Abstract
Redox signaling is important for embryogenesis, guiding pathways that govern processes crucial for embryo patterning, including cell polarization, proliferation, and apoptosis. Exposure to pro-oxidants during this period can be deleterious, resulting in altered physiology, teratogenesis, later-life diseases, or lethality. We previously reported that the glutathione antioxidant defense system becomes increasingly robust, including a doubling of total glutathione and dynamic shifts in the glutathione redox potential at specific stages during embryonic development in the zebrafish, Danio rerio. However, the mechanisms underlying these changes are unclear, as is the effectiveness of the glutathione system in ameliorating oxidative insults to the embryo at different stages. Here, we examine how the glutathione system responds to the model pro-oxidants tert-butylhydroperoxide and tert-butylhydroquinone at different developmental stages, and the role of Nuclear factor erythroid 2-related factor (Nrf) proteins in regulating developmental glutathione redox status. Embryos became increasingly sensitive to pro-oxidants after 72 h post-fertilization (hpf), after which the duration of the recovery period for the glutathione redox potential was increased. To determine whether the doubling of glutathione or the dynamic changes in glutathione redox potential are mediated by zebrafish paralogs of Nrf transcription factors, morpholino oligonucleotides were used to knock down translation of Nrf1 and Nrf2 (nrf1a, nrf1b, nrf2a, nrf2b). Knockdown of Nrf1a or Nrf1b perturbed glutathione redox state until 72 hpf. Knockdown of Nrf2 paralogs also perturbed glutathione redox state but did not significantly affect the response of glutathione to pro-oxidants. Nrf1b morphants had decreased gene expression of glutathione synthesis enzymes, while hsp70 increased in Nrf2b morphants. This work demonstrates that despite having a more robust glutathione system, embryos become more sensitive to oxidative stress later in development, and that neither Nrf1 nor Nrf2 alone appear to be essential for the response and recovery of glutathione to oxidative insults.
Keywords: Embryonic development, Glutathione, Oxidative stress, Redox, Zebrafish, Antioxidant
Highlights
-
•
Sensitivity of the embryo's glutathione system to pro-oxidants increases throughout development.
-
•
Nrf1a/b play significant roles in early embryonic redox homeostasis, while Nrf2a becomes important later in development.
-
•
Deficient Nrf signaling impacts expression of genes central to GSH synthesis and cytoprotection in the embryo.
1. Introduction
Oxidative stress is classically defined as the imbalance of the cellular environment towards a more oxidized, depolarized state. Under homeostatic conditions, the cell is in a balanced redox state, where the reducing power of the cell is able to mitigate or prevent the damage that would be done by oxidizing species. More recently, definitions of oxidative stress have been expanded to include disrupted redox signaling and control [1]. The endogenous antioxidant defense is provided by reduced glutathione (GSH) and other reduced thiols such as cysteine (Cys) and thioredoxin. These antioxidants serve multiple functions, contributing to the metabolism of potentially harmful agents and restoring the reducing power of the cell. The biosynthesis of many of these innate antioxidants is governed by the Cap ‘n’ Collar family of transcription factors, including the Nuclear factor erythroid 2 (NFE2)-related factor (Nrf) transcription factors. Nrf proteins bind to a specific DNA sequence, the antioxidant response element (ARE), found in the promoters of many chemoprotective genes, including those involved in the response to oxidative stress [2], [3]. The excessive generation of reactive oxygen species (ROS) may overwhelm innate antioxidant defenses, leading to pro-oxidizing cellular conditions, and in turn, cause lipid peroxidation, DNA damage, disruption of signaling cascades, irregular gene expression, or altered function and degradation of existing proteins.
Redox signaling plays a vital role in embryogenesis. During these developmental periods, shifts in embryonic cell redox potentials may guide cell fate towards proliferation in a reduced state, or towards differentiation, apoptosis or necrosis in increasingly oxidized states [4]. Many teratogens dysregulate development via oxidative stress and altered redox signaling during embryogenesis and organogenesis [4] and may decrease growth of tissues, alter tissue structure and patterning, decrease overall size, or increase embryo lethality. Untimely dysregulation of redox signaling may influence the mode of teratogenesis, as the susceptibility to oxidation of the GSH, Cys, and thioredoxin redox couples is independently regulated [4]. In this way, developmental exposure to chemicals may perturb different redox couples and their corresponding unique cellular response pathways, producing chemical-specific structural defects. Likewise, exposures to the same chemicals at specific developmental time points may also produce differing structural defects, as susceptibility to oxidative damage and ability to activate the endogenous antioxidant response pathways vary throughout development [4]. Thus, the timing of exposures and specificity of thiol targets are important factors in development.
Glutathione is the most abundant endogenous antioxidant in cells, present in millimolar concentrations. Once oxidized, GSH can form disulfide bonds with other oxidized thiols or it can dimerize to form glutathione disulfide (GSSG). The relative fractions of total glutathione (tGSH) that are reduced and oxidized can be used to quantitatively determine the redox potential (Eh). Eh is a more sensitive measure of cellular oxidative state than reduced/oxidized thiol ratios, accounting for the physiological state as well as the reducing power of the specific redox couple [5]. In a previous study, we reported that zebrafish embryonic tGSH is low through the first 24 hpf (zygote-segmentation), then rapidly increases by 30 hpf (pharyngula stage) and is maintained at that higher concentration through the end of the eleutheroembryo stage (96–120 hpf) [6]. The GSH/GSSG Eh, however, follows a different pattern. In the normal progression of embryonic development, the fertilized embryo is initially reduced, then becomes progressively oxidized between 3 and 48 hpf (blastula – hatching stage) before being restored to a reduced GSH/GSSG Eh by 72 hpf (protruding mouth stage) [6]. The increased GSH concentration, and the likely resultant reduced Eh, are the result of continual GSH recycling and biosynthesis [6].
The Nfe2 family of transcription factors is comprised of the Nfe2, Nfe2l1 (“Nrf1”), Nfe2l2 (“Nrf2”), and Nfe2l3 (“Nrf3”) proteins.1 Nrf1 upregulates the antioxidant response by increasing glutathione biosynthesis, and loss-of-function is embryo lethal in mice around mid-gestation [7], [8], [9]. Nrf2 is the most widely studied Nrf family member. Nrf2 is expressed across tissues and cell types throughout the animal kingdom, but unlike Nrf1, is not essential for viability [10]. Normally, the Nrf2 protein is found in the cellular cytosol bound to the Kelch Like ECH Associated Protein 1 (Keap1) repressor protein [11]. When bound, Nrf2 is targeted for ubiquitination and degradation [11], [12]. However, under oxidative conditions, Nrf2 translocates to the nucleus where it dimerizes with small Maf proteins, and this complex is able to bind to gene promoters that contain the ARE sequence [13]. AREs can be found in numerous gene promoters including many of those involved in xenobiotic response and Phase II metabolism, as well as GSH biosynthesis and recycling. Like Nrf2, Nrf1 and Nrf3 bind to the ARE to regulate expression of genes involved in the endogenous antioxidant response [14]. Nrf1 and Nrf2 paralogs were all found to be activated by oxidative stress in the zebrafish, and induced transcription of ARE targets [15], [16]. In human cells, Nrf3 is not activated by oxidative stress, and conversely, may repress transcription of ARE-regulated genes [17]. However, we do not yet understand the Nrf3 transcriptional response to oxidative stress in vertebrates. Unlike Nrf2, cytoplasmic localization of Nrf1 is independent of Keap1; instead, the N-terminus of Nrf1, in mammals, is bound to the membrane of the endoplasmic reticulum (ER) and nuclear translocation is typically indicative of ER stress [18].
A whole genome duplication occurred in the common ancestor of zebrafish and other teleost fish [19]. This duplication results in paralogs of some genes for which other vertebrates have only one copy, allowing for the partitioning of gene function. Nfe2 and Nrf3 have only single copy genes in the zebrafish, as in mammals. In contrast, zebrafish Nrf genes include duplicate paralogous copies of the genes encoding Nrf1 (Nrf1a, Nrf1b) and Nrf2 (Nrf2a, Nrf2b) [15], [20](reviewed in [16]). In the case of Nrf2, Nrf2a and Nrf2b have been subfunctionalized, where Nrf2a is a canonical activator of ARE targets and Nrf2b is a negative regulator of several crucial genes, including p53 and heme oxygenase 1 [20]. The functional partitioning of Nrf1 paralogs has not been explored, but the genes are expressed at different times in development, which may indicate separate functions [15]; a comprehensive examination of the redox roles of Nrf paralogs has yet to be conducted.
In this study, we address two key questions: 1) how does the glutathione system respond to oxidative challenges at different developmental stages, and 2) is the doubling of glutathione or the dynamic changes in glutathione redox potential mediated by either the Nrf1 or Nrf2 transcription factors (zebrafish co-ortholog genes nrf1a, nrf1b, nrf2a, nrf2b). This study also compares redox-sensitivity of specific stages of embryonic development, and constructs an ontogeny of glutathione redox consequences resulting from impaired Nrf1 and Nrf2 signaling in the zebrafish embryo model.
