Abstract
Mycobacterium tuberculosis secretes ESAT-6, a virulence factor that triggers cell-mediated immune responses and IFN-γ production during tuberculosis. ESAT-6 is transported across the bacterial envelope by a specialized secretion system with a FSD (FtsK-SpoIIIE domain) membrane protein. Although the presence of ESAT-6-like genes has been identified in the genomes of other microbes, the possibility that they may encode general virulence functions has hitherto not been addressed. Herein we show that the human pathogen Staphylococcus aureus secretes EsxA and EsxB, ESAT-6-like proteins, across the bacterial envelope. Staphylococcal esxA and esxB are clustered with six other genes and some of these are required for synthesis or secretion of EsxA and EsxB. Mutants that failed to secrete EsxA and EsxB displayed defects in the pathogenesis of S. aureus murine abscesses, suggesting that this specialized secretion system may be a general strategy of human bacterial pathogenesis.
Keywords: specialized secretion, Gram-positive, exoprotein, ess
Staphylococcus aureus and many other Gram-positive bacteria are known to secrete virulence factors or exotoxins into the extracellular milieu. These factors are secreted via the Sec translocon across the membrane by a mechanism that requires N-terminal signal peptides for recognition. Recent advances in Streptococcus pyogenes and Bacillus subtilis revealed that the translocon may be part of a larger complex (ExPortal), that organizes transport of signal peptide-bearing precursor proteins to dedicated sites within the bacterial cell envelope (1, 2). Sec-independent translocation of virulence factors has been observed in group A streptococci and other Gram-positive species (3). The mechanism of substrate recognition and transport for these “signal peptide less” polypeptides has thus far not been revealed (4). In group A streptococci, secretion across the plasma membrane and injection of polypeptides into the cytosol of host cells uses a cytolysin-mediated transport mechanism (5), that functionally resembles type III secretion of Gram-negative bacteria (6).
Mycobacteria are equipped with the typical Sec machinery. However, much attention has been drawn to the secretion of two virulence factors, ESAT-6 (early secreted antigen target 6 kDa) and CFP-10 (culture filtrate antigen 10 kDa), encoded by Mycobacterium tuberculosis H37Rv esxA (Rv3875) and esxB (Rv3874). M. tuberculosis variants lacking esxA display defects in the establishment of tuberculosis and EsxA induces a strong T-cell response during infection (7). Comparative genomics of mycobacteria related to M. tuberculosis H37Rv identified multiple regions of difference that have been deleted from both virulent and avirulent species (8–10). esxA (Rv3875) is located within region of difference 1 (RD1). RD1 is the only locus specifically deleted in the attenuated Mycobacterium bovis bacillus Calmette–Guérin (bacille Calmette-Guérin) vaccine strain and avirulent Mycobacterium microii strain (8, 10–13). Although RD1 is certainly not the only virulence trait of mycobacteria, it appears to be a prominent factor in attenuating vaccine strains (10, 14, 15).
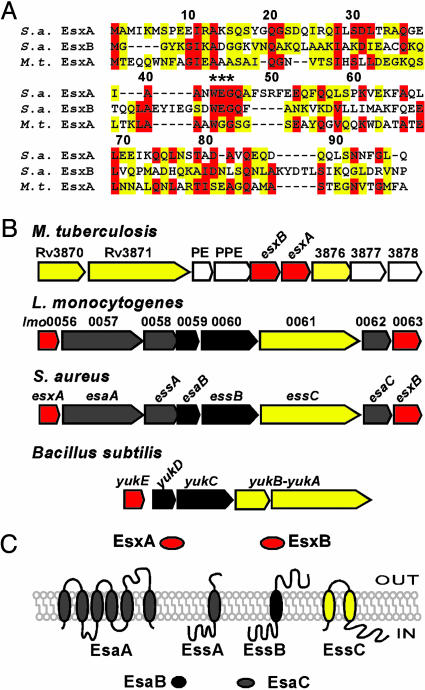
A survey of M. tuberculosis H37Rv genome sequence revealed at least 23 ESAT-6 homologues (9), 10 of which are encoded within five gene clusters that specify large soluble and membrane-bound ATPases with two or more FtsK/SpoIIIE-like domains (FSDs) (16, 17). Pallen (17) discovered ESAT-6 homologues in the sequenced genomes of other Gram-positive bacteria including B. subtilis, Bacillus anthracis, Clostridium acetobutylicum, Listeria monocytogenes, and S. aureus. The genes for the ESAT-6-like proteins are clustered with at least one gene encoding an FSD-type membrane protein (Fig. 1B), leading Pallen to propose that FSD ATPases may represent a universal portal for excretion of ESAT-6-like proteins (17).
Fig. 1.
S. aureus ess locus encoding ESAT-6-like proteins. (A) Protein sequence alignment of S. aureus (S.a.) EsxA and EsxB with M. tuberculosis (M.t.) EsxA. M. tuberculosis and S. aureus EsxA display 20.8% identity and 25% similarity, whereas S. aureus EsxB and M. tuberculosis EsxA are 17.8% identical and 35% similar. All three proteins contain the WXG motif. (B) Comparison of the M. tuberculosis H37Rv ESAT-6 locus with the S. aureus, L. monocytogenes ess loci and the B. subtilis yuk locus. Color of genes and proteins indicates FSD factors (yellow), ESAT-6-like (red), mycobacterial genes (light gray), and staphylococcal (also in listeria or bacilli, dark gray). (C) Membrane topology or soluble character of proteins encoded by the S. aureus ess locus.
