Short abstract
The Janus kinase (Jak) family, including Jak1, Jak2, Jak3 and Tyrosine kinase 2 (Tyk2), bind cytokine receptors through amino-terminal FERM domains and link them to other molecules, especially members of the signal transducer and activator of transcription (Stat) family.
Abstract
The Janus kinase (Jak) family is one of ten recognized families of non-receptor tyrosine kinases. Mammals have four members of this family, Jak1, Jak2, Jak3 and Tyrosine kinase 2 (Tyk2). Birds, fish and insects also have Jaks. Each protein has a kinase domain and a catalytically inactive pseudo-kinase domain, and they each bind cytokine receptors through amino-terminal FERM (Band-4.1, ezrin, radixin, moesin) domains. Upon binding of cytokines to their receptors, Jaks are activated and phosphorylate the receptors, creating docking sites for signaling molecules, especially members of the signal transducer and activator of transcription (Stat) family. Mutations of the Drosophila Jak (Hopscotch) have revealed developmental defects, and constitutive activation of Jaks in flies and humans is associated with leukemia-like syndromes. Through the generation of Jak-deficient cell lines and gene-targeted mice, the essential, nonredundant functions of Jaks in cytokine signaling have been established. Importantly, deficiency of Jak3 is the basis of human autosomal recessive severe combined immunodeficiency (SCID); accordingly, a selective Jak3 inhibitor has been developed, forming a new class of immunosuppressive drugs.
Gene organization and evolutionary history
Janus kinases (Jaks) are non-receptor tyrosine kinases and were discovered in searches for novel protein tyrosine kinases using PCR-based strategies or low-stringency hybridization [1-6]. In mammals, the family has four members, Jak1, Jak2, Jak3 and Tyrosine kinase 2 (Tyk2). In humans, the Jak1 gene is located on chromosome 1p31.3 and Jak2 is on 9p24; the Jak3 and Tyk2 genes are clustered together on chromosome 19p13.1 and 19p13.2, respectively. The murine genes are located on chromosomes 4 (Jak1), 19 (Jak2) and 8 (Jak3 and Tyk2). Since the sequencing of other vertebrate genomes has been completed, we know that there are four Jak family members in mammals, birds and fish (see the Additional data files for alignments).
Jaks have been identified in the primitive chordate Ciona; it is unclear, however, whether this species only has a single Jak or whether more will be found with further sequencing (see Additional data files). In Drosophila there is only one Jak kinase, Hopscotch (Hop) [7,8]. The ancestral Jak must therefore have arisen before the divergence of vertebrates and invertebrates. Nematode worms and slime molds lack the family, however, but they do express members of the signal transducer and activator of transcription (Stat) family of transcription factors - which in vertebrates interact with Jaks, among other proteins - suggesting that the Stats arose in evolution before the Jaks. It is of interest that the expansion of the Jak kinases in higher animals occurred at the same time as the evolution of innate and adaptive immune cells in fish; this is consistent with the multiple roles of Jaks in immune cells (see below). Thus, cytokine receptors acting via Jaks appear to have co-opted the Stat pathway for a variety of purposes, especially for host defense. The proximity of the Jak3 and Tyk2 genes suggests that one may have arisen from the other by gene duplication, but it is difficult to conclude which is the more ancestral. Jaks have approximately 20 exons; alternatively spliced forms of Jaks have been described but their functional significance is not known.
