Abstract
Gene therapy is a promising option for severe forms of genetic diseases. We previously provided evidence for the feasibility of trans-splicing, exon skipping, and gene replacement in a mouse model of hypertrophic cardiomyopathy (HCM) carrying a mutation in MYBPC3, encoding cardiac myosin-binding protein C (cMyBP-C). Here we used human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) from an HCM patient carrying a heterozygous c.1358-1359insC MYBPC3 mutation and from a healthy donor. HCM hiPSC-CMs exhibited ∼50% lower MYBPC3 mRNA and cMyBP-C protein levels than control, no truncated cMyBP-C, larger cell size, and altered gene expression, thus reproducing human HCM features. We evaluated RNA trans-splicing and gene replacement after transducing hiPSC-CMs with adeno-associated virus. trans-splicing with 5′ or 3′ pre-trans-splicing molecules represented ∼1% of total MYBPC3 transcripts in healthy hiPSC-CMs. In contrast, gene replacement with the full-length MYBPC3 cDNA resulted in ∼2.5-fold higher MYBPC3 mRNA levels in HCM and control hiPSC-CMs. This restored the cMyBP-C level to 81% of the control level, suppressed hypertrophy, and partially restored gene expression to control level in HCM cells. This study provides evidence for (1) the feasibility of trans-splicing, although with low efficiency, and (2) efficient gene replacement in hiPSC-CMs with a MYBPC3 mutation.
Keywords: hypertrophic cardiomyopathy, MYBPC3, trans-splicing, gene replacement, human induced pluripotent stem cell-derived cardiomyocytes
Introduction
In the last decade, several strategies have been developed to correct or remove gene mutations at the DNA or RNA level, including gene replacement by cDNA overexpression, CRISPR/Cas9 gene editing, exon skipping, spliceosome-mediated RNA trans-splicing (trans-splicing), and RNAi (as reviewed elsewhere1, 2, 3). Hypertrophic cardiomyopathy (HCM) is a myocardial disease with a revised estimated prevalence of 1:200 in the general population.4 It is mainly characterized by hypertrophy of the left ventricle, increased interstitial fibrosis, and diastolic dysfunction.5 Current therapies, including β-blockers and Ca2+-channel blockers, aim at the relief of symptoms, but they are not curative.6 HCM is caused by mutations in genes encoding sarcomeric proteins. Among them, MYBPC3, encoding cardiac myosin-binding protein C (cMyBP-C), is the most frequently mutated gene.5, 7 To date more than 350 HCM-causing MYBPC3 mutations have been reported in the literature.7 The majority of MYBPC3 mutations are truncating, leading to C-terminal-truncated cMyBP-C proteins, which were never detected in the myocardial tissue of HCM patients.8
cMyBP-C is a multidomain protein that plays a role in the regulation of cardiac function and in sarcomeric organization.9, 10, 11 A growing body of evidence indicates that double heterozygous, compound heterozygous, and homozygous mutations in sarcomeric genes are associated with severe forms of cardiomyopathies.12, 13, 14 Specifically, individuals carrying bi-allelic truncating MYBPC3 mutations presented already at birth with various forms of cardiomyopathies (hypertrophic, dilated, and/or left ventricular non-compaction), which quickly developed into systolic heart failure and death within the first year.15, 16, 17 For these infants, there is no curative therapy other than heart transplant. Recently, proof-of-concept studies reported the feasibility of exon skipping, trans-splicing, RNAi, or gene replacement as gene therapy options in HCM mouse models.18, 19, 20, 21 To move this concept toward clinical application, advantage could be taken from the use of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). These cells have already been used for disease modeling of long-QT syndrome,22, 23 dilated cardiomyopathy,24, 25, 26 and HCM,27, 28, 29 and they could be used as a platform for testing different gene therapy options. In the present study, we evaluated the feasibility and efficiency of MYBPC3 trans-splicing and gene replacement strategies in hiPSC-CMs obtained from a healthy donor and an HCM patient harboring a heterozygous truncating mutation in the MYBPC3 gene.
trans-splicing involves two separate mRNAs, the target endogenous mutant pre-mRNA and an exogenous wild-type (WT) pre-trans-splicing molecule (PTM; as reviewed elsewhere30, 31). After gene transfer of the PTMs, a splice event in trans can occur between the two molecules, resulting in a chimeric repaired mRNA, which is then translated into a corrected protein (Figure S1A). Beside the WT coding sequence, PTMs carry strong splice sequences and a binding domain for specific recognition of the target. Depending on the position of the mutation in the pre-mRNA, trans-splicing can occur in 5′ or 3′ mode, and PTMs will contain a splice donor or acceptor site, respectively. In the gene replacement approach, a correct copy of the defective gene, i.e., full-length WT cDNA, is provided to the cell in order to replace the non-functional and/or missing protein (Figure S1B; as reviewed elsewhere32, 33). In the present work, PTMs carrying each half of the MYBPC3 coding sequence and full-length WT MYBPC3 cDNA were packaged into the adenovirus-associated virus serotype 9-SRLSPPS (hereafter AAV),34 and analyses were done 7 days after AAV transduction.
Results
HCM hiPSC-CMs Show Hypertrophy and cMyBP-C Haploinsufficiency
At the time of septal myectomy, the patient had an interventricular septal thickness of 26 mm, ejection fraction of >60%, and a left ventricular outflow gradient of 85 mmHg. Genetic testing of genomic DNA with a panel of 19 HCM genes revealed an insertion of a C in exon 16 of the MYBPC3 gene (c.1358_1359insC; Figure S2) on one allele of the HCM patient. This variant was not found in the Exome Association Consortium (ExAC) Browser, which harmonized sequencing data from more than 60,000 unrelated individuals (http://exac.broadinstitute.org/), supporting its causal effect. The mutation induced a frameshift and a premature termination codon (PTC) in exon 16. At the protein level, an amino acid substitution at position 454 was followed by 21 new amino acids (p.Val454CysfsX21) and truncation in the C3 domain of cMyBP-C.
Dermal fibroblasts from the HCM patient and from a healthy donor (control) were reprogrammed into hiPSCs, followed by differentiation into CMs (see the Materials and Methods). Both HCM and control hiPSC-CMs were seeded in a confluent monolayer and cultured for 7 days. hiPSC-CMs of both cell lines exhibited spontaneous beating (data not shown). After fixation, immunofluorescence analysis with antibodies directed against cMyBP-C and α-actinin showed a cross-striated pattern, indicating proper formation of sarcomeres (Figure 1A). Cell size, as determined by automated analysis of large numbers of cells (see the legend of Figure 1B), was significantly higher in HCM than in control hiPSC-CMs (3,656 ± 201 μm2 versus 2,213 ± 145 μm2; Figure 1B). Total MYBPC3 mRNA level was 50% lower in HCM than in control cells (Figure 1C). Mutant MYBPC3 mRNA was not detected by Sanger sequencing or with a specific Taqman probe (data not shown). The amount of cMyBP-C protein normalized to α-actinin tended to be lower (p = 0.064) in HCM than in control hiPSC-CMs (Figures 1D and 1E). No truncated protein (∼52 kDa) was detected in HCM cells (data not shown). The disease phenotype of HCM hiPSC-CMs was further evaluated by gene expression profile of 49 proteins involved in the regulation of cardiac hypertrophy and contraction with the nanoString nCounter Elements technology (Figure S3; Table S1). Most of the proteins associated with hypertrophy, cardiomyopathy, and/or PI3K-Akt signaling exhibited higher mRNA levels in HCM than in control CMs (green bars), whereas many proteins involved in excitation-contraction coupling or adrenergic signaling had lower mRNA levels in HCM than in control CMs (red bars).
Figure 1.