Experimental support for this hypothesis was garnered when deletions of FSD-like genes (Rv3870, Rv3871) prevented the secretion of M. tuberculosis ESAT-6 and CFP-10 (14, 15, 18, 19). A third membrane protein encoded by Rv3877/snm4 was found to be required for secretion of ESAT-6 and CFP-10 (18). Therefore, in mycobacteria secretion of ESAT-6 and CFP-10 requires a specialized mechanism encoded by genes flanking esxA and esxB on the chromosome (Fig. 1B). This specialized secretion system has been referred to as Snm for secretion in mycobacteria (18). Most importantly, this dedicated secretion pathway may explain the absence of processing of ESAT-6 or CFP-10 during secretion (7).
Although it has been surmised that FSD proteins may represent a general Gram-positive secretion system (17), this compelling hypothesis is still in need of experimental verification. Here, the putative ESAT-6 secretion system (Ess) encoded by a cluster of eight genes in the Gram-positive pathogen S. aureus has been examined (Fig. 1B). Two ESAT-6-like proteins, Ess extracellular A (EsxA) and EsxB, were found to be transported across the bacterial envelope. Furthermore, the requirement of the FSD protein, EssC, as well as two other membrane proteins, EssA and EssB, for transport of EsxA and EsxB in S. aureus was demonstrated. The importance of EsxA and EsxB secretion during S. aureus infection was revealed by analyzing esxA, essC, and esxB mutants in a murine model of abscess formation.
Materials and Methods
Strains, Media, and Growth Conditions. Escherichia coli and S. aureus were grown in Luria–Bertani broth and tryptic soy broth at 37°C, respectively. Ampicillin and erythromycin were used at 100 mg·liter–1 and 10 mg·liter–1, respectively. essABC, esaAB, and esxB mutants were obtained from the Phoenix (ΦNΞ) library (20) (Table 1). Each Phoenix isolate is a derivative of the clinical isolate Newman (21) and carries the transposon bursa aurealis at a site on the chromosome that has been determined by DNA sequencing (20) and compared to S. aureus Mu50 genomic sequence (22). All bursa aurealis insertions were transduced into wild-type S. aureus Newman by using bacteriophage φ85 (23). Strain esxA24 lacking the EsxA gene product and strains overproducing EsxA and EsxB used for antibody production are described in the supporting information, which is published on the PNAS web site.
Table 1. S. aureus strains used in this study.
|
bursa aurealis insertion/mutation
|
Phenotype
|
|||||
|---|---|---|---|---|---|---|
| Strain | Genotype | nt position | AA position | Orientation | EsxA secretion | EsxB secretion |
| Newman | Clinical isolate | Yes | Yes | |||
| ΦNΞ187-01 | esxA24 | 24 | 8 | NA | No | No |
| ΦNΞ09349 | esaA::erm | 1110 | 370 | (+) | Yes | Yes |
| ΦNΞ03742 | essA::erm | 121 | 41 | (-) | No | No |
| ΦNΞ 13152 | esaB::erm | 43 | 15 | (+) | Yes | Yes |
| ΦNΞ 02411 | essB::erm | 199 | 67 | (+) | No | No |
| ΦNΞ04464 | essC::erm | 792 | 264 | (-) | No | No |
| ΦNΞ02191 | essC::erm | 1854 | 618 | (-) | No | No |
| ΦNΞ02832 | essC::erm | 2161 | 721 | (+) | Low | Low |
| ΦNΞ13038 | essC::erm | 3835 | 1,279 | (+) | Yes | Yes |
| ΦNΞ07912 | esxB::erm | 2 | 1 | (+) | No | No |
Culture Fractionation. Cultures were grown to an optical density of 0.8 at 660 nm (OD660), and 1.5 ml of culture was centrifuged at 10,000 × g for 4 min. Proteins in 1 ml of supernatant were precipitated with 7.5% trichloroacetic acid (TCA), using deoxycholic acid (0.2%) as a carrier, and sedimented by centrifugation 10,000 × g for 10 min (medium fraction). Culture (1.5 ml) was incubated in the presence of lysostaphin (100 μg/ml) for 30 min at 37°C. A 1-ml aliquot was precipitated with TCA (total culture). For extraction of proteins that are associated with the cell wall (loosely associated fraction), the cells of 1.5 ml of culture were washed three times with an equal volume of TSM buffer (50 mM Tris·HCl, pH 7.5/0.5 M sucrose/10 mM MgCl2). A 1-ml cell suspension was transferred to a fresh tube and precipitated with TCA. All TCA precipitates were washed with ice-cold acetone, solubilized in 50 μl of 0.5 M Tris·HCl (pH 8.0)/4% SDS and heated at 90°C for 10 min. Proteins were separated on SDS/PAGE and transferred to poly(vinylidene difluoride) membrane for immunoblot analysis with appropriate polyclonal antibodies. Immunoreactive signals were revealed by using a secondary antibody coupled to horseradish peroxidase and chemiluminescence.
Staphylococcal Fractionation. Cultures were centrifuged as described above and supernatants TCA precipitated in the presence of deoxycholic acid (medium). Cell pellets were suspended in 1 ml TSM buffer containing 100 μg/ml lystostaphin and incubated at 37°C for 30 min. Protoplasts were collected by centrifugation at 10,000 × g for 10 min, and the supernatant (cell wall fraction) was precipitated with TCA. The protoplasts were suspended in membrane buffer (0.1 M Tris·HCl, pH 7.5/0.1 M NaCl/10 mM MgCl2) and subjected to five rounds of freeze–thaw in a dry ice ethanol bath. Soluble proteins (cytoplasmic fraction) were separated from insoluble materials and membranes (membrane fraction) by centrifugation at 100,000 × g for 30 min. All samples were TCA-precipitated before immunoblotting.