Characteristic structural features
The three-dimensional structure of the Jaks is at present unknown. This is no doubt partly because they are relatively large proteins of more than 1,100 amino acids, with apparent molecular masses of 120-140 kDa; expressing and purifying them has been problematic. From the primary structure, putative domain structures have been recognized that are conserved between mammalian, avian, teleost and insect Jaks. Seven Jak homology (JH) domains have been identified, numbered from the carboxyl to the amino terminus (Figure 1). The JH1 domain at the carboxyl terminus has all the features of a typical eukaryotic tyrosine kinase domain. Interestingly, this domain is most closely related to the kinase domains of the epidermal growth factor family of receptor tyrosine kinases, suggesting that the Jak family may have arisen from this larger family of protein kinases [9]. Adjacent to the JH1 domain is a catalytically inactive pseudokinase or kinase-like domain (JH2), which is distantly related to other tyrosine kinase domains [9]. This tandem architecture of kinase domains is the hallmark of Jak kinases and gives them their name; just like the Roman god Janus, they are two-faced with respect to these domains. Although the pseudokinase domain lacks catalytic activity, it has an essential regulatory function. A number of patient-derived and artificial mutations within this domain abrogate kinase activity, underscoring its critical function [10,11]. Conversely, a mutation within this domain in Drosophila Hop activates the kinase and leads to transformation (discussed below) [12-14]. In mammalian Jak2, experimentally introduced mutations in this domain can also increase basal activity, but they abrogate ligand-dependent activation [11,15].
Figure 1.
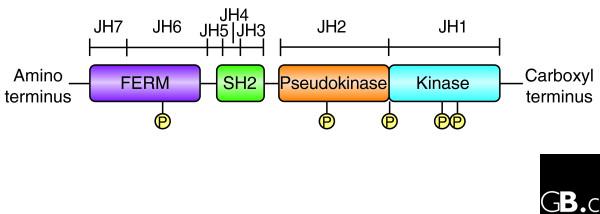
A schematic representation of the primary structure of Janus kinases (Jaks), which are made up of FERM, SH2-like, pseudokinase and kinase domains. An alternative nomenclature for the putative domains is as a series of Janus homology (JH) domains. The FERM domain mediates binding to cytokine receptors. Both the FERM and the pseudokinase domains regulate catalytic activity and appear to interact with the kinase domain. Jaks autophosphorylate at multiple sites (P), including two in the activation loop of the kinase domain, but the precise function of these modifications is just beginning to be understood.
The amino terminus of Jaks contains an SH2-like domain (JH3-JH4) and a Band-4.1, ezrin, radixin, moesin (FERM) homology domain (JH6-JH7). The FERM domain is 300 amino acids long and is implicated in mediating interactions with transmembrane proteins such as cytokine receptors; for some but not all cytokines, Jaks appear to be important in regulating cell-surface expression of the cognate receptors [16,17]. In addition, the FERM domain binds the kinase domain and positively regulates catalytic activity [18]. Unfortunately, the lack of crystal structures severely limits the understanding of the intramolecular interactions that involve Jaks. Binding partners for the Jak SH2 domain have not been identified.
Localization and function
In mammals Jak1, Jak2 and Tyk2 are ubiquitously expressed. In contrast, the expression of Jak3 is more restricted; it is predominantly expressed in hematopoietic cells and is highly regulated with cell development and activation [6,19,20]. At the cellular level, Jaks can be found in the cytosol when they are experimentally expressed in the absence of cytokine receptors, but, because of their intimate association with cytokine receptors, they ordinarily localize to endosomes and the plasma membrane, along with their cognate receptors [21,22]. The link between Jaks and cytokine signaling was first made using mutant cell lines that lacked responsiveness to interferon (IFN). One such cell line was shown to lack Tyk2, and adding back this kinase restored IFN signaling [16]. Shortly thereafter other Jaks were shown to be associated with various cytokine receptors [23-26], and subsequently Jak knockout mice have illustrated their essential and specific functions (see Table 1).
Table 1.
Functions of Jaks
| Gene | Phenotype of mouse knockout | Cytokines whose signaling requires this Jak |
| Jak1 | Viable but early postnatal lethal owing to neurological deficits; SCID | Families of receptor with the shared subunits γc or gp130; IFNs |
| Jak2 | Embryonic lethal owing to a defect of erythropoiesis | IL-3; family of receptors with the shared subunit gp130; IFN-γ hormone-like cytokines (EPO, GH, PRL, TPO) |
| Jak3 | SCID, viable and fertile | Family of receptor with the shared subunit γc |
| Tyk2 | Viable and fertile; susceptible to parasite infection; resistant to LPS; resistant to collagen-induced arthritis | IL-12; LPS |
Abbreviations: EPO, erythropoietin; γc, common γ chain; GH, growth hormone; IFN, interferon; IL, interleukin; LPS, bacterial lipopolysaccharide; PRL, prolactin; SCID, severe combined immunodeficiency; TPO, thrombopoietin.