2. Materials & methods
2.1. Chemicals & reagents
tert-Butyl hydroperoxide (tBOOH) was purchased from Alfa Aesar (Haverhill, MA). Tert-Butyl hydroquinone (tBHQ) was purchased from Acros Organics (Morris Plains, NJ). Iodoacetic acid, dansyl chloride, perchloric acid, GSH, GSSG, and γ-glutamyl glutamate were purchased from Sigma/Aldrich (St. Louis, MO). Methanol was obtained from Fisher Scientific (Pittsburgh, PA). Morpholino oligonucleotides (MOs) were purchased from Gene Tools, LLC (Philomath, OR). Morpholino sequences were previously published [15], [20].
2.2. Fish Husbandry
Zebrafish from the Tupfel/Long fin mutation wild-type strain (TL) were used throughout this study. Fish were maintained on a 14-h light/10-h dark cycle. Tank temperature was held at 28.5 °C, and water quality was monitored daily. Adult fish were fed a mixture of brine shrimp and 50:50 spirulina (Ocean Star International, Snowville, UT) and flake food (Lansy NRD 4/6 flake food, INVIE Aquaculture, Salt Lake City, UT) twice daily. Embryos were maintained in 0.3× Danieau's water (17 mM NaCl, 2 mM KCl, 0.12 mM MgSO4, 1.8 mM Ca(NO3)2, 1.5mMHEPES, pH 7.6) throughout the experiment. Animal procedures were performed at the Woods Hole Oceanographic Institution, University of Massachusetts Amherst (UMass), and Bates College under the approval of the Institution Animal Care and Use Committees.
2.3. Embryo Sampling
Breeding tanks were maintained in the WHOI, UMass or Bates College zebrafish facility, containing approximately 30 females and 15 males. Carefully timed embryo collections occurred within 20–30 min of fertilization, and maintained in 0.3x Danieau's solution. Any developmentally delayed or abnormal embryos were excluded, as determined by staging defined in [21].
2.4. Knockdown of Nrf protein
We used morpholino oligonucleotides to transiently knock down translation of Nrf1a, Nrf1b, Nrf2a, and Nrf2b proteins. Morpholinos to all these targets have been previously described, well vetted for non-specific effects, and morphant embryos phenocopy mutants where available (e.g. Nrf2a) [15], [20], [22], [23], [24]; oligonucleotide sequences are provided in Supplemental Table 1. Embryos were obtained at the 1–4 cell stage and injected with approximately 3 nl of 0.1 mM of MO. We have previously used and validated these morpholino concentrations in zebrafish embryos for loss of ARE-dependent gene regulation, and in vitro they produce reductions of 66% (Nrf1a), 68% (Nrf1b), 60% (Nrf2a), and 80% (Nrf2b) of protein expression [15], [20]. All MO were fluorescein-tagged at the 3′ end for visualization of distribution within embryonic tissues. At 24 hpf, embryos were examined for incorporation of MO, and only healthy embryos with uniform MO incorporation were utilized for this study. As a control, we employed a widely used standard negative control MO (Gene Tools), which targets a human beta-globin intron mutation but is without a specific target in zebrafish; we have used this in our previous studies of Nrf function [15], [20]. No significant differences were observed between non-injected and control-MO embryos for any measure, therefore all control measures reported are control-MO values.
While gene editing approaches were considered, there are several reasons why the use of knockouts or null mutations is not optimal for this particular study. Gene editing approaches can be used to generate homozygous germline mutants or to directly study the effect of the mutation in the injected embryo. In the case of the first approach, germline mutants exist currently only for Nrf2a but not for Nrf1a, Nrf1b, or Nrf2b. In addition, we have previously reported that eggs from homozygous Nrf2a mutant zebrafish have larger yolks [25], thus introducing a variable that may occlude changes in glutathione parameters. With respect to the second approach, the use of gene editing directly in embryos fails to affect maternally deposited RNAs, and as there are high levels of nrf2b that fall into this category [20], gene editing may not be effective for this target.
2.5. Exposures
Tert butyl hydroquinone (tBHQ) is a weak pro-oxidant, acutely generating ROS which can later upregulate the ARE-mediated antioxidant response [26], [27]. Triplicate pools of embryos and larvae were exposed at specified developmental windows to 5 μM tBHQ for 1 h at a density of 1 embryo/mL in glass petri dishes, in order to compare the sensitivity of the glutathione redox couple at different stages in development (Fig. 1A). Chorions were manually removed from any unhatched embryos at 72 hpf using watchmaker's forceps prior to exposure.
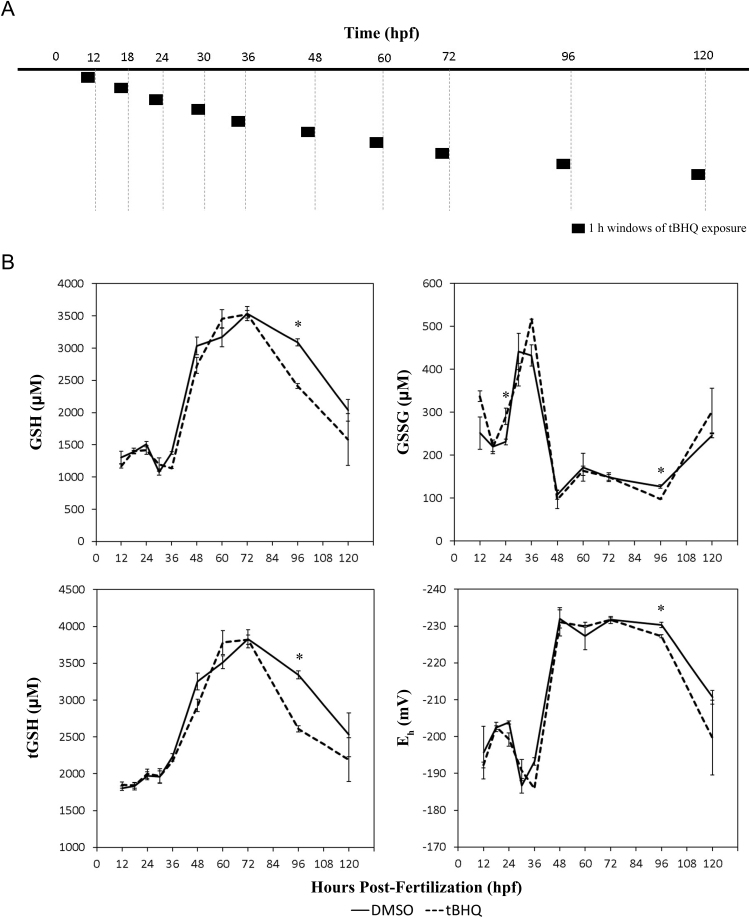
Fig. 1.
Acute treatment with pro-oxidant tBHQ is most oxidizing at 96 hpf. (A) Diagram of exposure and sampling timelines for experiments. Embryos were exposed to tBHQ starting at different developmental stages for 1 h prior to sampling. Vertical dashed lines represent sampling times. Black boxes represent tBHQ exposures for each sampling. (B) tBHQ exposures later in development affect embryonic reduced (GSH) and oxidized (GSSG) glutathione concentrations, and perturb glutathione redox potentials (Eh). Asterisks (*) indicate a statistically significant difference between control and tBHQ-treated embryos (p < 0.05). N = 3 pools of 30 embryos for all groups.
Exposures to the oxidant tert butyl hydroperoxide (tBOOH) were performed in order to monitor glutathione response and Eh recovery dynamics under oxidative stress and in the impaired Nrf expression scenarios. tBOOH (final concentration of 750 μM) was added to the Danieau's media in 1.5 mL Eppendorf tubes (recovery experiments; 1 mL total volume and 10 embryos per group) or a 24-well plate (MO knockdown experiments; 2 mL total volume and 10 embryos per group). Two-three experimental replicates were performed for each treatment and morpholino group. tBOOH exposures for each experiment were performed for specific periods of time, ranging from 1 min to 2 h before sample collection as described in the figure legends. New pools of embryos were used for each experiment at the different time points of sample collection.
2.6. Sample collection
At sampling time points, embryos were prepared for glutathione analysis and redox profiling as previously described [6]. Briefly, triplicate pools of 10–30 embryos were collected in 5% perchloric acid/boric acid solution containing γ-glutamylglutamate, an internal standard for thiol measurements. This preservation solution is formulated to immediately protect and preserve thiols and prevent any oxidation of degradation of the sample and has been shown to preserve GSH/GSSG ratios for more than two months at −80 °C [28]. Samples were snap frozen and stored at −80 °C until the time for glutathione measurements.
2.7. Quantification of GSH, GSSG, and Eh
Quantification of GSH and GSSG was performed using reverse phase High Performance Liquid Chromatography (HPLC) with fluorescence detection, as previously described in [6]. Samples were derivatized using dansyl chloride, using methods previously described in [28], [29], and previously performed in [6], [30].