Phenotypic Characterization of HCM hiPSC-Derived Cardiomyocytes
Patient-specific (HCM) and control (Ctrl) hiPSC-derived cardiomyocytes (hiPSC-CMs) were seeded in a confluent monolayer, cultured for 7 days, and prepared for subsequent analysis. (A) Representative immunofluorescence images of HCM and control hiPSC-CMs. Both cell lines were stained with antibodies directed against cMyBP-C (red), α-actinin (green), and with Hoechst33342 for nuclear staining (blue; scale bars, 50 μm). Higher magnification images of a few sarcomeres are shown on the right panels. (B) Quantification of CM cell size. Control and HCM hiPSC-CMs were seeded in 96-well plates at a density of 10,000 cells/well, cultured for 7 days, and stained with an antibody against α-actinin. Cell size was measured in 27–51 fields/well in both groups. Control hiPSC-CMs were evaluated from 33 wells of three independent experiments and HCM hiPSC-CMs from 21 wells of two independent experiments, for a total of 28,336 and 12,874 CMs, respectively. Images were taken with the Opera High-Content Screening System (PerkinElmer), and the analyses were performed with the Columbus Image Data Management and Analysis System. (C) Evaluation of MYBPC3 mRNA levels determined by qRT-PCR with SYBR Green in control and HCM hiPSC-derived CMs (n = 3–5, with n = number of wells from one transduction experiment). (D) Western blot of control and HCM hiPSC-CMs stained with antibodies directed against cMyBP-C. α-actinin was used as the loading control. (E) Quantification of cMyBP-C protein levels in control and HCM hiPSC-CMs normalized to α-actinin and related to control (n = 3–4, with n = number of wells from two independent transduction experiments). Values are expressed as mean ± SEM (*p < 0.05 and ***p < 0.001, unpaired Student’s t test). cMyBP-C, cardiac myosin-binding protein C; CMs, cardiomyocytes; Ctrl, control.
5′ trans-Splicing Is Feasible in hiPSC-CMs
We designed a 5′ PTM that would be able, in principle, to repair all MYBPC3 mutations contained in the first half of the gene. It carried the WT MYBPC3 cDNA sequence from exon 1 to exon 21 under the control of a CM-specific promoter (TNNT2, human cardiac troponin T; Figure 2A). The 5′ PTM included a binding domain for base pairing with a complementary sequence (120 nt) in intron 21 of the MYBPC3 gene, a canonical 5′ splice site sequence followed by a downstream intronic sequence enhancer element, which has been shown to markedly increase trans-splicing efficiency (Figure S4A).35 To allow specific detection of MYBPC3 trans-spliced mRNA and cMyBP-C protein, we included a FLAG-tag sequence at the N terminus of the coding sequence (Figure 2A). To prevent translation of the PTM, we removed the polyA signal as described previously.19 For gene transfer in hiPSC-CMs, 5′ PTMs were packaged into AAV. Beforehand, we tested the transduction efficiency of different MOIs of AAV-TNNT2-GFP in control hiPSC-CMs. The MOI of 10,000 proved to transduce >80% of hiPSC-CMs after 7 days of transduction (Figure S5).
Figure 2.
5′ trans-Splicing in Control hiPSC-Derived Cardiomyocytes
(A) Schematic representation of 5′ trans-splicing. 5′ pre-trans-splicing molecules (5′ PTMs) carry a 5′ FLAG-tagged wild-type (WT) MYBPC3 cDNA sequence from exon 1 to exon 21, conserved 5′ splice donor site (5′SS) sequences, and a binding domain (BD) targeting intron 21. Upon successful binding of 5′ PTM to the endogenous MYBPC3 pre-mRNA, 5′ trans-splicing can occur and results in a trans-spliced MYBPC3 mRNA. (B) Schematic illustration of primer pairs used to amplify either only trans-spliced or trans-spliced and/or endogenous (total) MYBPC3 mRNAs. The primer pair FLAG-F/E23-R was used to generate a 2,180-bp fragment corresponding to the trans-spliced MYBPC3 mRNA. trans-spliced and/or endogenous MYBPC3 mRNA was amplified either using the primer pair E1-F/E23-R (2,138-bp fragment) or the primer pair E15-F/E16-R (193-bp fragment). (C) hiPSC-derived cardiomyocytes (hiPSC-CMs) were transduced with AAV-5′ PTM or AAV-Mock (TNNT2 promoter without insert) at an MOI of 10,000 and cultured in 2D for 7 days prior to harvesting. Representative agarose gel of RT-PCR, using specific primer pairs for trans-spliced and total MYBPC3 mRNA, is shown (n = 5, from three independent transductions). (D) Determination of the percentage of trans-spliced MYBPC3 mRNA by qRT-PCR. In a first round of PCR, either trans-spliced (FLAG-F/E23-R primers) or total (E1-F/E23-R primers) MYBPC3 mRNAs were amplified from cDNA of AAV-5′ PTM-transduced hiPSC-CMs. PCR fragments were column-purified and used in qPCR, together with a standard dilution of a plasmid encoding full-length wild-type MYBPC3 cDNA, using a common primer pair (E15-F/E16-R primers). Data are expressed as mean ± SEM with n = 3 (three wells of one transduction experiment). AAV, adeno-associated virus; bp, base pair; CMs, cardiomyocytes; E, exon; F, forward; hiPSC, human induced pluripotent stem cell; NT, non-transduced; MOI, multiplicity of infection; R, reverse; RT, reverse transcriptase; WT, wild-type.
Control hiPSC-CMs were then transduced with AAV-5′ PTM or AAV-Mock (empty virus; both at an MOI of 10,000) and cultured for 7 days in 2D. Using PCR primers that amplify only trans-spliced MYBPC3 transcript (Figure 2B), we obtained a specific fragment only in AAV-5′ PTM-transduced hiPSC-CMs, but not in non-transduced or mock-transduced hiPSC-CMs (Figure 2C). After agarose gel extraction, sequencing of the 2,180-bp trans-spliced MYBPC3 fragment validated the presence of the FLAG-tag (Figure S6A). To evaluate the effect of trans-splicing on cis-splicing, total MYBPC3 mRNA ( = trans-spliced + endogenous) was amplified with primers in exon 1 and exon 23 (Figure 2B). A specific amplicon was obtained in all samples without a major intensity difference among them (Figure 2C). To estimate the efficiency of 5′ trans-splicing, cDNA from AAV-5′ PTM-transduced hiPSC-CMs was used to amplify either trans-spliced or total MYBPC3 mRNA by PCR (25 cycles). PCR fragments were column-purified and further analyzed by qPCR using a common primer pair for amplification of the same fragment in all MYBPC3 transcripts. The percentage of trans-spliced MYBPC3 mRNA was calculated using a standard dilution (2 × 109 − 2 × 101 copy number) of a plasmid encoding the full-length WT MYBPC3 cDNA. Samples and standards were amplified with the same primer pair, enabling us to specifically calculate a 5′ trans-splicing efficiency of 0.96% (Figure 2D). FLAG immunoprecipitation experiments to detect trans-spliced cMyBP-C protein were not successful (data not shown), probably due to low trans-splicing efficiency.
