Abstract
Patterned poly(oligo ethylene glycol) methyl ether methacrylate (POEGMEMA) brush structures may be formed by using a combination of atom-transfer radical polymerization (ATRP) and UV photopatterning. UV photolysis is used to selectively dechlorinate films of 4-(chloromethyl)phenyltrichlorosilane (CMPTS) adsorbed on silica surfaces, by exposure either through a mask or using a two-beam interferometer. Exposure through a mask yields patterns of carboxylic acid-terminated adsorbates. POEGMEMA may be grown from intact Cl initiators that were masked during exposure. Corrals, traps, and other structures formed in this way enable the patterning of proteins, vesicles, and, following vesicle rupture, supported lipid bilayers (SLBs). Bilayers adsorbed on the carboxylic acid-terminated surfaces formed by C–Cl bond photolysis in CMPTS exhibit high mobility. SLBs do not form on POEGMEMA. Using traps consisting of carboxylic acid-functionalized regions enclosed by POEGMEMA structures, electrophoresis may be observed in lipid bilayers containing a small amount of a fluorescent dye. Segregation of dye at one end of the traps was measured by fluorescence microscopy. The increase in the fluorescence intensity was found to be proportional to the trap length, while the time taken to reach the maximum value was inversely proportional to the trap length, indicating uniform, rapid diffusion in all of the traps. Nanostructured materials were formed using interferometric lithography. Channels were defined by exposure of CMPTS films to maxima in the interferogram, and POEGMEMA walls were formed by ATRP. As for the micrometer-scale patterns, bilayers did not form on the POEGMEMA structures, and high lipid mobilities were measured in the polymer-free regions of the channels.
Introduction
Lipid membranes play a central role in biology: they form the cellular membrane, separating the interior of the cell from its external environment, and they provide the means by which the interior of the cell is compartmentalized into discrete organelles.1 Understanding how biological systems use compartmentalization is a fundamental scientific challenge, and one that is also intricately connected with attempts to build biologically inspired nanosystems.2,3 However, native lipid membranes are difficult to study in situ. Supported lipid bilayers (SLBs) provide a convenient model for biological lipid membranes, facilitating direct interrogation by a plethora of techniques, including spectroscopic methods,4 quartz crystal microbalance measurements,5 surface plasmon resonance and atomic force microscopy.5 SLBs may be formed by the adsorption, fusion, and rupture of vesicles from an aqueous solution onto a clean oxide substrate.6,7 Although the precise mechanism for this process is not fully understood, it is thought that electrostatic interactions between the lipids and substrate play an important role.8 Clean silica substrates,9 or other inorganic surfaces such as mica, have been widely used; vesicles rupture readily on these surfaces to form continuous and highly mobile SLBs. There has also been much interest in forming SLBs on other surfaces, including polymers.10−12
The dynamical behavior of lipids and membrane components is important in controlling many biological processes.13 For example, bacterial photosynthesis is driven by a variety of membrane transport processes, including intramembrane transfer of charge, via diffusion of quinols, and transmembrane proton transport through the activity of cytochromebc1 and ATPsynthase.14,15 In eukaryotes, Groves and co-workers have demonstrated the importance of intramembrane transport in the immune system. Using “mazes” (collections of staggered lines) formed from 100 to 200 nm wide, 5.5 nm high chrome structures at 1.5–2 μm spacings, they were able to investigate the role of spatial organization in T-cell receptor signaling.16 It was found that the recognition of a peptide antigen by T cells involves coordinated movement of T cell receptors (TCRs) along with other costimulatory and signaling molecules, leading to the formation of immunological synapses, in which cluster size directly influences protein spatial positioning.17 However, in a review of bilayer patterning techniques, DeMond and Groves noted a wide range of significant experimental challenges. In particular, there are few reliable methods for control of bilayer organization,18 and substantial problems associated with the incorporation of transmembrane proteins into supported lipid bilayers.
