Abstract
The Nrf2 transcription factor belongs to the Cap’n’collar family, named after the founding member of this group, the product of the Drosophila cap’n’collar gene. The encoded protein, Cap’n’collar, abbreviated Cnc, offers a convenient and accessible model to study the structure, function and biology of Nrf2 transcription factors at organism, tissue, cell and molecular levels, using the powerful genetic, genomic and biochemical tools available in Drosophila. In this review we provide an account of the original identification of Cnc as a regulator of embryonic development. We will then describe the discovery of Nrf2 like-functions of Cnc and its role in acute stress signaling and aging. The establishment of Drosophila as a model organism in which the mechanisms and functions of Nrf2 signaling can be studied has led to several discoveries, the regulation of stem cell activity by an Nrf2-mediated redox mechanism, the interaction of Nrf2 with p62 and Myc in the control of tissue growth and unfolded protein response, and more. Several of these more recent lines of investigation will be highlighted. Model organisms like fly and worm remain powerful experimental platforms that can help to unravel the many remaining puzzles regarding the role of Nrf2 and its relatives in controlling the physiology and maintaining the health of multicellular organisms.
Introduction
Mammalian Cap’n’collar transcription factors, include the NFE2 Nrf 1, 2 and 3 proteins and their dimerization partners of the small Maf family and Bach family. These proteins, and most notably Nrf2, are the subject of intense scientific interest (Hayes and Dinkova-Kostova, 2014; Kensler et al., 2007; Sykiotis and Bohmann, 2010). Nrf2 has been characterized as a master regulator of gene expression programs that defend cells, tissues and organisms against oxidative stress and various types of chemical attack. A large number of studies have shown that Nrf2-mediated gene expression programs are protective against diseases that are caused or exacerbated by oxidative stress, including inflammatory diseases of the lung, liver, kidney and other organs, various cancer types, and neurodegenerative diseases, such as Parkinson’s, and Alzheimer’s (Hayes and McMahon, 2009; Osburn and Kensler, 2008). More recently, functions that appear not to be directly associated with chemical or oxidative stress have been ascribed to Nrf2. Such functions include the control of energy metabolism, or stem cell regulation (Hayes and Dinkova-Kostova, 2014; Tsai et al., 2013).
Much remains to be learned about the function of Nrf2 and the other Cap’n’collar family members: How do they respond to different signals and signaling pathways? How is their activity integrated with that of other signaling systems that affect stress response, metabolism and other functions? Can pharmacological manipulation of Nrf2 function have beneficial preventive or curative effects? How does Nrf2 influence the aging process and, conversely, what is the effect of aging on Nrf2 function?
One avenue to address these and other questions is offered by the use of simple, genetically tractable model organisms such as C. elegans (see chapter xx by Keith Blackwell) and Drosophila. Both flies and worms have signaling systems that are homologous to the mammalian Nrf2 pathway, and possibly the related Nrf1 pathway as well. In this chapter we will describe the Drosophila Nrf2 pathway, its discovery, its molecular constituents, and organismic functions. We will then highlight some of the discoveries that were made by applying the powerful experimental tools that are available in the fruitfly system to the study of Nrf2. We hope to make the reader familiar with the general features of the Drosophila Nrf2 pathway, but also to convey a sense of the unique experimental possibilities that Drosophila offers to the study of this important transcription factor.
History: Discovery of Cnc and its developmental functions
The Drosophila cnc gene was discovered in the early 1990s in a chromosome walk covering region 94E of the third chromosome, which had been conducted in order to find genes involved in Drosophila development (Mohler et al., 1991). Sequence analysis of the isolated cDNAs revealed that cnc encodes a putative transcription factor with a basic region / leucine zipper DNA-binding and dimerization domain and thus represents a distant relative of famous gene regulatory proteins such as Jun, Fos and C/EBP.
Cnc expression was detected in the embryonic blastoderm at a time when the identity of the body segments along the anterior-posterior axis is determined (for a schematic representation of Drosophila development refer to Figure 1). At this stage cnc mRNA is expressed in a unique pattern, consisting of a “cap” at the anterior end of the embryo and a stripe or “collar” surrounding the embryo at approximately 70% of its length. This expression pattern gave the gene, and with it the whole family, its name.
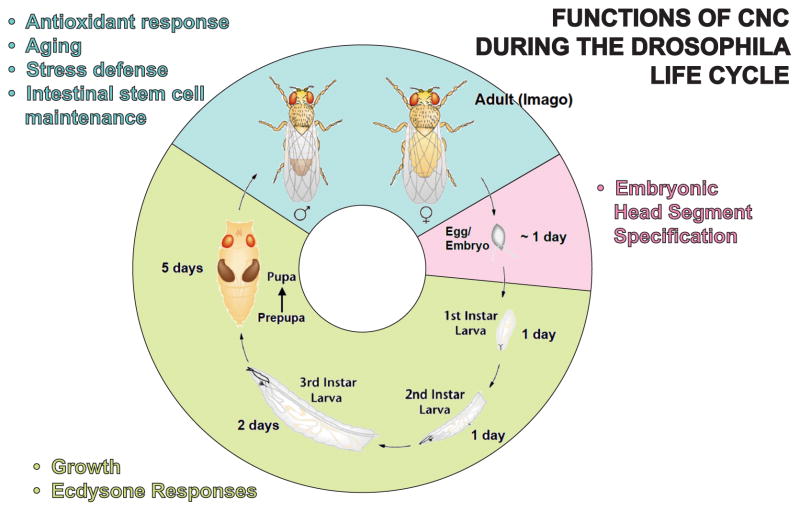
Figure 1. Functions of Cnc during the Drosophila life cycle.
The cartoon depicts the stages of Drosophila development (embryogenesis, pink; larval and pupal stages, green) and adulthood (blue). Females deposit fertilized eggs. Upon completion of embryogenesis the first larval stage (1. instar) hatches. Subsequent molting gives rise to second and third instar larvae. At the end of the last larval stage pupae are formed and metamorphosis to the adult form (imago) commences. The durations of these different developmental phases under optimal growth conditions are indicated. Cnc-regulated biological processes that occur at different stages of the Drosophila life cycle and are discussed in this review are listed.
The analysis of cnc mutants revealed that, consistent with the expression of the gene in the anterior region of the early embryo, it is required for head development (Harding et al., 1995; Mohler et al., 1995). It was found that Cnc genetically interacts with the homeobox protein Deformed (Dfd), which specifies the identity of the maxillary segment, a part of the developing head (McGinnis et al., 1998). In cnc mutants, the neighboring mandibular segment is transformed to a maxillary fate. Based on these observations it was concluded that Cnc contributes to embryonic pattern formation by suppressing Dfd function in the presumptive mandibular region.
Structure of the cnc locus, alternative gene products
Further molecular analysis by McGinnis and colleagues (Veraksa et al., 2000) showed that the cap’n’collar locus produces multiple alternatively spliced transcripts, giving rise to Cnc variant proteins of different sizes and domain composition. Three different isoforms were described in those early studies: CncA, CncB and CncC. All of these include the COOH-terminally located bZIP DNA-binding and dimerization domain (Fig. 2). Moving from the CncA to the CncC isoforms, the different splice variants are distinguished by the inclusion of progressively more NH2-terminal sequences. In this way, the 533 amino acid sequence of CncA is completely contained within the 805 amino acid CncB protein, and CncC is comprised of all these sequences plus an additional NH2-terminal extension that brings its total length to 1383 amino acids (Figs. 2 and 3). Of these three isoforms, CncB is the predominant gene product expressed during embryonic head development in the characteristic “cap and collar” pattern. Consistent with genetic evidence for an antagonistic role of Cnc towards Dfd during early embryonic development as mentioned above, this analysis demonstrated an inhibitory function of CncB on Dfd-dependent target gene activation (McGinnis et al., 1998).