3′ trans-Splicing Is Feasible in hiPSC-CMs
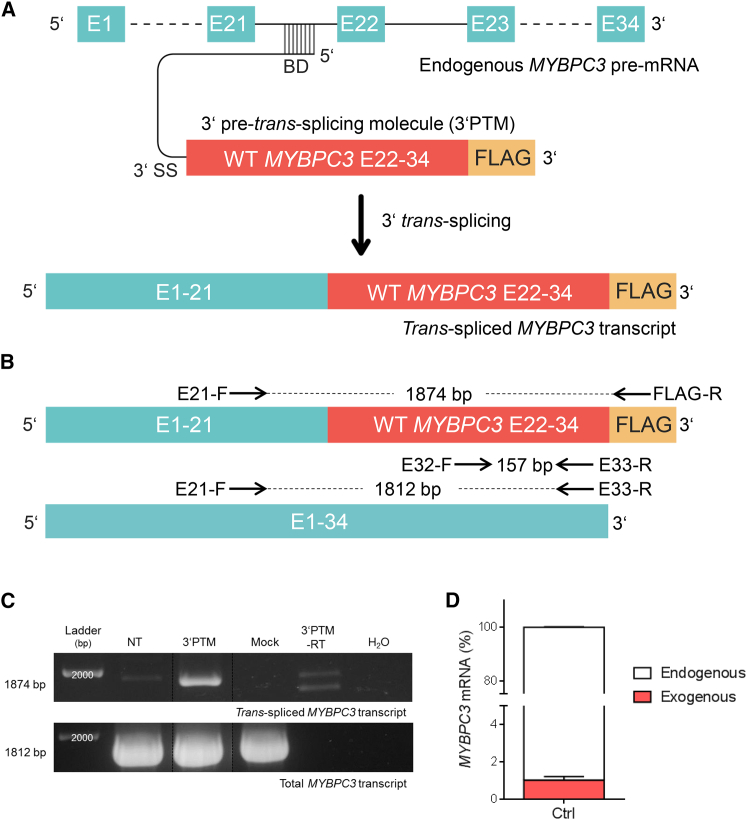
We designed a 3′ PTM, that would principally be able to repair all MYBPC3 mutations contained in the second half of the gene. It carried the WT MYBPC3 cDNA sequence from exon 22 to exon 34 under the control of the TNNT2 promoter (Figure 3A). The MYBPC3 cDNA sequence was FLAG-tagged at the C terminus to discriminate the trans-spliced MYBPC3 mRNA and cMyBP-C protein from endogenous ones. To prevent any translation of the 3′ PTM, no ATG sequence was introduced. To allow 3′ trans-splicing, we also inserted in the 3′ PTM a binding domain (same sequence as in 5′ PTM) targeting intron 21 followed by a linker sequence, a branch point, a polypyrimidine tract, and a 3′ splice site (Figure S4B). After packaging in AAV, control hiPSC-CMs were transduced with AAV-3′ PTM or AAV-Mock (MOI 10,000), and they were cultured for 7 days in 2D prior to collection. Primer pairs for detection of trans-spliced or total MYBPC3 mRNA are shown in Figure 4B. A specific 1,874-bp amplicon was obtained in the AAV-3′ PTM-transduced sample with primers designed to recognize trans-spliced MYBPC3 mRNA. Sequencing of this amplicon validated the presence of the FLAG sequence (Figures 3B and 3C; Figure S6B). A fragment of similar size was detected in the non-transduced sample, but sequencing did not reveal the FLAG sequence (data not shown). Similarly, PCR fragments also appeared in the AAV-3′ PTM-transduced but without reverse transcriptase sample, indicating nonspecific binding of the primers to the endogenous mRNA. RT-PCR with a primer pair that amplified total MYBPC3 mRNA showed a signal of similar intensity in non-transduced and in AAV-transduced samples (Figure 3C). As was done for 5′ trans-splicing, the efficiency of 3′ trans-splicing was determined by qRT-PCR with a standard dilution (2 × 109 − 2 × 101 copy number) of the same plasmid carrying the full-length WT MYBPC3 cDNA. The concentration of trans-spliced MYBPC3 mRNA reached 1% of the total (Figure 3D). Similar to 5′ trans-splicing, the trans-spliced cMyBP-C protein was not detected after FLAG immunoprecipitation (data not shown).
Figure 3.
3′ trans-Splicing in Control hiPSC-Derived Cardiomyocytes
(A) Schematic illustration of 3′ trans-splicing. 3′ pre-trans-splicing molecules (3′ PTMs) contain a BD complementary to MYBPC3 intron 21, conserved 3′ splice donor site sequences (3′SS), and a 3′-FLAG-tagged wild-type (WT) MYBPC3 cDNA sequence from exon 22 to exon 34. Upon successful binding of the 3′ PTM to the endogenous MYBPC3 pre-mRNA, 3′ trans-splicing can occur and results in trans-spliced MYBPC3 mRNA. (B) Schematic representation of primer pairs used to amplify either only trans-spliced MYBPC3 mRNA or trans-spliced and/or endogenous (total) MYBPC3 mRNA. The primer pair E21-F/FLAG-R amplifies a 1,874-bp fragment specific of trans-spliced MYBPC3 mRNA. The primer pairs E21-F/E33-R and E33-F/E33-R amplify in both trans-spliced and endogenous MYBPC3 mRNA a 1,812-bp and a 157-bp fragment, respectively. (C) Control hiPSC-CMs were transduced with AAV-3′ PTM or AAV-Mock (MOI 10,000) for 1 week prior to harvesting. Representative agarose gel of RT-PCR, using specific primer pairs for trans-spliced and total MYBPC3 mRNA, is shown (n = 9 from four independent transductions). (D) Determination of the percentage of trans-spliced MYBPC3 mRNA by qRT-PCR was performed as described in Figure 2D, with the common primer pair E33-F/E33-R. Data are expressed as mean ± SEM with n = 5 (five wells of one transduction experiment). bp, base pair; CMs, cardiomyocytes; E, exon; F, forward; hiPSC, human induced pluripotent stem cell; NT, non-transduced; MOI, multiplicity of infection; R, reverse; RT, reverse transcriptase; WT, wild-type.
Figure 4.
Proof-of-Concept of MYBPC3 Gene Replacement in Human iPSC-Derived Cardiomyocytes
(A) Schematic illustration of full-length wild-type (WT) exogenous and total MYBPC3 cDNA/mRNA sequence (exons 1–34). Location of primer pairs and size of amplicons are shown. The primer pair FLAG-F/E2-R amplifies exclusively the exogenous MYBPC3 sequence (amplicon size: 193 bp). The primer pair E1-F/E2-R is suitable for amplification of both exogenous and endogenous (total) MYBPC3 sequence (amplicon size: 151 bp). (B) Control and HCM hiPSC-CMs were transduced with AAV-FLAG-MYBPC3 (GR) or AAV-Mock (Mock) at an MOI of 10,000 and cultured in 2D for 7 days before harvesting. Representative RT-PCR using primers shown in (A) for specific amplification is shown (n = 3, one well each from three independent transductions). (C) Evaluation of MYBPC3 transcript levels in GR-transduced control and HCM hiPSC-CMs by qRT-PCR with a common primer pair (n = 3–5, with n = number of wells of one transduction experiment). (D) Determination of the percentage of exogenous MYBPC3 mRNA by qRT-PCR. In a first round of RT-PCR, either exogenous (FLAG-F/E2-R primers) or total (E1-F/E2-R primers) MYBPC3 transcripts were amplified in control and HCM GR samples. After column purification of PCR fragments, a qPCR with a common primer pair (E1-F/E2-R) was performed together with a standard dilution of a plasmid encoding full-length WT MYBPC3 cDNA (n = 3, three wells of one transduction experiment). (E) Western blot performed on pooled protein samples (n = 6, from three independent transduction experiments) of non-transduced (NT) or transduced (GR) control and single protein samples (n = 3, three wells of one transduction experiment) of HCM hiPSC-CMs with anti-cMyBP-C and anti-α-actinin antibodies. (F) Quantification of cMyBP-C level normalized to α-actinin and related to control NT pooled sample. Data are expressed as mean ± SEM (*p < 0.05, unpaired Student’s t test). E, exon; F, forward; NT, non-transduced; MOI, multiplicity of infection; R, reverse; WT, wild-type.
Gene Replacement in HCM hiPSC-CMs Partially Corrects cMyBP-C Haploinsufficiency and Reduces Cell Hypertrophy
To evaluate a gene replacement therapy option, the FLAG-tagged WT MYBPC3 cDNA under the control of the TNNT2 promoter was packaged in AAV (AAV-FLAG-MYBPC3). After transduction (MOI 10,000) of control and HCM hiPSC-CMs, cells were cultured for 7 days in 2D prior to harvesting. RT-PCR with specific primers for exogenous MYBPC3 transcript revealed a specific fragment in both control and HCM transduced hiPSC-CMs (Figures 4A and 4B). A signal of lower intensity was also obtained in AAV-transduced samples, which were not retrotranscribed, corresponding to the transgene. The transcription of the entire FLAG-MYBPC3 cDNA sequence was also validated (data not shown). Total MYBPC3 transcript was amplified using a common primer pair in all samples, transduced or not (Figures 4A and 4B). To quantify the overall level of MYBPC3 mRNA after transduction, a qRT-PCR was performed with a primer pair recognizing both exogenous and endogenous MYBPC3 mRNA transcripts (Figures 4A and 4C). About 2.5-fold overexpression of MYBPC3 was obtained in both control and HCM transduced hiPSC-CMs over the non-transduced cells (Figure 4C). To evaluate the efficiency of gene replacement, RT-PCR with specific primers to amplify either exogenous or total MYBPC3 transcripts was performed in control and HCM transduced samples. PCR fragments were column-purified and used further for qPCR with a common primer pair, together with a standard dilution curve of a plasmid carrying the full-length wild-type MYBPC3 cDNA sequence (2 × 109 − 2 × 101 copy number). In control and HCM hiPSC-CMs, exogenous MYBPC3 transcript level reached 70% and 57% of total, respectively (Figure 4D). The level of cMyBP-C protein was determined by western blot analysis in protein samples, normalized to α-actinin content and related to non-transduced control hiPSC-CM sample (Figures 4E and 4F). The total cMyBP-C protein level did not differ between transduced and non-transduced control hiPSC-CMs. In contrast, cMyBP-C protein level was 2.5-fold higher (p < 0.05, Student’s t test) in transduced than in non-transduced HCM hiPSC-CMs, reaching 81% of the cMyBP-C level of the non-transduced control sample (Figures 4E and 4F).
