigolactone responses
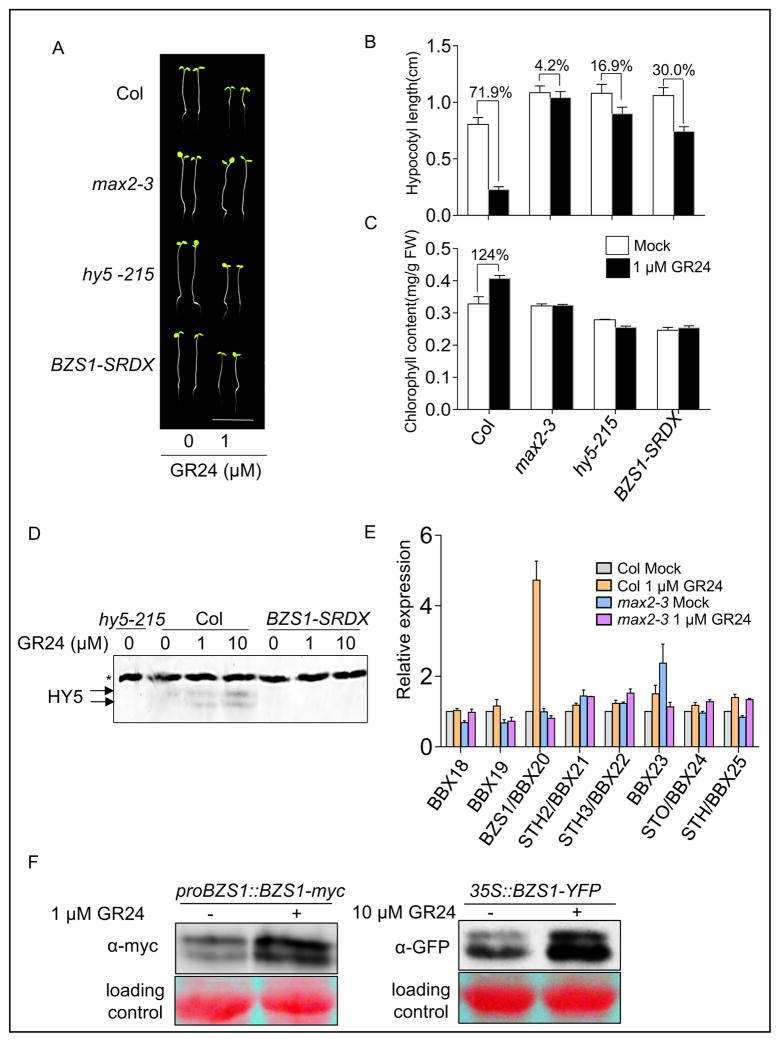
A previous study showed that HY5 is required for SL inhibition of hypocotyl elongation. The HY5 protein level is increased by SL treatment and the hypocotyl elongation of hy5 is partially insensitive to SL (Tsuchiya et al., 2010; Jia et al., 2014). Since BZS1’s function is dependent on HY5 in the light, we examined if BZS1 is also involved in SL signaling. As reported previously (Jia et al., 2014), treatment with 1 μM GR24, an analog of SL, dramatically inhibited the hypocotyl elongation of wild-type seedlings but had no effect on the SL insensitive mutant max2-3 (Fig. 4A and B). We found that the hypocotyl elongation of BZS1-SRDX seedlings was partially insensitive to GR24, similar to the hy5-215 mutant. The GR24 treatment decreased the hypocotyl length of wild-type seedlings by about 72% compared to the untreated control, but only by about 17% for hy5-215 and 30% for the BZS1-SRDX seedlings (Fig. 4A and B). GR24 also increased the chlorophyll content in wild-type plants by about 24%, but had no significant effect in max2-3, hy5-215 and BZS1-SRDX seedlings (Fig. 4C). Additionally, GR24 induced HY5 accumulation in wild-type background but not in the BZS1-SRDX seedlings (Fig. 4D). These results indicated that, like HY5, BZS1 also plays an important role in SL regulation of hypocotyl elongation and chlorophyll accumulation.