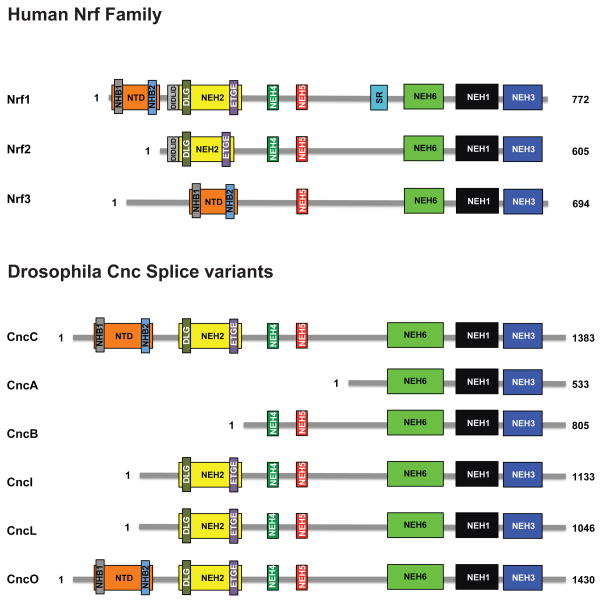
Figure 2. A representation of cap’n’collar transcription factors found in humans and Drosophila.
Structural domains were identified using MAFFT and T-COFFEE to compare publicly available amino acid sequences of human Nrf1, Nrf2, Nrf3, and different protein isoforms expressed from the Drosophila cnc locus (McWilliam et al., 2013). The N-terminal domains (NTD) present in Nrf1, Nrf3, CncC, and CncO contain the NHB1/2 subdomains that are believed to play an important role in targeting these proteins to the endoplasmic reticulum (Zhang et al., 2014b). The NEH2 domains contain the DIDLID/DLG and the ETGE motifs that are essential for Keap1 binding as well as Keap1-mediated degradation of these proteins (Fukutomi et al., 2014; Katoh et al., 2005). NEH4 and NEH5 are transactivation domains involved in the recruitment of the CREB binding protein, CBP (Katoh et al., 2001). The SR region is a serine-rich region of the Nrf1 protein. The highly conserved cap’n’collar (CNC) motif as well as the basic region and leucine zipper, all of which play a key role in DNA binding, are located within the NEH1 domain (Motohashi et al., 2004). The NEH3 domain is thought to be involved in transactivation by recruitment of CHD6, a chromo-ATPase/helicase DNA binding protein (Nioi et al., 2005). The NEH6 domain contains a putative repression site for β-TrCP and several conserved serine residues that may be phosphorylated by GSK-3 (Chowdhry et al., 2013).
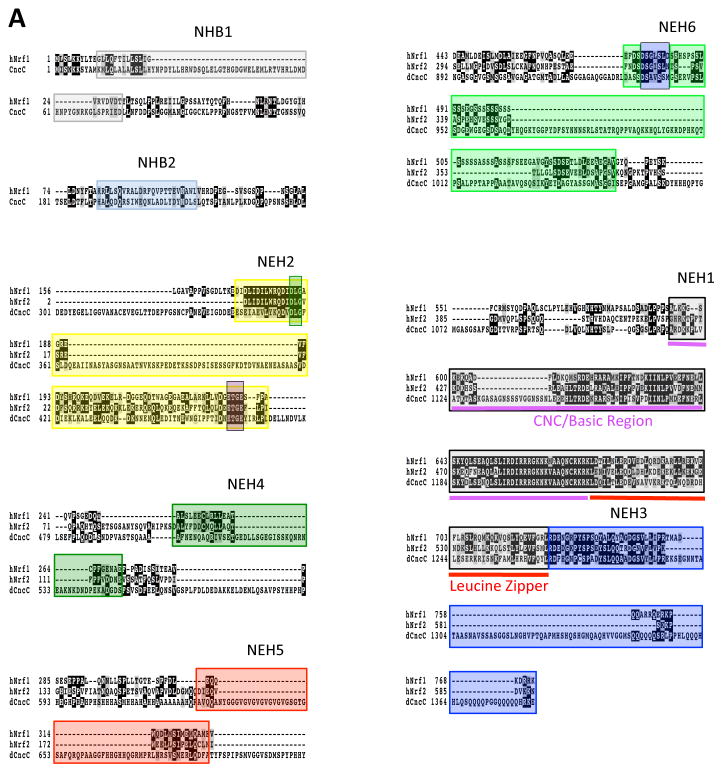
Figure 3. Amino acid sequence homology between Cnc, Keap1 and small Maf proteins of human and Drosophila.
A) Alignment of key functional elements in Nrf1, Nrf2, and CncC. Multisequence alignment using MAFFT, T-COFFEE, and BOXSHADE were used to compare publicly available amino acid sequences of human Nrf1, Nrf2, and CncC (McWilliam et al., 2013). Residues that are conserved between at least two of the sequences are shaded in black and residues with similar characteristics are shaded in gray. Domains and features described in Figure 1 are highlighted.
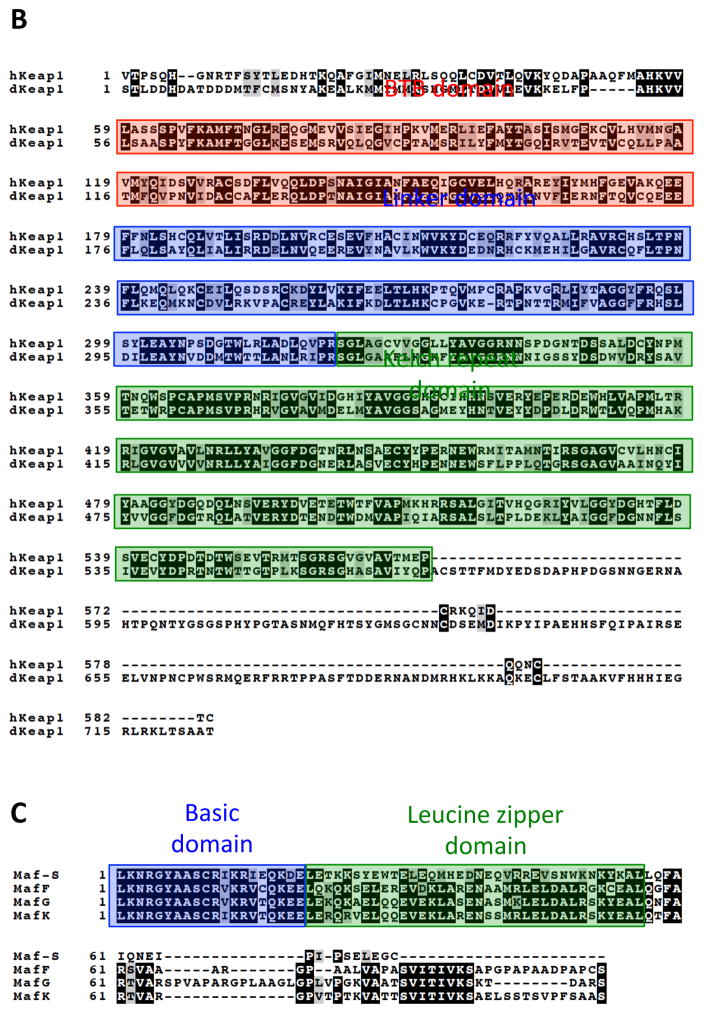
B) Alignment of Human and Drosophila Keap1. Multisequence alignment using MAFFT, T-COFFEE, and BOXSHADE were used to compare publicly available amino acid sequences of human Keap1 (hKeap1) and Drosophila Keap1 (dKeap1) (McWilliam et al., 2013). Residues that are conserved are shaded in black and residues with similar characteristics are shaded in gray. The BTB (Broad complex, Tramtrack, and Bric-a-Brac) domain contains key cysteine residues important for sensing oxidative stress (Zhang and Hannink, 2003). The linker domain plays a role in targeting Nrf2 for ubiquitination leading to its degradation as well as maintaining localization of the Nrf2-Keap1 complex in the cytoplasm (Zhang and Hannink, 2003). The Kelch repeat domain is responsible for Nrf2 interaction by interacting with the NEH2 regulatory domain of Nrf2 (Itoh et al., 1999).
C) Alignment of Mouse and Drosophila small Maf proteins. Multisequence alignment using MAFFT, T-COFFEE, and BOXSHADE were used to compare publicly available amino acid sequences of human Keap1 and Drosophila Keap1 (McWilliam et al., 2013). Residues that are conserved are shaded in black and residues with similar characteristics are shaded in gray. The basic-leucine zipper region is highlighted.