Murine Abscess Model. S. aureus strains were grown at 37°C overnight in tryptic soy broth, diluted 100-fold in fresh broth and incubated at 37°C until an OD660 of 0.4 was reached. Cells were washed and suspended in PBS, and 100 μl of bacterial suspension was injected intravenously into 10 6- to 8-week-old female BALB/c mice anesthetized by intraperitoneal injection of 100 mg/ml ketamine and 2 mg/ml xylazine. Viable staphylococci were enumerated by colony formation on tryptic soy agar to quantify the infection dose [≈106 colony forming units (cfu)/ml]. Four days after challenge, mice were killed by CO2 asphyxiation. Spleen, kidneys, and liver were removed, and organs were homogenized in 1 ml of 1% Triton X-100 in PBS. Dilutions of the homogenates were plated on agar for enumeration of viable staphylococci. The statistical analysis of the data were performed with Student's t test using analyze-it (Analyze-It Software, Leeds, England).
Results
A Cluster of S. aureus Genes Encoding ESAT-6-Like Proteins. S. aureus EsxA and EsxB correspond to the Mu50 ORFs SAV0282 and SAV0290 (22) and display 20.8% and 17.8% identity as well as 25% and 35% similarity to M. tuberculosis EsxA, respectively (Fig. 1 A). The peptide sequences of S. aureus EsxA and EsxB encompass the WXG motif, a signature sequence of ESAT-6-like proteins (17) (Fig. 1 A). EsxA and EsxB are encoded within a cluster comprised of eight predicted ORFs (esxA, esaA, essA, esaB, essB, essC, esxY, and esxB) as shown in Fig. 1B (esa, ESAT-6 secretion accessory gene). Beside esxA and esxB, the other genes in this cluster appear to be unique in the chromosome. The cluster is referred to as the Ess cluster. Genes located immediately downstream of esxB are not depicted in Fig. 1B. However, their conservation in other related clusters from low G+C organisms suggest that they may also be functionally associated with the Ess cluster. The deduced peptide sequence of one of these genes, essC, displays similarity with FSD proteins of the M. tuberculosis RD1 cluster (Fig. 1B). Because of a sequencing error, essC entry in the Mu50 genome appears as two ORFs (SAV0287–SAV0288) when it is, in fact, a single combined ORF (data not shown). Fig. 1C shows the membrane topology of four proteins, EsaA, EssA, EssB, and EssC, as predicted by the tmhmm algorithm (www.cbs.dtu.dk/services/TMHMM-2.0) and psort-b (24). Four other factors (EsxA, EsxB, EsaB, and EsaC) are predicted by the same computational methods to be soluble, and not membrane associated. A summary of the relevant features of proteins encoded by the ess locus is presented in Table 2, which is published as supporting information on the PNAS web site.
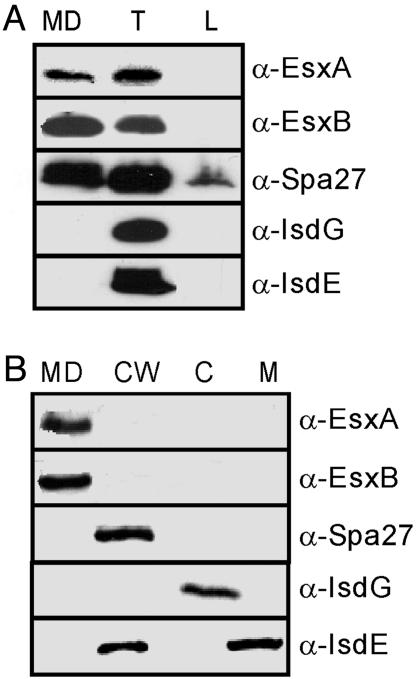
EsxA and EsxB Are Secreted into the Culture Medium of S. aureus. To study the location of EsxA and EsxB, cultures of S. aureus strain Newman were centrifuged and bacterial cells in the sediment were separated from the supernatant containing conditioned culture medium. Staphylococci were suspended in buffer and the cell wall was digested with lysostaphin, an endopeptidase that cleaves staphylococcal peptidoglycan (25). Proteins in the total lysate of staphylococci (T) or the culture medium (MD) were precipitated with TCA before separation on SDS/PAGE and immunoblotting. EsxA and EsxB were detected with specific rabbit antisera and were found in the total lysate and in the culture medium, indicating that S. aureus strain Newman secretes both polypeptides across the bacterial envelope (Fig. 2A). As a control, S. aureus IsdG, a cytoplasmic protein, and IsdE, a membrane lipoprotein, were found in total lysates but not in the bacterial culture medium (Fig. 2 A) (26). As a control for polypeptides that are covalently anchored to the cell wall, we also examined staphylococcal protein A (27). Cell wall degradation products, i.e., peptidoglycan fragments with linked polypeptide, are being released from the staphylococcal surface, and some protein A can therefore be found in both the total lysate and culture medium (28) (Fig. 2 A). Staphylococci were harvested from cultures by centrifugation and were boiled in hot SDS to release polypeptides that are only loosely associated with the cell wall (L). Boiling in hot SDS released small amounts of protein A, but not IsdE and IsdG. This is an expected result as the cell wall of staphylococci cannot be disintegrated by boiling in ionic detergents (29). EsxA and EsxB were not found to be associated with the staphylococcal surface after boiling bacteria in hot SDS, suggesting that staphylococci ESAT-6-like proteins are soluble in the extracellular milieu.