Fig. 4. BZS1 is involved in strigolactone signaling.
A: The hy5-215 and BZS1-SRDX mutants showed reduced sensitivity to GR24 treatment. Representative seedlings were grown on ½ MS medium containing 0 or 1 μM GR24 under red light for 4 days. Scale bar, 1 cm. B: Hypocotyl length measurement of seedlings described in (A). Error bars represent SD (n = 30). C: Chlorophyll content measurement of seedlings described in (A). Error bars represent SD from three biological repeats. D: BZS1-SRDX blocks GR24 induction of HY5 accumulation. Seedlings were grown on ½ MS medium containing 0, 1 or 10 μM GR24 under red light (20 μmol m–2s–1) for 4 days. Immunoblot was probed with an anti-HY5 antibody. The arrows indicate HY5, and the star marks a non-specific band that serves as a loading reference. E: Quantitative RT-PCR analysis of the B-BOX IV family genes. Seedlings were grown on ½ MS medium with or without 1 μM GR24 under constant white light for 5 days. Expression data are average of three biological repeats. The CACS (At5g46630) gene was used as an internal reference. Error bars represent SD. F: GR24 increases BZS1 protein level. The proBZS1::BZS1-myc or 35S::BZS1-YFP seedlings were grown on ½ MS medium containing indicated concentration of GR24 under red light or dark for 4 days, and total proteins were immunoblotted and detected with anti-myc or anti-GFP monoclonal antibody.
We then tested if SL regulates the expression of BZS1/BBX20 and its homologs. Real-time reverse transcription PCR (qRT-PCR) analysis showed that GR24 increased the expression level of BZS1/BBX20 mRNA in wild type, but not in the max2-3 mutant (Fig. 4E). Interestingly, expression levels of other members of BBX IV family, including STH2/BBX21, were not dramatically affected by GR24. Immunoblot analysis confirmed that GR24 treatment increased the levels of the BZS1-myc protein expressed from the BZS1 native promoter and the BZS1-YFP protein expressed from the constitutive 35S promoter, suggesting that SL regulates BZS1 at both transcriptional and posttranscriptional levels (Fig. 4F). These results indicated that BZS1 plays a positive role in SL signaling downstream of MAX2 at the early stage of seedling development.
DISCUSSION
Seedling development is crucial for establishment of life for a plant, and is thus highly responsive to a wide range of environmental and hormonal signals. The signaling pathways that transduce these signals are highly integrated at the molecular level to ensure coherent cellular responses and optimal growth according to environmental condition and endogenous physiology (Chaiwanon et al., 2016). This study uncovers additional mechanisms for such signal integration. Our quantitative proteomic analysis of the BZS1 complex reveals BZS1’s interaction with HY5, as well as provides direct evidence for in planta BZS1-COP1 interaction. Genetic analyses using overexpression and dominant negative loss-of-function transgenic plants demonstrate that BZS1 interacts with HY5 to activate gene expression and promote photomorphogenesis. Further, we find that BZS1 also mediates SL regulation of HY5 level and hypocotyl elongation. Together with previous finding of BZS1 function downstream of the BR pathway (Sun et al., 2010; Fan et al., 2012), our study establishes BZS1 as a key integrator of light, BR, and SL signals for regulating seedling morphogenesis.