Since the initial description of the alternatively spliced gene products of the cnc locus, CncA, CncB and CncC by McGinnis and colleagues (Veraksa et al., 2000), additional coding variants have been reported in FLYBASE (www.flybase.org) (McQuilton et al., 2012; Mohr et al., 2014) and are depicted in Fig. 2. Following the McGinnis nomenclature, these longer isoforms are designated CncD, CncE and so on. These splice variants were identified in large scale RNA-seq based experiments as part of the modENCODE project (www.modencode.org) (Li et al., 2014, modENCODE Consortium, 2010) and validated by specific RT PCR (M. Tian, personal communication). Further study of the function of these alternative gene products is warranted.
Analysis of conserved domains yielded more insight into the molecular functions of Cnc gene products. As already pointed out in the early papers by Mohler and McGinnis (Harding et al., 1995; Mohler et al., 1995), the bZIP DNA binding and dimerization region of Cnc proteins shows homology to the corresponding domains in the mammalian transcription factors NF-E2 and Nrf1, 2 and 3.
Drosophila Cnc products contain peptide sequences resembling a number of other domains that are conserved among mammalian cap’n’collar proteins. These include the NEH3, 4, 5 and 6 homology regions (Figs. 2 and 3) which have been implicated in transcriptional activation and interactions with various regulatory proteins. Careful structural and functional analyses to verify that the Cnc domains have similar molecular functions as their mammalian counterparts have not yet been completed. Interestingly, the long splice variant, CncC, shows conservation with additional functionally important domains of Nrf2 (Kobayashi et al., 2002). Notably, sequences resembling the ETGE and DLG motifs, which serve as docking sites for the Nrf2-specific negative regulator Keap1 (Baird et al., 2013; Dinkova-Kostova et al., 2002; McMahon et al., 2006; Wakabayashi et al., 2003; Zhang et al., 2004), are present in the longer splice variants including CncC. Experiments on the mammalian proteins had shown that the interaction of Nrf2 with Keap1 targets the transcription factor for ubiquitin-dependent proteasomal degradation. This proteolytic inhibition of Nrf2 activity is alleviated in conditions of oxidative stress. The subsequent stabilization and nuclear accumulation of Nrf2 accounts for the stress inducible transcription of Nrf2 target genes. It was later experimentally confirmed that CncC is a stress responsive transcription factor that functionally resembles Nrf2 (Sykiotis and Bohmann, 2008). Thus, in addition to their role in embryonic development and spatial patterning, cnc gene products were found to have Nrf2-like functions in stress defense and the control of redox homeostasis.
In addition to repression of CncC by Keap1, it is possible that Cnc isoforms are also repressed by glycogen synthase kinase-3 (GSK-3). Phosphorylation of the DSGIS motif found in the NEH6 domain of mammalian Nrf1/2 by GSK-3 leads to the recruitment of the E3 ubiquitin ligase adaptor, β-transducin repeat-containing protein (β-TrCP) at these sites. In mammalian cells β-TrCP was found to bind to two motifs, DSGIS and DSAPGS, respectively, found within the NEH6 domain of mammalian Nrf1 and Nrf2. Phosphorylation of the DSGIS motif is dependent upon GSK-3 activity and is required for recruitment of β-TrCP while recruitment of β-TrCP to the DSAPGS motif is GSK-3 independent. (Chowdhry et al., 2013; Rada et al., 2011). The NEH6 domains of Cnc isoforms contain a DSGIS-like motif, DSAVS, as well as several conserved Ser residues that are phosphorylated by GSK-3 in mammalian Nrf1/2 (Figure 3a). Interestingly, Cnc isoforms do not appear to have a conserved DSAPGS motif. These observations suggest that like Nrf1/2, Cnc isoforms may be regulated by the Drosophila GSK-3 ortholog, shaggy (sgg) and the β-TrCP ortholog, slimb (slmb) (Siegfried et al., 1990; Spencer et al., 1999). Thus, Drosophila can provide a system in which the mechanisms and the biological function of the crosstalk between GSK-3 and Nrf2 signaling can be further dissected.
Relationship of Cnc with Nrf2 and Nrf1
In spite of the presumably vital functions of Nrf2 in oxidative stress defense and redox homeostasis, mouse mutants lacking this factor are viable, albeit with increased sensitivity to oxidative stress and consequent pleiotropic phenotypes, notably susceptibility to a range of diseases including pulmonary, cardiovascular and neurodegenerative conditions (Chan et al., 1996; Osburn and Kensler, 2008). In contrast, Drosophila cncC loss-of-function mutants die during development (Harding et al., 1995; Mohler et al., 1995). One explanation for the comparatively mild, non-lethal phenotype of Nrf2 mutant mice would be a compensatory effect of the related Nrf1 and Nrf3 factors (Beyer et al., 2008; Derjuga et al., 2004). Some functions in stress response and redox regulation might be executed by these gene products. Indeed, Nrf1, like its more intensely studied cousin Nrf2, controls the transcription of a battery of antioxidant and stress defense genes. Nrf1 activity increases in response to oxidative stress and glucose depletion (Chen et al., 2003; Ohtsuji et al., 2008; Zhang et al., 2014b). In addition, signaling through the TorC1 complex can stimulate the expression of proteasome subunit genes by a mechanism that relies on Nrf1 but is Nrf2 independent (Zhang et al., 2014a; Zhang et al., 2014b). Nrf1 deficiency in mice causes developmental lethality (Chan et al., 1998; Chen et al., 2003).
Interestingly, the signal-dependent activation of Nrf1 relies on a mechanism that is strikingly different from the Keap1-mediated regulation of Nrf2. Nrf1 is attached to the ER membrane via its NH2-terminally localized N-terminal domain (NTD) (Wang and Chan, 2006; Zhang et al., 2006). As opposed to other membrane-tethered transcription factors such as SREBP or ATF6 the activation of Nrf1 does not appear to involve signal regulated intramembrane proteolysis, but rather a change in its posttranslational glycosylation status (Zhang et al., 2007; Zhang et al., 2014b). It has been suggested that deglycolsylation of Nrf1 changes the topology and the biochemical function of transcription activation domains ultimately resulting in target gene activation (Zhang et al., 2014b).
The emerging evidence thus suggests that Nrf1 and Nrf2 have similar but at least partially non-redundant functions in stress defense and maintenance of proteostasis. Examination of the different Cnc splicing and translation products has revealed that CncC contains peptide sequences with homology to the N-terminal domain of Nrf1 in addition to sequence homology to Nrf2 (Fig. 2) (Grimberg et al., 2011). Experimental evidence demonstrates that Drosophila CncC combines the functions of the mammalian Nrf1 and Nrf2 genes. Grimberg and colleagues described a role for CncC in both the antioxidant response resembling known Nrf2 function, as well as a role in the regulation of proteasomal activity much like Nrf1. The discovery that CncC seems to be homologous not only to Nrf2, but also to Nrf1 suggested that the Drosophila protein might resemble and ancestral form of the two mammalian transcription factors. The existence of a Drosophila Nrf1 homolog is intriguing because it offers the possibility of examining Cnc-C’s Nrf2 and Nrf1-like functions in using powerful fly genetics in order to gain understanding of shared and distinct roles of Nrf1 and Nrf2, as well as the ways in which these functions are regulated.
Negative regulation by Drosophila Keap1 (dKeap1)
The recognition of the CncC gene product as an Nrf2-related transcription factor led to the characterization of additional Drosophila proteins that are involved in Nrf2 signaling and target gene activation. The Drosophila gene CG3962 encodes a Kelch domain protein with sequence similarity to Keap1 (Sykiotis and Bohmann, 2008). A peptide sequence alignment of the Drosophila and mammalian Keap1 proteins is shown in Fig. 3B. Genetic and cell culture experiments confirmed that, like its mammalian counterparts, Drosophila Keap1 acts as an inhibitor of CncC. For example, overexpression of CncB or CncC in the developing Drosophila eye anlage causes malformation of the adult eye. Co-expression of Keap1 with CncC suppresses this phenotype, whereas the effect of CncB overexpression remains unaffected (Sykiotis and Bohmann, 2008). This genetic interaction between Keap1 and CncC is consistent with the presence of the Keap1-interacting ETGE and DLG motifs in CncC but not in CncB. The function of Drosophila Keap1 as a selective inhibitor of CncC has been confirmed in reporter gene studies in transgenic flies and in cell culture where Keap1 over-expression suppresses CncC dependent gene activation (Chatterjee and Bohmann, 2012). Finally co-immunoprecipitation and bimolecular fluorescence complementation (BiFC) experiments confirmed a physical interaction between CncC and Drosophila Keap1 ((Deng and Kerppola, 2014), M. Tian and D. Bohmann, unpublished observations).