A large number of cytokines are dependent upon Jak1, including a family that use a shared receptor subunit called common γ chain (γc), which includes interleukin (IL)-2, IL-4, IL-7, IL-9, IL-15 and IL-21. These cytokines are also dependent upon Jak3, because Jak3 binds γc. Jak1 is also essential for another family that uses the shared receptor subunit gp130 (IL-6, IL-11, oncostatin M, leukemia inhibitory factor (LIF), ciliary neurotrophic factor (CNF)) as well as granulocyte colony-stimulating factor (G-CSF) and IFNs. Jak2 is essential for the hormone-like cytokines such as growth hormone (GH), prolactin (PRL), erythropoietin (EPO), thrombopoietin (TPO) and the family of cytokines that signal through the IL-3 receptor (IL-3, IL-5 and granulocyte-macrophage colony-stimulating factor, GM-CSF). Jak2 is also important for cytokines that use the gp130 receptor and for some IFNs.
Tyk2 was the first Jak to be implicated in IFN signaling, but subsequent studies indicate that Tyk2 is essential for IL-12 signaling but not for IFN-α/β signaling or for cytokines that use gp130 [27,28]. Tyk2-/- mice also have defective responses to lipopolysaccharide (LPS, a component of the outer membrane of Gram-negative bacteria), but whether this is a direct or indirect effect has not been defined. In particular, a role for Tyk2 in signaling through the Toll receptor, which mediates the response to LPS, has not been established [29,30].
Jak1 knockout mice have a perinatal lethal phenotype, probably related to the neurological defects that prevent them from suckling [30] (Table 1). These mice also have defective lymphoid development and function as a result of defective signaling by cytokines through Jak1. Jak2 deficiency results in embryonic lethality at embryonic day 12.5 as a result of a failure in definitive erythropoiesis [31,32]. Interestingly, Jak3 deficiency was first identified in humans with autosomal recessive severe combined immunodeficiency (SCID) [33,34]. We now know that Jak3 binds to γc and that deficiency of either Jak3 or γc abrogates signaling by the family of cytokines using this receptor subunit. Not surprisingly, this has devastating consequences in terms of immune-cell development and function. Together, mutations in the receptor for IL-7, γc and Jak3 account for two-thirds to three-quarters of cases of SCID [35]. Jak3-/- mice were subsequently generated, and they too exhibit SCID but notably do not have non-immune defects [36-38]; this is notable because it suggests that an inhibitor of Jak3 would have restricted effects in vivo ([35]; see below).
The Jak/Stat pathway has been extensively studied in Drosophila and has been demonstrated to be involved in stem-cell maintenance, ovarian-cell migration and sex determination [13,39]. In development, this pathway is important for embryonic segmentation and larval hematopoiesis as well as for development of the eye, wing, trachea, hindgut and limb [14,40-44]. A gain-of-function mutation in Hop has been identified that results in a leukemia-like phenotype in the affected flies; this is designated tumorous lethal (Hoptum-l) [12,45,46]. In human leukemias, chromosomal translocations result in fusion proteins of the Tel transcription factor with Jaks. This creates a constitutively active Jak, which has also been documented to be transforming [47,48]. In human cells transformed with T-cell leukemia virus-1, Jak3 and Stat5 are constitutively activated [49]. Constitutive activation of Stats is very common in many other types of tumors, although the mechanisms underlying this activation have yet to be defined.