IP-MS is a powerful method for identification of interacting proteins, which has been widely used in dissecting signal transduction pathways (Wang et al., 2013; Ni et al., 2014). With increased sensitivity of modern mass spectrometers, IP-MS tends to identify not only specific interacting proteins but also large numbers of non-specific proteins. Under our experimental conditions, over 300 proteins were identified in each IP-MS analysis. Distinguishing specific from non-specific interactors is challenging without quantitative measurement. SILIA-IP-MS provides an ideal quantitative method for this purpose, as the sample and negative control can be mixed at an early step of the immunoprecipitation experiment to avoid technical variations. Indeed, among the large numbers of proteins identified by mass spectrometry, only 29 showed enrichment by the BZS1-YFP fusion protein, and thus were considered BZS1-associated proteins. The interactions of BZS1 with HY5, COP1, and its homolog STH2/BBX21 were confirmed by yeast two-hybrid or co-immunoprecipitation assays. Consistent with COP1-mediated ubiquitination of BZS1, our BZS1-interactome data includes ubiquitin and one proteasome activating protein PA200 (Table S2).
In theory, the ratio between sample and negative control should be infinite for proteins that specifically interact with the bait protein in SILIA-IP-MS. However, due to background signals in the control samples, either from non-specific binding of proteins in immunoprecipitation or interfering signals in MS1, the ratios actually distribute within a wide range. For example, Hubner et al. (2010) observed that pull-down with Aly-GFP leads to only moderate enrichment because Aly itself binds to control beads as well. In our study, only 2 of the 254 proteins identified in the YFP sample were enriched over 2× median, suggesting that even 2-fold cutoff yields low false discovery rate when two reverse-labeled replicates are used.
Our genetic analyses support that BZS1 interacts with HY5 to activate gene expression and promote photomorphogenesis. First, comparison of genome-wide data shows that BZS1 tends to activate, rather than repress, HY5 direct target genes (Fig. 2A). Second, dominant inactivation of BZS1 causes similar phenotypes as the hy5-215 mutant (Fig. 2B–D), supporting that BZS1 and HY5 act in the same or overlapping pathway(s). Third, the phenotypes of BZS1-YFP plants are suppressed by hy5-215 (Fig. 2B–D), confirming that BZS1 functions in a HY5-dependent manner. These results together provide strong evidence for a model that BZS1 interacts with HY5 to activate HY5-bound target genes.
BBX proteins contain one or two B-box zinc finger motifs in their N-terminal regions, and are organized into five subfamilies (Khanna et al., 2009). The fourth subfamily includes eight B-box proteins (BBX18–BBX25) containing two tandem B-boxes without CCT domain (CO, COL, TOC1). Our study together with previous studies show that five members of the BBX subfamily IV (BZS1/BBX20, STH2/BBX21, LZF1/STH3/BBX22, STO/BBX24, and STH/BBX25) interact with COP1 and HY5 (Datta et al., 2007; Datta et al., 2008; Jiang et al., 2012; Gangappa et al., 2013; Gangappa and Botto, 2014). Thus, interaction with HY5 seems to be a common mechanism for these B-box proteins to regulate gene expression. Interestingly, BZS1/BBX20, STH2/BBX21 and LZF1/STH3/BBX22 are positive regulators in photomorphogenesis, while BBX19, STO/BBX24 and STH/BBX25 are negative regulators (Bowler et al., 2013; Gangappa and Botto, 2014; Wang et al., 2015). Our finding of STH2/BBX21 and STO/BBX24 as interactors of BZS1/BBX20 suggests that these factors form hetero-dimers. The dominant negative effect of the BZS1-SRDX fusion indicates that BZS1/BBX20 normally functions as a transcription activator, which is consistent with previous finding that STH2/BBX21 functions as a transcription activator (Datta et al., 2007). It has been reported that STO/BBX24 and STH/BBX25 interact with HY5 and most likely inhibit HY5 function by forming inactive heterodimers (Jiang et al., 2012; Gangappa et al., 2013). Our identification of STO/BBX24 as a BZS1-associated protein suggests another possibility that STO/BBX24 may form a non-functional hetero-dimer with BZS1/BBX20 and hence inhibit BZS1/BBX20 activity.