Fig. 2.
Subcellular location of EsxA and EsxB. (A) S. aureus strain Newman cultures were separated into medium (MD), total (T), and loosely wall-associated (L) fractions. Proteins were precipitated with TCA, separated on SDS/PAGE, and detected by immunoblotting with specific antibodies [α-EsxA, α-EsxB, α-Spa (protein A), α-IsdG, and α-IsdE]. (B) S. aureus strain Newman cultures were fractionated into medium (MD), cell wall (CW), cytoplasm (C), and membrane (M) compartments. TCA-precipitated proteins in each compartment were revealed by SDS/PAGE and immunoblotting.
Subcellular Localization of EsxA and EsxB. To examine the subcellular localization of EsxA and EsxB in staphylococci, cultures of S. aureus strain Newman were separated into cytoplasm, membrane, cell wall, and medium fractions (Fig. 2B). Proteins in all fractions were revealed by immunoblotting with specific antibodies. EsxA and EsxB were found exclusively in the culture medium. As a control, protein A was detected in the cell wall fraction, whereas IsdG and IsdE resided in the cytoplasm and the plasma membrane or cell wall of staphylococci, respectively (Fig. 2B). Together, these results demonstrate that S. aureus strain Newman secretes EsxA and EsxB across the bacterial envelope into the culture medium.
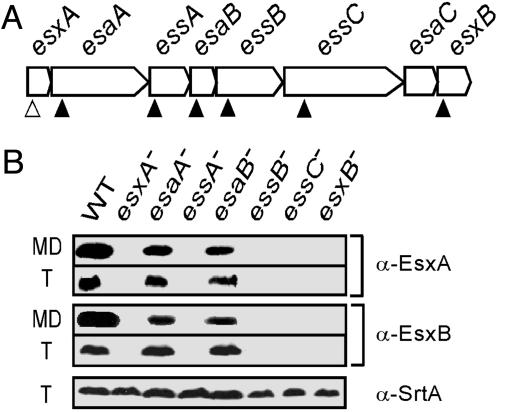
Factors Affecting the Production and Secretion of EsxA and EsxB. We wondered whether the factors encoded by the S. aureus ess cluster regulate production or promote secretion of EsxA and EsxB. Previous work from our laboratory generated the Phoenix (ΦNΞ) library, a collection of bursa aurealis insertion mutants in S. aureus strain Newman that have been mapped by inverted PCR and DNA sequencing (20). S. aureus strain Newman variants ΦNΞ09349, ΦNΞ03742, ΦNΞ13152, ΦNΞ02411, ΦNΞ04464, and ΦNΞ07912 carry bursa aurealis insertions in esaA, essA, esaB, essB, essC, or esxB, respectively (Table 1 and Fig. 3A). S. aureus Newman variant ΦNΞ187-01 carries a stop codon at position 8 of esxA coding sequence. The ability of these mutant strains to synthesize and secrete EsxA and EsxB was examined by immunoblotting of culture medium and total lysostaphin lysate samples as described in the legend of Fig. 2 A.
Fig. 3.
Factors affecting the production and secretion of EsxA and EsxB. (A) Schematic drawing of the ess cluster with the position of bursa aurealis insertions (filled triangles) and esxA24 mutation (open triangles). (B) Medium (MD) and total protein fractions (T) were separated on SDS/15% PAGE and immunoblotted with α-EsxA and α-EsxB. α-SrtA is used as a control for total protein loaded.
Bursa aurealis insertions at nucleotide positions 1,110 (codon 370) of esaA, or 43 (codon 15) of esaB did not affect the production or secretion of EsxA and EsxB when examined by immunoblot analysis (Fig. 3B). However, bursa aurealis insertions at nucleotide positions 121 (codon 41) of essA, 328 (codon 110) of essB, or 792 (codon 264) of essC abolished production and thus secretion of both EsxA and EsxB polypeptides as immunoreactive species could not be detected in either the medium or total culture lysate of strains ΦNΞ03742, ΦNΞ02411, and ΦNΞ04464, respectively (Fig. 3B). One bursa aurealis insertion in the Phoenix library mapped to nucleotide 2 (codon 1) of esxB (mutant ΦNΞ07912), a mutation that abolished the expression of all EsxB immunoreactive species (Fig. 3B). The inability of strain ΦNΞ07912 (esxB::erm) to produce EsxB abolished all EsxA production and, therefore, secretion. Conversely, the inability of strain ΦNΞ187–01 (esxA24) to produce EsxA abolished all EsxB production as well (Fig. 3B). None of the mutants used here abolished the transcription of either esxA or esxB as judged by RT-PCR (see Supporting Text, which is published as supporting information on the PNAS web site), suggesting that the phenotypes observed were not caused by polar effects of mutations or to transcriptional down-regulation.
EsaA is a 1,009-aa polytopic membrane protein with six predicted transmembrane helices that is conserved in staphylococcal and listerial species but absent from mycobacteria (Fig. 1B). essA and esaB specify 152- and 80-aa long protein products, respectively. Bioinformatic prediction of subcellular localization suggests that EssA contains one transmembrane domain, whereas EsaB may be soluble as it lacks a canonical signal peptide (Table 1). EssB and EsaB share sequence homology with listerial genes located in a similar ess cluster, and with B. subtilis YukC and YukD, respectively (30). The function of YukC and YukD is unknown, but their corresponding genes map to a chromosomal locus with yukA, yukB, and yukE (30) (Fig. 1B). YukE bears the WXG motif typical of ESAT-6-like proteins.
