Abstract
Although reduced fetal growth in response to hypoxia has been appreciated for decades, we have a poor understanding of the effects of hypoxia on embryonic development and the underlying cellular and molecular mechanisms. Here we show that hypoxia treatment not only resulted in embryonic growth retardation but also caused significant delay in developmental speed and the timing of morphogenesis in vital organs of zebrafish. Hypoxia strongly induced the expression of insulin-like growth factor (IGF)-binding protein (IGFBP)-1, a secreted protein that binds IGFs in extracellular environments. Hypoxia did not change the expression levels of IGFs, IGF receptors, or other IGFBPs. The hypothesis that elevated IGFBP-1 mediates hypoxia-induced embryonic growth retardation and developmental delay by binding to and inhibiting the activities of IGFs was tested by loss- and gain-of-function approaches. Knockdown of IGFBP-1 significantly alleviated the hypoxia-induced growth retardation and developmental delay. Overexpression of IGFBP-1 caused growth and developmental retardation under normoxia. Furthermore, reintroduction of IGFBP-1 to the IGFBP-1 knocked-down embryos restored the hypoxic effects on embryonic growth and development. When tested in vitro with cultured zebrafish embryonic cells, IGFBP-1 itself had no mitogenic activity, but it inhibited IGF-1- and IGF-2-stimulated cell proliferation. This inhibitory effect was abolished when IGF-1 or IGF-2 was added in molar excess, suggesting that IGFBP-1 inhibits embryonic growth and development by binding to and inhibiting the activities of IGFs. The induction of IGFBP-1 expression may be a conserved physiological mechanism to restrict the IGF-stimulated growth and developmental process under hypoxic stress.
Keywords: zebrafish, heart development, craniofacial skeleton, morphogenesis, oxygen
Hypoxic stress causes major metabolic changes in all organisms requiring oxygen. Hypoxic stress also influences fetal growth and development and the pathogenesis of several human diseases, including intrauterine growth restriction (IUGR) (1–3). Reduced birth weight is also observed at high altitudes (4). IUGR not only increases fetal and neonatal morbidity and mortality, but also increases the risk of having adult diseases, such as cardiovascular disease, type-2 diabetes, obesity, and hypertension (5).
Recent evidence suggests that hypoxia may influence fetal growth through its connection to the insulin-like growth factor (IGF) signaling system. IGFs are well known fetal growth factors (6–9). Several groups have reported that the circulating level of IGF-binding protein (IGFBP)-1, a secreted protein that binds to IGF in extracellular environments, is elevated in IUGR fetuses (10–12) and that there is a striking inverse correlation between IGFBP-1 levels and fetal size (13). In addition, higher maternal serum IGFBP-1 levels are found at higher altitude (14). In vitro studies with cultured human cells and in vivo studies with mammalian animal models suggest that IGFBP-1 gene expression is elevated in hypoxic conditions (15–17) and that this up-regulation is mediated through the hypoxia-inducible factor (HIF)-1 pathway (16). Because IGFBP-1 binds IGFs with high affinity and inhibits IGF activities on cell growth in vitro (18, 19) and because IGFBP-1-overexpressing transgenic mice had reduced birth weight (20–22), it was postulated that the elevated IGFBP-1 plays a major role in hypoxia-caused IUGR by binding fetal IGFs and inhibiting their growth-promoting activities (3, 16, 17). This appealing model, however, has not been directly tested in vivo, and a causative relationship between the elevated IGFBP-1 expression and IUGR has not been established. Moreover, the impact of hypoxia on early developmental processes, such as morphogenesis, is poorly understood, and the role of IGFBP-1, if any, in mediating the hypoxic effects on embryonic development is unknown.
The zebrafish has now become an informative vertebrate model organism for the study of IGF signaling in early development (23). Zebrafish embryos develop externally, eliminating the complication of maternal compensation. Fast developing and transparent zebrafish embryos make it possible to manipulate environmental factors and observe the phenotypic changes in organ formation in real time. Furthermore, major components of the zebrafish IGF-signaling pathway, including IGF ligands, receptors, IGFBPs, and intracellular signal transduction network components, have been characterized, and they are highly conserved (23–26). Therefore, information gained from studying this unique animal model will not only provide new insight into zebrafish developmental biology but will also deepen our understanding of growth and developmental regulation in vertebrates in general.
The objective of this study was to determine the effects of hypoxia on embryonic development and growth and to examine the role of IGFBP-1 in mediating these hypoxic effects by using zebrafish embryos. Our studies suggest that hypoxia causes embryonic growth retardation and developmental delay and that IGFBP-1 plays a key role in mediating these hypoxic effects.
Materials and Methods
Materials. All chemicals and reagents were purchased from Fisher Scientific (Pittsburgh, PA) unless noted otherwise. Human IGF-1 and IGF-2 were purchased from GroPep (Adelaide, Australia). RNA polymerase and RNase-free DNase were purchased from Promega (Madison, WI). Restriction endonucleases were purchased from New England BioLabs (Beverly, MA). Superscript II reverse transcriptase and oligonucleotide primers were purchased from Invitrogen Life Technologies. Morpholino-modified oligonucleotides (MOs) were purchased from Gene Tools (Corvalis, OR).
Experimental Animals and Procedures. Wild-type zebrafish (Danio rerio) were maintained on a 14-h light/10-h dark cycle at 28°C and fed twice daily. Embryos were obtained by natural cross. Fertilized eggs were raised in embryo medium at 28.5°C and staged according to Kimmel et al. (27). To inhibit embryo pigment formation, embryo medium was supplemented with 0.003% (wt/vol) 2-phenylthiourea. The dissolved oxygen level of the system water under ambient conditions (normoxia) was ≈6.5 ± 0.5 mg/liter. For the hypoxia treatment, oxygen level was reduced to 0.6 ± 0.1 mg/liter (hypoxia) by bubbling nitrogen gas into water. After embryos of various stages were transferred, the container was sealed and the dissolved oxygen levels were monitored by a dissolved oxygen meter (SonTek/YSI model 58, Fisher Scientific). After 18–24 h of hypoxia treatment, the embryos were fixed in 4% paraformaldehyde in 1× PBS and stored at –20°C in 100% methanol or frozen in liquid nitrogen and stored at –80°C for further analysis.