In addition to direct interaction between BZS1 and HY5 proteins in regulating target gene expression, BZS1 and HY5 also regulate each other’s expression level. BZS1 positively regulates the RNA and protein levels of HY5 (Fig. 3A and B). Recent studies have shown that HY5 binds to its own promoter to regulate its own level (Abbas et al., 2014; Binkert et al., 2014), thus BZS1 may regulate HY5 transcription through interaction with HY5 protein. In contrast, the BZS1 protein level is increased in hy5-215, suggesting a negative regulation by HY5 at the protein level. HY5 may promote BZS1 degradation by interacting with COP1. Similarly, a previous study showed that the degradation of BBX22 is also promoted by both COP1 and HY5 (Chang et al., 2011), whereas BBX22 transcription is directly activated by HY5 and repressed by BBX24 (Gangappa et al., 2013). Such positive and negative regulation between interacting partners potentially contributes to the signaling dynamics during dark-to-light transition and fluctuating light intensities.
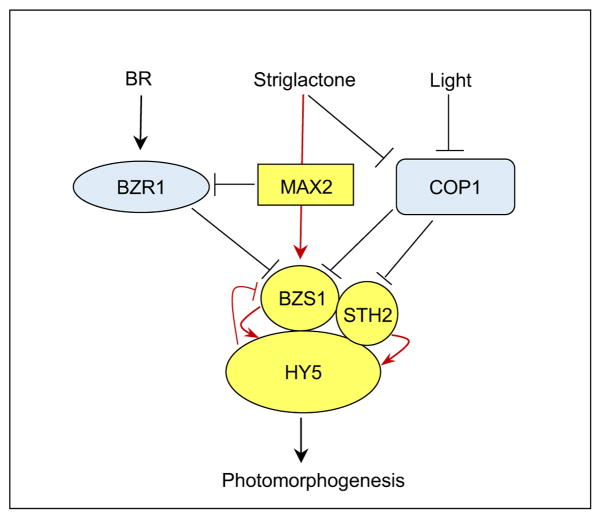
Our study uncovers a major role for BZS1 in SL response. Previous studies have shown that SL promotes photomorphogenesis by increasing HY5 level (Tsuchiya et al., 2010). However, the molecular links from SL signaling to HY5 regulation have remained unclear. Our results show that BZS1 mediates SL regulation of HY5 level and photomorphogenesis. Similar to hy5-215, BZS1-SRDX seedlings are partially insensitive to GR24 treatment under light (Fig. 4A–D), which indicates that BZS1 plays a positive role in SL regulation of seedling morphogenesis. Actually, BZS1 is the only member in the subfamily IV of B-box protein family that is regulated by SL (Fig. 4E), suggesting that BZS1 plays a unique role in SL regulation of photomorphogenesis. As BZS1 increases HY5 level, SL activation of BZS1 expression would contribute, together with inactivation of COP1 (Tsuchiya et al., 2010), to the SL-induced HY5 accumulation. On the other hand, the BZS1-SRDX plants showed normal branching phenotypes (data not shown), which suggests that BZS1 is only involved in SL regulation of HY5 activity and seedling photomorphogenesis but not shoot branching. Our finding of BZS1 function in SL response further supports a key role for BZS1 in integration of light, BR and SL signals to control seedling photomorphogenesis (Fig. 5).
Fig. 5. A model for BZS1/BBX20 function in integrating light and hormone pathways.
Negative and positive regulators for photomorphogenesis are marked in blue and yellow, respectively. Previous known pathways and new connections identified in this study are marked by black and red lines, respectively.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana accession Col-0 was used as a background in all experiments of this study. Arabidopsis seeds were surface sterilized with 70% ethanol plus 0.1% Triton X-100 for 5 min, followed by one-time wash of anhydrous ethanol. After drying on a sterile filter paper, seeds were sown on half-strength Murashige and Skoog (MS) medium with 0.8% Phytoblend (Caisson Laboratories, USA). The plates were stratified at 4°C in dark for 2 days and followed by white light treatment at 22°C for 4 h to promote germination. White light (about 100 μmol m–2s–1) was provided by fluorescence light source in a growth room. Red light treatment (about 20 μmol m–2s–1) was carried out in an LED light chamber (E-30LEDL3, Percival). Hypocotyl length was measured with Image J software (Schneider et al., 2012).