Nrf2 and small Maf proteins in Drosophila
Mammalian Nrf2 proteins bind DNA as a heterodimer with one of several members of the small Maf protein family (Blank, 2008). Drosophila Cnc proteins similarly require a small Maf as a dimerization partner to bind to target gene promoters and activate transcription. Veraksa and colleagues first identified the product of the lone member of the small Maf family in Drosophila, Maf-S, as an obligate dimerization partner of CncB (Veraksa et al., 2000). Maf-S is a 132 amino acid polypeptide mostly comprised of a bZIP domain. The experiments by Veraksa et al. showed that Maf-S is required for the developmental and gene regulatory functions of CncB. An alignment of Drosophila Maf-S with mammalian small Maf proteins is shown in Figure 2C.
More recent experiments by Rahman et al. confirmed that Maf-S is also required for the Nrf2-like functions of CncC (Rahman et al., 2013). The Maf-S protein dimerizes with CncC and mediates both base level and inducible expression of target genes. Maf-S binds to the promoters of CncC target genes even when they are quiescent and appears to facilitate the activation of these genes in response to stress or other signals (Rahman et al., 2013). A similar function has been described for MafK, a mammalian small Maf protein and Nrf2 dimerization partner. MafK was found to occupy the ARE in the 5′ regulatory region of the nqo1 gene in an Nrf2- and stress-independent manner (Nioi et al., 2003). In this way the protein was reported to maintain the nqo1 gene at a low activity in basal conditions and to facilitate transcriptional activation by Nrf2 in response to stress. Functions of pre-bound small Maf proteins, namely to mark specific binding sites in the transcription regulatory regions of target genes and thereby facilitate the specific and efficient binding of Nrf2 in response to appropriate signals, might therefore be conserved between insects and mammals.
It is not clear whether in the absence of nuclear CncC Maf-S binds to AREs as a homodimer or whether it might have a different dimerization partner. Two-hybrid experiments identify no leucine zipper proteins other than Cnc as MafS binding partners (http://www.droidb.org). Furthermore, mammalian small Mafs, are able to bind DNA as a homodimers, at least in vitro (Blank, 2008; Kimura et al., 2007; Motohashi et al., 1997). Thus, it is plausible that in flies in which Nrf2 signaling is not active, ARE elements are occupied by Maf-S homodimers. This speculation however still awaits experimental validation. Interestingly, Maf-S function seems to get lost in older adults resulting in a decline of inducible CncC target gene expression, an effect that appears to contribute to the loss of fitness with increasing age (Rahman et al., 2013).
As outlined above, the conservation of Nrf2 signaling between flies and mammals extends to the small Maf and Keap1 proteins, thereby making Drosophila an attractive model to study functions of the pathway in mechanistic detail. Interestingly, the Skn1 pathway in C. elegans, while clearly sharing many of the characteristic functions with the Nrf2 and CncC systems (see chapter xx by Keith Blackwell), operates without small Mafs or Keap1 homologs. Skn1 lacks a leucine zipper and binds to DNA as a monomer, without the assistance of a dimerization partner such as MafS (Blackwell et al., 1994). A homolog of Keap1 has not been identified in the worm, even though the WD40 domain protein WDR-23, might functionally replace Keap1 (Choe et al., 2009).
Drosophila CncC in stress response and aging
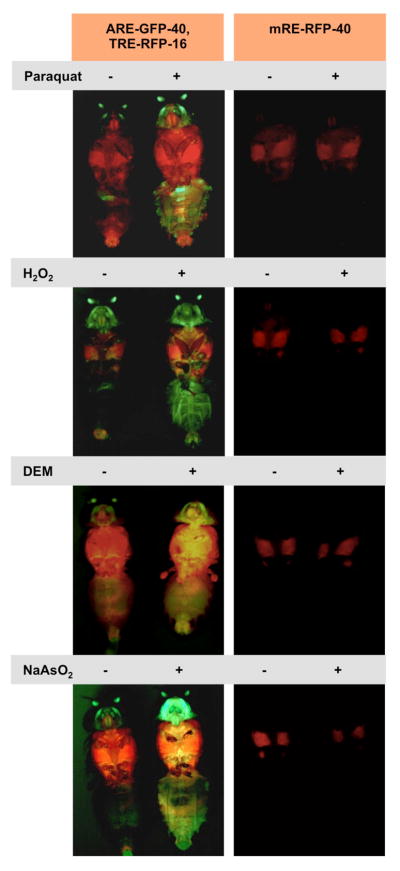
As mentioned above, sequence similarities between Nrf2 and CncC raised the suggestion that the Drosophila protein might be a functional homolog of the mammalian transcription factor and serve as an inducer of stress responsive, protective gene expression programs. (Misra et al., 2011; Sykiotis and Bohmann, 2008). Drosophila CncC can selectively bind to Nrf2 binding sites, or AREs (antioxidant response elements) to activate the transcription of nearby genes. The AREs found in Drosophila are strikingly similar to those found in mammalian systems, indicating that the sequence specificity of DNA binding is conserved between mammalian Nrf2 portions and CncC (Chatterjee and Bohmann, 2012). CncC regulated genes included many homologs of well-established Nrf2 target genes such as those encoding glutathione S-transferases, thioredoxin and enzymes in the glutathione synthesis pathway. CncC transcriptional activity increased when Keap1 expression was genetically diminished or when flies where exposed to acute xenobiotic stress. CncC-dependent target gene activation was, for example, observed upon oral administration of paraquat, an organic compound that promotes mitochondrial ROS production or DEM, a glutathione inhibitor, H2O2 and also in response to heavy metals such as arsenate (Sykiotis and Bohmann, 2008). (see Fig. 4) Interestingly, flies also respond to cancer chemo preventive agents such as oltipraz and sulforaphane with up regulation of CncC (Sykiotis and Bohmann, 2008. G.P. Sykiotis, unpublished results). These drugs can stimulate Nrf2 activity without causing harmful levels of stress and are therefore well tolerated. They can confer protection against chemically induced cancer in rodent models, an effect that is dependent on Nrf2 activity (Kensler et al., 1999; Zhang and Gordon, 2004). Nrf2-activating drugs can also be beneficial in the treatment of other diseases that are associated with oxidative stress, including neurodegenerative and inflammatory conditions. Indeed, an Nrf2 inducer, dimethyl fumarate, marketed under the brand name Tecfidera®, has recently gained FDA approval for treatment of multiple sclerosis (English and Aloi, 2015; Linker et al., 2011). The possibility of modeling the effect Nrf2 inducing drugs in Drosophila may contribute to a better understanding of the orgasmic functions and the mechanism of action of this promising class of therapeutics.
Figure 4. Activation of Drosophila Nrf2 reporter genes by different stressors.
As in mammals, Nrf2 activity can be activated in Drosophila in response to a variety of different types of cells stress. Flies carrying a CncC responsive ARE-GFP reporter and, for comparison, a separate AP-1 (Fos & Jun) responsive TRE-GFP GFP reporter (Chatterjee and Bohmann, 2012) were exposed to dietary Paraquat, H2O2, DEM, and sodium aresenite. Controls treated with matching solvents and photographed under the same exposure are shown. A mutated, Nrf2 unresponsive reporter (mRE-RFP) is not affected by these treatment conditions.