Jaks are constitutively associated with the membrane-proximal regions of cytokine receptors, although in some cases interaction between the Jak and the receptor is increased upon ligand binding (Figure 2). It has been proposed that ligand binding promotes a conformational change in the receptor, which promotes Jak activation through reciprocal interaction of two juxtapositioned Jak kinases and auto- and/or trans-phosphorylation of tyrosine residues on the activation loop of the Jak kinase domain.
Figure 2.
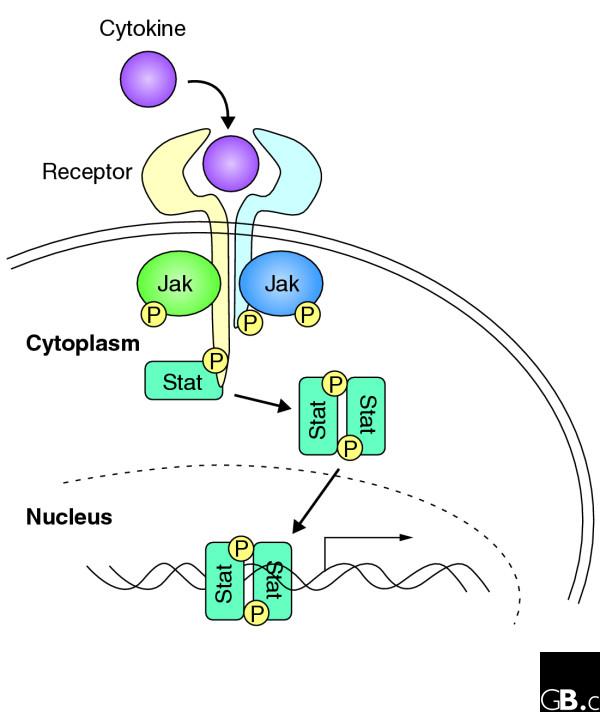
An overview of cytokine signaling. Cytokines bind to homodimeric or heterodimeric receptors, which are constitutively bound to Jaks. Jaks are thought to be activated by a conformational change in the receptor that allows trans- and/or auto-phosphorylation of the two bound Jaks. These in turn phosphorylate the cytokine receptors. Stat proteins bind the phosphorylated receptor chains, allowing the Jaks to phosphorylate the Stats. Phosphorylated Stats form dimers and translocate and accumulate in the nucleus, where they regulate gene expression.
Like other tyrosine kinases, Jaks undergo autophosphorylation, but the importance of this modification in Jak-dependent signaling is not very well understood. Autophosphorylation within the activation loop positively regulates kinase activity; in Jak3, however, phosphorylation in this region can enhance or inhibit catalytic activity, depending upon the site of phosphorylation [50] (Figure 1). Other sites of autophosphorylation have recently been identified. For instance, a conserved residue in Jak2 and Jak3 that resides in the hinge region between JH1 and JH2 is a prominent site of autophosphorylation (Tyr813 in Jak2 and Tyr785 in Jak3) [51]. This site serves to recruit the adapter protein SH2-Bβ, which positively regulates Jak2 activity. Other sites of autophosphorylation in Jak2 include Tyr221 and Tyr570 [52].
Frontiers
Despite intensive studies during the past decade that have generated the model shown in Figure 2, the exact molecular mechanisms of Jak activation have largely remained elusive. It is clear that much more detailed structural information pertaining to Jaks and the Jak-cytokine-receptor complex is needed to enhance our understanding of the mechanism of Jak activation. Also, the exact mechanism and functional relevance of autophosphorylation at different sites in Jaks is not known but will be an interesting area for future research.