There has been interest in the formation of patterned SLBs for use in studies of electrophoresis. By applying an electric field in the plane of the SLB, charged components such as lipids could be moved. This was first demonstrated by Sackman et al, who used electrophoresis to determine the mobility and diffusion coefficients of lipids in an SLB.19 Yoshina-Ishii and Boxer continued this work by showing that it was possible to manipulate lipids within membrane arrays.20 More recently, Cheetham and Roth and co-workers published a series of papers in which ratchet structures were fabricated by photolithography and microcontact printing for the movement and concentration of both lipids and membrane proteins within SLBs.21−24
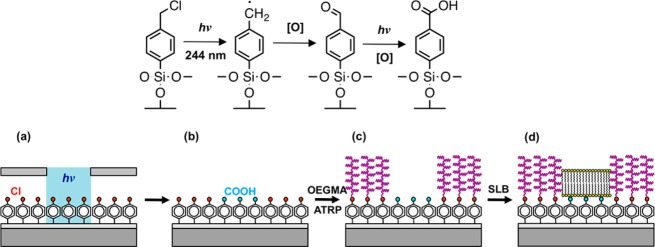
The present work reports a new approach to the fabrication of structures for the investigation of dynamic phenomena in SLBs (Figure 1). The method is effective across a wide range of length scales, from hundreds of micrometers to tens of nanometers, and relies upon simple chemistry. When 4-(chloromethyl)phenyltrichlorosilane (CMPTS) is exposed to UV light, photolysis of the C–Cl bond occurs to create first an aldehyde and then a carboxylic acid (Figure 1b).25 This rapid process enables the definition of hydrophilic, anionic regions in which SLBs may be formed. Lipid mobilities on such surfaces are comparable to those observed on glass. To contain lipid diffusion, “walls” are grown from unmodified regions of the sample by atom-transfer radical polymerization (ATRP)26,27 of oligo(ethylene glycol) methyl ether methacrylate (OEGMEMA), using intact Cl as the initiator (Figure 1c); SLBs are then deposited by standard methods into the carboxylate regions (Figure 1d). Trap structures were formed and used in studies of electrophoresis. ATRP of OEGMEMA28−31 and of zwitterionic monomers such as 2-methacryloloxyethyl phosphorylcholine,32,33 sulfobetaine methacrylates34 and amino acid methacrylates35 has been shown to be a very effective means of passivating surfaces against adsorption of biological molecules.36,37 By carrying out exposure using a two-beam interferometer, nanostructured polymers were also formed that enclosed nanostructured lipid channels and were used to study diffusional transport in confined environments.
Figure 1.
Top: reaction scheme for the photochemical oxidation of CMPTS. Bottom: schematic diagram showing the fabrication of SLBs confined by poly(oligoethylene glycol)methacrylate brushes.
Experimental Section
Silicon wafers (test grade, B-doped, < 100>, 380 μm thick) were supplied by Pi-KEM (Peterbrough, UK). Copper electron microscope grids (1000–2000 mesh) were obtained from Agar Scientific (Stanstead, UK). 4-(Chloromethyl)phenyltrichlorosilane was obtained from Alfa Aesar (Heysham, UK). Oligo(ethylene glycol) methyl ether methacrylate (Mn 475), 2,2′-bipyridyl (Bipy, > 99%), copper(I) bromide (99%), and copper(II) bromide (99.5%) were obtained from Sigma-Aldrich (Poole, UK). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) were purchased from Avanti Polar Lipids (Alabaster, AL). Atto 590-labeled 1, 2-dioleoyl-sn-glycero-3-phosphoehanomamine (Atto590-DOPE) and Atto 488-labeled 1, 2-dioleoyl-sn-glycero-3-phosphoehanomamine (Atto488-DOPE) were purchased from Atto-TEC (Siegen, Germany).
To prepare polymer brushes by ATRP, samples were placed in carousel tubes, sealed, degassed, and placed under nitrogen. In a round-bottom flask, water (10 mL) and methanol (10 mL) were added to the monomer, and the solution was degassed for 30 min. To the monomer solution, 0.37g of copper(I) bromide and 0.81 g of 2,2′-bipyridyl were added, and the solution was degassed for a further 5 min and sonicated. 1–2 mL of the monomer–catalyst solution was added to the carousel tubes, and the samples were left to polymerize for various times (to control the brush thickness). Once the polymerization was complete, the samples were sonicated in water, rinsed with ethanol, and blown dry with nitrogen.