The protective function of CncC against acute oxidative stress and the longstanding notion that progressive increase of oxidative damage contributes to the degenerative and ultimately fatal effects of aging led to the idea that CncC function might affect the aging process. This hypothesis was tested by measuring the effects of CncC activity on Drosophila lifespan. The decreased gene dose of keap1 in a flystock that is heterozygous for a loss of function allele causes a mild increase of CncC activity. Males of this genotype showed a moderate, but statistically significant lifespan extension compared to otherwise isogenic wild type stocks (Sykiotis and Bohmann, 2008). This result indicated a role of Nrf2 proteins in the regulation of aging. Further support for this conclusion came from experiments in C. elegans that showed the gain of function conditions for the worm ortholog of Nrf2, Skn1, can promote longevity (Tullet et al., 2008). In addition, it was found that Skn1 expression in a single pair of sensory neurons, called ASI, is essential for the lifespan extending effects of calorie restriction (Bishop and Guarente, 2007). Still, much has to be learned about the signals and pathways, including Nrf2, that control the aging process. Flies and worms offer attractive and tractable genetic models to address these questions experimentally.
Recent advances
The identification of CncC as the functional and structural homolog of Nrf2, and possibly Nrf1, in the fly has opened the field to the experimental possibilities available in the Drosophila model system. Recent discoveries made using Drosophila have expanded our understanding of the role played by Cnc-family transcription factors in aging, cancer, development, and pesticide resistance, all of which could have profound implications on human health and diseases.
Aging and Proteostasis
Aging is characterized by the progressive decline in an organism’s ability to maintain homeostasis and correlates with an increased risk of death and disease (Lopez-Otin et al., 2013). Although a number of theories have been proposed to mechanistically explain aging, the precise causal mechanisms remain unclear (Moskalev et al., 2014; Rattan, 2006). A common thread in many theories of aging is that accumulation of damaged biomolecules is a driving force of the aging process (Gems and Partridge, 2013; Harman, 1956; Ziegler et al., 2015). In addition to preventing such damage by directing the expression of antioxidants, the cap’n’collar protein family has been shown to play a key role in the breakdown and clearance of damaged proteins by regulating the transcriptional activation of molecular chaperones and proteolytic systems that include the autophagosome and proteasome (Arlt et al., 2009; Pickering et al., 2012; Radhakrishnan et al., 2010; Tsakiri et al., 2013b; Zhang et al., 2014a). The central proteolytic component of the ubiquitin-proteasome system (UPS), the 26S proteasome, plays an important role in maintaining protein homeostasis (or proteostasis) by degrading mutated, damaged or misfolded proteins, as well as targeting normally short lived proteins for degradation (Bedford et al., 2009; Totta et al., 2014). The 26S proteasome is comprised of one or two regulatory 19S particles and a 20S core particle that contains the proteolytically active sites (da Fonseca and Morris, 2008). Recent work by Tsakiri and colleagues has examined the role of Cap’n’collar transcription factors in proteostasis and the regulation of the proteasome in different tissues (Tsakiri et al., 2013a). In this study they compared mechanisms that maintain proteostasis in somatic and in germ line tissues at different ages. They discovered that, despite showing elevated CncC and antioxidant activity, the abundance of damaged proteins, as indicated by increases in protein carbonylation and advanced glycation end product (AGE), increased with advancing age in somatic tissues. This increase in oxidatively damaged protein abundance correlated with decreases in the peptidase activity of isolated proteasomes and was attributed to down regulation of both proteins and mRNA of all major 19S and 20S proteasome subunits in a variety of somatic tissues tested. In striking contrast, gonadal tissue isolated from aged animals maintained proteasome expression and activity levels similar to those observed in young animals. In addition, protein carbonylation and AGE accumulation were virtually undetectable in these tissues. Further comparison of somatic versus germ line tissue revealed that expression and activity of the proteasome, as well as antioxidant responses, remained stable throughout life in the germ line tissue, but declined with age in somatic tissues. It was also observed that gonadal tissue expressed higher levels of CncC and Keap1 when compared to the soma, suggesting that germline-specific maintenance of CncC activity may underlie the persistence of the proteasome function. Consistent with this interpretation, tissue specific genetic activation of the Nrf2 pathway by keap1RNAi in the soma restored youthful proteasome activity to somatic tissues while inactivation of the pathway by cncCRNAi in the gonads led to decline in proteasome activity similar to what was observed in aging somatic tissue. These findings demonstrate that the Nrf2 pathway plays a key role in the regulation of proteostasis in somatic and reproductive tissues, that this function can decline during the aging process, and that it may even contribute to it. It is interesting to note that this prolonged protective function of CncC in the germline compared to the soma is consistent with a “dispensable soma theory” which postulates that more energy is spent protecting the germ line in order to safeguard the genetic material being passed on to the next generation at the cost of sacrificing the less essential somatic tissue (Kirkwood, 2005).
Autophagy
Autophagy is a process that, like UPS, is critical for the maintenance of proteostasis and intracellular homeostasis. In the course of autophagy, proteins, lipids, and organelles are captured within double-membrane organelles called autophagosomes and targeted either for lysosomal degradation or for recycling (Mizushima, 2007). Autophagy plays an important role in maintaining integrity and energy balance in eukaryotic cells. An important function of autophagy is the clearing of damaged proteins and organelles from the cell. However, autophagy can also support the survival and growth of malignant tumors and excessive autophagic activity can cause cell death. The process therefore must be tightly controlled (Kimmelman, 2011; White, 2012). Myc is a protein required for the expression of a variety of genes involved in cell growth and proliferation (Carroll et al., 2015; Eilers and Eisenman, 2008). Deregulation of Myc is observed in a large number of different cancer types and is considered a classic oncogenic event (Dang, 2012; Land et al., 1983). A study performed by Nagy and colleagues explored the relationship between Myc and autophagy using Drosophila as a model organism. The authors discovered a novel relationship between Myc-induced overgrowth and the Nrf2 pathway (Nagy et al., 2013). Depletion of Myc using RNAi correlated with reduced cell size within the fat body in a cell-autonomous manner and stabilization of the autophagic cargo, p62 (a commonly used marker for estimating autophagic degradation) (Nezis et al., 2008; Pircs et al., 2012). Conversely, overexpression of Myc in the fat body led to increases in cell growth as well as increases in lipidation of the autophagosome-associated protein, Atg8a-II. The latter is indicative of increased autophagic activity. Additionally, they showed that overexpression of Myc caused an increase in flux through the autophagy pathway. Together these observations demonstrated that Myc plays an important role in the regulation of both the induction of autophagy, as well as the degradation steps of the autophagy response. Nagy and colleagues also observed that overexpression of Myc led to the accumulation of p62 and of ubiquitinated proteins. Previous work had shown that p62 plays an important role in tumorigenesis by promoting the persistent activation of antioxidant responses via disrupting the interaction between Keap1 and Nrf2 (Komatsu et al., 2010). Furthermore, overexpression of Myc activated the Nrf2 transcriptional reporters (gstD-lacZ and gstD-GFP) and knockdown of p62 or Cnc prevented the activation of these reporters, suggesting that Myc was activating the antioxidant response through the p62/Nrf2 signaling pathway. Finally, both autophagy and the antioxidant response were found to be activated simultaneously by Myc deregulation and both are required to maintain the Myc-induced overgrowth in Drosophila. Non-oncogene addiction is described as any cellular process that is required for cancer cell growth but is (at least to some degree) dispensable in normal cells (Luo et al., 2009). Nrf2 knockout mice are hypersensitive to oxidants, but are able to survive into adulthood, and tissue specific knockout of autophagy genes (Atg) in mouse models are viable with adverse effects occurring in aged animals (Chan et al., 1996; Mizushima and Komatsu, 2011). These pathways may provide potential non-oncogene addiction targets for future cancer therapies aimed at Myc-dependent human cancers.