Another important topic for future studies is to define the mechanisms of crosstalk between Jaks and other pathways. For instance, the receptor Notch has been reported to promote Stat3 activation, and the Notch effectors Hes1 and Hes5 have been found to associate directly with Jak2 and Stat3 [53]. Evidence for cooperation between the Jak/Stat and Notch pathways has also been provided by work from Drosophila [54] and genetic screens in Drosophila have identified additional potential modifiers of the Jak/Stat pathway [55]. Jaks have also been reported to be activated by a variety of structurally diverse receptors, beyond the cytokine receptors. Examples include receptor tyrosine kinases, death receptors (such as CD40) and G-protein-coupled receptors (such as chemokine receptors). Many of the studies have employed overexpression or putatively specific inhibitors to implicate the Jaks, but we now know that these inhibitors are not specific, so the essential function of Jaks for non-cytokine receptors remains uncertain. This is clearly another critical area for future work.
Finally, because of the crucial role of Jak3 in cytokine signaling through γc and because of its limited tissue expression, the inhibition of Jak3 activity has emerged as a promising strategy for immunosuppression. A highly selective and potent Jak3 inhibitor (CP-69O, 550) has recently been developed that has nanomolar potency against Jak3 in vitro, with much less potency against other Jak family members. Consequently, CP-690, 550 was both very efficacious and well-tolerated in animal models of organ transplantation [56]. One might anticipate that this drug will help to overcome the unwarranted side effects often seen in patients under current immunosuppressive therapy. Thus, the drug could be useful in blocking transplant rejection and in the treatment of autoimmune diseases. Conceivably, it might also be useful in treating those hematological malignancies that exhibit constitutive Jak3 activation. Targeting Tyk2 with specific drugs would also be logical, given its restricted role; presumably a Tyk2 inhibitor would be useful in some immune-mediated diseases. Whether a Jak2 inhibitor would be useful in malignancies is also worthy of consideration.
Additional data files
Protein sequence alignments in text and jpeg format are available for orthologs of Jak1 (Additional data files 1 and 6), Jak2 (Additional data files 2 and 7), Jak3 (Additional data files 3 and 8), Tyk2 (Additional data files 4 and 9), and undefined members of the family (Additional data files 5 and 10), and a key for the alignments (Additional data file 11).
Supplementary Material
Protein sequence alignments in text format for orthologs of Jak1
Protein sequence alignments in text format for orthologs of Jak2
Protein sequence alignments in text format for orthologs of Jak3
Protein sequence alignments in text format for orthologs of Tyk2
Protein sequence alignments in text format for undefined members of the family
Protein sequence alignments in jpeg format for orthologs of Jak1
Protein sequence alignments in jpeg format for orthologs of Jak2
Protein sequence alignments in jpeg format for orthologs of Jak3
Protein sequence alignments in jpeg format for orthologs of Tyk2
Protein sequence alignments in jpeg format for undefined members of the family
A key for the alignments
References
- Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, Krolewski JJ. Tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene. 1990;5:1329–1336. This paper and [2-6] were the first studies to report the cloning of Jaks. [PubMed] [Google Scholar]
- Wilks AF. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:1603–1607. doi: 10.1073/pnas.86.5.1603. See [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks AF. Cloning members of protein-tyrosine kinase family using polymerase chain reaction. Methods Enzymol. 1991;200:533–546. doi: 10.1016/0076-6879(91)00169-w. See [1] [DOI] [PubMed] [Google Scholar]
- Harpur AG, Andres AC, Ziemiecki A, Aston RR, Wilks AF. JAK2, a third member of the JAK family of protein tyrosine kinases. Oncogene. 1992;7:1347–1353. See [1] [PubMed] [Google Scholar]