Samples were injected and peaks were quantified using a Waters 2695 separations module fitted with a Supelcosil LC-NH2 column. These were coupled to a Waters 2475 fluorescence detector, and analyzed using the Waters Empower software. Excitation and emission wavelengths were set for 335 and 518 nm, respectively. Flow rate was 1.0 mL/min, using a gradient method for two mobile phases: A) 80% methanol and 20% water, and B) 62.5% methanol, 12.5% glacial acetic acid, and 214 mg/mL sodium acetate trihydrate in water.
The Nernst equation was utilized (pH 7.4) to calculate redox potential: Eh = E0 + (RT/nF) * log([GSSG]/[GSH]2), where E0 = −264 mV and (RT/nF) = 30. These calculations were normalized to estimate cellular volume and sample protein concentration determined by BCA assay [31].
2.8. RNA Isolation and Reverse Transcription
RNA was isolated from Control-MO, nrf1a-MO, nrf1b-MO, nrf2a-MO, and nrf2b-MO embryos at 96 hpf in order to examine expression of genes related to glutathione synthesis and induction of the antioxidant response. Briefly, 10 embryos were pooled and collected into RNAlater (Fisher Scientific), and stored at −80 °C until use. A total of 3 samples were collected for each morpholino group, across 3 experimental replicates. RNA was isolated using the GeneJET RNA Purification Kit (Fisher Scientific), following manufacturer instructions. RNA concentrations were quantified using the BioDrop µLITE spectrophotometer (BioDrop), and 500 ng of RNA were reverse transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Sample cDNA was stored at −20 °C until use.
2.9. Quantitative PCR
Experiments were conducted in compliance with the Minimum Information for Publication of Quantitative Real-Time PCR Experiments [32]. All samples were blinded to remove experimental bias. cDNA was diluted to 2.5 ng/µl working stocks for use in reactions. Analysis of nrf1a-MO, nrf1b-MO, and their respective Control-MO samples was performed on an Agilent Mx3000 qPCR system (Agilent, Santa Clara, CA) using Brilliant II SYBR Master-mix as described in Williams et al. [33]. Quantitative PCR analyses of nrf2a-MO, nrf2b-MO, and respective Control-MO samples were performed using a Bio-Rad CFX Connect Real-Time PCR Detection System. All reaction volumes were 20 µl, containing 10 µl of 2x iQ SYBR Green Supermix (Bio-Rad), 7 µl water, 5 pM of each primer, and 5 ng (2 µl) of cDNA template. Primers used in this study are provided in Supplemental Table 2. All designed primers were exon-spanning to avoid amplification of any contaminating genomic DNA. Melting curves were conducted to control for primer stability, and NTCs were utilized to confirm lack of contamination. Data was analyzed using the Bio-Rad CFX Manager software, and fold changes for gene expression were calculated using the ΔΔCT method [34]. Beta-2-macroglobulin (b2m) was used as the housekeeping gene [35], and expression did not change between morphant groups. A second housekeeping gene, b-actin, was also run and confirmed the findings calculated with b2m.
2.10. Statistical analyses
Data was analyzed using Microsoft Excel 2013 and IBM SPSS Statistics. Non-parametric ANOVA with a Games-Howell post hoc test were performed, assuming a confidence level of 95% (α = 0.05). Welch's t-tests were used to compare between two groups, without assuming equal variances or normal distributions (α = 0.05). Values presented are means ± standard error of the mean, and N represents the number of pooled samples. Pools contained 10–30 embryos, depending on the experiment.
3. Results
3.1. Acute sensitivity of the glutathione redox couple to oxidation changes during development
In order to compare the sensitivity of the glutathione redox couple to exogenous oxidative stress at different stages of embryogenesis, embryos were exposed to a nontoxic concentration of the Nrf2-activator, tBHQ (1 µM for 1 h) prior to sampling at specific stages of embryonic development following exposure (Fig. 1A). The 1 h exposure time was selected based on the amount of time expected to observe significant changes in the reduced GSH, oxidized GSSG, and Eh at these stages. We previously characterized the endogenous GSH, GSSG, tGSH, and redox potentials for embryos throughout this same window of development [6], and showed that glutathione concentrations doubled after 36 hpf. Here, this finding was replicated (Fig. 1B). However, no significant changes in GSH, GSSG, or Eh due to tBHQ treatment were observed until the 96 hpf time point (protruding mouth stage). At 96 hpf, tBHQ-treated embryos had significantly depleted GSH, GSSG, and tGSH (p = 0.003, p = 0.017, and p = 0.002 respectively), and Eh was significantly oxidized (p = 0.041). However, at 120 hpf, there were not statistically significant effects of tBHQ on the glutathione system, although there was high variability among treated embryos.
3.2. The embryonic response following pro-oxidant exposure is stage- and time-dependent
The redox consequences of several pro-oxidant exposures during development have been characterized, but a quantitative examination of response and recovery dynamics has yet to be performed. Here, we assessed the temporal response to pro-oxidant insults by quantifying the glutathione redox response within 1 h of exposure at discrete windows of development. Age-matched embryos at specific stages of embryonic and larval development were collected following 1, 10, or 60 min of a low tBOOH exposure in order to elucidate the amount of time it takes for the embryo to recover to a control-matched redox status. Exposures used tBOOH instead of tBHQ because tBOOH is a direct oxidant and can more easily and quickly penetrate membranes than tBHQ. Not only does tBOOH act as a faster oxidant than tBHQ, but it is also a limited reaction, unlike tBHQ which can produce electrophilic interactions and metabolites capable of undergoing redox cycling. Glutathione dynamics after tBOOH treatment were assessed by measuring GSH and GSSG, and calculating tGSH (GSH + 2*GSSG), and glutathione redox potentials (Eh) (Fig. 2).
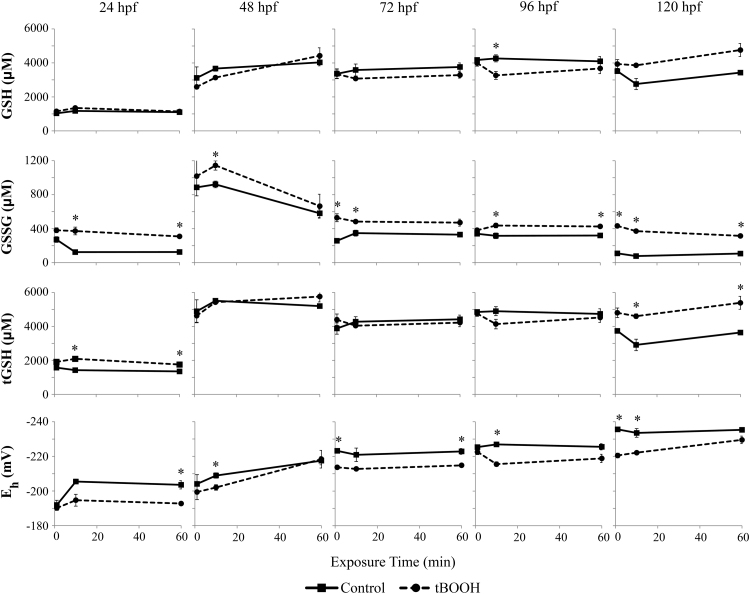
Fig. 2.
GSH, GSSG, Total glutathione, and redox potentials change during development, and recovery from oxidation is stage- and time-dependent. Embryos were exposed to 750 µM tBOOH at specific times of development (24, 48, 72, 96, 120 hpf) and sampled 1, 10, or 60 min following initial exposure. Reduced (GSH) and oxidized (GSSG) glutathione were quantified and total glutathione content and resulting redox potentials were calculated. Asterisks (*) indicate a statistically significant difference between control and tBOOH-treated embryos (p < 0.05). N = 3 pools of 15 embryos for all groups.
At 24 hpf, no immediate changes within 1 min were observed, likely due to the presence of the chorion which inhibits solute uptake. After 10 min of tBOOH exposure, GSSG and tGSH were both significantly elevated (p = 0.029 and p < 0.001, respectively). After 60 min, GSSG and tGSH remained elevated in tBOOH-exposed samples compared to controls (p = 0.004 and p = 0.008), but these changes also resulted in a significantly oxidized redox potential (p = 0.031). No changes in GSH were observed.
At 48 hpf, as at 24 hpf, no immediate changes within 1 min were observed, and no changes in GSH were observed due to tBOOH exposure after any exposure time. Following 10 min of exposure, GSSG was significantly elevated (p = 0.033) and glutathione Eh was significantly oxidized (p = 0.035). However, these effects were attenuated by 60 min post-treatment. GSSG across control and tBOOH-treated embryos was elevated compared to all other embryonic ages.
After 72 hpf, tBOOH exposure had several immediate effects on the hatched eleutheroembryos. GSSG was increased following 1 min of exposure (p = 0.015), and glutathione Eh was significantly oxidized (p = 0.005). Eh remained oxidized throughout the 60 min exposure period, also significantly oxidized at 60 min post-treatment (p = 0.021). GSSG was also significantly increased after 10 min exposure (p = 0.039), and remained elevated after 60 min.
At 96 hpf, no immediate changes were observed. Following 10 min tBOOH exposure, GSH was decreased (p = 0.034), GSSG was increased (p = 0.048), and Eh was significantly oxidized (p = 0.003). GSSG remained significantly elevated after 60 min (p = 0.005).