Plasmids construction
The coding sequence of BZS1, HY5 or STH2 were amplified from wild-type cDNA and cloned into gateway ENTRY vector pENTR/SD/D-TOPO or pENTRY-SRDX (for BZS1-SRDX) and then recombined into different destination vectors according to manufacturer’s protocol (Invitrogen, USA). For yeast two-hybrid assay, pGADT7 or pGBKT7 vectors were used; for BZS1-SRDX overexpression lines, pEarleyGate 100 vector was used; for STH2-myc-His overexpression, pG7MH1 vector was used (Wang et al., 2013).
For construction of proBZS1::BZS1-myc transgenic lines, a 1793-bp genomic fragment containing promoter region of BZS1 was amplified from wild-type genomic DNA and cloned into the vector Native-pG7MH (native promoter) by Gateway Cloning system. Native-pG7MH vector is modified from pCAMBIA1390. Different from pG7MH1, 35S promotor was not included for native promotor insertion (Wang et al., 2013)
SILIA-IP-MS assay
To generate 15N-labeled seeds, Arabidopsis plants were grown hydroponically (Bindschedler et al., 2012) in diluted Hoagland solution (Hoagland’s No. 2 Basal Salt Mixture, Caisson Laboratories) containing 10 mM K15NO3 (Cambridge Isotope Laboratories,USA). One-eighth diluted Hoagland medium was used at seedling stage and 1/4 Hoagland medium was used when plant started to bolt. After the siliques were fully developed, 1/8 Hoagland medium was used till seeds were fully mature.
For SILIA-IP-MS assay, the 14N- or 15N-labeled seeds were grown on Hoagland medium containing 10 mM K14NO3 or K15NO3, respectively, for 5 days under constant white light. The seedlings were harvested and ground to fine powder in liquid nitrogen. Five grams each of 14N-labeled BZS1-YFP or YFP and 15N-labeled wild-type tissue power were mixed and total proteins were extracted using extraction buffer (20 mM HEPES, pH7.5, 40 mM KCl, 1 mM EDTA, and protease inhibitor cocktail tablets from Roche). After removing the cell debris by centrifugation, 20 μL GFP-Trap®_MA Beads (ChromoTek GmbH, Germany) were added to the supernatant and then incubated in the cold room for 2 h with constant rotating. The beads were washed three times with IP wash buffer (20 mM HEPES, pH7.5, 40 mM KCl, 0.1% Triton X-100). The proteins were eluted twice using 50 μL 2 × SDS sample loading buffer by incubating at 95°C for 10 min. The isotope labels were switched in repeat experiments.
The eluted proteins were separated by NuPAGE® Novex 4–12% Bis-Tris Gel (Thermo Fisher Scientific, USA). After Colloidal Blue staining (Thermo Fisher Scientific), the gel was cut into five fractions for trypsin digestion. In-gel digestion procedure was performed according to Tang et al. (2008). Extracted peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS). The LC separation was performed using an Eksigent 425 NanoLC system on a C18 trap column (5 mm × 0.3 mm, 4.6 μm) and a C18 analytical column (75 μm × 15 cm, 4.6 μm). Solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in acetonitrile. The flow rate was 300 nL/min. The MS/MS analysis was conducted with a Thermo Scientific Q Exactive mass spectrometer in positive ion mode and data dependent acquisition mode to automatically switch between MS and MS/MS acquisition. The identification and quantification were done by pFind and pQuant softwares (Liu et al., 2014; Chi et al., 2015) in an open search mode. The parameters of software were set as follows: parent mass tolerance, 15 ppm; fragment mass tolerance, 0.6 Da. The FDR of the pFind analysis was 1% for peptides. Arabidopsis TAIR10 database was used for data search.