- Krolewski JJ, Lee R, Eddy R, Shows TB, Dalla-Favera R. Identification and chromosomal mapping of new human tyrosine kinase genes. Oncogene. 1990;5:277–282. See [1] [PubMed] [Google Scholar]
- Kawamura M, McVicar DW, Johnston JA, Blake TB, Chen YQ, Lal BK, Lloyd AR, Kelvin DJ, Staples JE, Ortaldo JR, et al. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc Natl Acad Sci USA. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. See [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Mahowald AP. I(1)hopscotch, a larval-pupal zygotic lethal with a specific maternal effect on segmentation in Drosophila. Dev Biol. 1986;118:28–41. doi: 10.1016/0012-1606(86)90070-9. This paper and [8] report the first identification of the Drosophila Jak. [DOI] [PubMed] [Google Scholar]
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. See [7] [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. An excellent discussion of the all the kinases present in the human genome. [DOI] [PubMed] [Google Scholar]
- Chen M, Cheng A, Candotti F, Zhou YJ, Hymel A, Fasth A, Notarangelo LD, O'Shea JJ. Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: evidence for interactions between the kinase and pseudokinase domains. Mol Cell Biol. 2000;20:947–956. doi: 10.1128/MCB.20.3.947-956.2000. This paper and [11-15] report analyses of the function of the Jak pseudokinase domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/MCB.20.10.3387-3395.2000. See [10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanratty WP, Dearolf CR. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol Gen Genet. 1993;238:33–37. doi: 10.1007/BF00279527. See [10] [DOI] [PubMed] [Google Scholar]
- Jinks TM, Polydorides AD, Calhoun G, Schedl P. The JAK/STAT signaling pathway is required for the initial choice of sexual identity in Drosophila melanogaster. Mol Cell. 2000;5:581–587. doi: 10.1016/S1097-2765(00)80451-7. See [10] [DOI] [PubMed] [Google Scholar]
- Johansen KA, Iwaki DD, Lengyel JA. Localized JAK/STAT signaling is required for oriented cell rearrangement in a tubular epithelium. Development. 2003;130:135–145. doi: 10.1242/dev.00202. See [10] [DOI] [PubMed] [Google Scholar]
- Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. See [10] [DOI] [PubMed] [Google Scholar]
- Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-L. The first evidence that Jaks are critical for cytokine signaling. The study also indicates that Jaks may regulate cytokine receptor expression. [DOI] [PubMed] [Google Scholar]
- Huang LJ, Constantinescu SN, Lodish HF. The N-terminal domain of Janus kinase 2 is required for Golgi processing and cell surface expression of erythropoietin receptor. Mol Cell. 2001;8:1327–1338. doi: 10.1016/S1097-2765(01)00401-4. Further support for a role of Jaks in regulating intracellular trafficking of cytokine receptors. [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Chen M, Cusack NA, Kimmel LH, Magnuson KS, Boyd JG, Lin W, Roberts JL, Lengi A, Buckley RH, et al. Unexpected effects of FERM domain mutations on catalytic activity of Jak3: structural implication for Janus kinases. Mol Cell. 2001;8:959–969. doi: 10.1016/S1097-2765(01)00398-7. Evidence that the Jak FERM domain has two important functions: mediating receptor association and regulating kinase activity. [DOI] [PubMed] [Google Scholar]
- Musso T, Johnston JA, Linnekin D, Varesio L, Rowe TK, O'Shea JJ, McVicar DW. Regulation of JAK3 expression in human monocytes: phosphorylation in response to interleukins 2, 4, and 7. J Exp Med. 1995;181:1425–1431. doi: 10.1084/jem.181.4.1425. This article and [20] demonstrate the tissue-specific and activation-dependent expression of Jak3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortolani PJ, Lal BK, Riva A, Johnston JA, Chen YQ, Reaman GH, Beckwith M, Longo D, Ortaldo JR, Bhatia K, et al. Regulation of JAK3 expression and activation in human B cells and B cell malignancies. J Immunol. 1995;155:5220–5226. See [19] [PubMed] [Google Scholar]