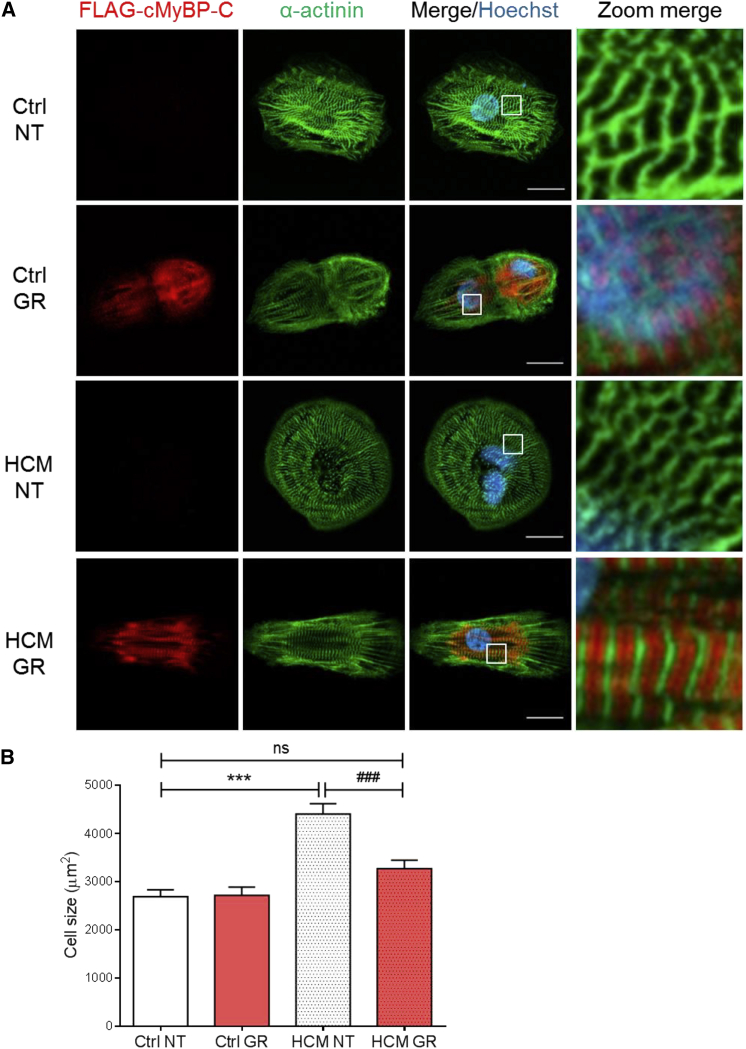
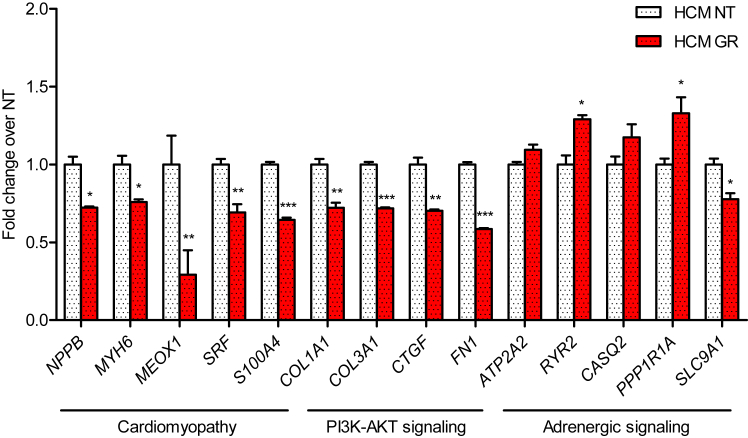
Sarcomere organization after transduction of control and HCM hiPSC-CMs with AAV-FLAG-MYBPC3 was evaluated by immunofluorescence analysis with anti-FLAG and anti-α-actinin antibodies. FLAG-cMyBP-C protein was properly organized in doublets at the A-band of the sarcomere, well alternating with α-actinin located at the Z-disk (Figure 5A). Finally, we evaluated the impact of MYBPC3 gene transfer on cell size. Cell size was significantly higher in non-transduced HCM than in control hIPSC-CMs (Figure 5B). MYBPC3 gene transfer did not affect cell size in control hiPSC-CMs (2,715 ± 168 μm2 versus 2,665 ± 120 μm2 in non-transduced control). In contrast, cell size was significantly lower in MYBPC3-transduced than in non-transduced HCM hiPSC-CMs (3,205 ± 139 μm2 versus 4,409 ± 217 μm2), and it did not differ from non-transduced control hiPSC-CMs (Figure 5B). MYBPC3 gene replacement lowered mRNA levels of several proteins involved in cardiomyopathy and PI3K-Akt signaling, and it increased mRNA levels of calcium-handling proteins in HCM hiPSC-CMs (Figure 6).
Figure 5.
Phenotypic Characterization of Control and HCM hiPSC-Derived Cardiomyocytes after MYBPC3 Gene Transfer
(A) Representative images of non-transduced (NT) and transduced (GR) control (Ctrl) and HCM hiPSC-CMs after immunofluorescence staining with anti-FLAG and anti-α-actinin antibodies. Images were taken with a Zeiss LSM 800 microscope (scale bars, 20 μm). Higher magnification images of a few sarcomeres are shown on the right panel. (B) Quantification of CM cell size. NT and GR control and HCM hiPSC-CMs were seeded in 96-well plates at a density of 10,000 cells/well, cultured for 1 week, and stained with antibodies against FLAG-tag and α-actinin. Low-resolution images of >100 cells in each group from six wells of one experiment each (Ctrl NT n = 108; Ctrl GR n = 110; HCM NT n = 114; HCM GR n = 133) were taken with the Zeiss LSM 800 microscope and analyzed with Fiji software (ImageJ). Data are expressed as mean ± SEM (***p < 0.001, two-way ANOVA plus Bonferroni post-test; ###p < 0.001, unpaired Student’s t test).
Figure 6.
Gene Expression Analysis of HCM hiPSC-Derived Cardiomyocytes in the Absence of or after MYBPC3 Gene Transfer
The mRNA levels of transduced (GR) and non-transduced (NT) HCM hiPSC-CMs were determined with the nanoString nCounter Elements technology, normalized to housekeeping genes, and related to NT CMs. Data are expressed as mean ± SEM (*p < 0.05, **p < 0.01, and ***p < 0.001, unpaired Student’s t test; n = 3, with n = number of wells from one transduction experiment).