Co-immunoprecipitation assay and western blot analysis
Three-day-old Arabidopsis seedlings expressing BZS1-YFP or YFP alone were grown under constant light and used for BZS1-COP1 co-immunoprecipitation assay. For the BZS1, HY5 and STH2 co-immunoprecipitation assay, about one-month-old healthy Nicotiana benthamiana leaves were infiltrated with Agrobacterium tumefaciens GV3101 harboring corresponding plasmids. The plants were then grown under constant light for 48 h and infiltrated leaves were collected.
The harvested materials were frozen and ground into fine powder in liquid nitrogen. Total proteins from 0.3 g tissue powder were extracted with 0.6 mL extraction buffer (20 mM HEPES, pH7.5, 40 mM KCl, 1 mM EDTA, 1% TritonX-100 and protease inhibitor cocktail tablets from Roche). The lysate was pre-cleared by centrifugation twice at 20,000 g for 10 min at 4°C, and then diluted with equal volume of extraction buffer without Triton X-100. Twenty microliter of Pierce Protein A Magnetic Beads (Thermo Fisher Scientific) coupled with 10 μg anti-GFP polyclonal antibody (custom made) were added to each protein extract and incubated at 4°C for 1 h with rotation. The beads were then collected by DynaMag™-2 Magnet (Thermo Fisher Scientific) and washed three times with wash buffer (20 mM HEPES, pH7.5, 40 mM KCl, 0.1% Triton X-100). The bonded proteins were eluted with 50 μL 2 × SDS loading buffer by incubating at 95°C for 10 min.
For western blot analysis, proteins were separated by SDS-PAGE electrophoresis and transferred onto a nitrocellulose membrane (Merck Millipore Corporation, Germany) by semi-dry transfer cell (Bio-Rad Laboratories, USA). The membrane was blocked with 5% none-fat milk followed by primary and secondary antibodies. Chemiluminescence signal was detected using SuperSignal™ West Dura Extended Duration Substrate (Thermo Fisher Scientific) and FluorChem™ Q System (Protein Simple, USA). Monoclonal GFP antibody (JL-8) was purchased from Clontech, USA. Myc antibody (9B1) and ubiquitin antibody (P4D1) were from Cell Signaling Technology, USA) HY5 and COP1 antibodies were from Dr. Hongquan Yang’s lab. Secondary antibodies goat anti-mouse-HRP or goat anti-rabbit-HRP were from Bio-Rad Laboratories.
Anthocyanin and chlorophyll measurements
Five-day-old seedlings grown under white light were used for anthocyanin measurement. Anthocyanin extraction and determination was according to Datta et al. (2007). Four-day-old seedlings grown under red light were used for chlorophyll determinations. The protocol for extracting chlorophyll was modified based on the previous report (Ni et al., 2009). Fresh leaves were frozen and ground into fine power in liquid nitrogen, and then 50 mg tissue powder was extracted with 1 mL of 80% acetone at 4°C and incubated for 15 min in the dark. The extract was then centrifuged at 4°C for 15 min, and the chlorophyll in supernatant was measured by spectrophotometer at the wavelengths of 645 and 663 nm, respectively. The chlorophyll concentration was calculated as follows: Concentration: a+b (mg/g) = [8.02 × A663 + 20.20 × A645] ×V/1000×W (Chlorophyll a+b), where V = volume of the extract (mL), and W = weight of fresh leaves (g).
Total RNA extraction and real-time PCR analysis
Total RNA was extracted using RNeasy Plant Mini kit (QIAGEN, Germany) and reverse transcription was performed using PrimeScriptTM RT reagent Kit (Takara Biotechnology, Japan) with gDNA Eraser (Takara Biotechnology, Japan). Real-time PCR was performed using TAKARA SYBR Premix Ex TaqTM reagent (Takara Biotechnology) and carried out on ABI7500 machine (Applied Biosystems, USA). The expression level was normalized to actin2 or CACS (At5g46630) controls. Primers are listed in Table S3. Three biological repeats were performed for each sample.