- Hofmann SR, Lam AQ, Frank S, Zhou YJ, Ramos HL, Kanno Y, Agnello D, Youle RJ, O'Shea JJ. Jak3-independent trafficking of the common gamma chain receptor subunit: chaperone function of Jaks revisited. Mol Cell Biol. 2004;24:5039–5049. doi: 10.1128/MCB.24.11.5039-5049.2004. This paper and [22] present analyses of the intracellular trafficking of Jaks and their cognate receptors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragimbeau J, Dondi E, Alcover A, Eid P, Uze G, Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. See [21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argetsinger LS, Campbell GS, Yang X, Witthuhn BA, Silvennoinen O, Ihle JN, Carter-Su C. Identification of JAK2 as a growth hormone receptor-associated tyrosine kinase. Cell. 1993;74:237–244. doi: 10.1016/0092-8674(93)90415-M. This paper and [24-26] are the studies that first demonstrated that cytokine receptors couple to Janus kinases. [DOI] [PubMed] [Google Scholar]
- Witthuhn BA, Quelle FW, Silvennoinen O, Yi T, Tang B, Miura O, Ihle JN. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-L. See [23] [DOI] [PubMed] [Google Scholar]
- Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur AG, Barbieri G, Witthuhn BA, Schindler C, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature. 1993;366:129–135. doi: 10.1038/366129a0. See [23] [DOI] [PubMed] [Google Scholar]
- Silvennoinen O, Ihle JN, Schlessinger J, Levy DE. Interferon-induced nuclear signalling by Jak protein tyrosine kinases. Nature. 1993;366:583–585. doi: 10.1038/366583a0. See [23] [DOI] [PubMed] [Google Scholar]
- Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, Yamashita M, Numata A, Takase K, Kobayashi S, et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–571. doi: 10.1016/S1074-7613(00)00055-8. This report and [28,29] on the generation of Tyk2 knockout mice reveal its restricted role. [DOI] [PubMed] [Google Scholar]
- Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/S1074-7613(00)00054-6. See [27] [DOI] [PubMed] [Google Scholar]
- Karaghiosoff M, Steinborn R, Kovarik P, Kriegshauser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. See [27] [DOI] [PubMed] [Google Scholar]
- Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, et al. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/S0092-8674(00)81166-6. A description of the Jak1 knockout mice and its essential roles in cytokine signaling. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/S0092-8674(00)81168-X. This paper and [32] describe the embryonic lethality associated with Jak2 deficiency. [DOI] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/S0092-8674(00)81167-8. See [31] [DOI] [PubMed] [Google Scholar]
- Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O'Shea JJ, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature. 1995;377:65–68. doi: 10.1038/377065a0. This paper and [34] report the first identification of humans with Jak mutations, which result in severe combined immunodeficiency. [DOI] [PubMed] [Google Scholar]
- Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. See [33] [DOI] [PubMed] [Google Scholar]
- O'Shea JJ, Pesu M, Borie DC, Changelian PS. A new modality for immunosuppression: targeting the JAK/STAT pathway. Nat Rev Drug Discov. 2004;3:555–564. doi: 10.1038/nrd1441. A comprehensive review of Jak3 antagonists. [DOI] [PubMed] [Google Scholar]
- Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. This paper and [37,38] are the first reports of Jak3 knockout mice and their resultant immunodeficiency. [DOI] [PubMed] [Google Scholar]
- Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. See [36] [DOI] [PubMed] [Google Scholar]
- Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. See [36] [DOI] [PubMed] [Google Scholar]
- Sorrentino RP, Melk JP, Govind S. Genetic analysis of contributions of dorsal group and JAK-Stat92E pathway genes to larval hemocyte concentration and the egg encapsulation response in Drosophila. Genetics. 2004;166:1343–1356. doi: 10.1534/genetics.166.3.1343. This paper and [40-45] give evidence for the broad functions of the Jak/Stat pathway in regulating insect development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Asha H, Kockel L, Parke T, Mlodzik M, Dearolf CR. The Drosophila Jak kinase hopscotch is required for multiple developmental processes in the eye. Dev Biol. 1999;213:432–441. doi: 10.1006/dbio.1999.9390. See [39] [DOI] [PubMed] [Google Scholar]
- Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. See [39] [DOI] [PubMed] [Google Scholar]