Whole-Mount in Situ Hybridization and RT-PCR Analysis. Whole-mount in situ hybridization with digoxigenin-labeled RNA riboprobe was performed as reported in ref. 25. For RT-PCR analysis, total RNA was extracted by using TriReagent (Molecular Research Center, Cincinnati). After DNase treatment (Invitrogen), the RNA (2.5 μg) was reverse-transcribed with random hexamer primers and SuperScript II reverse transcriptase (Invitrogen). Sequences of primers used for RT-PCR are shown in Table 1, which is published as supporting information on the PNAS web site. PCR cycles and amounts of templates were optimized for each primer set in pilot experiments. PCR cycles were as follows: 94°C for 2 min, 32 cycles (IGFBP-5 and IGF-2) or 35 cycles (IGFBP-1, IGFBP-2, IGFBP-3, IGF-1, and IGF-1 receptor-a) of 94°C for 30 sec, 57°C for 30 sec, and 72°C for 1 min. PCR products were analyzed by electrophoresis followed by ethidium bromide staining. The levels of ornithinedecarboxylase mRNA was measured and used as an internal control (28).
Determining the IGFBP-1 Gene Structure. The zebrafish IGFBP-1 genomic structure was determined by searching the zebrafish genome database and PCR analysis. Genomic DNA isolated from adult zebrafish (AB-line) was used for genomic PCR analysis. The amplified PCR product was subcloned to PCR2.1-TOPO vector (Invitrogen) and sequenced.
Morpholino Knockdown. Two MOs were designed for targeting IGFBP-1 exon1/intron1 and exon3/intron 3 boundaries, and a standard control MO was used in all experiments (Table 2, which is published as supporting information on the PNAS web site). The MOs were dissolved in 1× Danieau Solution (27) and injected into 1–2 cell stage embryos by using a PV830 pneumatic pico pump and glass pipettes (World Precision Instruments, Sarasota, FL). To determine the efficiency and specificity of MO knockdown, total RNA was extracted from the injected embryos at various time points and RT-PCR was conducted as described above. The IGFBP-1 PCR products were subcloned into PCR2.1-TOPO vector (Invitrogen) and sequenced.
Overexpression of IGFBP-1. A DNA fragment containing the entire ORF of zebrafish IGFBP-1 was generated by PCR with a primer set (5′-GGC GGATCC GCC GCC ACC ATG AAC AGA CTG CTT CTG-3′/5′-CCC GGA TCC GTG GTT GAG TTC CTC GGG-3′). This DNA fragment was subcloned into the pCS 2+ vector to generate the IGFBP-1–EGFP fusion protein. The construct was verified by DNA sequencing. To express IGFBP-1–EGFP, linearized plasmid (75 pg) was injected into embryos with or without 2 ng of IGFBP-1 MO1. Injected embryos were raised to 36 h postfertilization (hpf) and homogenized for Western immunoblotting and ligand blotting as described in refs. 24 and 29.
Cell Culture and BrdUrd Incorporation Assay. Zebrafish embryonic (ZF4) cells, obtained from American Type Culture Collection, were grown in a 1:1 mixture of Ham's F12 medium and DMEM with penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% FBS at 28.5°C. A BrdUrd incorporation assay was performed as reported in ref. 30.
Statistics. Values are expressed as means ± SE. Differences among groups were analyzed by one- or two-way ANOVA followed by Fisher's protected least significant difference by using statview software (SAS Institute, Cary, NC). Significance was accepted at P < 0.05.
Results
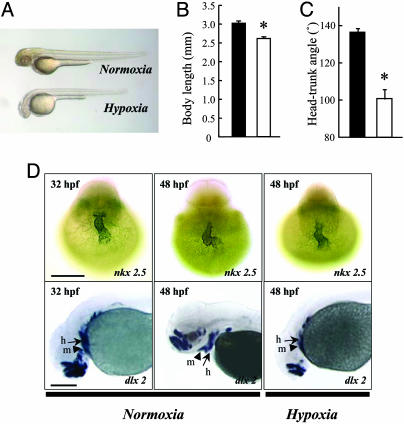
Hypoxia Causes Growth Retardation and Developmental Delay in Zebrafish Embryos. As shown in Fig. 1 A and B, 24-h hypoxia treatment significantly decreased the body length. Hypoxia treatment also caused a significant reduction in the development rate, as indicated by the reducing head–trunk angles (HTA) (Fig. 1C), which is a quantitative parameter for determining the developmental stage 24–48 hpf (i.e., during the pharyngula period) (27). Based on the HTA data, 48-hpf embryos in the hypoxia group were developmentally equivalent to control embryos at 32 hpf. Similar experiments were carried out by using 6-hpf embryos. Hypoxia treatment for 18 h significantly reduced the somite number from 30.2 ± 1.0 (n = 38) of the control group to 21.4 ± 0.7 (n = 42, P < 0.001) of the hypoxia group. The somite number is used for staging embryos 10–24 hpf (i.e., during the segmentation period) (27). Embryos at 24 hpf in the hypoxia group were therefore developmentally equivalent to control embryos at 19.5 hpf.
Fig. 1.