Dried lipids (DOTAP:POPC:Atto590-DOPE = 24.9:74.6:0.5) were dissolved in a 50:50 mixture of HPLC-grade chloroform and methanol and transferred to glass vials. The lipids were dried under a flow if nitrogen for 1 h and rehydrated using phosphate buffer (a 10 mM mixture of sodium dihydrogen phosphate and disodium hydrogen phosphate in deionized water, adjusted to pH 7.1 with NaOH or HCl. Vesicle solutions (1.0 mg mL–1) were prepared by vortex mixing for 1 min (Vortex Genie2, Jencons Ltd., Leighton Buzzard, UK) to create multilamellar vesicles as a cloudy suspension. Small unilamellar vesicles were prepared by tip sonication of the aforementioned solution (Branson Sonifer 750, Branson Ultrasonics Corp, Danbury, CT) at 4 °C for 30 min, during which time the suspension became clear. The suspension was centrifuged (Heraeius Fresco 17, Thermo Fisher Scientific, Loughborough, UK) for 1 min at 14 500g, after which the Ti precipitate (formed at the surface of the tip of the sonicator during the tip-sonication process) was removed and the supernatant was retained. The suspension was diluted with phosphate buffer to 0.5 mg mL–1 prior to use and stored at 4 °C in the dark for no longer than 5 days.
Bilayer formation was carried out in a custom-built flow cell. For bare glass substrates, the vesicles were injected and incubated for 1 h at 22 °C. The samples were rinsed subsequently for 20 min with degassed, deionized water at a flow rate of 2.6 mL min–1. For the polymer brush patterns, the samples were first soaked in buffer solution for 10 min, followed by injection of vesicles, incubation and rinsing.
Photopatterning was carried out using a Coherent Innova 300C frequency-doubled argon ion laser (Coherent UK, Cambridge, UK) emitting at 244 nm. Micropatterns were formed by carrying out the exposure through a mask. Interferometric lithography was carried out as described previously using a Lloyd’s mirror interferometer in combination with the same laser. The laser beam was directed at a sample stage and mirror held at an angle 2θ relative to each other, such that half the beam struck the sample and the other half struck the mirror from where it was reflected onto the sample to interfere with the first half of the beam. The resulting interferogram had a sinusoidal cross-section with a period of λ/2sin θ.
X-ray photoelectron spectroscopy (XPS) was carried out using a Kratos Axis Ultra X-ray photoelectron spectrometer equipped with a monochromatized X-ray source operating at a power of 150 W and emission current of 8 mA. Samples were mounted using double-sided adhesive tape, and an electron flood was used to compensate for sample charging. Electron energy analyzer pass energies of 160 and 20 eV were used to acquire wide (survey) spectra and high resolution spectra, respectively. Data were analyzed using CasaXPS software (Casa, http://www.casaxps.com, UK).
Secondary ion mass spectrometry (SIMS) was carried out using an IonToF SIMS V imaging secondary ion mass spectrometer (IonToF, Münster, Germany), equipped with a bismuth cluster source and a single-stage reflectron time-of-flight mass analyzer. A minimum of 2 spectra per sample and multiple samples were analyzed. High mass-resolution images were obtained by using high-current bunched mode, with Bi2+ as the primary projectile and a target current of 0.1 pA. The data were analyzed using the SurfaceLab 6 software (IonToF).
Fluorescence microscopy was carried out using an epifluorescence microscope (Nikon Instruments Europe, B.V., Kingston, UK). Fluorescence images were captured using a 12-bit greyscale digital camera, Orca-ER (Hammamatsu Photonics UK Ltd., Welwyn Garden City, UK).
Atomic force microscopy was carried out using a Digital Instruments Nanoscope IV Multimode instrument (Veeco, Santa Barbara, USA) equipped with a ‘J’ scanner (0–125 μm). In contact mode, silicon nitride nanoprobes with nominal force constants of 0.06 or 0.12 N m–1 and tip radii in the range 20–60 nm were used (Bruker, Coventry, UK). In tapping mode, silicon probes with spring constants between 20 and 80 N m–1 were used (Bruker). Prior to analysis, samples were washed with ethanol and dried under a stream of nitrogen. Samples were then secured to a metal disc using double-sided adhesive tape.
Fluorescence recovery after photobleaching (FRAP) was carried out using an epifluoroescence microscope (E600 Nikon, USA). A small amount of Atto590-DOPE was introduced to the lipid mixture, and the sample was illuminated and bleached by a high pressure mercury arc lamp. The bleached spot radius was 14 μm when using a 40× objective lens. Fluorescence images were collected using a Zyla sCMOS CCD (Andor Technology Ltd., Belfast, UK) with 2 × 2 binning, and recorded on NIS elements software. Images were collected until complete fluorescence recovery was observed. The Axelrod method of analysis38 was employed, which provides both the diffusion coefficient and the mobile fraction.