Cap’n’collar, Keap1 and endocrine signaling
Whereas the role of the Nrf2 pathway in the antioxidant response is fairly well characterized, its function in other biological processes is less understood. Experiments in the Kerppola lab have unraveled an unexpected connection between CncC and the hormonal control of development (Deng and Kerppola, 2013). Drosophila metamorphosis involves the coordinated endocrine regulation of transcription throughout the entire organism. A key regulator of metamorphosis and development in insects like Drosophila is the hormone, ecdysone (Ables et al., 2015; Ohhara et al., 2015). Several points during metamorphosis, which mark the transitions between the different larval and pupal stages of development, are controlled by the release of ecdysone pulses. The resulting ecdysone-mediated regulation of transcriptional activity manifests itself in the form of chromosomal “puffs” at specific regions of polytene chromosomes found within the salivary gland (Ashburner and Lemeunier, 1972). Polytene chromosomes are oversized versions of normal chromosomes found in the salivary gland of several insect species including Drosophila. They result from endoreplication, DNA replication without cell division, in these cells. Hundreds of thus formed chromosomal DNA strands align to form structures that are readily observable by light microscopy and accessible to cytological analysis such as immunohistochemistry to localize the position of specific DNA bound proteins. Recent work from the Kerppola group (Deng and Kerppola, 2013) demonstrated that both CncC and Keap1 play an important role in the coordination of ecdysone-mediated metamorphosis. By examining the localization of CncC and Keap1 on polytene chromosome they observed that CncC and Keap1 bind directly to a majority of previously identified ecdysone-regulated genes. In apparent contrast to their well-established opposing effects on antioxidant signaling, where a loss of Keap1 causes target gene activation in a CncC dependent manner, depletion of both CncC or Keap1 in the salivary glands led to the reduction of many of the same ecdysone regulated transcripts. This suggested a previously unrecognized nuclear function for Keap1 where both CncC and Keap1 support the expression of ecdysone responsive genes in a manner that is distinct from their roles in the antioxidant response pathway. It was also observed that depletion of CncC or Keap1 led to a delayed pupation phenotype caused by decreases in the molting hormone 20-hydroxyecdysone (20E). The timing of pupation and production of 20E involves the Ras signaling pathway as well as the neuropeptide prothoracictropic hormone (PTTH) (Rewitz et al., 2009). To determine the relationship between Ras and CncC the effects of CncC depletion in a constitutively active Ras (RasVal12) mutant were explored in the prothoracic gland (PG), an endocrine gland that secretes ecdysone. As previously reported, constitutive activation of Ras caused early pupation and smaller pupae (Caldwell et al., 2005). Depletion of CncC in the PG restored pupation timing and size to levels similar to wild type. Further examination of CncC binding to polytene chromosomes in the RasVal12 mutant revealed increases in both the level as well as the number of loci occupied by CncC suggesting that Ras influences the efficiency and specificity of CncC binding to polytene chromosomes. Together these results suggest that CncC plays an active role in the regulation of pupation by mediating the effects of the Ras signaling pathway and controlling the transcription of ecdysone target genes. Ecdysone and other ecdysteroids appear to be primarily found in invertebrates, but steroid hormones are found in nearly all higher organisms. This raises the possibility that Nrf2 and/or Keap1 may play a previously unappreciated role in other biological processes such as human development by coordinating the responses to external and internal cues in order to fine tune progression through this process.
CncC as regulators of adult stem cell maintenance
Regulation of the intracellular redox balance has previously been shown to play a key role in the maintenance of stem cell populations. Elevated levels of ROS can reduce the self-renewal capacity of stem cells by causing hyperproliferation in a number of progenitor populations in mammalian systems (L’Honore et al., 2014; Wang et al., 2015). Similarly, it has been observed that the redox state of intestinal stem cells (ISCs) found within the midgut of Drosophila is a driving force behind stem cell maintenance. Nrf2-Keap1 signaling is a major regulator of redox balance within these cells (Hochmuth et al., 2011). Given the prominent role of ROS signaling in the maintenance of stem cell pluripotency, Hochmuth and colleagues examined CncC activity in ISCs using CncC target gene reporter (gstD1::lacZ) and found it to be highly activated in stem cell populations. Interestingly, they observed this elevated reporter activity in the absence of external stress. Surprisingly, however, and contrary to what is observed in differentiated cells, under oxidative stress conditions gstd1::lacZ activity was repressed in ISCs. Overexpression of CncC or knockdown of Keap1 by RNAi prior to exposure to oxidative stress reduced the proliferative potential of ISCs as well as gstd1::lacZ activity in these cells. To determine if CncC might function as a general inhibitor of stem cell proliferation, the effects of overexpression of CncC under conditions previously observed to increase stem cell proliferation (JNK activation, overexpression of the insulin receptor (InR), NotchRNAi) was explored. Under all three of these conditions coexpression of CncC inhibited ISC proliferation suggesting that CncC functions as a negative regulator of proliferation in this stem cell population. To examine the role that CncC signaling has on intracellular redox state of ISCs following oxidative stress, they used redox-sensitive dye, dihydro-ethidium (DHE). Compared to differentiated cells, the stem cells demonstrated significantly reduced oxidation of DHE. Loss of CncC by CncCRNAi lead to increases in intracellular oxidation and stem cell proliferation suggesting that CncC is responsible for responding to changes in ROS levels that are involved in promoting stem cell proliferation. It had previously been reported that overproliferation of ISCs leads to loss of epithelial homeostasis in aged Drosophila (Biteau et al., 2008). To determine if CncC plays a role in this loss of homeostasis Hochmuth and colleagues aged flies that overexpressed CncC or had knocked down levels of CncC by RNAi. Flies that were aged expressing the CncCRNAi construct displayed increased stem cell proliferation and accelerated loss in tissue homeostasis compared to their wild type counterparts. In contrast, flies that overexpressed CncC had lower ISC proliferation rates and reduced loss of tissue homeostasis. Together these observations suggest a model in which the Nrf2/CncC pathway exhibits a “reverse” stress response in ISCs. Elevated levels of CncC helps to maintain stem cell populations by keeping intracellular ROS levels low that in turn keeps ISCs in a quiescent state. Upon insult or injury CncC is downregulated creating a state that is permissive for stem cell expansion, differentiation, and regeneration. These results provide further evidence that redox state is a major factor in the maintenance of stem cell populations and that the Nrf2/Keap1 signaling module plays an integral role in maintaining stem cell quiescence by modulating redox balance.
Pesticide resistance
The development of pesticide resistance in insect populations poses a huge challenge in controlling insect-borne human diseases, as well as in protection of agricultural resources targeted by insects (Heckel, 2012). There are two primary mechanisms thought to underlie the development of pesticide resistance. Mutations that result in a decrease in the ability of the pesticide to bind or block the activity of the target is known as target site resistance (Edi et al., 2014). Alternatively, upregulation of detoxification pathways to eliminate or remove the pesticide from the animal is known as metabolic resistance. Metabolic resistance can also involve mutations that cause constitutive activation of detoxification pathways (Mulamba et al., 2014). Drosophila has long been used as a model for studying the mechanisms and the development of insecticide resistance (Morton, 1993; Pittendrigh et al., 1997). Misra and colleagues used two independently established insecticide resistant Drosophila lines (91R and RDDTR) to better understand the mechanisms surrounding metabolic resistance (Misra et al., 2013). They observed that both of these resistant lines showed elevated levels of several CncC target genes suggesting that the Nrf2 pathway might be upregulated in these strains and may contribute to their enhanced insecticide resistance. Using a CncC reporter construct (Cyp6a8-lacZ) they demonstrated that the reporter activity was elevated in both resistant lines and that the reporter activity was dependent upon CncC binding to the target gene promoter. In addition, intercrossed F1 animals containing one set of 91R chromosomes and one set of the RDDTR chromosomes had an additive effect on Cyp6a8-lacZ reporter activity. Previous work has established that upregulation of cytochrome P450 detoxification and CncC target genes (Cyp6a2 and Cyp6a8) in several insecticide resistant backgrounds involves gene(s) located on the third chromosome (Maitra et al., 2000). Following outcrossing, fly lines carrying the third chromosome from either the 91R or RDDTR backgrounds caused constitutive activation of the CncC reporter, indicating that the factor(s) involved in the upregulation of the Nrf2 pathway is located on the third chromosome. Furthermore, disruption of CncC activity (by CncCRNAi or Keap1 overexpression) caused a significant decrease in the upregulation of Cyp6a2 and Cyp6a8 in both resistant lines providing further evidence that the Nrf2 pathway is playing a critical role in upregulating the proteins involved in the detoxification process. Given the conserved detoxification function of the Nrf2 pathway, constitutive activation of the pathway may be a common mechanism of resistance found in other insecticide resistant insect populations and therefore may provide a potential target for novel insecticides and/or improve the efficacy of current methods of insect control. Furthermore, this system may be used to explore the varying degrees of misregulation of the Nrf2 and other detoxification pathways as an underlying cause for variation in the susceptibility as well as efficacy of drugs in disease states such as cancer (Jaramillo and Zhang, 2013; Wang et al., 2010).