EssC is homologous to two genes of M. tuberculosis, Rv3870 (snm1) and Rv3871 (snm2), and to B. subtilis YukA–YukB. Recent sequence analysis revealed that yukA–yukB represents a single ORF (31). EssC, Rv3870, Rv3871, and YukA sequences encompass one or more FSDs (17). Gene SA0276 (essC, in the N315 genome) is annotated as “conserved hypothetical protein, similar to diarrheal toxin.” The annotation derives from similarity to the Bacillus cereus gene bceT and appears to be erroneous (32). Bioinformatic analysis predicted two transmembrane helices at the N terminus of the 1,479-aa EssC polypeptide (Table 1 and Fig. 1C). The N and C termini of EssC are presumed to reside in the staphylococcal cytoplasm based on the assumption that the FSDs hydrolyze ATP in this compartment.
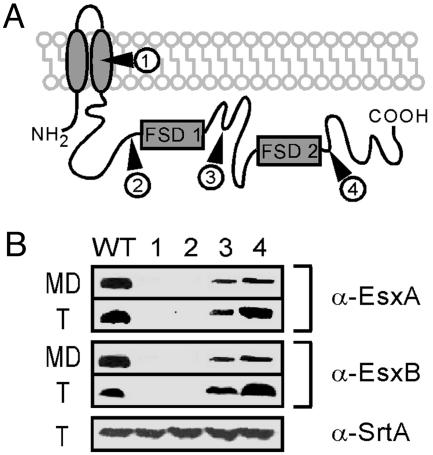
Phenotypic Analysis of Various Insertion Mutants in essC. The Phoenix library contains several strains carrying bursa aurealis insertions in the essC gene (Table 1 and Fig. 4A). Four essC mutants were used to analyze the effect of EssC truncations on the secretion of EsxA and EsxB (Fig. 4B). As shown in Fig. 3B, strain ΦNΞ 04464 carries a transposon insertion at nucleotide 792 (codon 264, mutant 1) (Fig. 4A), whereas strains ΦNΞ 02191, ΦNΞ 02832, and ΦNΞ 13038 carry bursa aurealis insertions at nucleotide positions 1854 (codon 618, mutant 2), 2161 (codon 721, mutant 3), and 3835 (codon 1,279, mutant 4), respectively (Fig. 4A). None of these transposon insertions affected the transcription of the two genes downstream (data not shown). Transposon insertion at codons 264 and 618 of essC abolished all production and secretion of EsxA and EsxB. In contrast, the synthesis and secretion of EsxB or EsxA occurred when bursa aurealis inserted at codons 721 and 1279 (Fig. 4B). These results imply that a single FSD domain is sufficient for EssC activity, and suggest that EsxA and EsxB secretion may be fueled by EssC-mediated ATP hydrolysis.
Fig. 4.
Role of EssC in EsxA and EsxB secretion. (A) Predicted topology of EssC with insertion sites of bursa aurealis in each strain (filled triangles). EssC contains two FstK/SpoIIIE-like domains (FSD). (B) Immunoblotting of total cellular protein (T) or culture medium (MD) with α-EsxA and α-EsxB antibodies. Insertional mutations at codon 264 (mutant 1) and codon 618 (mutant 2) prevented production/secretion of EsxA and EsxB. Mutants 1–4 are strains ΦNΞ 04464 (insertion at codon 264), ΦNΞ 02191 (insertion at codon 618), ΦNΞ 02832 (insertion at codon 721), and ΦNΞ 13038 (insertion at codon 1279), respectively. α-SrtA was used as a control for the total protein loaded.
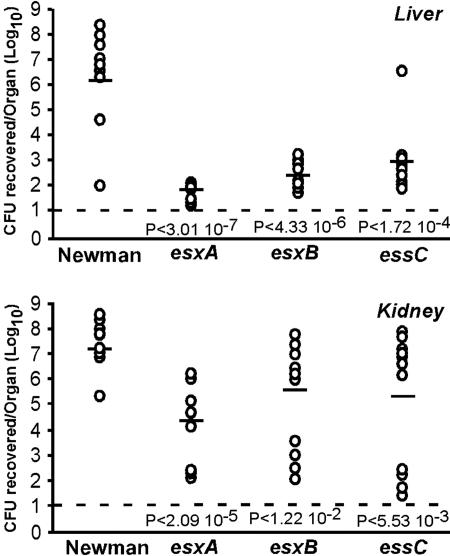
esxA, esxB, and essC Mutants Are Defective in the Pathogenesis of Staphylococcal Abscesses. S. aureus host infection and dissemination within organ tissues depends on staphylococcal synthesis and secretion of a wide variety of virulence factors (33). Because of the astonishing armament of staphylococci with virulence factors, the pathogenesis of S. aureus infections is considered multifactorial (33). Thus, with the exception of α-hemolysin (hla) mutants (34), strains carrying mutations that abrogate the expression of individual exoproteins typically do not display significant defects in the pathogenesis of S. aureus infections (35–37). We wondered whether the Ess secretion pathway might be involved in the establishment of staphylococcal disease. S. aureus Newman variants ΦNΞ187–01 (esxA24), ΦNΞ07912 (esxB::erm), and ΦNΞ04464 (essC::erm) do not synthesize EsxA and EsxB. These strains were chosen for experimental measurement of bacterial virulence. Viable staphylococci of mutant or wild-type Newman strains (≈106 staphylococci) were administered intravenously via retro-orbital injection into mice. The animals were killed 4 days after infection. Internal organs were removed, inspected for abscess formation, and then homogenized and spread on agar medium to quantify staphylococcal replication in host tissues by colony formation (38). Mutations in esxA, esxB, and essC genes caused a significant reduction in the ability of S. aureus Newman to establish kidney or liver abscesses in infected mice (Fig. 5 and data not shown). Kidneys of mice infected with Newman carried a mean value of 3.89 × 107 cfu per organ, whereas those of mice infected with strains lacking esxA, esxB, or essC harbored a mean value of 3.00 × 104 cfu per organ (P < 2.09 × 10–5), 6.17 × 105 cfu per organ (P < 1.2 × 10–2), and 2.51 × 105 cfu per organ (P < 5.53 × 10–3), respectively. It should be noted that half of the animals infected with mutant bacteria had cleared most staphylococci from the kidneys after 4 days, suggesting that complete clearing may occur by day 5 after infection with the mutant strains. The formation of liver abscesses in infected animals was even more affected when staphylococci did not produce EsxA and EsxB. Homogenized livers of mice infected with Newman were found to contain a mean value of 2.09 × 106 cfu per organ of staphylococci, whereas the livers of mutants lacking esxA, esxB, or essC contained a mean value of 9 × 101 cfu per organ (P < 3.01 × 10–7), 4.17 × 102 cfu per organ (P < 4.33 × 10–6), and 1.35 × 103 cfu per organ (P < 1.72 × 10–4), respectively.