Supplementary Material
Table. S1. Proteins identified in BZS1-YFP and YFP by SILIA-IP-MS assays.
Table. S2. BZS1-associated proteins identified in both repeated SILIA-IP-MS experiments.
Table. S3. Primer sequences used in this study.
Fig. S1. Workflow of identifying BZS1 protein complex by SILIA-IP-MS in Arabidopsis.
Fig. S2. Immunoprecipitation of BZS1 protein complex in Arabidopsis.
Fig. S3. Genotyping BZS1-SRDX genetic background by PCR.
Acknowledgments
We thank Dr. Xingwang Deng for providing hy5-215 seeds and Dr. Hongquan Yang for providing anti-HY5 and anti-COP1 antibodies. We also thank Drs. Chao Liu, Hao Chi, and Shuolei Bu for their kind assistance with mass spectrometry data analyses. This study was supported by a grant from National Institute of Health (NIH, R01GM066258) to Z.-Y. Wang and “One-hundred Talents Project” of Hebei province, China (E2013100004) to Y. Sun. C.-Q. Wang was supported by the China Scholarship Council.
Footnotes
Author contributions
C-Q.W. and Z-Y.W. together designed the experiments. C-Q.W. performed BZS1 SILIA-IP-MS and other experiments. C-Q.W., L-F. A. C-W. C and K.H.L. analysis the data under the supervision of A.L.B. and Z-Y.W.. J.Z. and Z-Z.Z. generated the BZS1-Myc transgenic plants. C-Q.W., Y.S. and Z-Y.W. wrote the manuscript.
References
- Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S. Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell. 2014;26:1036–1052. doi: 10.1105/tpc.113.122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Shang J, Oh E, Fan M, Bai Y, Zentella R, Sun T, Wang Z. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nature Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, Mills DJS, Cramer R. Hydroponic isotope labeling of entire plants and high-performance mass spectrometry for quantitative plant proteomics. In: Marcus K, editor. Quantitative Methods in Proteomics. Humana Press; Totowa, NJ: 2012. pp. 155–173. [DOI] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognár L, Terecskei K, De Veylder L, Nagy F, Ulm R. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell. 2014;26:4200–4213. doi: 10.1105/tpc.114.130716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotech. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- Bowler C, Botto J, Deng XW. Photomorphogenesis, B-Box transcription factors, and the legacy of Magnus Holm. Plant Cell. 2013;25:1192–1195. doi: 10.1105/tpc.113.250412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J, Wang W, Zhu JY, Oh E, Wang ZY. Information integration and communication in plant growth regulation. Cell. 2016;164:1257–1268. doi: 10.1016/j.cell.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CSJ, Maloof JN, Wu SH. COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 2011;156:228–239. doi: 10.1104/pp.111.175042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H, He K, Yang B, Chen Z, Sun RX, Fan SB, Zhang K, Liu C, Yuan ZF, Wang QH, Liu SQ, Dong MQ, He SM. pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J Proteomics. 2015;125:89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007;19:3242–3255. doi: 10.1105/tpc.107.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-Box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008;20:2324–2338. doi: 10.1105/tpc.108.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, Wei CQ, Zhu SW, Sun Y, Chong K, Wang ZY. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant. 2012;5:591–600. doi: 10.1093/mp/sss041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C. Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol. 2015;34:46–53. doi: 10.1016/j.conb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19:460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell. 2013;25:1243–1257. doi: 10.1105/tpc.113.109751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodriguezfranco M. Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 2007;51:563–574. doi: 10.1111/j.1365-313X.2007.03162.x. [DOI] [PubMed] [Google Scholar]