Discussion
In this study, we report the principal feasibility of two gene therapy options for HCM in hiPSC-CMs from a healthy donor and an HCM patient carrying a MYBPC3 truncating mutation. While our data suggest that trans-splicing efficiency is too low to be a therapeutic option to treat severe forms of HCM, the MYBPC3 gene replacement strategy looks promising, particularly for its ability to circumvent haploinsufficiency of cMyBP-C (restoration of a correct amount of protein) and to reduce CM hypertrophy. Numerous studies have used hiPSC-CMs as a tool for disease modeling of different cardiac diseases, including HCM with MYBPC3 mutations.22, 23, 24, 25, 26, 27, 28, 29, 36, 37, 38, 39 However, so far only one study used the gene replacement strategy to correct a phospholamban (PLN) mutation associated with dilated cardiomyopathy in vitro.39
We believe that the truncating p.Val454CysfsX21 MYBPC3 mutation, described here for the first time, is HCM causing for the following reasons. First, truncating MYBPC3 mutations were reported to have a >110-fold higher odds ratio to cause HCM.40 Second, this specific mutation was not found in the population of about 60,000 unrelated individuals of the ExAC Browser (http://exac.broadinstitute.org/). HCM hiPSC-CMs revealed larger cell size, higher mRNA levels of proteins associated with hypertrophy, such as four and a half LIM domains 1 (FHL1), S100 calcium-binding protein A4 (S100A4), and connective tissue growth factor (CTGF), and lower mRNA levels of proteins involved in calcium handling, such as PLN, SERCA2A (ATP2A2), ryanodine receptor (RYR2), and inhibitor 1 (PPP1R1A) than control CMs, all hallmarks of HCM. This is in agreement with previous findings using HCM hiPSC-CMs or human embryonic stem cell (hESC) carrying MYBPC3 mutations.28, 29, 37, 41 While Dambrot et al.37 described that the supplementation of serum in culture medium masks the hypertrophic phenotype in 2D culture of hiPSC-CMs, in the present study the enlarged cell size was evident already in the presence of serum, as also reported by Ojala et al.28
In addition to hypertrophy, the HCM CMs used in the present work presented another hallmark of the MYBPC3-causing HCM, namely haploinsufficiency (for review, see Marston et al.42). Indeed, the amounts of MYBPC3 mRNA and cMyBP-C protein were ∼50% of the control hiPSC-CMs. Furthermore, the mutant nonsense mRNA was not detected, suggesting that it is degraded by the nonsense-mediated mRNA decay (NMD), as previously shown in Mybpc3-targeted knockin HCM mice.43 The location of the PTC at >50–55 nt upstream of the most 3′ exon-exon junction perfectly matches the NMD rules,44 therefore supporting its NMD-mediated degradation. The expected 52-kDa truncated cMyBP-C protein was also not detected by western blot, in agreement with previous observations in human myocardial samples with truncating MYBPC3 mutations.8, 45 Thus, the combination of a unique truncating MYBPC3 mutation, cMyBP-C haploinsufficiency, and CM hypertrophy makes this hiPSC cell line a good test bed for the application of different treatment options for HCM.
In the last years, mRNA trans-splicing has been evaluated both in vitro and in vivo as an alternative option for currently incurable genetic diseases, such as Duchenne muscular dystrophy and spinal muscular atrophy (as reviewed elsewhere30, 31). In the case of HCM-associated MYBPC3 mutations, the trans-splicing approach was appealing because by generating only two PTMs covering the first and the second half of the MYBPC3 mRNA, one could principally repair all the mutations and, therefore, treat 40%–60% of all HCM patients.6, 7 In addition, and in contrast to exon skipping mediated by antisense oligonucleotides (AONs), trans-splicing is expected to result in a repaired full-length and functional cMyBP-C protein. Furthermore, FDA or EMA authorization for marketing of two PTMs as new medicinal products would be easier and quicker than for several AONs.
In the present study, we showed the feasibility of both 5′ and 3′ trans-splicing in control hiPSC-CMs. However, we did not test these approaches in HCM hiPSC-CMs, since the efficiency of the process was low and we were unable to detect repaired cMyBP-C protein. This supports our previous data obtained in vivo in Mybpc3-targeted knockin mice, in which the low amount of repaired cMyBP-C protein produced by 5′ trans-splicing was not sufficient to prevent the development of the cardiac disease phenotype.19 Efficient trans-splicing depends on different factors, such as efficient delivery of PTMs to desired cell type and especially PTM design.30 Increasing the number of viral particles used for transduction could be difficult, since at a certain point AAV turned to be toxic for the cells (data not shown). On the other hand, PTM design could be improved, particularly for the binding domain. To have approximately the same packaging size for the 5′ PTM and 3′ PTM, we chose to target intron 21 in the MYBPC3 gene. The binding domain covered 120 nt (of 246) and was located roughly in the middle. However, we included highly conserved splice donor and acceptor sites in the 5′ PTM and 3′ PTM, respectively, which could be even stronger than the corresponding sequences in MYBPC3 intron 21. We are aware that additional systematic testing of different lengths and complementary regions might have led to higher binding affinities for the endogenous pre-mRNA and, therefore, to higher trans-splicing efficiencies. Further optimization should be done in the future. Nevertheless, this is the first proof of concept of trans-splicing in hiPSC-CMs.
Gene replacement by cDNA overexpression is another very attractive treatment option for sarcomeric cardiomyopathies. Indeed, we recently demonstrated that AAV-mediated delivery of full-length WT Mybpc3 cDNA in homozygous Mybpc3-targeted knockin mice not only prevented the development of cardiac hypertrophy and dysfunction, by increasing the amount of cMyBP-C protein, but also suppressed the expression of the endogenous mutant alleles.21 Exogenous expression of a sarcomeric protein is expected to replace in part or completely the endogenous counterpart, since the sarcomere is a tightly regulated system with a preserved stoichiometry of all structural components.46 This is the case in this study for the control cell line since MYBPC3 gene transfer resulted in a 2.4-fold higher level of MYBPC3 mRNA without change in the level of full-length cMyBP-C protein. Similarly, MYBPC3 gene transfer resulted in a 2.6-fold higher amount of MYBPC3 mRNA in HCM cells. Importantly and because the basal level was lower than in control cell lines, the cMyBP-C amount after gene transfer in HCM CMs reached 81% of the control cell line. In this condition, we expected at least a partial correction of the disease phenotype, since we previously showed that restoration of 80% of the cMyBP-C level by MYBPC3 gene transfer partially restores the disease phenotype of engineered heart tissue derived from Mybpc3-deficient mice.47 Indeed, the partial restoration of cMyBP-C haploinsufficiency was sufficient to suppress hypertrophy in HCM CMs. Exogenous FLAG-cMyBP-C proteins were properly incorporated into the sarcomere in both cell lines. MYBPC3 gene replacement had a positive effect on mRNA levels of proteins associated with hypertrophy or calcium handling. For instance, serum response factor (SRF) and S100A4, which are known to be higher in cardiomyopathies,48, 49 were significantly reduced after 7 days of MYBPC3 therapy. In addition, levels of mRNAs encoding proteins belonging to the PI3K-Akt Kyoto Encyclopedia of Genes and Genome (KEGG) pathway, such as CTGF, collagens (COL1A1 and COL3A1), and fibronectin (FN1), were significantly reduced. The mechanism by which the latter are regulated by gene therapy is not certain, but it could be related to paracrine factors mediating the crosstalk between CMs and fibroblasts (as reviewed elsewhere50, 51). Finally, gene therapy increased mRNA levels of calcium-handling proteins, suggesting improvement of the cardiac contraction. We are aware that a weakness of the findings reported here concerns the comparison of the diseased line to an unrelated control and not to a corrected isogenic hiPSC line. However, in line with our findings, adenoviral-mediated MYBPC3 gene delivery for 7 days restored the amount of cMyBP-C protein toward WT level and prevented hypertrophy in CMs derived from hESCs.41
In conclusion, our findings support gene replacement by overexpression of MYBPC3 cDNA as a promising therapeutic option, particularly for infants with bi-allelic truncating MYBPC3 mutations.15, 16, 17 This causal therapy could prolong and improve quality of life of these affected infants for whom no other therapy exists except heart transplantation. The evaluation of the efficacy of therapeutic options in a human cellular model, such as hiPSC-CMs, represents an intermediate step toward clinical application for these infants. Further studies with large animal models of severe forms of cardiomyopathy are still required to test AAV doses and delivery before going to first-in patient.
Materials and Methods
Vector Design
The 5′ PTM was obtained by fusion PCR of MYBPC3 exon 1–21 coding sequence (PCR1) and the binding domain in intron 21 (PCR2) with partially overlapping primers. The list of primers is given in Table S2. The amplicon of PCR1 was obtained with a forward primer (F5-1) containing the NheI restriction site, the FLAG sequence, and the first 16 coding nt of MYBPC3 exon 1. The reverse primer (R5-1) for PCR1 contained the BamHI restriction site, the downstream intronic splicing enhancer (DISE) element sequence, followed by the 5′ canonical splice donor site and the last 15 nt of MYBPC3 exon 21. The MYBPC3 coding sequence was amplified from the pSPORT-CMV-MYBPC3, clone BC151211.1 (Thermo Scientific). The amplicon of PCR2 was obtained with a forward primer (F5-2) containing a few nucleotides of the DISE, a BamHI restriction site, and 18 nt of intron 21. The reverse primer (R5-2) comprised an NotI restriction site and 18 nt of intron 21. Amplicons obtained in PCR1 and PCR2 were then used as templates for a third (fusion) PCR with primers F5-1 and R5-2. The 3′ PTM was obtained by fusion PCR of three different PCRs. PCR1 was performed to amplify the binding domain in intron 21 using a forward primer (F3-1) containing an NheI restriction site followed by 18 nt of MYBPC3 intron 21 and a reverse primer (R3-1) comprising 14 nt of the linker sequence, a BamHI restriction site, and 18 nt of MYBPC3 intron 21. PCR2 was performed to amplify the 3′ splicing sequences with forward primer F3-2 (same sequence as R3-1) and reverse primer (R3-2) carrying 10 nt of MYBPC3 exon 22, 3′ canonical splice acceptor site sequence, and the polypyrimidine tract. PCR3 was performed to amplify the MYBPC3 coding sequence from exon 22 to exon 34 with a forward primer F3-3 (same sequence as R3-2) and a reverse primer R3-3 carrying an NotI restriction site, the stop codon TGA, the FLAG sequence, and 12 nt of exon 34. PCR1 and PCR2 were then used as templates in PCR4 with primers F3-1 and R3-2 and finally PCR4 and PCR3 were fused in PCR5 with primers F3-1 and R3-3.