Hypoxia causes growth retardation and developmental delay. (A) Morphology of wild-type zebrafish embryos at 48 hpf after 24 h of normoxia or hypoxia treatment. (B and C) Effect of hypoxia on embryonic growth and development. Embryos at 24 hpf were transferred to normal (filled bars) or hypoxic (open bars) water. After 24 h, the total body length (B) and HTA (C) were measured. Values are expressed as means ± SE. (n = 28–36). *, P < 0.05. (D) Effect of hypoxia treatment on heart and head skeleton morphogenesis. Embryos raised in water with normal oxygen (normoxia) were fixed at 32 and 48 hpf. A subset of embryos was subjected to 24-h hypoxia exposure beginning at 24 hpf. Whole-mount in situ hybridization was performed by using nkx 2.5 (Upper) and dlx 2 (Lower) riboprobes. h, hyroid arch (arrows); m, mandibular arch (arrow heads). (Scale bar, 200 μm.)
We next investigated the effect of hypoxia on heart morphogenesis by examining the mRNA expression pattern of nkx 2.5, a cardiac marker (31). In zebrafish, the shape of the heart changes from a tube-like structure to a looped structure between 24 and 48 hpf under normoxia (Fig. 1D Upper). Under hypoxia, this looping process was significantly delayed. As shown in Fig. 1D, embryos exposed to hypoxia have yet to complete the cardiac looping, and their hearts remained a tube structure at 48 hpf, resembling that of the control embryos at 32 hpf. The appearance of pharyngeal arches is another well timed morphogenesis event in the pharyngula period and can be monitored by the mRNA expression patterns of dlx 2, a homeobox transcription factor (27). Under normoxia, dlx 2 expression was detected in the primordial region of the budding mandibular (Fig. 1D Lower, arrow heads) and hyroid arches (Fig. 1D Lower, arrows) at 32 hpf. At 48 hpf, embryos developed distinct branchial arches. In comparison, the mandibular arch and hyroid of embryos on the hypoxia group were located far posterior to the forebrain at 48 hpf, similar to embryos of the normoxia group at 32 hpf. These results suggest that hypoxia not only results in growth retardation but also causes significant delay in embryonic development and the timing of heart and head skeleton morphogenesis.
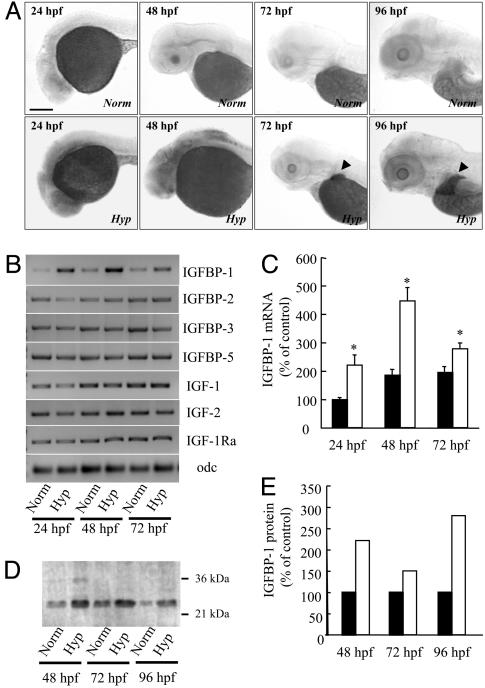
Hypoxia Significantly Increases IGFBP-1 mRNA and Protein Levels. The IGFBP-1 mRNA expression levels were low under normoxia in all examined stages (Fig. 2A). Hypoxia treatment increased IGFBP-1 expression in different spatial domains at different developmental stages. In early embryos (24 and 48 hpf), hypoxia stimulated ubiquitous expression of IGFBP-1. In advanced embryos (72 and 96 hpf), the hypoxia-induced and basal IGFBP-1 expression became liver-specific (Fig. 2A Lower). Semiquantitative RT-PCR analysis showed that hypoxia significantly increased IGFBP-1 mRNA levels at all of the time points examined (Fig. 2 B and C). Ligand blotting analysis revealed that the increase in IGFBP-1 mRNA levels was accompanied by increased functional IGFBP-1 protein levels (Fig. 2 D and E). No significant changes in the expression levels of IGFBP-2, IGFBP-3, IGFBP-5, IGF-1, IGF-2, and IGF-1 receptor-a were detected (Fig. 2B). These data suggest that hypoxia treatment significantly increased IGFBP-1 mRNA and protein levels but not those of other components of the IGF-signaling system.
Fig. 2.
Hypoxia induces IGFBP-1 expression. (A) Hypoxia increased IGFBP-1 mRNA expression in many tissues in early embryonic stages (24 and 48 hpf) and in the liver (indicated by arrow heads) in advanced stages (72 and 96 hpf). Zebrafish embryos at various stages were transferred to normal or hypoxic water. After 24 h, the embryos were fixed and subjected to in situ hybridization analysis. (Scale bar, 200 μm.) (B) RT-PCR analysis of IGFBP-1, IGFBP-2, IGFBP-3, IGFBP-5, IGF-1, IGF-2, IGF-1 receptor-a (IGF-1Ra), and ornithinedecarboxylase (odf) in embryos of the indicated stages. Norm, normoxia; Hyp, hypoxia. (C) Relative IGFBP-1 mRNA levels normalized by the ornithinedecarboxylase mRNA level. Filled bars represent the normoxia group, and open bars represent the hypoxia group. Values are expressed as means ± SE. (n = 6). *, P < 0.05 compared with the corresponding normoxia group. (D) Western ligand blotting analysis. Wild-type embryos at various indicated stages were subjected to 24 h of hypoxia or normoxia treatment. The levels of IGFBP-1 protein were determined by ligand blotting with digoxigenin-labeled IGF-1. (E) Densitometric analysis results of D. Filled bars represent the normoxia group, and open bars represent the hypoxia group.