- Jia KP, Luo Q, He SB, Lu XD, Yang HQ. Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol Plant. 2014;7:528–540. doi: 10.1093/mp/sst093. [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li QF, Björn LO, He JX, Li SS. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012;22:1046–1057. doi: 10.1038/cr.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. Curr Top Dev Biol. 2010;91:29–66. doi: 10.1016/S0070-2153(10)91002-8. [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. The Arabidopsis B-box zinc finger family. Plant Cell. 2009;21:3416–3420. doi: 10.1105/tpc.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N. Quantitative measurement of phosphopeptides and proteins via stable isotope labeling in Arabidopsis and functional phosphoproteomic strategies. Methods Mol Biol. 2012;876:17–32. doi: 10.1007/978-1-61779-809-2_2. [DOI] [PubMed] [Google Scholar]
- Liu C, Song CQ, Yuan ZF, Fu Y, Chi H, Wang LH, Fan SB, Zhang K, Zeng WF, He SM, Dong MQ, Sun RX. pQuant improves quantitation by keeping out interfering signals and evaluating the accuracy of calculated ratios. Anal Chem. 2014;86:5286–5294. doi: 10.1021/ac404246w. [DOI] [PubMed] [Google Scholar]
- Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, Cheng M, Hao Y, Oh E, Tian M, Liu L, Zhang M, Xie Q, Chong K, Wang ZY. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, Chalkley RJ, Pham TN, Guan S, Maltby DA, Burlingame AL, Wang ZY, Quail PH. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell. 2013;25:2679–2698. doi: 10.1105/tpc.113.112342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim E, Chen Z. Chlorophyll and starch assays. Protocol Exchange 2009 [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J, Wang Z. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, Patil S, Kim TW, Ji H, Wong WH, Rhee SY, Wang ZY. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Deng Z, Oses-Prieto JA, Suzuki N, Zhu S, Zhang X, Burlingame AL, Wang ZY. Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol Cell Proteomics. 2008;7:728–738. doi: 10.1074/mcp.M700358-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, Lamond A. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P. A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol. 2010;6:741–749. doi: 10.1038/nchembio.435. [DOI] [PubMed] [Google Scholar]
- Von Arnim A, Deng XW. Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Wang CQ, Sarmast MK, Jiang J, Dehesh K. The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell. 2015;27:1128–1139. doi: 10.1105/tpc.15.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shang JX, Chen QX, Oses-Prieto JA, Bai MY, Yang Y, Yuan M, Zhang YL, Mu CC, Deng Z, Wei CQ, Burlingame AL, Wang ZY, Sun Y. Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol Cell Proteomics. 2013a;12:3653–3665. doi: 10.1074/mcp.M113.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bai MY, Wang ZY. The brassinosteroid signaling network—a paradigm of signal integration. Curr Opin Plant Biol. 2014;21:147–153. doi: 10.1016/j.pbi.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Bai M, Oh E, Zhu J. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Waters MT, Smith SM. KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol Plant. 2013;6:63–75. doi: 10.1093/mp/sss127. [DOI] [PubMed] [Google Scholar]
- Yang Z, Guo G, Zhang M, Liu CY, Hu Q, Lam H, Cheng H, Xue Y, Li J, Li N. Stable isotope metabolic labeling-based quantitative phosphoproteomic analysis of Arabidopsis mutants reveals ethylene-regulated time-dependent phosphoproteins and putative substrates of constitutive triple response 1 kinase. Mol Cell Proteomics. 2013;12:3559–3582. doi: 10.1074/mcp.M113.031633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table. S1. Proteins identified in BZS1-YFP and YFP by SILIA-IP-MS assays.
Table. S2. BZS1-associated proteins identified in both repeated SILIA-IP-MS experiments.
Table. S3. Primer sequences used in this study.
Fig. S1. Workflow of identifying BZS1 protein complex by SILIA-IP-MS in Arabidopsis.
Fig. S2. Immunoprecipitation of BZS1 protein complex in Arabidopsis.
Fig. S3. Genotyping BZS1-SRDX genetic background by PCR.