Electrophoresis was carried out in a home-built flow cell, which served to maintain the membrane in an aqueous environment and facilitate the connection of external electrodes to the on-substrate interdigitated electrodes. An arbitrary waveform generator (Thurlby Thandar Instruments Ltd., Huntingdon, UK) and a home-built amplifier were used to generate the electrical signal for the experiments. Currents of 10–100 μA between the electrodes were monitored using a Keithley picoammeter (Keithley Instruments Ltd., Theale, UK). A constant flow of degassed deionized water at 0.75 mL min–1 was maintained for the duration of the experiment to reduce Joule heating generated by the electric current, maintain a constant temperature, and remove bubbles generated by redox processes at the electrode surfaces.
Results and Discussion
POEGMEMA Patterning
A detailed investigation of the mechanism of dehalogenation of CMPTS films was reported previously by Sun et al.,25 who reported a substantial decline in contact angle following exposure of films to UV light. To confirm that the dehalogenation reaction was occurring as required, the change in contact angle was measured as a function of the UV exposure (Figure 2a). The contact angle of the virgin film was 68°, and this declined to 10° after an exposure of 2.7 J cm–2. Thereafter, no significant change in contact angle was measured. XPS Cl 2p spectra were acquired before and after exposure of films to 4 J cm–2 of UV light (Figure 2b). It can be seen that at this exposure, Cl is undetectable by XPS. An exposure of 4 J cm–2 was deemed suitable for all of the subsequent patterning experiments.
Figure 2.
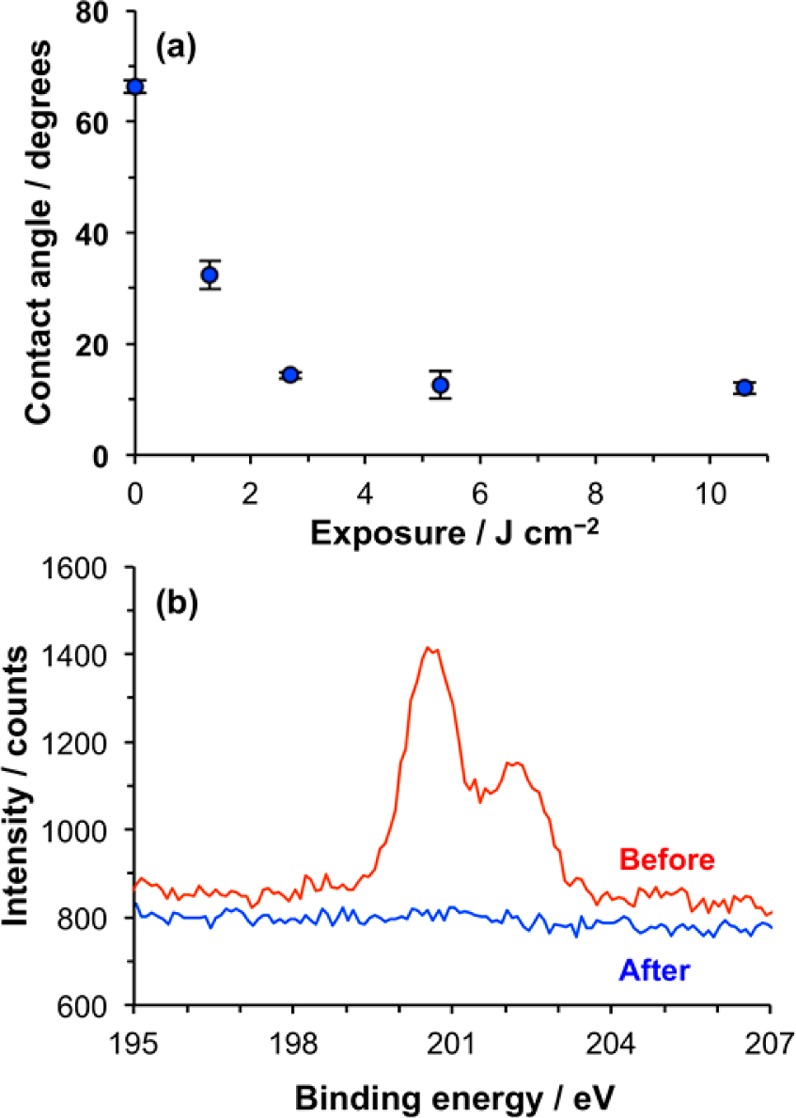
(a) Variation in the advancing water contact angle with UV exposure for CMPTS films. (b) XPS Cl 2p spectra recorded before and after exposure of samples to 4 J cm–2 of UV irradiation.