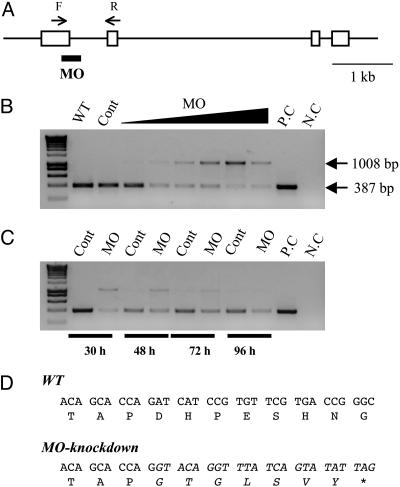
Knockdown of IGFBP-1 Abrogates Hypoxia-Induced Growth Retardation and Developmental Delay. Splice-site-targeting MOs have been proven to be an efficient and specific way to “knock down” targeted genes in zebrafish embryos by altering premRNA splicing (28). This approach has the advantage of being verifiable by RT-PCR and thus eliminates the need for specific antibodies. To design specific MOs, we first determined the zebrafish IGFBP-1 gene structure by searching the zebrafish genome database followed by genomic PCR and sequencing analysis. The zebrafish IGFBP-1 gene spans 5 kb in the genome (Fig. 3A) and is composed of four exons (exon 1, 465 bp; exon 2, 158 bp; exon 3, 129 bp, and exon 4, 254 bp) and three introns (intron 1, 621 bp; intron 2, ≈3,199 bp; and intron 3, 212 bp).
Fig. 3.
Knockdown of functional IGFBP-1 with splice-site-targeting MOs. (A) Structure of the zebrafish IGFBP-1 gene. Exons are shown as boxes, and introns are shown as lines. The splice donor site targeted by the IGFBP-1 MO is underlined. The primer set used for RT-PCR is shown as arrows. F, forward primer; R, reverse primer. (Scale bar, 1 kb.) (B) Dose-dependent effect after 0.5, 1, 2, 4, 8, or 16 ng MO1 was injected into embryos. The specific primer set amplified a 387-bp spliced form of IGFBP-1, whereas MO knockdown caused an aberrant splice form (1,008 bp). WT, wild-type; Cont, control MO; P.C., positive control of PCR; N.C., negative control of PCR. (C) Time-course effect. MO1 at dose of 2 ng was injected into embryos, and the knockdown effect was analyzed by RT-PCR at 30, 48, 72, and 96 h after the injection. (D) Sequence analysis of the IGFBP-1 cDNAs isolated from the wild-type and MO1-injected embryos. The sequence of the aberrantly spliced form is italicized. Note that the aberrant splicing form results in a coding-frame shift and a premature stop codon.
Two MOs were designed to target the IGFBP-1 exon 1/intron 1 and exon 3/intron 3 boundaries, respectively. Each MO was injected into embryos at various doses (0.5, 1, 2, 4, 8, and 16 ng) and the successful knockdown was confirmed by RT-PCR. The results showed that injection of IGFBP-1 MO1 (the exon 1/intron 1 MO) at doses >1 ng resulted in the reduction of endogenous IGFBP-1 (387 bp) and an aberrant splice form (1,008 bp) (Fig. 3B). The knockdown effect lasted until at least 96 hpf (Fig. 3C). The exon 3/intron 3 MO2 also efficiently altered splicing, although a higher dose was required (data not shown). Zebrafish IGFBP-1 consists of 262 amino acids, including a 25-aa signal peptide and a 237-aa mature protein that can be divided into a N-terminal domain, middle linker domain, and C-terminal domain (26). Sequence analysis of the altered splicing product revealed an insertion of 621 bp into intron 1, which caused a reading-frame shift and introduced a premature stop codon (Fig. 3D). As a result, the encoded protein is 125 aa and therefore lacks part of the middle linker domain and all of the C-terminal domain. This truncated IGFBP-1 is predicted to be nonfunctional, because deletion of the C-terminal 20 aa or residue changes in this region completely abolish IGF binding (32). This MO knockdown of IGFBP-1 is highly specific given that RT-PCR analysis revealed that there were no aberrant splice forms nor significant changes in the abundance of other IGFBPs, including IGFBP-2, IGFBP-3, and IGFBP-5 (Fig. 6, which is published as supporting information on the PNAS web site).
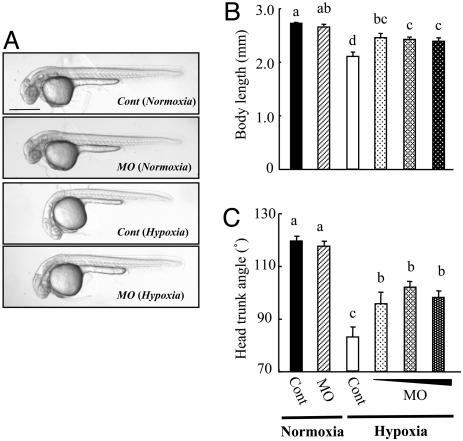
To test whether the elevated IGFBP-1 plays any role in mediating hypoxia-induced growth and developmental retardation, the IGFBP-1 MO1 or control MO was injected into embryos at the 1- to 2-cell stage. As shown in Fig. 4A, the hypoxia treatment caused severe growth retardation and developmental delay indicated by the smaller body size and reduced HTA. Knockdown of IGFBP-1 alleviated these hypoxic effects by 43–54% and 48–65%, based on the body length and HTA data (Fig. 4 B and C). Injection of MO2 had similar effects. Embryos in the IGFBP-1 knockdown group under normoxia were morphologically indistinguishable from those of the control group under normoxia (Fig. 4 B and C), suggesting that knockdown of IGFBP-1 had little effect in growth and development under normoxia. This observation also indicates that the IGFBP-1 MO at the concentration used had little, if any, side effect.
Fig. 4.
Knockdown of IGFBP-1 partially abrogates hypoxia-caused growth retardation and developmental delay. (A) Morphology of control MO- (Cont) or IGFBP-1 MO-injected embryos at 36 hpf. After raised to 12 hpf in normoxic water, the injected embryos were transferred to normal (normoxia) or hypoxic (hypoxia) water for 24 h. (Scale bar, 500 μm.) (B and C) Knockdown of IGFBP-1 attenuated hypoxia-induced growth and developmental retardations but had no effect under normoxia. Embryos injected with 2 ng of control MO or 1, 2, or 4 ng of MO1 were raised to 12 hpf and then transferred to normal or hypoxic water. After 24 h, their body lengths (B) and HTA (C) were measured. Values are expressed as means ± SE (n = 20–30). Groups with the same letters (a, b, c, or d) are not significantly different from each other (P < 0.05).