Production and Titration of AAV Particles
For production of AAV vectors, HEK293T cells were triple transfected with modified rep2/cap9-plasmid p5E18-VD-2/9-SLRSPPS,34 plasmid pDGΔVP containing adenovirus helper functions, and one of the ITR-containing plasmids (scAAV-TNNT2-EGFP,52 pGG2-TNNT2-FLAG-MYBPC3, pGG2-TNNT2-5′ PTM, or pGG2-TNNT2-3′ PTM, with FLAG-MYBPC3, 5′ PTM, and 3′ PTM inserted by NheI and NotI). Transfection, harvesting, and purification were performed as described previously.34 In brief, cells were transfected with PEI (Polysciences), harvested after 48–72 hr cells by Trypsin-EDTA, and lysed in cell lysis buffer (50 mM Tris-Cl [pH 8.5], 150 mM NaCl, and 5 mM MgCl2). Cell lysates were sonicated with a Sonorex TK device for 1 min at 48 W (Bandelin) and treated with 100 U Benzonase (Sigma-Aldrich) per milliliter of lysate for 30 min at 37°C. AAV particles were then purified by iodixanol density gradient ultracentrifugation, desalted (ZebaSpin desalting columns, recommended for processing compounds >7,000 Da [7 MWCO]; Thermo Fisher Scientific) from 40% iodixanol to 1× PBS, followed by concentration using Vivaspin6 columns (10 MWCO, Sartorius).
Generation of Patient-Specific hiPSC Line
The HCM patient was recruited in the outpatient clinic at the University Heart Center Hamburg and provided written informed consent for the use of fibroblasts. A skin biopsy was taken, washed in PBS, minced, and placed in a six-well plate in fibroblast medium (DMEM with 10% fetal bovine serum [FBS, PAA], 2 mM L-glutamine, and 0.5% penicillin and streptomycin; all Life Technologies). Dermal fibroblasts growing out of the explants were collected for passaging or cryopreservation and used for subsequent reprogramming at passage 5. The reprogramming was performed according to previously published protocols with retroviruses encoding the human transcription factors OCT3/4, SOX2, KLF4, and L-MYC.53, 54, 55
CM Differentiation, AAV Transduction, and Culture
CM differentiation from hiPSCs was performed following a three-step protocol with generation of embryoid bodies (EBs) in spinner flasks as described.56, 57 After dissociation with collagenase 2, beating CMs were transduced in suspension for 1 hr at 37°C with AAV at an MOI of 10,000. Transduced and non-transduced CMs were plated on Geltrex-coated (1:100, Gibco) 12-well or 96-well plates at a density of 440,000 cells/well or 10,000 cells/well, respectively. CMs were mantained in culture as a monolayer in DMEM (10% heat inactivated fetal calf serum [FCS, Gibco], 0.1% insulin [Sigma-Aldrich], and 0.5% penicillin/streptomycin [Gibco]) for 7 days at 37°C and 10% CO2 prior to further analyses.
Immunofluorescence Staining of hiPSC-CMs
Human iPSC-CMs were cultured for 7 days in 96-well plates (μclear, Greiner), then rinsed once with pre-warmed 1× PBS and fixed with Histofix (Carl Roth) for 20 min at 4°C. After washing two times in cold 1× PBS, hiPSC-CMs were incubated with primary antibodies directed against the M-motif of cMyBP-C (1:200, custom made), FLAG (1:800, Sigma), and α-actinin (1:800, Sigma); diluted in permeabilization buffer (1× PBS [Gibco], milk powder 3% [w/v, Carl Roth], and Triton X-100 0.1% [Carl Roth]); and incubated overnight at 4°C under gentle agitation. After washing two times in cold 1× PBS, hiPSC-CMs were incubated with secondary antibodies anti-mouse Alexa Fluor 488 (1:800, Life Technologies) and anti-rabbit Alexa Fluor 546 (1:800, Life Technologies), diluted in permeabilization buffer, and incubated for 1–2 hr at room temperature under gentle agitation and protected from light. In a final step, Hoechst 33342 (1:2,500, Thermo Fisher Scientific) diluted in 1× PBS was added to the wells and incubated for an additional 20 min. After washing two times in 1× PBS, hiPSC-CMs were ready for subsequent analysis in the Opera High-Content Screening System (PerkinElmer) or by confocal microscopy using a Zeiss LSM 800 microscope with Airyscan technology.
Measurement of CM Size
For quantification of hiPSC-CMs and for cell size analysis, the Opera High-Content Screening System and the Zeiss LSM 800 with Airyscan technology system were used. For Opera analysis, stained hiPSC-CMs in 96-well plates were loaded into the microscope, and, depending on how many wells were included in the analysis, 500–2,500 images were taken in 10× magnification. Those images were transferred in the Columbus Image Data Management and Analysis System (PerkinElmer). Columbus delivers pre-tested scripts, which were modified according to the characteristics of the used hiPSC-CMs. In this process, single building blocks were defined and accustomed to the hiPSC-CMs. Using the customized script, bulk analyses of the experiments were done and cell parameters of interest were obtained. For confocal microscopy, hiPSC-CMs were stained in 96-well plates and >100 images per sample were recorded, prepared according to the Opera analysis. Cell sizes from confocal images were measured by using Fiji software (ImageJ). Quality criteria for hiPSC-CM inclusion were set for single cells with well-formed sarcomeres.
RNA Isolation and Expression Analysis
Total RNA was extracted from hiPSC-CMs using TRIzol Reagent (Life Technologies), following the manufacturer’s protocol. The conversion of cDNA was performed with SuperScript III First-Strand Synthesis System (Invitrogen), according to the manufacturer’s instructions. For reverse transcription, oligo(dT) primers supplied in the kit and 200 ng extracted RNA were used. Touchdown RT-PCR to detect different MYBPC3 transcripts in 5′ or 3′ trans-splicing experiments (63°C–58°C and 67°C–62°C, respectively) was performed using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific) in a total volume of 20 μL for 35 cycles, according to the instructions of the manufacturer’s protocol. The qRT-PCR was performed in triplicates with SYBR-Green (Fermentas) according to the manufacturer’s instructions. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene. The target sequences were amplified during 40 cycles in an AbiPrism7000HT cycler (Applied Biosystems). For expression analysis with the nanoString nCounter Elements technology, a total amount of 50 ng RNA was hybridized with a customized nanoString Gene Expression CodeSet (Table S1) and analyzed using the nCounter Sprint Profiler. The mRNA levels were normalized to five housekeeping genes (ABCF1, CLTC, GAPDH, PGK1, and TUBB) and expressed as fold change in HCM over control CMs.
Western Blot
Western blot analysis was performed on total crude protein lysates from cultured hiPSC-CMs in the different conditions. Same amount of proteins (20 μg/lane) of single samples or pooled samples (n = 5–6) were separated on a 10% SDS-polyacrylamide (29:1) mini-gels (Bio-Rad) and transferred by wet-electroblotting to nitrocellulose membranes. Membranes were stained with the primary antibodies directed against the M-motif of cMyBP-C (polyclonal, 1:10,000, custom made) and α-actinin (monoclonal, 1:10,000, Sigma). Peroxidase-conjugated secondary antibodies against mouse (1:20,000, Dianova) or against rabbit (1:20,000, Sigma) were used. Proteins were visualized using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare), and the signals were detected on Amersham Hyperfilm MP (GE Healthcare). Signals were quantified with GeneTools image analyzing software (Syngene).