Concluding Remarks
Since its initial description almost a quarter century ago, the Drosophila cnc gene has been a remarkable research subject that has helped to advance several scientific disciplines. In addition to the insights into principles of early embryonic morphogenesis, pattern formation and developmental gene regulation that came out of the initial studies, cnc provided a simple, but powerful model to investigate the mechanisms and functions of Nrf2 signaling. In this chapter we have described the components mediating Nrf2 signaling in Drosophila and their relationship to human and worm homologs. Several examples were presented to illustrate how the experimental assets available in the fly system have been used to generate new information on the biological functions of Nrf2 and to develop concepts that will fuel future research. Other interesting projects have expanded our knowledge of Nrf2 biology but could not be covered in this chapter. These include the demonstration that CncC can protect flies from neurodegeneration and loss of motor function in a Parkinson’s Disease model (Barone et al., 2011; Trinh et al., 2010). As the experimental tools to assess and manipulate Cnc function in the fly system become more and more refined and widely accessible, we can expect further discoveries on specific biological functions of the cap’n’collar transcription factors.
The cap’n’collar family remains a subject of intense scientific interest and many important questions still need to be experimentally addressed: What is the connection between antioxidant and metabolic regulation by these factors? More generally, how does the regulation of cellular redox balance by Nrf2 and similar factors modulate the activities and readouts of other signaling pathways and how does this interplay affect cell regulation and organism function? Are the biological effects of short-term Nrf2 stimulation, for example in response to acute stress different from prolonged changes in activity? What are the relevant upstream signaling molecules mediating such different responses? What is the role of Nrf2 and its relatives in controlling aging, and, conversely what is the effect of age on the protective function of these transcription factors? How do epigenetic factors, small RNA or drugs affect the function of the Nrf2 in an intact, aging organism? Genetically tractable, comparatively simple organisms like Drosophila and C. elegans offer facile, rapid and economic avenues towards addressing these important questions. Research employing these model organisms will continue to complement studies in mammalian and in vitro systems in productive and innovative ways for the foreseeable future.
Acknowledgments
This work was supported by R01 AG039753 to DB and a fellowship from T32 5T90DE021985-04 to AP. The authors would like to thank Catherine Ovitt for helpful comments on the manuscript and Nirmalya Chatterjee for contributions to Figure 4.
References
- Ables ET, Bois KE, Garcia CA, Drummond-Barbosa D. Ecdysone response gene E78 controls ovarian germline stem cell niche formation and follicle survival in Drosophila. Developmental biology. 2015 doi: 10.1016/j.ydbio.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt A, Bauer I, Schafmayer C, Tepel J, Muerkoster SS, Brosch M, Roder C, Kalthoff H, Hampe J, Moyer MP, et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2) Oncogene. 2009;28:3983–3996. doi: 10.1038/onc.2009.264. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Lemeunier F. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VII. Homology of puffing patterns on chromosome arm 3L in D. melanogaster and D. yakuba, with notes on puffing in D. teissieri. Chromosoma. 1972;38:283–295. doi: 10.1007/BF00290926. [DOI] [PubMed] [Google Scholar]
- Baird L, Lleres D, Swift S, Dinkova-Kostova AT. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone MC, Sykiotis GP, Bohmann D. Genetic activation of Nrf2 signaling is sufficient to ameliorate neurodegenerative phenotypes in a Drosophila model of Parkinson’s disease. Dis Model Mech. 2011;4:701–707. doi: 10.1242/dmm.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Paine S, Rezvani N, Mee M, Lowe J, Mayer RJ. The UPS and autophagy in chronic neurodegenerative disease: six of one and half a dozen of the other--or not? Autophagy. 2009;5:224–227. doi: 10.4161/auto.5.2.7389. [DOI] [PubMed] [Google Scholar]
- Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. The EMBO journal. 2008;27:212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- Caldwell PE, Walkiewicz M, Stern M. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release. Current biology : CB. 2005;15:1785–1795. doi: 10.1016/j.cub.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Carroll PA, Diolaiti D, McFerrin L, Gu H, Djukovic D, Du J, Cheng PF, Anderson S, Ulrich M, Hurley JB, et al. Deregulated Myc Requires MondoA/Mlx for Metabolic Reprogramming and Tumorigenesis. Cancer cell. 2015;27:271–285. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. The EMBO journal. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Bohmann D. A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PloS one. 2012;7:e34063. doi: 10.1371/journal.pone.0034063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kwong M, Lu R, Ginzinger D, Lee C, Leung L, Chan JY. Nrf1 is critical for redox balance and survival of liver cells during development. Molecular and cellular biology. 2003;23:4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Molecular and cellular biology. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. The Journal of biological chemistry. 2008;283:23305–23314. doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Kerppola TK. Regulation of Drosophila metamorphosis by xenobiotic response regulators. PLoS genetics. 2013;9:e1003263. doi: 10.1371/journal.pgen.1003263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Kerppola TK. Visualization of the Drosophila dKeap1-CncC interaction on chromatin illumines cooperative, xenobiotic-specific gene activation. Development (Cambridge, England) 2014;141:3277–3288. doi: 10.1242/dev.110528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derjuga A, Gourley TS, Holm TM, Heng HH, Shivdasani RA, Ahmed R, Andrews NC, Blank V. Complexity of CNC transcription factors as revealed by gene targeting of the Nrf3 locus. Molecular and cellular biology. 2004;24:3286–3294. doi: 10.1128/MCB.24.8.3286-3294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edi CV, Djogbenou L, Jenkins AM, Regna K, Muskavitch MA, Poupardin R, Jones CM, Essandoh J, Ketoh GK, Paine MJ, et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS genetics. 2014;10:e1004236. doi: 10.1371/journal.pgen.1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc’s broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English C, Aloi JJ. New FDA-Approved Disease-Modifying Therapies for Multiple Sclerosis. Clinical therapeutics. 2015 doi: 10.1016/j.clinthera.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Fukutomi T, Takagi K, Mizushima T, Ohuchi N, Yamamoto M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Molecular and cellular biology. 2014;34:832–846. doi: 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annual review of physiology. 2013;75:621–644. doi: 10.1146/annurev-physiol-030212-183712. [DOI] [PubMed] [Google Scholar]
- Grimberg KB, Beskow A, Lundin D, Davis MM, Young P. Basic leucine zipper protein Cnc-C is a substrate and transcriptional regulator of the Drosophila 26S proteasome. Molecular and cellular biology. 2011;31:897–909. doi: 10.1128/MCB.00799-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding KW, Gellon G, McGinnis N, McGinnis W. A screen for modifiers of Deformed function in Drosophila. Genetics. 1995;140:1339–1352. doi: 10.1093/genetics/140.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in biochemical sciences. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends in biochemical sciences. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Heckel DG. Ecology. Insecticide resistance after Silent spring. Science. 2012;337:1612–1614. doi: 10.1126/science.1226994. [DOI] [PubMed] [Google Scholar]
- Hochmuth CE, Biteau B, Bohmann D, Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell. 2011;8:188–199. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes & development. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes & development. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Iida K, Kang MI, Kobayashi A, Mizukami M, Tong KI, McMahon M, Hayes JD, Itoh K, Yamamoto M. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch Biochem Biophys. 2005;433:342–350. doi: 10.1016/j.abb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes to cells : devoted to molecular & cellular mechanisms. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Groopman JD, Sutter TR, Curphey TJ, Roebuck BD. Development of cancer chemopreventive agents: oltipraz as a paradigm. Chem Res Toxicol. 1999;12:113–126. doi: 10.1021/tx980185b. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & development. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Yamamoto T, Zhang J, Itoh K, Kyo M, Kamiya T, Aburatani H, Katsuoka F, Kurokawa H, Tanaka T, et al. Molecular basis distinguishing the DNA binding profile of Nrf2-Maf heterodimer from that of Maf homodimer. The Journal of biological chemistry. 2007;282:33681–33690. doi: 10.1074/jbc.M706863200. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature cell biology. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- L’Honore A, Commere PH, Ouimette JF, Montarras D, Drouin J, Buckingham M. Redox regulation by Pitx2 and Pitx3 is critical for fetal myogenesis. Developmental cell. 2014;29:392–405. doi: 10.1016/j.devcel.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Li JJ, Huang H, Bickel PJ, Brenner SE. Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data. Genome research. 2014;24:1086–1101. doi: 10.1101/gr.170100.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker RA, Lee DH, Ryan S, van Dam AM, Conrad R, Bista P, Zeng W, Hronowsky X, Buko A, Chollate S, et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain : a journal of neurology. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S, Dombrowski SM, Basu M, Raustol O, Waters LC, Ganguly R. Factors on the third chromosome affect the level of cyp6a2 and cyp6a8 expression in Drosophila melanogaster. Gene. 2000;248:147–156. doi: 10.1016/s0378-1119(00)00129-3. [DOI] [PubMed] [Google Scholar]