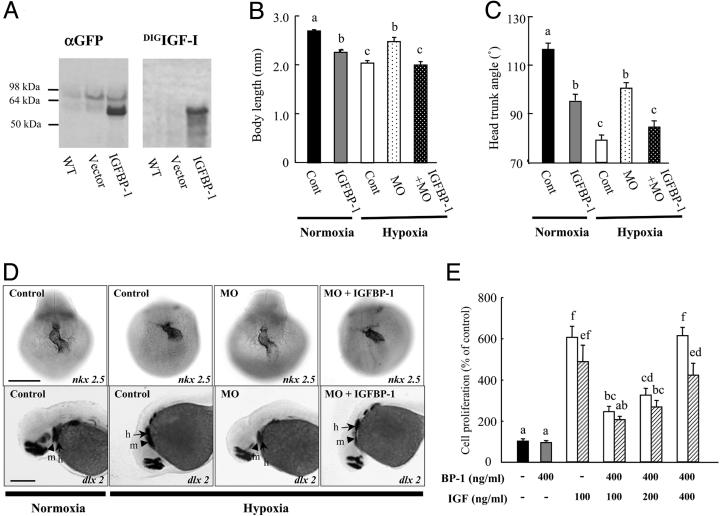
Overexpression of IGFBP-1 Inhibits Embryonic Growth and Development Under Normoxia and Restores the Hypoxic Effects in the Knockdown Background. To further test the hypothesis through a gain-of-function approach, we generated an IGFBP-1–EGFP expression construct. The transcript resulting from this construct is intronless and therefore resistant to the IGFBP-1 MOs. Western immunoblotting analysis with a GFP antibody detected a major band at 55 kDa, corresponding to the IGFBP-1–EGFP fusion protein in these embryos (Fig. 5A Left). This protein was not detected in wild-type or vector-injected embryos. Western ligand blotting analysis indicated that IGFBP-1–EGFP is a functional IGFBP (Fig. 5A Right). This binding is specific because the band was completely displaced by the addition of an excess amount of unlabeled IGF-1 (data not shown).
Fig. 5.
IGFBP-1 inhibits embryonic growth and development by inhibiting IGF actions. (A) Expression of a functional IGFBP-1 fusion protein. Embryos were injected with the IGFBP-1–EGFP construct (IGFBP-1) or empty vector (vector). At 36 hpf, the embryos were subjected to immunoblot (Left) and ligand blot analysis (Right). WT, wild-type embryos; vector, empty-vector-injected embryos; IGFBP-1, embryos injected with the IGFBP-1–EGFP construct. (B and C) Effect of IGFBP-1 overexpression under normoxia and hypoxia. Embryos injected with control MO (Cont), IGFBP-1 MO1 (MO), or IGFBP-1 MO plus IGFBP-1–EGFP overexpression vector at 12 hpf were transferred to normal (normoxia) or hypoxic water (hypoxia). The body length (B) and HTA (C) were measured 24 h later. Values are expressed as means ± SE (n = 20–34). Groups with common letters are not significantly different from each other (P < 0.05). (D) MO knockdown abrogates and IGFBP-1 overexpression restores hypoxia-induced delay in head skeleton and heart morphogenesis. Embryos injected with control MO, IGFBP-1 MO, or IGFBP-1 MO plus IGFBP-1–EGFP overexpression vector were transferred to normal or hypoxic water for 24 h and subjected to whole-mount in situ hybridization with nkx 2.5 (Upper) and dlx 2 (Lower). h, hyroid arch (arrows); m, mandibular arch (arrow heads). (Scale bar, 200 μm.) (E) IGFBP-1 inhibits IGF-stimulated zebrafish embryonic cell proliferation. Serum-starved, confluent cells were exposed to IGFBP-1 (400 ng/ml) with or without various concentrations of IGF-1 (open bars) or IGF-2 (shadow bars) in the presence of BrdUrd (20 μM) for 24 h. BrdUrd-labeled cells were detected by immunostaining. Values are expressed as means ± SE of two independent experiments, each of which was performed in duplicates. Groups with the same letters (a, b, c, d, e, or f) are not significantly different from each other (P < 0.05).
To determine whether elevated IGFBP-1 mediates the hypoxia effect, a series of gain-of-function studies were performed. As shown in Fig. 5B, overexpression of IGFBP-1 reduced embryo size to 83% of the control under normoxia. Overexpression of IGFBP-1 also slowed down the developmental rate, as indicated by the significantly reduced HTA (Fig. 4C). The body length in the IGFBP-1-plus-MO group was indistinguishable from that of the control group under hypoxia and significantly smaller than the MO group in hypoxia. Likewise, IGFBP-1 overexpression reversed the effects of MO in development as indicated by the reduction in HTA (Fig. 5C).
We also examined the effects of loss and gain of IGFBP-1 on heart and head skeleton development. As shown in Fig. 5D, the looping of the developing heart and the positions of the mandibular and hyroid arches in the MO group under hypoxia were similar to those of the control group under normoxia. Overexpression of IGFBP-1–EGFP restored these delays. The positions of the mandibular and hyroid arches and the heart structure of the IGFBP-1-plus-MO group were similar to those of the control group in hypoxia.