Poly(oligo(ethylene glycol) methyl ether methacrylate) (POEGMEMA) brushes may be grown from halogenated surfaces by ATRP to yield thick, highly protein-resistant surfaces.29,31,37 Growth is slower from chlorinated surfaces than from the more commonly used bromine initiators,39 but is nevertheless substantial. Patterned brushes were fabricated by first exposing CMPTS films to UV irradiation (4 J cm–2 at 244 nm) through a 2000 mesh electron microscope grid, and then subsequently carrying out ATRP. Because a grid was used as a mask, a large number of features were fabricated close together, enabling the uniformity of the patterning process to be evaluated. Figure 3a shows an AFM tapping mode topographical image of a typical sample. The dark squares correspond to regions that were exposed to UV light; here the Cl has been removed by C–Cl bond photolysis and no polymer grows. The bars (bright contrast) correspond to regions that were masked during UV exposure. Here polymer molecules have grown from surface-immobilized Cl initiators. Line sections (Figure 3b shows a representative example) indicated that the mean thickness of the brush layer, measured as the height difference between the masked and exposed regions, was 104 nm.
Figure 3.
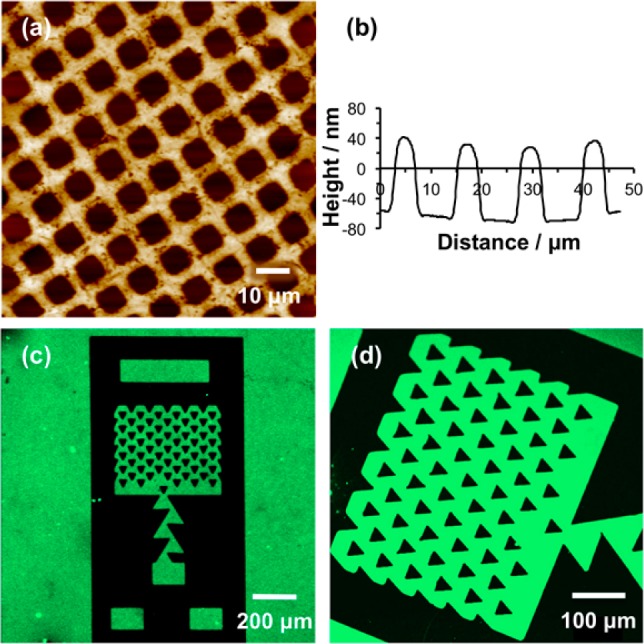
(a) AFM topographical image of a patterned POEGMEMA brush formed by UV exposure of a CMPTS film through a mask, followed by ATRP. (b) Line section through image a. (c,d) Fluorescence microscopy images of trap structures formed by photopatterning of CMPTS combined with ATRP after immersion in a solution of GFP. GFP adsorbs to the carboxylic acid terminated regions of the pattern formed by UV exposure (bright contrast) but not the POEGMEMA brushes (dark contrast).
To further test the effectiveness of the polymer patterning, samples were immersed in solutions of green fluorescent protein (GFP). GFP is not expected to adsorb to POEGMEMA, which exhibits strong resistance to protein adsorption,31,40 but it is expected to adsorb to POEGMEMA-free regions defined by dehalogenation of the CMPTS film. Figure 3c,d shows fluorescence microscopy images of a trap structure formed by UV exposure of CMPTS through a mask, followed by ATRP of OEGMEMA and immersion in GFP solution. Dark contrast is observed from regions that were masked during exposure (for example, the triangular features in Figure 3d). However, bright contrast is observed on regions that were exposed to UV light. The contrast difference between the masked and exposed regions is abrupt, indicating that the patterning has been effective.
To achieve mobile SLBs of high quality, it is essential that there be low rates of polymer growth from the exposed regions of the patterns. Because high molecular weights may be achieved via ATRP, defects are effectively amplified. To assess polymer growth from residual Cl “defects” in the exposed regions, imaging secondary ion mass spectrometry (SIMS) was used to characterize trap structures similar to the one in Figure 3c,d. SIMS enables retrospective mass spectral imaging at high spatial resolution. Figure 4a shows a region of the negative ion SIMS spectrum of an unpatterned POEGMEMA brush. The spectrum exhibits a plethora of oxygen-containing fragment ions that may be used to differentiate the brush from the surrounding surface. Patterned samples were imaged by mapping the intensity of the C2H3O– species (m/z 43). Figure 4b shows a secondary ion image formed for a trap structure similar to the one shown in Figure 3c,d. The regions that exhibited dark contrast in Figure 3c,d are thought to be occupied by polymer brushes, and this is confirmed by the C2H3O– image. By contrast, the SIMS image exhibits dark contrast in regions corresponding to those that displayed bright contrast in Figure 3c,d. This confirms that the polymer is largely absent from the exposed regions of the sample; if polymers are formed from low densities of Cl “defects”, they are present at levels too small to be readily detectable by SIMS.