Fig. 5.
S. aureus esxA, esxB, and essC mutants are defective in the pathogenesis of murine abscesses. Ten 6-week-old BALB/c mice were injected retro-orbitally with 106 cfu for each strain. Mice were killed 4 days after infection. Kidneys and liver were removed and homogenized. Viable bacteria were counted after dilution and colony formation on tryptic soy agar. Statistical significance was examined with the Student t test, and P values were recorded. The limit of detection (solid line) was determined to be 10 cfu (101).
Discussion
S. aureus as well as several other staphylococcal species are physiological commensals of the human skin and nares (39). Breaches in local defense, for example, a skin cut or hair follicle trauma, provide a bacterial opportunity for replication and spread in underlying tissues (39). Successful implementation of this strategy requires S. aureus secretion of exoproteins, factors that are transported across the bacterial envelope via the Sec secretion system (40). The establishment of staphylococcal abscesses with liquefaction necrosis can thus be viewed as the sum of all pathogenic events implemented by exoproteins and the corresponding host responses (41). These infectious lesions provide further opportunity for invading pathogens to enter deeper tissues or to spread via blood circulation, whereupon a reiterative cycle of secretion and destruction will be initiated (33, 41).
Previous work showed that S. aureus secretes 30 or more exoproteins (42) and mutations that abrogate the expression and secretion of only one of these factors typically do not cause significant defects in pathogenesis when measured in animal models of disease (35, 37). Nevertheless, some exceptions to this unifying view have been revealed. S. aureus mutants with hla deletions display defects in the pathogenesis of skin or mammary abscess formation in a murine model (42). Panton–Valentine leucocidin (PVL), a bacteriophage-encoded heterodimeric toxin, is believed to inflict immune cell necrosis by selective interaction with host membranes (43). S. aureus strains lysogenic for PVL bacteriophage are known to cause necrotizing pneumonia in children and healthy adults, a frequently lethal disease (44). Staphylococcal toxinoses depend on the secretion and functional role of specific toxins, but not on staphylococcal tissue invasion as the deciding parameter of pathological outcome (41).
Herein we examined the ESAT-6 system of S. aureus, a specialized secretion pathway that contributes to the pathogenesis of blood borne infections. EsxA and EsxB, two proteins with sequence identity to M. tuberculosis ESAT-6 (EsxA), are secreted by S. aureus Newman and are expressed by other staphylococcal strains (data not shown). EsxA and EsxB do not contain a classical N-terminal signal sequence and the polypeptides do not appear to be processed upon secretion (data not shown). Furthermore, EsxB is required for EsxA synthesis as well as secretion and vice versa. In M. tuberculosis, EsxA physically interacts with EsxB both in vitro and in vivo (45, 46). The data obtained herein support the notion that staphylococcal EsxA and EsxB could also form a complex. Although molecular determinants for the presumed dimerization and secretion EsxA and EsxB are not yet known, the WXG motif or the predicted coiled-coil structure of ESAT-6-like proteins represent obvious candidates for such functions. Knowledge of the presumed EsxA/EsxB dimerization will be important for future design of experiments that seek to examine these factors in pathogenesis studies.
Mutations in essA, essB, or essC abolished synthesis and therefore secretion of EsxA and EsxB without affecting transcription of esxA and esxB. Because EssA, EssB, and EssC are predicted transmembrane proteins, it is tempting to speculate that these proteins may represent a secretion apparatus. Without secretion, EsxA and EsxB may be rapidly degraded in the bacterial cytoplasm or posttranscriptional feedback inhibition may reduce esxA and esxB expression. Although experimental distinction between these possibilities has not yet been achieved, it seems noteworthy that many secretion systems, including type III and sec pathways, regulate secretion substrate gene expression via posttranscriptional mechanisms until the machinery is fully assembled and functional (47).