IGFBP-1 Inhibits IGF Action in Cultured Embryonic Cells. The mode of action of IGFBP-1 was examined in vitro by using cultured zebrafish embryonic cells. As shown in Fig. 5E, IGF-1 or IGF-2 (100 ng/ml) resulted in a 6.0 ± 0.5- and 4.9 ± 0.8-fold increases in the number of divining cells over the control, respectively (P < 0.05). Addition of purified fish IGFBP-1 at an equimolar concentration (400 ng/ml) inhibited IGF-1- or IGF-2-stimulated cell proliferation by 60% and 58%, respectively. These inhibitory effects were statistically significant (P < 0.05). IGFBP-1 itself had no effect on cell proliferation (Fig. 5E), suggesting that IGFBP-1 does not directly regulate zebrafish cell proliferation. We postulated that IGFBP-1 might act by binding to and sequestering the actions of IGFs. To test this idea, IGF and IGFBP-1 were added to the cells at a 2:1 and 4:1 molar ratio. As shown in Fig. 5E, the inhibitory effect of IGFBP-1 on cell proliferation was diminished when IGF-1 or IGF-2 was added to the cells at a 4:1 molar ratio.
Discussion
In this study, we show that chronic hypoxia treatment results in significant embryonic growth retardation and developmental delay in zebrafish, concomitant with a significant increase in IGFBP-1 mRNA and protein levels. Overexpression of IGFBP-1 reduced the growth and development rate under normoxia conditions, and targeted knockdown of IGFBP-1 abrogated the hypoxia-caused growth retardation and developmental delay. Furthermore, reintroduction of a MO-resistant IGFBP-1 to the IGFBP-1 knockdown embryos restored the hypoxia effects. These findings demonstrate a clear causative relationship between elevated IGFBP-1 expression and hypoxia-induced embryonic growth and developmental retardation and have provided unequivocal evidence supporting the hypothesis that IGFBP-1 plays a key role in mediating the hypoxic effects on embryonic growth and development.
Our data show that oxygen availability has a profound impact on embryonic growth. Hypoxia exposure for 18–24 h significantly reduced body size, as it does to human and other mammalian fetuses. Our results further reveal that hypoxia also causes significant delays in embryonic development. This conclusion is supported by the reduced somite number and HTA and by the delayed heart and craniofacial skeleton morphorgenesis. Whole-mount in situ hybridization suggests that hypoxia delays the morphogenesis of these organs but does not cause patterning abnormality. In fact, the hypoxia-caused growth and developmental retardation is a reversible process. After transferring back to normal oxygen environment, the embryos can “catch up” and become nearly indistinguishable from the control group (S.K. and C.D., unpublished data). This catch-up growth pattern is also observed in IUGR infants and rodent models, and the catch-up growth is often associated with higher serum IGF-1 levels (33, 34). In contrast, zebrafish embryos enter suspended animation, and the developmental processes and cell division are stopped under anoxia (35).
In developing zebrafish embryos, IGFBP-1 mRNA is expressed at very low levels in many tissues in earlier stages but become liver-specific in advanced embryos after the liver primordium is formed (26). The induction of IGFBP-1 expression by hypoxia is operative at both stages: first in multiple embryonic tissues and then only in the liver. The induction appears to be specific to IGFBP-1 because all of the other IGF-signaling system components examined, including IGF-1, IGF-2, IGFBP-2, IGFBP-3, IGFBP-5, and IGF-1 receptor, show little change under hypoxia. These findings are consistent with previous studies performed in mammalian and teleost species (15–17, 26, 36). No significant changes were found in serum levels of IGF-1 and IGF-2 under hypoxia in ovine fetus (15). Although immunoreactive IGF-1 and IGF-2 were detected in teleost embryonic tissues by immunohistochemistry (37, 38), it is unclear whether hypoxia changes the levels of IGF peptides. A well characterized pathway of hypoxia-induced gene expression is the transcription factor HIF-1 and its close relatives HIF-2 and HIF-3 (39). To date, many hypoxia-regulated genes have been reported in mammals, and the majority of these genes appear to be under HIF-1 control (39). Tazuke et al. (16) reported that the human IGFBP-1 gene contains three consensus HIF response elements (HRE) in its intron 1, one of which is functional and responsible for its hypoxia-induced up-regulation when tested in cultured human hepatoma (HepG2) cells. Zebrafish IGFBP-1 promoter contains several putative HIF response elements (40). In addition, treatment of zebrafish embryos with CoCl2, a chemical inducer of HIF-1, significantly increased IGFBP-1 mRNA levels (S.K. and C.D., unpublished data), suggesting that the HIF pathway may be involved in the induction of IGFBP-1 expression by hypoxia in zebrafish embryos.
Our gain- and loss-of-function analysis results provide strong evidence supporting the theory that IGFBP-1 plays an important role in mediating hypoxia-caused growth and developmental retardation. Overexpression of a functional IGFBP-1 fusion protein reduced the embryo size to 83% of the control size under normoxia. This result is similar to the reduced birth size in IGFBP-1 overexpressing mice (20, 21). Overexpression of IGFBP-1 also slowed down the developmental rate to a degree similar to that of the hypoxia treatment. Therefore, IGFBP-1 is sufficient to induce growth retardation and developmental delay. Furthermore, targeted knockdown of IGFBP-1 significantly alleviated the hypoxia-induced growth retardation and developmental delay. To our knowledge, this study is the first to provide in vivo evidence for a requirement of IGFBP-1 in mediating hypoxia-caused growth and developmental retardation in a vertebrate embryo. Knockdown of IGFBP-1 significantly but only partially (43–65%) alleviated the hypoxia-caused growth retardation and developmental delay. This observation indicates that IGFBP-1 is probably one of many genes involved in hypoxia-induced embryonic growth and developmental retardation. In support of this view, a recent microarray analysis study reports a number of genes known to be important in cell cycle (cyclin G1 and histone 3), protein synthesis (ribosomal proteins), and metabolism regulation (ATP synthase) are regulated by hypoxia in zebrafish embryos (41).