- McGinnis N, Ragnhildstveit E, Veraksa A, McGinnis W. A cap ‘n’ collar protein isoform contains a selective Hox repressor function. Development. 1998;125:4553–4564. doi: 10.1242/dev.125.22.4553. [DOI] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. The Journal of biological chemistry. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J, FlyBase C. FlyBase 101--the basics of navigating FlyBase. Nucleic acids research. 2012;40:D706–714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic acids research. 2013;41:W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra JR, Horner MA, Lam G, Thummel CS. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes & development. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra JR, Lam G, Thummel CS. Constitutive activation of the Nrf2/Keap1 pathway in insecticide-resistant strains of Drosophila. Insect biochemistry and molecular biology. 2013;43:1116–1124. doi: 10.1016/j.ibmb.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes & development. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- modENCODE Consortium. Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler J, Mahaffey JW, Deutsch E, Vani K. Control of Drosophila head segment identity by the bZIP homeotic gene cnc. Development. 1995;121:237–247. doi: 10.1242/dev.121.1.237. [DOI] [PubMed] [Google Scholar]
- Mohler J, Vani K, Leung S, Epstein A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mechanisms of development. 1991;34:3–9. doi: 10.1016/0925-4773(91)90086-l. [DOI] [PubMed] [Google Scholar]
- Mohr SE, Hu Y, Kim K, Housden BE, Perrimon N. Resources for functional genomics studies in Drosophila melanogaster. Genetics. 2014;197:1–18. doi: 10.1534/genetics.113.154344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton RA. Evolution of Drosophila insecticide resistance. Genome / National Research Council Canada = Genome / Conseil national de recherches Canada. 1993;36:1–7. doi: 10.1139/g93-001. [DOI] [PubMed] [Google Scholar]
- Moskalev AA, Aliper AM, Smit-McBride Z, Buzdin A, Zhavoronkov A. Genetics and epigenetics of aging and longevity. Cell cycle. 2014;13:1063–1077. doi: 10.4161/cc.28433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic acids research. 1997;25:2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulamba C, Riveron JM, Ibrahim SS, Irving H, Barnes KG, Mukwaya LG, Birungi J, Wondji CS. Widespread pyrethroid and DDT resistance in the major malaria vector Anopheles funestus in East Africa is driven by metabolic resistance mechanisms. PloS one. 2014;9:e110058. doi: 10.1371/journal.pone.0110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Varga A, Pircs K, Hegedus K, Juhasz G. Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS genetics. 2013;9:e1003664. doi: 10.1371/journal.pgen.1003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. The Journal of cell biology. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. The Biochemical journal. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Molecular and cellular biology. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhara Y, Shimada-Niwa Y, Niwa R, Kayashima Y, Hayashi Y, Akagi K, Ueda H, Yamakawa-Kobayashi K, Kobayashi S. Autocrine regulation of ecdysone synthesis by beta3-octopamine receptor in the prothoracic gland is essential for Drosophila metamorphosis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:1452–1457. doi: 10.1073/pnas.1414966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. The Journal of biological chemistry. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering AM, Linder RA, Zhang H, Forman HJ, Davies KJ. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. The Journal of biological chemistry. 2012;287:10021–10031. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PloS one. 2012;7:e44214. doi: 10.1371/journal.pone.0044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh B, Reenan R, ffrench-Constant RH, Ganetzky B. Point mutations in the Drosophila sodium channel gene para associated with resistance to DDT and pyrethroid insecticides. Molecular & general genetics : MGG. 1997;256:602–610. doi: 10.1007/s004380050608. [DOI] [PubMed] [Google Scholar]
- Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Molecular and cellular biology. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Molecular cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Sykiotis GP, Nishimura M, Bodmer R, Bohmann D. Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging cell. 2013 doi: 10.1111/acel.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattan SI. Theories of biological aging: genes, proteins, and free radicals. Free radical research. 2006;40:1230–1238. doi: 10.1080/10715760600911303. [DOI] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, Gilbert LI, O’Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326:1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Perkins LA, Capaci TM, Perrimon N. Putative protein kinase product of the Drosophila segment-polarity gene zeste-white3. Nature. 1990;345:825–829. doi: 10.1038/345825a0. [DOI] [PubMed] [Google Scholar]
- Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes & development. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Stress-Activated Cap’n’collar Transcription Factors in Aging and Human Disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totta P, Pesiri V, Marino M, Acconcia F. Lysosomal function is involved in 17beta-estradiol-induced estrogen receptor alpha degradation and cell proliferation. PloS one. 2014;9:e94880. doi: 10.1371/journal.pone.0094880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh K, Andrews L, Krause J, Hanak T, Lee D, Gelb M, Pallanck L. Decaffeinated coffee and nicotine-free tobacco provide neuroprotection in Drosophila models of Parkinson’s disease through an NRF2-dependent mechanism. J Neurosci. 2010;30:5525–5532. doi: 10.1523/JNEUROSCI.4777-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JJ, Dudakov JA, Takahashi K, Shieh JH, Velardi E, Holland AM, Singer NV, West ML, Smith OM, Young LF, et al. Nrf2 regulates haematopoietic stem cell function. Nature cell biology. 2013;15:309–316. doi: 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri EN, Sykiotis GP, Papassideri IS, Gorgoulis VG, Bohmann D, Trougakos IP. Differential regulation of proteasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013a;27:2407–2420. doi: 10.1096/fj.12-221408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri EN, Sykiotis GP, Papassideri IS, Terpos E, Dimopoulos MA, Gorgoulis VG, Bohmann D, Trougakos IP. Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging cell. 2013b;12:802–813. doi: 10.1111/acel.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JMA, Hertweck M, An JH, Baker J, Oliviera RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity promoting factor SKN-1 by Insulinlike signaling in C. elegans. Cell. 2008;132 doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraksa A, McGinnis N, Li X, Mohler J, McGinnis W. Cap ‘n’ collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127:4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. The Journal of biological chemistry. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhang Y, Lu W, Liu K. Mitochondrial reactive oxygen species regulate adipocyte differentiation of mesenchymal stem cells in hematopoietic stress induced by arabinosylcytosine. PloS one. 2015;10:e0120629. doi: 10.1371/journal.pone.0120629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chorley BN, Pittman GS, Kleeberger SR, Brothers J, 2nd, Liu G, Spira A, Bell DA. Genetic variation and antioxidant response gene expression in the bronchial airway epithelium of smokers at risk for lung cancer. PloS one. 2010;5:e11934. doi: 10.1371/journal.pone.0011934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Molecular and cellular biology. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Molecular and cellular biology. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. The Biochemical journal. 2006;399:373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gordon GB. A strategy for cancer prevention: stimulation of the Nrf2-ARE signaling pathway. Mol Cancer Ther. 2004;3:885–893. [PubMed] [Google Scholar]
- Zhang Y, Lucocq JM, Yamamoto M, Hayes JD. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. The Biochemical journal. 2007;408:161–172. doi: 10.1042/BJ20070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014a;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ren Y, Li S, Hayes JD. Transcription factor Nrf1 is topologically repartitioned across membranes to enable target gene transactivation through its acidic glucose-responsive domains. PloS one. 2014b;9:e93458. doi: 10.1371/journal.pone.0093458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler DV, Wiley CD, Velarde MC. Mitochondrial effectors of cellular senescence: beyond the free radical theory of aging. Aging cell. 2015;14:1–7. doi: 10.1111/acel.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]