At 120 hpf, GSSG was significantly elevated (p < 0.001) and Eh was significantly oxidized (p = 0.002) following 1 min of tBOOH exposure. These effects remained following 10 min of exposure (p = 0.003 and p = 0.044, respectively), and were complemented by a significant increase of tGSH (p = 0.025). GSSG and tGSH were still significantly elevated after 60 min of tBOOH exposure (p < 0.001 and p = 0.044).
3.3. Nrf1a and Nrf1b morpholino knockdown reveals variable glutathione redox responses
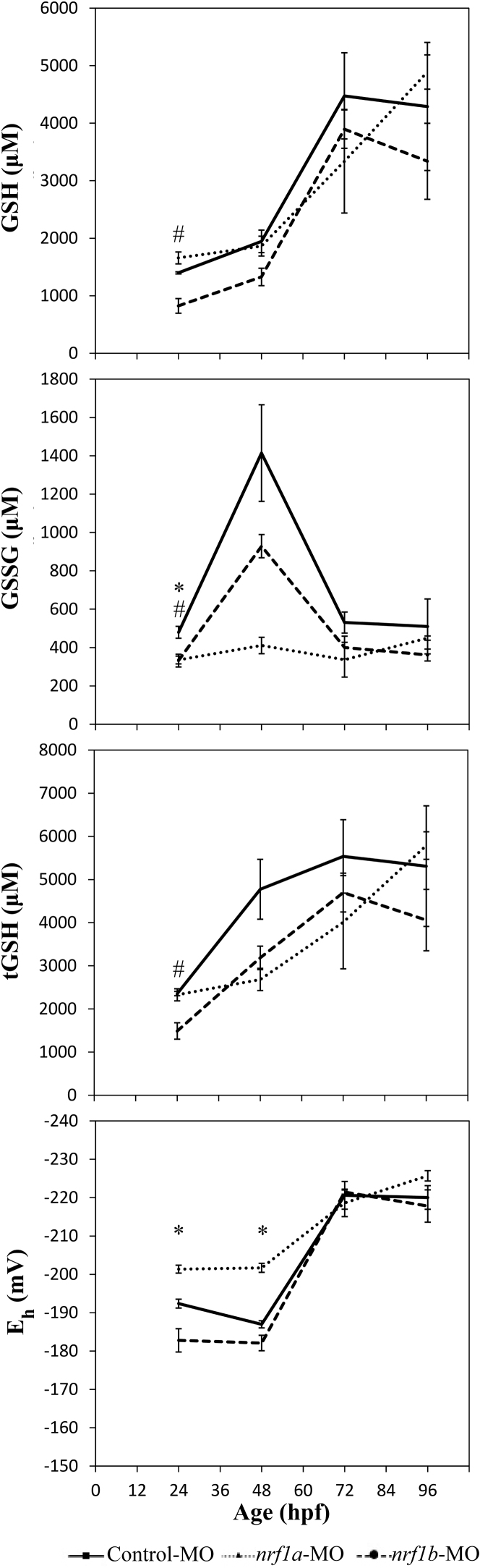
Nrf1 has been previously shown to contribute to the maintenance of GSH and GSSG levels during embryonic development in mouse fetal liver tissue [36], but the specific role (and potential subfunctionalization) of zebrafish paralogs Nrf1a and Nrf1b in the glutathione redox system during embryonic development required clarification. Here, embryos were injected with control, nrf1a, or nrf1b morpholinos in order to knock down expression of each protein and examine glutathione dynamics between 24 and 96 hpf (Fig. 3). At 24 hpf, Nrf1b morphants had decreased GSH compared to controls (p = 0.043), but Nrf1a morphants did not differ from controls. GSSG was significantly lower in both Nrf1a and Nrf1b morphants compared to controls (p = 0.024 and p = 0.032, respectively). Nrf1b morphants had decreased tGSH (p = 0.038). Glutathione Eh in Nrf1a morphants was more reduced compared to controls (p = 0.005).
Fig. 3.
Nrf1a and Nrf1b morpholino knockdown perturbs embryonic glutathione redox profiles. Wild type embryos were injected with a control-MO, nrf1a-MO, or nrf1b-MO within 1 h of fertilization (at or before the 4-cell stage). Asterisks (*) indicate a statistically significant difference between control and nrf1a-MO injected embryos (p < 0.05). Octothorpes (#) indicate a statistically significant difference between control and nrf1b-MO injected embryos (p < 0.05). N = 3 pools of 10–15 embryos for all groups.
At 48 hpf, no significant changes of GSH or tGSH were observed. Though not statistically significant, GSSG appeared elevated in control and Nrf1b morphants, but not Nrf1a morphants, at 48 hpf. For this reason, Nrf1a morphants had more reduced glutathione Eh compared to controls (p = 0.001). No changes in GSH, GSSG, tGSH, or Eh were observed due to Nrf1a or Nrf1b deficiency at 72 or 96 hpf.
3.4. Consequences of Nrf2a and Nrf2b knockdown differ temporally for glutathione
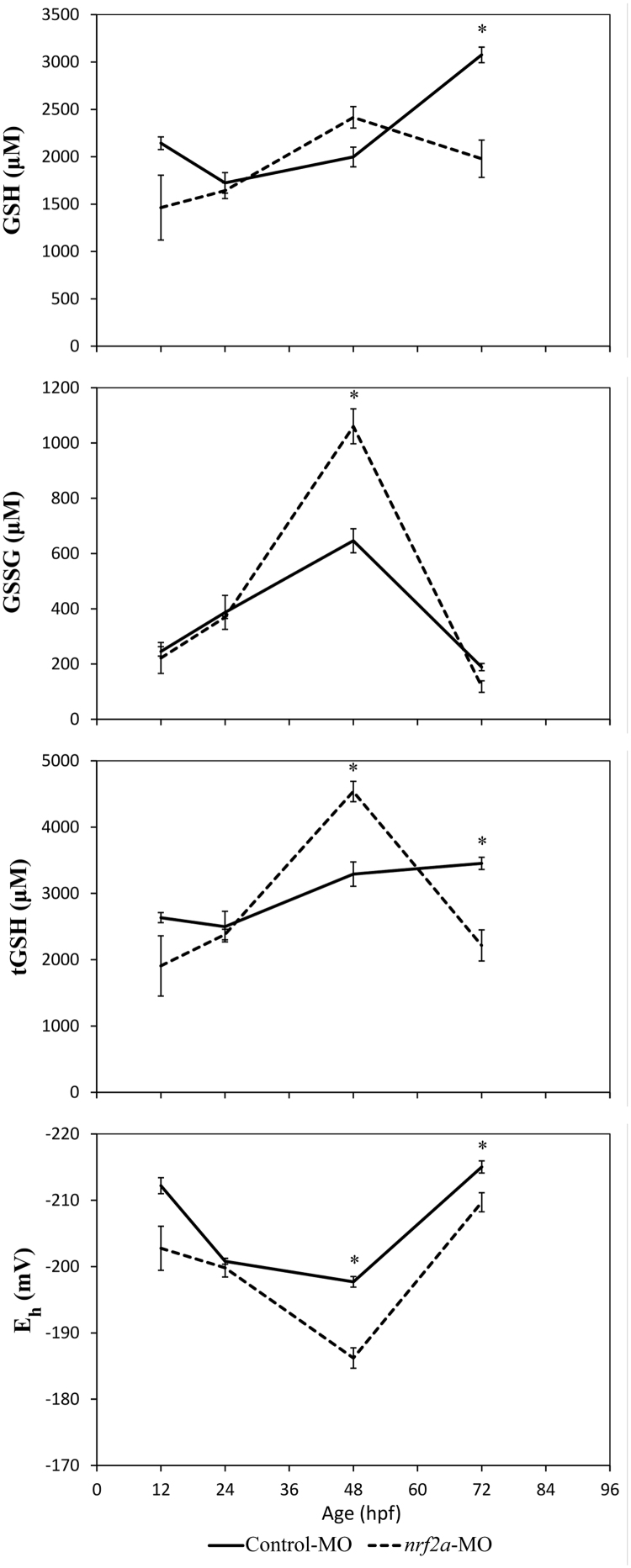
To build upon this redox characterization of Nrf1a and Nrf1b, we examined how Nrf2a and Nrf2b impact glutathione throughout development. Nrf2a morphants were analyzed for GSH, GSSG, tGSH, and Eh from 12 to 72 hpf (Fig. 4). No statistically significant changes in any measures were observed until 48 hpf, when GSSG and tGSH were both significantly elevated (p = 0.008 and p = 0.007, respectively) and Eh was oxidized (p = 0.007) in Nrf2a morphants. Though not statistically significant, GSH also appeared slightly elevated at 48 hpf in Nrf2a morphants (p = 0.054). By 72 hpf, GSH, GSSG, and tGSH were all depleted in Nrf2a morphants (p = 0.019, p = 0.056, and p = 0.022, respectively), and Eh remained oxidized (p = 0.045).
Fig. 4.