Numerous in vitro studies have shown that IGFBP-1 inhibits IGF actions in cultured human cells and in transgenic mice (18–22, 30). In this study, we show that fish IGFBP-1 inhibited IGF-stimulated zebrafish embryonic cell proliferation, although fish IGFBP-1 itself had no mitogenic activity. This inhibitory effect was abolished when IGF-1 or IGF-2 was added in molar excess. Because the IGF-1 receptor mediates the biological actions of IGFs and because IGFBP-1 bind to IGFs with high affinity, the inhibitory effects of IGFBP-1 can be rationally attributed to its competition for the ligand with the IGF-1 receptor (42). The lack of ligand-independent activity in fish IGFBP-1 is consistent with the fact that fish IGFBP-1 does not contain an RGD motif (26, 43). In contrast, human IGFBP-1 contains a functional RGD motif, which can interact with integrin and is responsible for its IGF-independent activity on cell migration (44).
It is of interest to note that knock down of IGFBP-1 did not alter the growth and development rate under normoxia condition. This observation is consistent with recent reports showing that IGFBP-1 knockout mice exhibited normal growth and development under a normal oxygen environment (45, 46). This lack of effect is in agreement with the observation that IGFBP-1 mRNA and protein levels are low under normal oxygen conditions. Therefore, the growth and developmental inhibition by IGFBP-1 may be in an “off” mode when there is ample oxygen, thus favoring fast growth and development. The induction of IGFBP-1 expression by hypoxia may be an evolutionally conserved physiological mechanism to restrict the IGF-stimulated growth and developmental process under stressful conditions. In addition to hypoxia, the IGFBP-1 gene is also highly responsive to food deprivation, malnutrition, stress, and chronic diseases in a wide variety of vertebrate species ranging from teleosts to humans (18, 47). Because these stressful and catabolic conditions lead to adaptive changes in metabolic reorganization, such as the activation of the anaerobic ATP-generating pathway (glycolysis), the adaptive significance of the IGFBP-1 up-regulation may be to divert important energy resources away from growth and development toward those metabolic processes essential for survival.
Supplementary Material
Acknowledgments
We thank H. Hirata and M. Shimizu for suggestions and discussions; P. S. Schlueter, E. J. Clowney, B. Beckman, and K. Cadigan for critical reading of the manuscript; and M. Ekker (University of Ottawa, Ottawa ON, Canada), V. Prince (University of Chicago, Chicago), and S. Lynes (University of Michigan, Ann Arbor) for providing reagents. This work was supported by National Science Foundation Grant IBN 0110864 (to C.D.). S.K. was supported by Japan Society for Promotion of Science Fellowship 11824.
Author contributions: S.K. and C.D. designed research; S.K. performed research; S.K., K.A., and C.D. analyzed data; and S.K. and C.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IUGR, intrauterine growth restriction; IGF, insulin-like growth factor; IGFBP, IGF-binding protein; HIF, hypoxia-inducible factor; MO, morpholino oligonucleotide; HTA, head–trunk angle; hpf, hours postfertilization.
References
- 1.Semenza, G. L. (2000) Gene Dev. 14, 1983–1991. [PubMed] [Google Scholar]
- 2.Le, Q. T., Denko, N. C. & Giaccia, A. J. (2004) Cancer Metastasis Rev. 23, 293–310. [DOI] [PubMed] [Google Scholar]
- 3.Giudice, L.C. (2002) J. Clin. Invest. 110, 307–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore, L. G. (2003) High Alt. Med. Biol. 4, 141–156. [DOI] [PubMed] [Google Scholar]
- 5.Ong, K. K. & Dunger, D. B. (2002) Best Pract. Res. Clin. Endocrinol. Metab. 16, 191–207. [DOI] [PubMed] [Google Scholar]
- 6.Woods, K. A., Camacho-Hubner, C., Savage, M. O. & Clark, A. J. L. (1996) N. Engl. J. Med. 335, 1363–1367. [DOI] [PubMed] [Google Scholar]
- 7.Abuzzahab, M. J., Schneider, A., Goddard, A., Grigorescu, F., Lautier, C., Keller, E., Kiess, W., Klammt, J., Kratzsch, J., Osgood, D., et al. (2003) N. Engl. J. Med. 349, 2211–2222. [DOI] [PubMed] [Google Scholar]
- 8.Liu, J. P., Baker, J., Perkins, A. S., Robertson, E. J. & Efstratiadis, A. (1993) Cell 75, 59–72. [PubMed] [Google Scholar]
- 9.Baker, J., Liu, J. P., Robertson, E. J. & Efstratiadis, A. (1993) Cell 75, 73–82. [PubMed] [Google Scholar]
- 10.Unterman, T., Simmons, R. A., Glick, R. P. & Ogata, E. S. (1993) Endocrinology 132, 327–336. [DOI] [PubMed] [Google Scholar]
- 11.Unterman, T., Lascon, R., Gotway, M. B., Oehler, D., Gounis, A., Simmons, R. A. & Ogata, E. S. (1990) Endocrinology 127, 2035–2037. [DOI] [PubMed] [Google Scholar]
- 12.Giudice, L. C., de Zegher, F., Gargosky, S. E., Dsupin, B. A., de las Fuentes, L., Crystal, R. A., Hintz, R. L. & Rosenfeld, R. G. (1995) J. Clin. Endocrinol. Metab. 80, 1548–1555. [DOI] [PubMed] [Google Scholar]