ESAT-6 (EsxA) and CFP-10 (EsxB) of M. tuberculosis are known to play an important role in virulence (10), causing macrophage subversion (18) and host cell lysis (15). To study the phenotype of esxA, esxB and essC mutations in S. aureus, a murine model of staphylococcal blood-borne dissemination and abscess formation was chosen (38). In these experiments, inoculated staphylococci must resist phagocytic killing, adhere to organ tissues, and mount an invasive strategy that leads to bacterial replication and abscess formation. Our results indicate that bursa aurealis insertion in esxA, esxB, or essC caused a 2- to 4-log reduction in the ability of mutant staphylococci to establish abscesses in infected mice, depending on the organ system examined (Fig. 5). Therefore, secretion of EsxA and EsxB represents an important virulence strategy. In fact, only S. aureus sortase A mutants seem to display a similarly strong phenotype with 2- to 3-log reduction in virulence in this model (23, 48).
It is still unclear whether S. aureus EsxA and EsxB function as toxins. For example, type III pathways of Gram-negative microbes are characterized by a conserved secretion machinery and chaperone genes; however, genes encoding effectors, i.e., proteins that implement virulence strategies by interacting with the host, are typically not conserved beyond species boundaries. Considering the conservation of esxAB and essC in genomes of several different bacteria, one could argue that the effectors of staphylococcal ESAT-6 may not yet have been identified. Indeed, preliminary analysis of the spectrum of secreted polypeptides in esxA and esxB mutant cultures suggests that the ESAT-6 pathway may transport or somehow affect secretion of several exoproteins (data not shown). Therefore, it seems plausible that ESAT-6 effectors may have evolved to perform specific functions in different microorganism. One could ask further whether immune responses to ESAT-6 factors can protect hosts against staphylococcal disease or whether mutant variants lacking ESAT-6 determinants can be exploited for live vaccine design with attenuated S. aureus strains. Future work in this area promises to be fertile, revealing EsxA/EsxB secretion mechanisms and a comprehensive view of EssABC substrates, or perhaps pointing to novel strategies that may prevent the plethora of human diseases caused by S. aureus, in particular drug-resistant strains with resistance to all known antibiotics (49).
Supplementary Material
Acknowledgments
We thank L. Ang for technical assistance, E. Skaar (University of Chicago, Chicago) for providing the anti-IsdG and anti-IsdE antibodies, members of our laboratory for discussion, and O. Schneewind (University of Chicago, Chicago) for providing the anti-SrtA antibody as well as critical reading of the manuscript. This study was supported in part by University of Chicago seed projects awards and start-up funds from the Department of Microbiology (to D.M.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FSD, FtsK/SpoIIIE-like domain; Ess, ESAT-6 secretion system; TCA, trichloroacetic acid; cfu, colony-forming unit.
References
- 1.Rosch, J. & Caparon, M. (2004) Science 304, 1513–1515. [DOI] [PubMed] [Google Scholar]
- 2.Campo, N., Tjalsma, H., Buist, G., Stepniak, D., Meijer, M., Veenhuis, M., Westermann, M., Muller, J. P., Bron, S., Kok, J., et al. (2004) Mol. Microbiol. 53, 1583–1599. [DOI] [PubMed] [Google Scholar]
- 3.Chaatwal, G. S. (2002) Trends Microbiol. 10, 205–208. [DOI] [PubMed] [Google Scholar]
- 4.Pancholi, V. & Fischetti, V. A. (1992) J. Exp. Med. 176, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madden, J. C., Ruiz, N. & Caparon, M. (2001) Cell 104, 143–152. [DOI] [PubMed] [Google Scholar]
- 6.Galan, J. E. & Collmer, A. (1999) Science 284, 1322–1333. [DOI] [PubMed] [Google Scholar]
- 7.Sorensen, A. L., Nagai, S., Houen, G., Andersen, P. & Andersen, A. B. (1995) Infect. Immun. 63, 1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahairas, G. G., Sabo, P. J., Hickey, M. J., Singh, D. C. & Stover, C. K. (1996) J. Bacteriol. 178, 1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537–544. [DOI] [PubMed] [Google Scholar]
- 10.Pym, A. S., Brodin, P., Brosch, R., Huerre, M. & Cole, S. T. (2002) Mol. Microbiol. 46, 709–717. [DOI] [PubMed] [Google Scholar]
- 11.Calmette, A. (1927) in La Vaccination Préventive contre la Tuberculose, (Mason-et-Cie, Paris), p. 251.