Knockdown of Nrf2a oxidizes the embryonic glutathione redox potential. Control or Nrf2a morpholinos were injected into the yolk sacs of wild-type eggs within 1 h of fertilization (before the 4-cell stage). Asterisks (*) indicate a statistically significant difference between control and nrf2a-MO injected embryos (p < 0.05). N = 3 pools of 10–15 embryos for all groups. A 96-hpf time point was not included because the nrf2a MO has declining efficacy at this time (unpublished data).
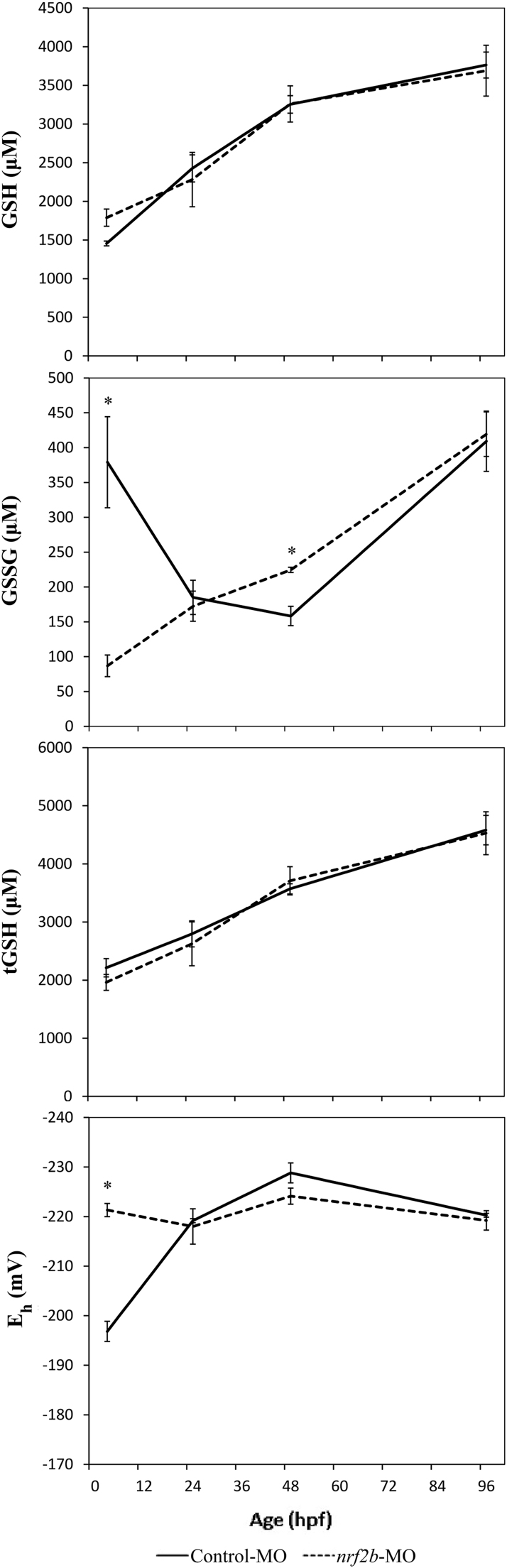
Nrf2b morphants were assessed for glutathione dynamics between 3 and 96 hpf (Fig. 5). No changes in GSH or tGSH were observed in Nrf2b morphants at any timepoint. At 3 hpf, GSSG was significantly decreased in Nrf2b morphants (p = 0.040). As a result, Eh was significantly reduced (p = 0.001). These effects were attenuated by 24 hpf. At 48 hpf, GSSG was significantly elevated in Nrf2b morphants (p = 0.034), though no other parameters were altered. At 96 hpf, all glutathione measures were not different between Nrf2b morphants and controls.
Fig. 5.
Knockdown of Nrf2b perturbs GSSG and glutathione redox state of embryos early in development. Control or Nrf2b morpholinos were injected into embryos prior to 1 hpf. Asterisks (*) indicate a statistically significant difference between control and nrf2b-MO injected embryos (p < 0.05). N = 3 pools of 10–15 embryos for all groups.
3.5. Nrf2 may play a moderate role in embryonic recovery from oxidative insults
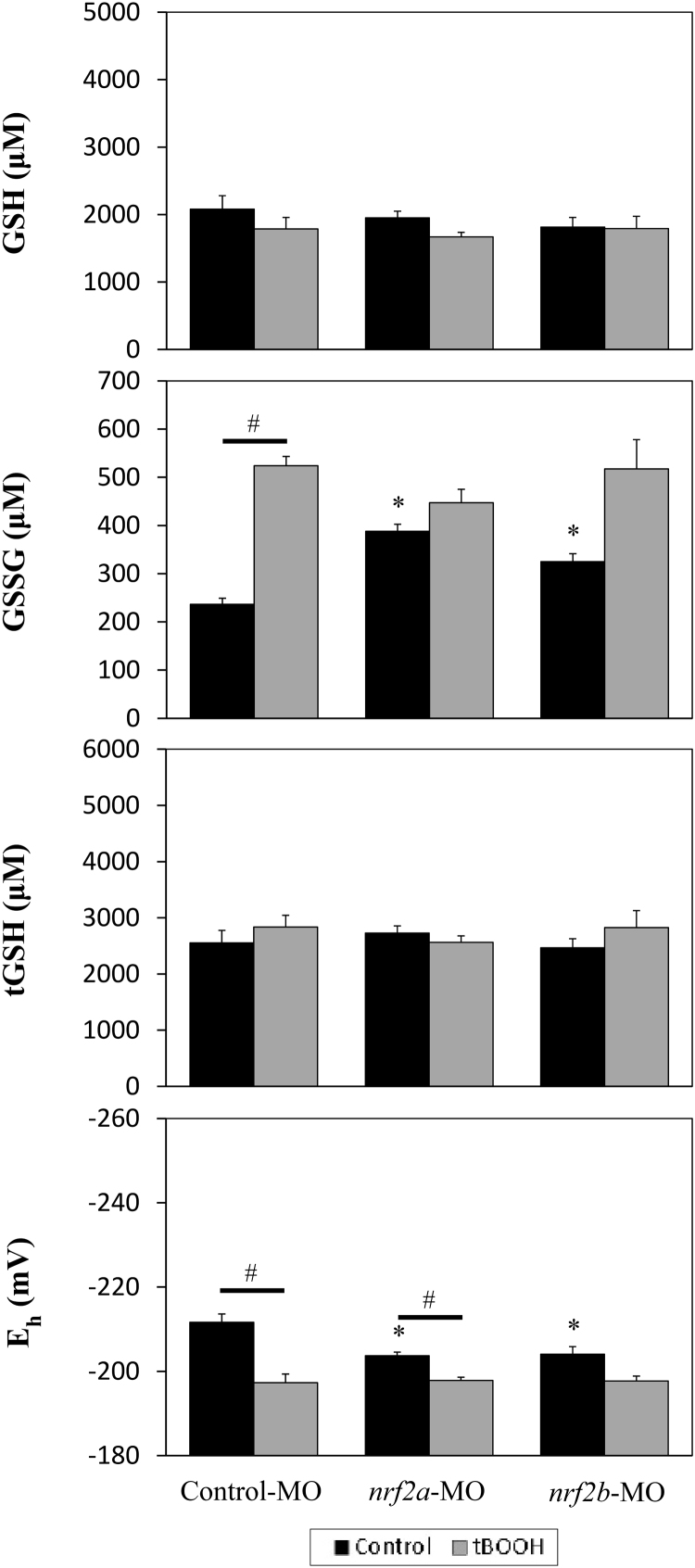
To biochemically assess whether embryos deficient in Nrf2a, Nrf2b, or combined Nrf2a and Nrf2b are more responsive and susceptible to pro-oxidant insults, glutathione concentrations and redox potentials were analyzed in Nrf2a or Nrf2b morphant zebrafish embryos at 26 hpf following 2 h exposure to tBOOH (Fig. 6). There were no significant changes in GSH or tGSH caused by tBOOH exposure for any of the control or morpholino groups. tBOOH exposure significantly increased GSSG (p < 0.001) and oxidized Eh (p = 0.007) in control-MO embryos. nrf2a morphants treated with tBOOH also had oxidized Eh compared to untreated nrf2a morphants (p = 0.006). Untreated nrf2a, nrf2b, and combined nrf2a+nrf2b morphants had elevated GSSG compared to untreated controls (p = 0.001, p = 0.013, and p = 0.003, respectively). All morphants also had oxidized Eh compared to Control-MO embryos (nrf2a-MO p = 0.020, nrf2b-MO p = 0.045, nrf2a+nrf2b-MO p = 0.020).
Fig. 6.
Nrf2 knockdown impacts glutathione signaling in control embryos but does not confer additional sensitivity to pro-oxidants at 26 hpf. Embryos were injected with Control, nrf2a, or nrf2b morpholinos within 1 hpf. Embryos from each morpholino group were either maintained in media (Control treatment) or tBOOH was added to media (tBOOH treatment) for 2 h prior to sampling. Asterisk (*) indicates a significant difference between untreated control and untreated morpholino-injected embryos (p < 0.05). Octothorpes (#) indicate a statistically significant difference between tBOOH-treated embryos are their matched morpholino untreated controls (p < 0.05). N = 3–5 pools of 10–15 embryos for all groups.