- 13.Fant, M., Salafia, C., Baxter, R. C., Schwander, J., Vogel, C., Pezzullo, J. & Moya, F. (1993) Regul. Pept. 48, 29–39. [DOI] [PubMed] [Google Scholar]
- 14.Krampl, E., Kametas, N. A., McAuliffe, F., Cacho-Zegarra, A. M. & Nicolaides, K. H. (2002) Obstet. Gynecol. 99, 594–598. [DOI] [PubMed] [Google Scholar]
- 15.McLellan, K. C., Hooper, S. B., Bocking, A. D., Delhanty, P. J. D., Phillips, I. D., Hill, D. J. & Han, V. K. M. (1992) Endocrinology 131, 1619–1628. [DOI] [PubMed] [Google Scholar]
- 16.Tazuke, S. I., Mazure, N. M., Sugawara, J., Carland, G., Faessen, G. H., Suen, L. F., Irwin, J. C., Powell, D. R., Giaccia, A. J. & Giudice, L. C. (1998) Proc. Natl. Acad. Sci. USA 95, 10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popovici, R. M., Lu, M., Bhatia, S., Faessen, G. H., Giaccia, A. J. & Giudice, L. C. (2001) J. Clin. Endocrinol. Metab. 86, 2653–2659. [DOI] [PubMed] [Google Scholar]
- 18.Clemmons, D. R. (2001) Endocr. Rev. 22, 800–817. [DOI] [PubMed] [Google Scholar]
- 19.Firth, S. M. & Baxter, R. C. (2002) Endocr. Rev. 23, 824–854. [DOI] [PubMed] [Google Scholar]
- 20.Rajkumar, K., Barron, D., Lewitt, M. S. & Murphy, L. J. (1995) Endocrinology 136, 4029–4034. [DOI] [PubMed] [Google Scholar]
- 21.Gay, E., Seurin, D., Babajko, S., Doublier, S., Cazillis, M. & Binoux, M. (1997) Endocrinology 138, 2937–2947. [DOI] [PubMed] [Google Scholar]
- 22.Crossey, P. A., Pillai, C. C. & Miell, J. P. (2002) J. Clin. Invest. 110, 41–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan, C., Ding, J., Schlueter, P. J., Li, Y., Zhang, J. & Royer, T. (2003) Acta Zool. Sinica 49, 421–431. [Google Scholar]
- 24.Duan, C., Ding, J., Li, Q., Tsai, W. & Pozios, K. C. (1999) Proc. Natl. Acad. Sci. USA 96, 15274–15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maures, T., Chan, S. J., Xu, B., Sun, H., Ding, J. & Duan, C. (2002) Endocrinology 143, 1858–1871. [DOI] [PubMed] [Google Scholar]
- 26.Maures, T. & Duan, C. (2002) Endocrinology 143, 2722–2731. [DOI] [PubMed] [Google Scholar]
- 27.Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. (1995) Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- 28.Draper, B. W., Morcos, P. A. & Kimmel, C. B. (2001) Genesis 30, 154–156. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu, M., Swanson, P., Fukada, H., Hara, A. & Dickhoff, W. W. (2000) Gen. Comp. Endocrinol. 119, 26–36. [DOI] [PubMed] [Google Scholar]
- 30.Bauchat, J. R., Busby, W. H., Jr., Garmong, A., Swanson, P., Moore, J., Lin, M. & Duan, C. (2001) J. Endocrinol. 170, 619–628. [DOI] [PubMed] [Google Scholar]
- 31.Lee, K. H., Xu, Q. & Breitbart, R. E. (1996) Dev. Biol. 180, 722–731. [DOI] [PubMed] [Google Scholar]
- 32.Brinkman, A., Kortleve, D. J., Zwarthoff, E. C. & Drop, S. L. (1991) Mol. Endocrinol. 5, 987–994. [DOI] [PubMed] [Google Scholar]
- 33.Thieriot-Prevost, G., Boccara, J. F., Francoual, C., Badoual, J. & Job, J. C. (1988) Pediatr. Res. 24, 380–383. [DOI] [PubMed] [Google Scholar]
- 34.Albertsson-Wikland, K., Boguszewski, M. & Karlberg, J. (1998) Horm. Res. 49, 7–13. [PubMed] [Google Scholar]
- 35.Padilla, P. A. & Roth, M. B. (2001) Proc. Natl. Acad. Sci. USA 98, 7331–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gracey, A. Y., Troll, J. V. & Somero, G. N. (2001) Proc. Natl. Acad. Sci. USA 98, 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrot, V., Moiseeva, E. B., Gozes, Y., Chan, S. J., Ingleton, P. & Funkenstein, B. (1999) Gen. Comp. Endocrinol. 116, 445–460. [DOI] [PubMed] [Google Scholar]
- 38.Radaelli, G., Patruno, M., Maccatrozzo, L. & Funkenstein, B. (2003) J. Endocrinol. 178, 285–299. [DOI] [PubMed] [Google Scholar]
- 39.Wenger, R. H. (2002) FASEB J. 16, 1151–1162. [DOI] [PubMed] [Google Scholar]
- 40.Kajimura, S., Aida, K. & Duan, C. (2004) Proceedings of the Fifth International Congress of Asia and Oceania Society of Comparative Endocrinology (Asia Oceania Soc. Comp. Endocrinol., Nara, Japan), in press.
- 41.Ton, C., Stamatiou, D. & Liew, C. C. (2003) Physiol. Genomics 16, 97–106. [DOI] [PubMed] [Google Scholar]
- 42.Duan, C. (2002) J. Endocrinol. 175, 41–54. [DOI] [PubMed] [Google Scholar]
- 43.Funkenstein, B. I., Tsai, W., Maures, T. & Duan, C. (2002) Gen. Comp. Endocrinol. 128, 112–122. [DOI] [PubMed] [Google Scholar]
- 44.Jones, J. I., Gockerman, A., Busby, W. H., Jr., Wright, G. & Clemmons, D. R. (1993) Proc. Natl. Acad. Sci. USA 90, 10553–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leu, J. I., Crissey, M. A. & Taub, R. (2003) J. Clin. Invest. 111, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leu, J. I., Crissey, M. A., Craig, L. E. & Taub, R. (2003) Mol. Cell. Biol. 23, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley, K. M., Schmidt, K. E., Berg, L., Sak, K., Galima, M. M., Gillespie, C., Balogh, L., Hawayek, A., Reyes, J. A. & Jamison, M. (2002) J. Endocrinol. 175, 3–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.