- 12.Harboe, M., Oettinger, T., Wiker, H. G., Rosenkrands, I. & Andersen, P. (1996) Infect. Immun. 64, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philipp, W. J., Nair, S., Guglielmi, G., Lagranderie, M., Gicquel, B. & Cole, S. T. (1996) Microbiology 142, 3135–3145. [DOI] [PubMed] [Google Scholar]
- 14.Pym, A. S., Brodin, P., Majlessi, L., Brosch, R., Demangel, C., Williams, A., Griffiths, K. E., Marchal, G., Leclerc, C. & Cole, S. T. (2003) Nat. Med. 9, 533–539. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, T., Hingley-Wilson, S. M., Chen, B., Chen, M., Dai, A. Z., Morin, P. M., Marks, C. B., Padiyar, J., Goulding, C., Gingery, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12420–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gey Van Pittius, N. C., Gamieldien, J., Hide, W., Brown, G. D., Siezen, R. J. & Beyers, A. D. (2001) Genome Biol. 2, research0044.1–0044.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pallen, M. J. (2002) Trends Microbiol. 10, 209–212. [DOI] [PubMed] [Google Scholar]
- 18.Stanley, S. A., Raghavan, S., Hwang, W. W. & Cox, J. S. (2003) Proc. Natl. Acad. Sci. USA 100, 13001–13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guinn, K. M., Hickey, M. J., Mathur, S. K., Zakel, K. L., Grotzke, J. E., Lewinsohn, D. M., Smith, S. & Sherman, D. R. (2004) Mol. Microbiol. 51, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae, T., Banger, A. K., Wallace, A., Glass, E. M., Åslund, F., Schneewind, O. & Missiakas, D. M. (2004) Proc. Natl. Acad. Sci. USA 101, 12312–12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duthie, E. S. & Lorenz, L. L. (1952) J. Gen. Microbiol. 6, 95–107. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., Cui, L., Oguchi, A., Aoki, K., Nagai, Y., et al. (2001) Lancet 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 23.Mazmanian, S. K., Liu, G., Jensen, E. R., Lenoy, E. & Schneewind, O. (2000) Proc. Natl. Acad. Sci. USA 97, 5510–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardy, J. L., Spencer, C., Wang, K., Ester, M., Tusnady, G. E., Simon, I., Hua, S., deFays, K., Lambert, C., Nakai, K. & Brinkman, F. S. L. (2003) Nucleic Acids Res. 31, 3613–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindler, C. A. & Schuhardt, V. T. (1964) Proc. Natl. Acad. Sci. USA 51, 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson, I. M., Mazmanian, S. K., Schneewind, O., Bremell, T. & Tarkowski, A. (2003) Microb. Infect. 5, 775–780. [DOI] [PubMed] [Google Scholar]
- 27.Schneewind, O., Fowler, A. & Faull, K. F. (1995) Science 268, 103–106. [DOI] [PubMed] [Google Scholar]
- 28.Navarre, W. W. & Schneewind, O. (1999) Microbiol. Mol. Biol. Rev. 63, 174–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneewind, O., Model, P. & Fischetti, V. A. (1992) Cell 70, 267–281. [DOI] [PubMed] [Google Scholar]
- 30.Oudega, B., Vandenbol, M. & Koningstein, G. (1997) Microbiology 143, 1489–1491. [DOI] [PubMed] [Google Scholar]
- 31.Sao-Jose, C., Baptista, C. & Santos, M. A. (2004) J. Bacteriol. 186, 8337–8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen, B. M., Hoiby, P. E., Jensen, G. B. & Hendriksen, N. B. (2003) FEMS Microbiol. Lett. 223, 21–24. [DOI] [PubMed] [Google Scholar]
- 33.Novick, R. P. (2003) Mol. Microbiol. 48, 1429–1449. [DOI] [PubMed] [Google Scholar]
- 34.Bhakdi, S. & Tranum-Jensen, J. (1991) Microbiol. Rev. 55, 733–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Reilly, M., de Azavedo, J. C., Kennedy, S. & Foster, T. J. (1986) Microb. Pathog. 1, 125–138. [DOI] [PubMed] [Google Scholar]
- 36.Jonsson, P., Lindberg, M., Haraldsson, I. & Wadstrom, T. (1985) Infect. Immun. 49, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, A. H., Nowlan, P., Weavers, E. D. & Foster, T. (1987) Infect. Immun. 55, 3103–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albus, A., Arbeit, R. D. & Lee, J. C. (1991) Infect. Immun. 59, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Archer, G. L. & Climo, M. W. (2001) N. Engl. J. Med. 344, 55–56. [DOI] [PubMed] [Google Scholar]
- 40.Archer, G. L. (1998) Clin. Infect. Dis. 26, 1179–1181. [DOI] [PubMed] [Google Scholar]
- 41.Dinges, M. M., Orwin, P. M. & Schlievert, P. M. (2000) Clin. Microbiol. Rev. 13, 16–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Recsei, P., Kreiswirth, B., O'Reilly, M., Schlievert, P., Gruss, A. & Novick, R. (1986) Mol. Gen. Genet. 202, 58–61. [DOI] [PubMed] [Google Scholar]
- 43.Prevost, G., Cribier, B., Couppie, P., Petiau, P., Supersac, G., Finck-Barbacon, V., Monteil, H. & Piemont, Y. (1995) Infect. Immun. 63, 4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillet, Y., Issartel, B., Vanhems, P., Fournet, J. C., Lina, G., Bes, M., Vandenesch, F., Piemont, Y., Brousse, N., Floret, D. & Etienne, J. (2002) Lancet 359, 753–759. [DOI] [PubMed] [Google Scholar]
- 45.Renshaw, P. S., Panagiotidou, P., Whelan, A., Gordon, S. V., Hewinson, R. G., Williamson, R. A. & Carr, M. D. (2002) J. Biol. Chem. 277, 21598–21603. [DOI] [PubMed] [Google Scholar]
- 46.Okkels, L. M. & Andersen, P. (2004) J. Bacteriol. 186, 2487–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramamurthi, K. S. & Schneewind, O. (2002) Annu. Rev. Cell Dev. Biol. 18, 107–133. [DOI] [PubMed] [Google Scholar]
- 48.Jonsson, I. M., Mazamanian, S. K., Schneewind, O., Vendrengh, M., Bremell, T. & Tarkowski, A. (2002) J. Infect. Dis. 185, 1417–1424. [DOI] [PubMed] [Google Scholar]
- 49.Weigel, L. M., Clewell, D. B., Gill, S. R., Clark, N. C., McDougal, L. K., Flannagan, S. E., Kolonay, J. F., Shetty, J., Killgore, G. E. & Tenover, F. C. (2003) Science 302, 1569–1571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.