3.6. Nrf1 and Nrf2 paralogs differentially affect glutathione-related gene expression
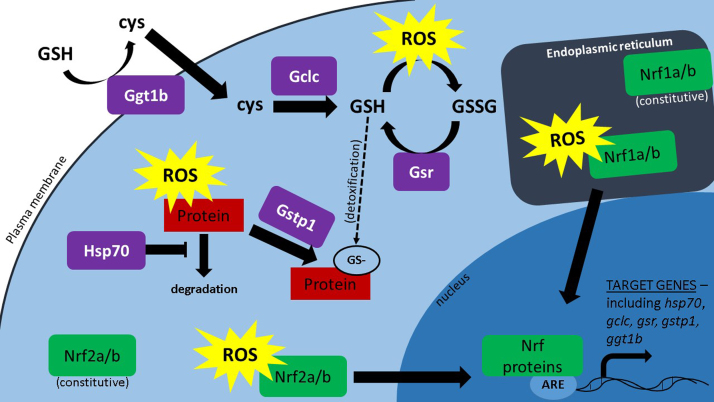
To examine the consequences of impaired Nrf signaling on glutathione signaling and the antioxidant response, gene expression of Nrf targets was analyzed. The function of these enzymes (shown in purple) is depicted in Fig. 7. Genes encoding each of these enzymes and subunits are targets of Nrf transcription factors, and play a role in the protection of cells against ROS. We have previously characterized the expression ontogeny for these genes in zebrafish embryos [6]. Gamma-glutamyltransferase 1b (Ggt1b) and glutamate-cysteine ligase catalytic subunit (Gclc) are enzymes that increase cellular cysteine supply and catalysis for glutathione synthesis. Glutathione-disulfide reductase (Gsr) activity also increases GSH by recycling oxidized GSSG into reduced GSH. Glutathione S-transferase pi 1 (Gstp1) and heat shock protein 70 (Hsp70) aid cellular detoxification by catalyzing the S-glutathionylation of proteins and stabilizing protein structure and function during excessive intracellular oxidative stress. Previous studies have identified gstp1 expression, but not hsp70, as a reliable biomarker of Nrf2a activation in zebrafish [20], [37].
Fig. 7.
Nrf proteins and target proteins maintain glutathione and redox signaling. Nrf proteins (green) are redox ‘sensors’ in the endoplasmic reticulum (Nrf1) and cytosol (Nrf2). In the presence of excessive ROS, Nrf proteins can translocate to the nucleus where they serve as transcription factors for a myriad of genes involved in glutathione synthesis and redox signaling (shown in purple). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Gene expression of ggt1b, gclc, and gstp1 was unchanged in Nrf1a, Nrf2a, and Nrf2b morphants at 96 hpf (Fig. 8). In Nrf1b morphants, ggt1b, gclc, and gstp1 expression was decreased by 75% (p = 0.024), 57% (p = 0.037), and 93% (p = 0.020), respectively. Expression of gsr was not significantly altered in any morphant group. Gene expression of hsp70 was unchanged in Nrf1a, Nrf1b, and Nrf2a morphants, but was increased by 121% in Nrf2b morphants (p = 0.044).
Fig. 8.
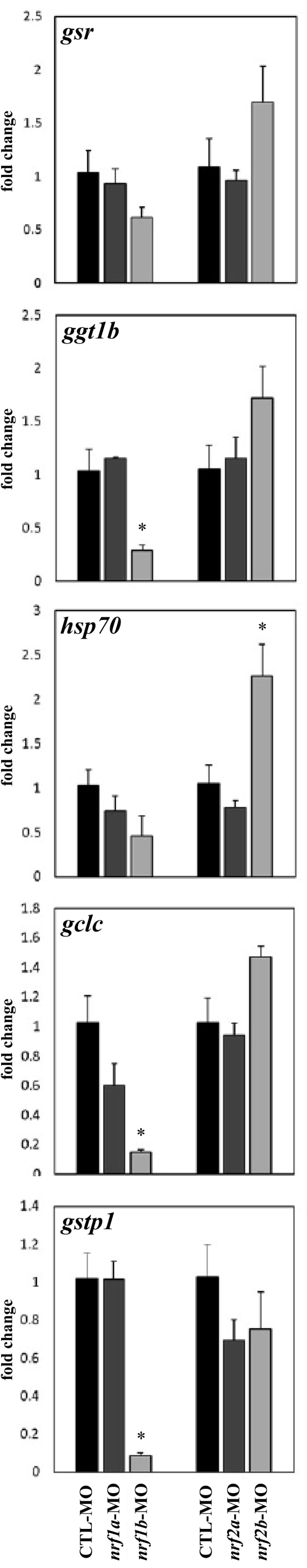
Deficiencies of Nrf1 and Nrf2 distinctly impact the expression of genes involved in glutathione synthesis and redox signaling. Embryos were injected with Control, nrf1a, nrf1b, nrf2a, or nrf2b morpholinos within 1 hpf and allowed to grow until RNA sampling at 96 hpf. Fold change refers to expression relative to the respective Control-MO control in each set of samples. Asterisk (*) indicates a significant difference between respective control and morpholino-injected embryos (p < 0.05). N = 3 samples of 10 embryos for all groups.
4. Discussion
Glutathione is the most abundant endogenous antioxidant in the developing embryo, and plays a critical role in embryonic development. We previously presented an ontogeny of GSH dynamics and related gene expression in the developing zebrafish embryo, identifying critical windows of rapid glutathione synthesis and redox shifts between oxidized and reduced cellular states [6]. Here, we further characterized the function of the embryonic glutathione redox system by quantifying the response to, and recovery from, pro-oxidant exposures at different developmental stages. Further, we tested the significance of Nrf expression in these responses by knocking down Nrf1a, Nrf1b, Nrf2a, and Nrf2b. This is the first study quantifying the timing and magnitude of the GSH response to pro-oxidants at different developmental stages in the zebrafish embryo, and elucidating the role of Nrf1 and Nrf2 paralogs in this dynamic system.
Redox regulation of embryogenesis and organogenesis is a well-controlled process. The response to and recovery from oxidative insults is essential in the embryo, and impaired function of the glutathione system may result in teratogenesis [4]. We previously showed that basal tGSH increased in the embryo around 36 hpf [6], a finding replicated in control embryos in the current study. Here, we also show tGSH became depleted in tBHQ-treated embryos compared to controls at 96 hpf. This decreased tGSH later in development is attributed to decreases in both GSH and GSSG. It is possible that the decreased GSH and concurrent decrease in GSSG indicates that glutathione may be utilized and bound by other proteins in the embryo. An examination of S-glutathionylation would provide a quantifiable estimate of these contributions to the tGSH pool, and also may identify protein targets that are susceptible to oxidative modification during this phase of organogenesis.
We previously found that the basal embryonic Eh was initially reduced in newly fertilized embryos, then almost immediately became oxidized until Eh recovery after 48 hpf [6]. This was validated here, as Eh reached approximately −240 mV around 48 hpf and remained highly reduced until 120 hpf. These findings are concordant with other published studies of glutathione redox parameters in embryos from zebrafish, rat, and mouse (Supplemental Table 3). As reported previously, the 48 hpf time point is highly variable with respect to glutathione parameters in the zebrafish embryo [6]. During the hatching window, which begins at 48 hpf, the glutathione profile can shift quite dramatically based on the embryo's initiation of hatching [6]. However, if we examine earlier (24 hpf) and later timepoints (96 hpf), we see that the values are more consistent, indicating that the period of hatching is one of high natural variation.
At 72 hpf, the GSH/GSSG Eh became relatively oxidized in tBHQ-treated embryos compared to controls. This oxidation suggests that the embryo became increasingly susceptible to pro-oxidant insults as development progressed, despite a more reduced GSH/GSSG redox environment. This increased susceptibility could either occur because pro-oxidants can more drastically perturb a reduced redox environment, or more likely, because of the activities of other redox couples in the embryo. In this study, we characterized the glutathione redox couple, because GSH is the most abundant antioxidant species in the embryo. However, other thiols and antioxidants, including the cysteine/cysteine and thioredoxin redox systems, comprise cellular redox signaling and produce other readouts of cellular Eh in the embryo [1], [4]. The Eh readout of these other redox couples may follow a different pattern during development, and could be selectively oxidized during oxidative stress events. Nrf proteins likely play a role in the differentiation of these responses. For example, Nrf1 activation suppresses the xCT antiporter, decreasing cystine uptake, which would further oxidize cysteine/cystine Eh. [38]. However, depleted cystine reduces the amount of free cysteine for glutathione synthesis, effectively depleting intracellular GSH [39]. Therefore, these redox couples are distinctly regulated and their collective examination may be required to understand the complete oxidative stress response.