- Chen X, Oh SW, Zheng Z, Chen HW, Shin HH, Hou SX. Cyclin D-Cdk4 and cyclin E-Cdk2 regulate the Jak/STAT signal trans-duction pathway in Drosophila. Dev Cell. 2003;4:179–190. doi: 10.1016/S1534-5807(03)00024-8. See [39] [DOI] [PubMed] [Google Scholar]
- O'Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109(Suppl):S121–S131. doi: 10.1016/S0092-8674(02)00701-8. See [39] [DOI] [PubMed] [Google Scholar]
- Hombria JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569–R575. doi: 10.1016/S0960-9822(02)01057-6. See [39] [DOI] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. See [39] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters P, Raynaud SD, Cools J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. This article and [47-49] give support for the role of Jaks in malignant transformation. [PubMed] [Google Scholar]
- Cools J, Peeters P, Voet T, Aventin A, Mecucci C, Grandchamp B, Marynen P. Genomic organization of human JAK2 and mutation analysis of its JH2-domain in leukemia. Cytogenet Cell Genet. 1999;85:260–266. doi: 10.1159/000015308. See [46] [DOI] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard OA. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. See [46] [DOI] [PubMed] [Google Scholar]
- Migone TS, Lin JX, Cereseto A, Mulloy JC, O'Shea JJ, Franchini G, Leonard WJ. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79–81. doi: 10.1126/science.7604283. See [46] [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Hanson EP, Chen YQ, Magnuson K, Chen M, Swann PG, Wange RL, Changelian PS, O'Shea JJ. Distinct tyrosine phosphorylation sites in JAK3 kinase domain positively and negatively regulate its enzymatic activity. Proc Natl Acad Sci USA. 1997;94:13850–13855. doi: 10.1073/pnas.94.25.13850. This study and [51,52] describe the functional significance of Jak autophosphorylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzer JH, Argetsinger LS, Zhou YJ, Kouadio JL, O'Shea JJ, Carter-Su C. Tyrosine 813 is a site of JAK2 autophosphorylation critical for activation of JAK2 by SH2-B beta. Mol Cell Biol. 2004;24:4557–4570. doi: 10.1128/MCB.24.10.4557-4570.2004. See [50] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feener EP, Rosario F, Dunn SL, Stancheva Z, Myers MG., Jr Tyrosine phosphorylation of Jak2 in the JH2 domain inhibits cytokine signaling. Mol Cell Biol. 2004;24:4968–4978. doi: 10.1128/MCB.24.11.4968-4978.2004. See [50] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. This paper and [54,55] provide emerging evidence of cross-talk between the Jak/Stat pathway and other signaling pathways. [DOI] [PubMed] [Google Scholar]
- Josten F, Fuss B, Feix M, Meissner T, Hoch M. Cooperation of JAK/STAT and Notch signaling in the Drosophila foregut. Dev Biol. 2004;267:181–189. doi: 10.1016/j.ydbio.2003.11.016. See [53] [DOI] [PubMed] [Google Scholar]
- Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila Janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–1166. doi: 10.1093/genetics/165.3.1149. See [53] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changelian PS, Flanagan ME, Ball DJ, Kent CR, Magnuson KS, Martin WH, Rizzuti BJ, Sawyer PS, Perry BD, Brissette WH, et al. Prevention of organ allograft rejection by a specific Janus kinase 3 inhibitor. Science. 2003;302:875–878. doi: 10.1126/science.1087061. A report of the first Jak inhibitor, which is efficacious as an immunosuppressant in a primate model of transplant rejection. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein sequence alignments in text format for orthologs of Jak1
Protein sequence alignments in text format for orthologs of Jak2
Protein sequence alignments in text format for orthologs of Jak3
Protein sequence alignments in text format for orthologs of Tyk2
Protein sequence alignments in text format for undefined members of the family
Protein sequence alignments in jpeg format for orthologs of Jak1
Protein sequence alignments in jpeg format for orthologs of Jak2
Protein sequence alignments in jpeg format for orthologs of Jak3
Protein sequence alignments in jpeg format for orthologs of Tyk2
Protein sequence alignments in jpeg format for undefined members of the family
A key for the alignments