Statistical Analysis
Data are presented as mean ± SEM. Statistical analyses were performed by Student’s t test and two-way ANOVA with Bonferroni post-test using the GraphPad Prism 6.0 software. A value of p < 0.05 was considered significant.
Author Contributions
Conceptualization, M. Prondzynski, G.M., and L.C.; Methodology, M. Prondzynski, G.M., and L.C.; Investigation, M. Prondzynski, E.K., and G.M.; Formal Analysis, M. Prondzynski, G.M., and L.C.; Visualization, M. Prondzynski; Resources, S.D.L., A.S., A.H., F.F., O.P., O.J.M., J.M., C.R., M. Patten, T.E., and L.C.; Writing – Original Draft, M. Prondzynski and G.M.; Writing – Review & Editing, M. Prondzynski, T.E., G.M., and L.C.; Project Administration, M. Prondzynski, G.M., and L.C.; Supervision, G.M. and L.C.; Funding Acquisition, T.E., G.M., and L.C.
Conflicts of Interest
L.C., T.E., O.J.M., and G.M. are co-authors of a provisional European patent application EP13164212, filed April 17, 2013, Gene-therapy vectors for treating cardiomyopathy, followed by an international patent US20160108430, application US 14/785,188, publication date April 21, 2016 (filing date April 17, 2014). The remaining authors declare no conflicts of interest.
Acknowledgments
We are thankful to Alessandra Moretti (Department of Cardiology, Klinikum rechts der Isar, Technische Universität München) for providing the control hiPSC cell line. We would like to thank the cardiac differentiation team, especially Ingra Mannhardt-Vollert, Tessa Werner, and Bärbel Ulmer for precious help (UKE-Pharmacology) and Andreas Jungmann for support with AAV productions (Internal Medicine III, University Hospital Heidelberg). We also want to thank Oliver Keminer for his help and technical assistance with the Opera High-Content Screening System and the Columbus Image Data Management and Analysis System (Fraunhofer IME Screening-Port). Finally, we would like to thank Suellen Lopes Oliveira for graphic design (http://www.suelopes.com) and visualization. This work was supported by the DZHK (German Centre for Cardiovascular Research, the German Ministry of Research Education (BMBF), and by the Seventh Framework Program of the European Union (Health-F2-2009–241577; Big-Heart project).
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.05.008.
Contributor Information
Giulia Mearini, Email: g.mearini@uke.de.
Lucie Carrier, Email: l.carrier@uke.de.
Supplemental Information
References
- 1.Hammond S.M., Wood M.J. Genetic therapies for RNA mis-splicing diseases. Trends Genet. 2011;27:196–205. doi: 10.1016/j.tig.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Wang D., Gao G. State-of-the-art human gene therapy: part I. Gene delivery technologies. Discov. Med. 2014;18:67–77. [PMC free article] [PubMed] [Google Scholar]
- 4.Semsarian C., Ingles J., Maron M.S., Maron B.J. New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2015;65:1249–1254. doi: 10.1016/j.jacc.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Elliott P.M., Anastasakis A., Borger M.A., Borggrefe M., Cecchi F., Charron P., Hagege A.A., Lafont A., Limongelli G., Mahrholdt H., Authors/Task Force members 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur. Heart J. 2014;35:2733–2779. doi: 10.1093/eurheartj/ehu284. [DOI] [PubMed] [Google Scholar]
- 6.Behrens-Gawlik V., Mearini G., Gedicke-Hornung C., Richard P., Carrier L. MYBPC3 in hypertrophic cardiomyopathy: from mutation identification to RNA-based correction. Pflugers Arch. 2014;466:215–223. doi: 10.1007/s00424-013-1409-7. [DOI] [PubMed] [Google Scholar]
- 7.Carrier L., Mearini G., Stathopoulou K., Cuello F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene. 2015;573:188–197. doi: 10.1016/j.gene.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marston S., Copeland O., Jacques A., Livesey K., Tsang V., McKenna W.J., Jalilzadeh S., Carballo S., Redwood C., Watkins H. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ. Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 9.Craig R., Lee K.H., Mun J.Y., Torre I., Luther P.K. Structure, sarcomeric organization, and thin filament binding of cardiac myosin-binding protein-C. Pflugers Arch. 2014;466:425–431. doi: 10.1007/s00424-013-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sequeira V., Witjas-Paalberends E.R., Kuster D.W., van der Velden J. Cardiac myosin-binding protein C: hypertrophic cardiomyopathy mutations and structure-function relationships. Pflugers Arch. 2014;466:201–206. doi: 10.1007/s00424-013-1400-3. [DOI] [PubMed] [Google Scholar]
- 11.Moss R.L., Fitzsimons D.P., Ralphe J.C. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ. Res. 2015;116:183–192. doi: 10.1161/CIRCRESAHA.116.300561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marziliano N., Merlini P.A., Vignati G., Orsini F., Motta V., Bandiera L., Intrieri M., Veronese S. A case of compound mutations in the MYBPC3 gene associated with biventricular hypertrophy and neonatal death. Neonatology. 2012;102:254–258. doi: 10.1159/000339847. [DOI] [PubMed] [Google Scholar]
- 13.Ho C.Y., Lever H.M., DeSanctis R., Farver C.F., Seidman J.G., Seidman C.E. Homozygous mutation in cardiac troponin T: implications for hypertrophic cardiomyopathy. Circulation. 2000;102:1950–1955. doi: 10.1161/01.cir.102.16.1950. [DOI] [PubMed] [Google Scholar]
- 14.Maron B.J., Maron M.S., Semsarian C. Double or compound sarcomere mutations in hypertrophic cardiomyopathy: a potential link to sudden death in the absence of conventional risk factors. Heart Rhythm. 2012;9:57–63. doi: 10.1016/j.hrthm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Wessels M.W., Herkert J.C., Frohn-Mulder I.M., Dalinghaus M., van den Wijngaard A., de Krijger R.R., Michels M., de Coo I.F., Hoedemaekers Y.M., Dooijes D. Compound heterozygous or homozygous truncating MYBPC3 mutations cause lethal cardiomyopathy with features of noncompaction and septal defects. Eur. J. Hum. Genet. 2015;23:922–928. doi: 10.1038/ejhg.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lekanne Deprez R.H., Muurling-Vlietman J.J., Hruda J., Baars M.J., Wijnaendts L.C., Stolte-Dijkstra I., Alders M., van Hagen J.M. Two cases of severe neonatal hypertrophic cardiomyopathy caused by compound heterozygous mutations in the MYBPC3 gene. J. Med. Genet. 2006;43:829–832. doi: 10.1136/jmg.2005.040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin B., Puffenberger E., Tumbush J., Bockoven J.R., Wang H. Homozygosity for a novel splice site mutation in the cardiac myosin-binding protein C gene causes severe neonatal hypertrophic cardiomyopathy. Am. J. Med. Genet. A. 2007;143A:2662–2667. doi: 10.1002/ajmg.a.31981. [DOI] [PubMed] [Google Scholar]
- 18.Gedicke-Hornung C., Behrens-Gawlik V., Reischmann S., Geertz B., Stimpel D., Weinberger F., Schlossarek S., Précigout G., Braren I., Eschenhagen T. Rescue of cardiomyopathy through U7snRNA-mediated exon skipping in Mybpc3-targeted knock-in mice. EMBO Mol. Med. 2013;5:1128–1145. doi: 10.1002/emmm.201202168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mearini G., Stimpel D., Krämer E., Geertz B., Braren I., Gedicke-Hornung C., Précigout G., Müller O.J., Katus H.A., Eschenhagen T. Repair of Mybpc3 mRNA by 5′-trans-splicing in a Mouse Model of Hypertrophic Cardiomyopathy. Mol. Ther. Nucleic Acids. 2013;2:e102. doi: 10.1038/mtna.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang J., Wakimoto H., Seidman J.G., Seidman C.E. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mearini G., Stimpel D., Geertz B., Weinberger F., Krämer E., Schlossarek S., Mourot-Filiatre J., Stoehr A., Dutsch A., Wijnker P.J. Mybpc3 gene therapy for neonatal cardiomyopathy enables long-term disease prevention in mice. Nat. Commun. 2014;5:5515. doi: 10.1038/ncomms6515. [DOI] [PubMed] [Google Scholar]