To better understand subtle temporal differences in susceptibility to pro-oxidant challenges, embryos were acutely challenged with tBOOH for 1, 10, or 60 min at specific stages of embryonic development. Regardless of exposure time, embryos exposed to tBOOH at any age had elevated GSSG compared to control embryos. However, this was not always concurrent with a decrease in GSH. Though GSH levels were fairly similar between controls and tBOOH-treated embryos through 48 hpf, GSH concentrations were decreased when embryos were treated with tBOOH at 72 or 96 hpf. Interestingly, this effect was reversed at 120 hpf, as control embryos had depleted GSH. The tGSH was similar for control and tBOOH-treated embryos for all time points until 120 hpf, when controls had significantly lower tGSH. Despite these changes, Eh for tBOOH-treated embryos was consistently oxidized compared to control embryos at all ages and response timepoints. In all, the data suggest that the embryo is susceptible to oxidation across the entire embryonic development window.
The Nrf family of transcription factors regulates the innate antioxidant defense pathway and GSH synthesis. We have previously characterized the expression of nrf genes during zebrafish embryogenesis and organogenesis [15], [20]. Following fertilization, gene expression of nrf1a and nrf2b was low through 48 hpf. Expression of nrf1b was initially elevated, but resembled that of nrf1a and nrf2b beginning at 6 hpf. nrf2a expression, however, rapidly increased and became expressed 3–fold higher than the other Nrf family members by 48 hpf. Here, embryos deficient in Nrf signaling exhibited temporal differences in their glutathione concentrations and redox potentials. While these differences may be pronounced due to each Nrf having distinct functions, they may also be attributed to differences in basal expression, which fluctuate throughout development [15]. Furthermore, the cytoplasmic tethering of Nrf2 protein to Keap1 and the targeted localization of Nrf1 to the endoplasmic reticulum may produce spatial or compartmental differences in redox signaling and response to oxidative insults.
Compared to control embryos, the GSH/GSSG Eh values in Nrf1b morphants were relatively oxidized, though they followed temporal fluctuations similar to those of the controls. In Nrf1a morphants, however, this measurement deviated from these temporal fluctuations, and ultimately had a more reduced GSH/GSSG Eh. Nrf2b morphant embryos exhibited time-dependent effects, initially more reduced like Nrf1a morphants until 24 hpf when they begin to resemble control embryos. Nrf2a morphants showed more canonical effects, exhibited oxidized GSH/GSSG Eh compared to control values. Nrf2a morphants also had significantly increased GSSG at 48 hpf. This data suggests that disruptions to Nrf signaling during embryonic development can perturb the glutathione redox system, even moderately in the absence of additional oxidative stress.
To assess the importance of Nrf2 in response to oxidative stress, we challenged Nrf2a and Nrf2b morphants with tBOOH for 2 h at 24 hpf. Because Nrf2 is primarily responsible for the upregulation of genes that control GSH biosynthesis and recycling, we expected Nrf2a and Nrf2b morphants to be more severely affected by tBOOH exposures. While there was relatively little change of GSH or tGSH due to tBOOH treatment, GSSG and Eh followed some interesting trends. GSSG was elevated in both Nrf2a and Nrf2b untreated morphants compared to untreated controls, and this also corresponded with oxidized Eh in untreated morphants. GSSG was also increased by tBOOH treatment in control embryos, and increased slightly in Nrf2a and Nrf2b morphants by tBOOH treatment. Cells have the ability to export GSSG [40], [41], and these exporters were measured in the embryo [42], [43]. In this study, we only examined the response to pro-oxidant challenge 2 h after initial tBOOH exposure, allowing for potential recovery. It is possible that the initial response to tBOOH was severe and that compensatory mechanisms were initiated, though these responses often require periods longer than 2 h to exhibit strong changes. Further examination of time-dependent aspects of recovery is necessary to elucidate the related mechanisms.
In order to examine the compensatory mechanisms which could be influencing glutathione concentrations and redox state, we examined gene expression of important glutathione-related enzymes in Nrf morphants at 96 hpf. All of these enzymes not only contribute to glutathione-related redox signaling, but also are inducible by Nrf transcription factors (Fig. 7). Gene expression of ggt1b and gclc was significantly decreased in Nrf1b morphants (Fig. 8). These genes encode enzymes which support glutathione biosynthesis by increasing rate-limiting intracellular cysteine and catalyzing its incorporation into GSH. Expression of gsr, the enzyme which converts oxidized GSSG into reduced GSH, was also slightly decreased, though not statistically significant. Therefore, it is possible that downregulation of these genes at 96 hpf, as well as decreased GSH, GSSG, and tGSH concentrations in Nrf1b morphants (Fig. 3), suggest depletion of GSH substrates such as rate-limiting cysteine. Gene expression of gstp1, and enzyme crucial for GSH-mediated detoxification through S-glutathionylation of oxidized cellular proteins, was also downregulated in Nrf1b morphants (Fig. 8). We had previously shown that Nrf2a morphants had significantly reduced gstp1 expression at 52 hpf [20]. Because nrf2a gene expression increases throughout development [20], it is likely that this is a temporal effect, and that Nrf2a activity or translocation may become less sensitive prior to 96 hpf.
These studies were performed with the use of morpholinos, decreasing translation of the target Nrf proteins. However, morpholinos provide a knock-down, not a knockout, and thus some minimal translation of the proteins is maintained. Also, the efficacy of morpholinos becomes diminished as development progresses [44]. Generation of loss-of-function mutant strains overcomes these limitations; however, germ-line mutants for nrf1a, nrf1b, and nrf2b do not yet exist. Importantly, it is unknown how parental deficiencies affect unrelated measures of egg quality, and therefore could introduce other variables to this embryonic study. There are suggestions that genomic loss-of-function mutations may lead to functional compensation via alternative mechanisms [45]. Because we wanted to control for confounding variables such as parental RNA deposition into the egg, morpholinos were appropriate. Here, we examined how knockdown of Nrf transcripts impacts glutathione dynamics from a functional, pragmatic perspective. It is unknown whether the small amount of activity maintained is enough to sustain increased GSH synthesis, as Nrf proteins can be exported from the nucleus and potentially recycled within the cell several times before degradation [46], [47]. An examination of Nrf-target protein concentrations after knock-down of Nrf function would allow for analysis of the remaining Nrf activity.
In conclusion, this study is a novel investigation into the embryonic response and recovery to oxidative stress conditions during embryogenesis and organogenesis. We have compared developmental windows of susceptibility to pro-oxidant challenges, and established the delay in recovery to these challenges well into the late organogenesis phase of development. Deficient Nrf1 and Nrf2 signaling altered GSH concentrations, redox potentials, and antioxidant gene expression in the embryo, though these effects were moderate in magnitude. This study builds upon our existing foundation of glutathione dynamics during development, to elucidate the consequences and responses of the embryo to oxidative stress during these sensitive phases of development.
Funding
This research was supported by several NIH grants, including F32ES028085 (to KES), F32ES017585 (to ART-L), F32ES019832 (to LMW), P20GM103423 (to LMW), R01ES025748 (to ART-L), R01ES015912 (JJS), and R01ES016366 (MEH). Additional research support was provided by the J. Seward Johnson Fund at WHOI and the WHOI Postdoctoral Scholar Award with funding from Walter A. and Hope Noyes Smith (to ART-L).
Acknowledgements
The authors kindly acknowledge the excellent fish care provided by Gale Clark and Brandy Joyce at the Woods Hole Oceanographic Institution and Mary Hughes at Bates College.