- 22.Moretti A., Bellin M., Welling A., Jung C.B., Lam J.T., Bott-Flügel L., Dorn T., Goedel A., Höhnke C., Hofmann F. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 23.Lahti A.L., Kujala V.J., Chapman H., Koivisto A.P., Pekkanen-Mattila M., Kerkelä E., Hyttinen J., Kontula K., Swan H., Conklin B.R. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis. Model. Mech. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinson J.T., Chopra A., Nafissi N., Polacheck W.J., Benson C.C., Swist S., Gorham J., Yang L., Schafer S., Sheng C.C. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun N., Yazawa M., Liu J., Han L., Sanchez-Freire V., Abilez O.J., Navarrete E.G., Hu S., Wang L., Lee A. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gramlich M., Pane L.S., Zhou Q., Chen Z., Murgia M., Schötterl S., Goedel A., Metzger K., Brade T., Parrotta E. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol. Med. 2015;7:562–576. doi: 10.15252/emmm.201505047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan F., Lee A.S., Liang P., Sanchez-Freire V., Nguyen P.K., Wang L., Han L., Yen M., Wang Y., Sun N. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ojala M., Prajapati C., Pölönen R.P., Rajala K., Pekkanen-Mattila M., Rasku J., Larsson K., Aalto-Setälä K. Mutation-Specific Phenotypes in hiPSC-Derived Cardiomyocytes Carrying Either Myosin-Binding Protein C Or α-Tropomyosin Mutation for Hypertrophic Cardiomyopathy. Stem Cells Int. 2016;2016:1684792. doi: 10.1155/2016/1684792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka A., Yuasa S., Mearini G., Egashira T., Seki T., Kodaira M., Kusumoto D., Kuroda Y., Okata S., Suzuki T. Endothelin-1 induces myofibrillar disarray and contractile vector variability in hypertrophic cardiomyopathy-induced pluripotent stem cell-derived cardiomyocytes. J. Am. Heart Assoc. 2014;3:e001263. doi: 10.1161/JAHA.114.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger A., Maire S., Gaillard M.C., Sahel J.A., Hantraye P., Bemelmans A.P. mRNA trans-splicing in gene therapy for genetic diseases. Wiley Interdiscip. Rev. RNA. 2016;7:487–498. doi: 10.1002/wrna.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wally V., Murauer E.M., Bauer J.W. Spliceosome-mediated trans-splicing: the therapeutic cut and paste. J. Invest. Dermatol. 2012;132:1959–1966. doi: 10.1038/jid.2012.101. [DOI] [PubMed] [Google Scholar]
- 32.Wang D., Gao G. State-of-the-art human gene therapy: part II. Gene therapy strategies and clinical applications. Discov. Med. 2014;18:151–161. [PMC free article] [PubMed] [Google Scholar]
- 33.Collins M., Thrasher A. Gene therapy: progress and predictions. Proc. Biol. Sci. 2015;282:20143003. doi: 10.1098/rspb.2014.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varadi K., Michelfelder S., Korff T., Hecker M., Trepel M., Katus H.A., Kleinschmidt J.A., Müller O.J. Novel random peptide libraries displayed on AAV serotype 9 for selection of endothelial cell-directed gene transfer vectors. Gene Ther. 2012;19:800–809. doi: 10.1038/gt.2011.143. [DOI] [PubMed] [Google Scholar]
- 35.Lorain S., Peccate C., Le Hir M., Garcia L. Exon exchange approach to repair Duchenne dystrophin transcripts. PLoS ONE. 2010;5:e10894. doi: 10.1371/journal.pone.0010894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma D., Wei H., Lu J., Ho S., Zhang G., Sun X., Oh Y., Tan S.H., Ng M.L., Shim W. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 2013;34:1122–1133. doi: 10.1093/eurheartj/ehs226. [DOI] [PubMed] [Google Scholar]
- 37.Dambrot C., Braam S.R., Tertoolen L.G., Birket M., Atsma D.E., Mummery C.L. Serum supplemented culture medium masks hypertrophic phenotypes in human pluripotent stem cell derived cardiomyocytes. J. Cell. Mol. Med. 2014;18:1509–1518. doi: 10.1111/jcmm.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karakikes I., Stillitano F., Nonnenmacher M., Tzimas C., Sanoudou D., Termglinchan V., Kong C.W., Rushing S., Hansen J., Ceholski D. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat. Commun. 2015;6:6955. doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh R., Thomson K.L., Ware J.S., Funke B.H., Woodley J., McGuire K.J., Mazzarotto F., Blair E., Seller A., Taylor J.C. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monteiro da Rocha A., Guerrero-Serna G., Helms A., Luzod C., Mironov S., Russell M., Jalife J., Day S.M., Smith G.D., Herron T.J. Deficient cMyBP-C protein expression during cardiomyocyte differentiation underlies human hypertrophic cardiomyopathy cellular phenotypes in disease specific human ES cell derived cardiomyocytes. J. Mol. Cell. Cardiol. 2016;99:197–206. doi: 10.1016/j.yjmcc.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marston S., Copeland O., Gehmlich K., Schlossarek S., Carrier L. How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J. Muscle Res. Cell Motil. 2012;33:75–80. doi: 10.1007/s10974-011-9268-3. [DOI] [PubMed] [Google Scholar]
- 43.Vignier N., Schlossarek S., Fraysse B., Mearini G., Krämer E., Pointu H., Mougenot N., Guiard J., Reimer R., Hohenberg H. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ. Res. 2009;105:239–248. doi: 10.1161/CIRCRESAHA.109.201251. [DOI] [PubMed] [Google Scholar]
- 44.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 45.van Dijk S.J., Dooijes D., dos Remedios C., Michels M., Lamers J.M., Winegrad S., Schlossarek S., Carrier L., ten Cate F.J., Stienen G.J., van der Velden J. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 46.Tardiff J.C., Carrier L., Bers D.M., Poggesi C., Ferrantini C., Coppini R., Maier L.S., Ashrafian H., Huke S., van der Velden J. Targets for therapy in sarcomeric cardiomyopathies. Cardiovasc. Res. 2015;105:457–470. doi: 10.1093/cvr/cvv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wijnker P.J., Friedrich F.W., Dutsch A., Reischmann S., Eder A., Mannhardt I., Mearini G., Eschenhagen T., van der Velden J., Carrier L. Comparison of the effects of a truncating and a missense MYBPC3 mutation on contractile parameters of engineered heart tissue. J. Mol. Cell. Cardiol. 2016;97:82–92. doi: 10.1016/j.yjmcc.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Doroudgar S., Quijada P., Konstandin M., Ilves K., Broughton K., Khalafalla F.G., Casillas A., Nguyen K., Gude N., Toko H. S100A4 protects the myocardium against ischemic stress. J. Mol. Cell. Cardiol. 2016;100:54–63. doi: 10.1016/j.yjmcc.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X., Azhar G., Chai J., Sheridan P., Nagano K., Brown T., Yang J., Khrapko K., Borras A.M., Lawitts J. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H1782–H1792. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 50.Mooren F.C., Viereck J., Krüger K., Thum T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am. J. Physiol. Heart Circ. Physiol. 2014;306:H557–H563. doi: 10.1152/ajpheart.00711.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda N., Manabe I. Cellular Interplay between Cardiomyocytes and Nonmyocytes in Cardiac Remodeling. Int. J. Inflamm. 2011;2011:535241. doi: 10.4061/2011/535241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werfel S., Jungmann A., Lehmann L., Ksienzyk J., Bekeredjian R., Kaya Z., Leuchs B., Nordheim A., Backs J., Engelhardt S. Rapid and highly efficient inducible cardiac gene knockout in adult mice using AAV-mediated expression of Cre recombinase. Cardiovasc. Res. 2014;104:15–23. doi: 10.1093/cvr/cvu174. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi K., Okita K., Nakagawa M., Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 55.Ohnuki M., Takahashi K., Yamanaka S. Generation and characterization of human induced pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 2009;Chapter 4:Unit 4A.2. doi: 10.1002/9780470151808.sc04a02s9. [DOI] [PubMed] [Google Scholar]
- 56.Mannhardt I., Breckwoldt K., Letuffe-Brenière D., Schaaf S., Schulz H., Neuber C., Benzin A., Werner T., Eder A., Schulze T. Human Engineered Heart Tissue: Analysis of Contractile Force. Stem Cell Reports. 2016;7:29–42. doi: 10.1016/j.stemcr.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breckwoldt K., Letuffe-Brenière D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.