Footnotes
We adhere to the gene and protein nomenclature guidelines established by the Zebrafish Nomenclature Committee, outlined on the ZFIN Zebrafish Nomenclature website. Human genes and proteins are designated using all capitals and italics, e.g. NRF2 and NRF2, respectively. Zebrafish genes are designated nrf2a and Nrf2a for genes and proteins, respectively.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.05.023.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Jones D.P. Redefining oxidative stress. Antioxid. Redox Signal. 2006;8(9–10):1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 2.Wasserman W.W., Fahl W.E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. 1997;94(10):5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen T., Nioi P., Pickett C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009;284(20):13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen J.M., Harris C. Redox control of teratogenesis. Reprod. Toxicol. 2013;35(0):165–179. doi: 10.1016/j.reprotox.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Jones D.P. Redox potential of GSH/GSSG couple: assay and biological significance. In: Helmut S., Lester P., editors. Methods in Enzymology. Academic Press; 2002. pp. 93–112. [DOI] [PubMed] [Google Scholar]
- 6.Timme-Laragy A.R., Goldstone J.V., Imhoff B.R., Stegeman J.J., Hahn M.E., Hansen J.M. Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic. Biol. Med. 2013;65(0):89–101. doi: 10.1016/j.freeradbiomed.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.Y., Kwong M., Lu R., Chang J., Wang B., Yen T.S., Kan Y.W. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17(6):1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L.Y., Kwong M., Lu R.H., Ginzinger D., Lee C., Leung L., Chan J.Y. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell Biol. 2003;23(13):4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong M., Kan Y.W., Chan J.Y. The CNC basic leucine zipper factor, Nrf1, Is essential for cell survival in response to oxidative stress-inducing agents: role for Nrf1 in y-gcsL and gss expression in mouse fibroblasts. J. Biol. Chem. 1999;274(52):37491–37498. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- 10.Chan K., Lu R., Chang J.C., Kan Y.W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA. 1996;93(24):13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon M., Itoh K., Yamamoto M., Hayes J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 13.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnsen O., Murphy P., Prydz H., Kolsto A.B. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26(2):512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams L.M., Timme-Laragy A.R., Goldstone J.V., McArthur A.G., Stegeman J.J., Smolowitz R.M., Hahn M.E. Developmental expression of the Nfe2-related factor (Nrf) transcription factor family in the zebrafish, Danio rerio. PLoS ONE. 2013;8(10):e79574. doi: 10.1371/journal.pone.0079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn M.E., Timme-Laragy A.R., Karchner S.I., Stegeman J.J. Nrf2 and Nrf2-related proteins in development and developmental toxicity: insights from studies in zebrafish (Danio rerio) Free Radic. Biol. Med. 2015;88:275–289. doi: 10.1016/j.freeradbiomed.2015.06.022. (Part B) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sankaranarayanan K., Jaiswal A.K. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H: quinone oxidoreductase1 gene. J. Biol. Chem. 2004;279(49):50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 18.Wang W., Chan J.Y. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J. Biol. Chem. 2006;281(28):19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 19.Jaillon O., Aury J.M., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Nicaud S., Jaffe D., Fisher S., Lutfalla G., Dossat C., Segurens B., Dasilva C., Salanoubat M., Levy M., Boudet N., Castellano S., Anthouard V., Jubin C., Castelli V., Katinka M., Vacherie B., Biemont C., Skalli Z., Cattolico L., Poulain J., De Berardinis V., Cruaud C., Duprat S., Brottier P., Coutanceau J.P., Gouzy J., Parra G., Lardier G., Chapple C., McKernan K.J., McEwan P., Bosak S., Kellis M., Volff J.N., Guigo R., Zody M.C., Mesirov J., Lindblad-Toh K., Birren B., Nusbaum C., Kahn D., Robinson-Rechavi M., Laudet V., Schachter V., Quetier F., Saurin W., Scarpelli C., Wincker P., Lander E.S., Weissenbach J., Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 20.Timme-Laragy A.R., Karchner S.I., Franks D.G., Jenny M.J., Harbeitner R.C., Goldstone J.V., McArthur A.G., Hahn M.E. Nrf2b, novel zebrafish paralog of oxidant-responsive transcription factor NF-E2-related factor 2 (NRF2) J. Biol. Chem. 2012;287(7):4609–4627. doi: 10.1074/jbc.M111.260125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 22.Timme-Laragy A.R., Van Tiem L.A., Linney E.A., Di Giulio R.T. Antioxidant responses and NRF2 in synergistic developmental toxicity of PAHs in zebrafish. Toxicol. Sci. 2009;109(2):217–227. doi: 10.1093/toxsci/kfp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima H., Nakajima-Takagi Y., Tsujita T., Akiyama S., Wakasa T., Mukaigasa K., Kaneko H., Tamaru Y., Yamamoto M., Kobayashi M. Tissue-restricted expression of Nrf2 and its target genes in zebrafish with gene-specific variations in the induction profiles. PLoS One. 2011;6(10):e26884. doi: 10.1371/journal.pone.0026884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukaigasa K., Nguyen L.T., Li L., Nakajima H., Yamamoto M., Kobayashi M. Genetic evidence of an evolutionarily conserved role for Nrf2 in the protection against oxidative stress. Mol. Cell Biol. 2012;32(21):4455–4461. doi: 10.1128/MCB.00481-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rousseau M.E., Sant K.E., Borden L.R., Franks D.G., Hahn M.E., Timme-Laragy A.R. Regulation of Ahr signaling by Nrf2 during development: effects of Nrf2a deficiency on PCB126 embryotoxicity in zebrafish (Danio rerio) Aquat. Toxicol. 2015;167:157–171. doi: 10.1016/j.aquatox.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinkus R., Weiner L.M., Daniel V. Role of oxidants and antioxidants in the induction of AP-1, NF-kappaB, and glutathione S-transferase gene expression. J. Biol. Chem. 1996;271(23):13422–13429. doi: 10.1074/jbc.271.23.13422. [DOI] [PubMed] [Google Scholar]
- 27.Hahn M.E., McArthur A.G., Karchner S.I., Franks D.G., Jenny M.J., Timme-Laragy A.R., Stegeman J.J., Woodin B.R., Cipriano M.J., Linney E. The transcriptional response to oxidative stress during vertebrate development: effects of tert-butylhydroquinone and 2,3,7,8-tetrachlorodibenzo-p-dioxin. PLoS One. 2014;9(11):e113158. doi: 10.1371/journal.pone.0113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones D.P., Carlson J.L., Samiec P.S., Sternberg P., Jr, Mody V.C., Jr, Reed R.L., Brown L.A.S. Glutathione measurement in human plasma: evaluation of sample collection, storage and derivatization conditions for analysis of dansyl derivatives by HPLC. Clin. Chim. Acta. 1998;275(2):175–184. doi: 10.1016/s0009-8981(98)00089-8. [DOI] [PubMed] [Google Scholar]
- 29.Harris C., Hansen J. Oxidative stress, thiols, and redox profiles. In: Harris C., Hansen J.M., editors. Developmental Toxicology. Humana Press; 2012. pp. 325–346. [Google Scholar]
- 30.Harris C., Shuster D.Z., Roman Gomez R., Sant K.E., Reed M.S., Pohl J., Hansen J.M. Inhibition of glutathione biosynthesis alters compartmental redox status and the thiol proteome in organogenesis-stage rat conceptuses. Free Radic. Biol. Med. 2013;63(0):325–337. doi: 10.1016/j.freeradbiomed.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirlin W.G., Cai J., Thompson S.A., Diaz D., Kavanagh T.J., Jones D.P. Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic. Biol. Med. 1999;27(11–12):1208–1218. doi: 10.1016/s0891-5849(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 32.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 33.Williams L.M., Lago B.A., McArthur A.G., Raphenya A.R., Pray N., Saleem N., Salas S., Paulson K., Mangar R.S., Liu Y., Vo A.H., Shavit J.A. The transcription factor, Nuclear factor, erythroid 2 (Nfe2), is a regulator of the oxidative stress response during Danio rerio development. Aquat. Toxicol. 2016;180:141–154. doi: 10.1016/j.aquatox.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.McCurley A.T., Callard G.V. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008;9(1):102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L., Kwong M., Lu R., Ginzinger D., Lee C., Leung L., Chan J.Y. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell. Biol. 2003;23(13):4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T., Takagi Y., Osanai H., Li L., Takeuchi M., Katoh Y., Kobayashi M., Yamamoto M. Pi class glutathione S-transferase genes are regulated by Nrf 2 through an evolutionarily conserved regulatory element in zebrafish. Biochem. J. 2005;388(Pt 1):65–73. doi: 10.1042/BJ20041860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsujita T., Peirce V., Baird L., Matsuyama Y., Takaku M., Walsh S.V., Griffin J.L., Uruno A., Yamamoto M., Hayes J.D. Transcription factor Nrf1 negatively regulates the cystine/glutamate transporter and lipid-metabolizing enzymes. Mol. Cell Biol. 2014;34(20):3800–3816. doi: 10.1128/MCB.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X., Long Y.C. Crosstalk between cystine and glutathione is critical for the regulation of amino acid signaling pathways and ferroptosis. Sci. Rep. 2016;6:30033. doi: 10.1038/srep30033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keppler D. Export pumps for glutathione S-conjugates. Free Radic. Biol. Med. 1999;27(9–10):985–991. doi: 10.1016/s0891-5849(99)00171-9. [DOI] [PubMed] [Google Scholar]
- 41.Ballatori N., Krance S.M., Marchan R., Hammond C.L. Plasma membrane glutathione transporters and their roles in cell physiology and pathophysiology. Mol. Asp. Med. 2009;30(1–2):13–28. doi: 10.1016/j.mam.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin J., Yang J.M., Zhang F., Miao P., Lin Y., Chen M.L. Individual and joint toxic effects of cadmium sulfate and alpha-naphthoflavone on the development of zebrafish embryo. J. Zhejiang Univ. Sci. B. 2014;15(9):766–775. doi: 10.1631/jzus.B1400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosnjak I., Uhlinger K.R., Heim W., Smital T., Franekic-Colic J., Coale K., Epel D., Hamdoun A. Multidrug efflux transporters limit accumulation of inorganic, but not organic, mercury in sea urchin embryos. Environ. Sci. Technol. 2009;43(21):8374–8380. doi: 10.1021/es901677r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisen J.S., Smith J.C. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135(10):1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 45.Rossi A., Kontarakis Z., Gerri C., Nolte H., Holper S., Kruger M., Stainier D.Y.R. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 46.Sun Z., Zhang S., Chan J.Y., Zhang D.D. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell Biol. 2007;27(18):6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velichkova M., Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol. Cell Biol. 2005;25(11):4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material