Figure 4.
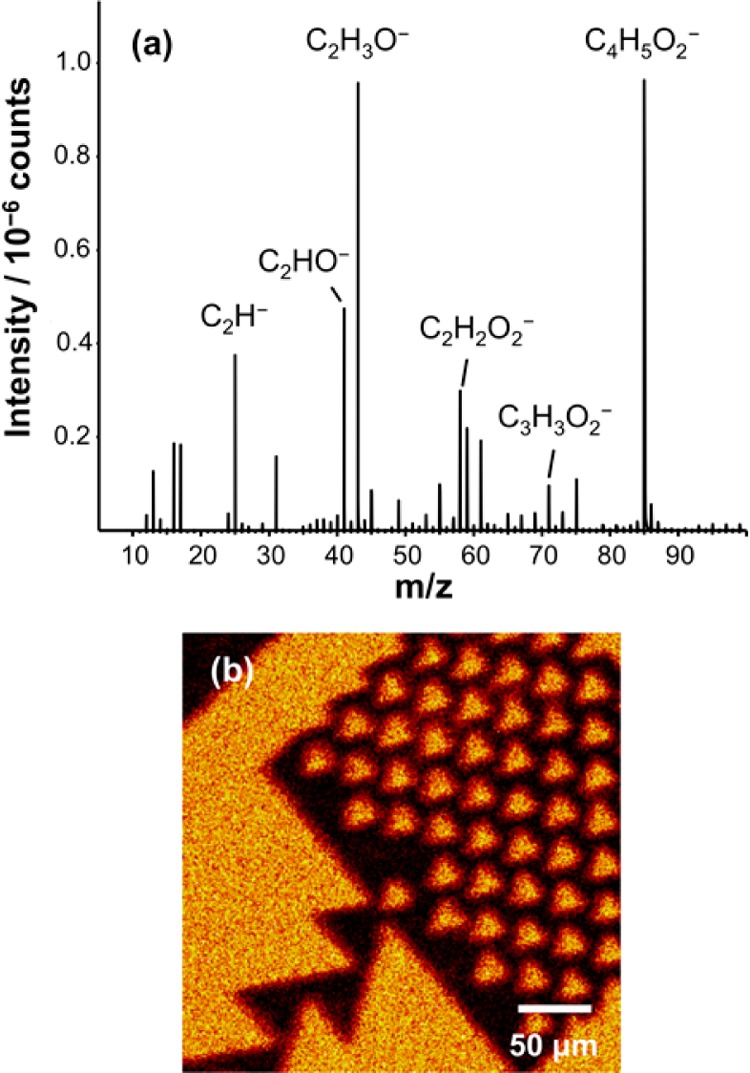
(a) Negative ion SIMS spectrum of an unpatterned POEGMEMA brush. (b) SIMS image of a POEGMEMA trap structure formed by mapping the intensity of the C2H3O– species.
SLB Formation
To test the effectiveness of POEGMEMA as a means of confining SLBs, trap structures were fabricated as described above and incubated in suspension containing vesicles formed using a 24.9:74.6:0.5 DOTAP:POPC:Atto590-DOPE mixture. The DOTAP is positively charged and is expected to have a favorable electrostatic interaction with carboxylate groups formed at the photomodified CMPTS surface, aiding vesicle rupture.
To evaluate the efficacy of confinement of the SLB by the POEGMEMA brushes, trap structures were formed and characterized by fluorescence microscopy after deposition of vesicles (Figure 5a,b). It can be seen that the lipids are confined to the carboxylic acid-terminated regions formed during UV exposure: the pattern of fluorescence from the lipid layer in Figure 5a matches the distribution of intensity due to GFP in Figure 3c. The high magnification image (Figure 5b) displays a clear contrast difference between the triangular lipid-free regions (dark) and the surrounding SLB (bright). These data confirm that POEGMEMA brushes resist the formation of an SLB, and are a highly effective and convenient means to organize SLBs into patterns.
Figure 5.
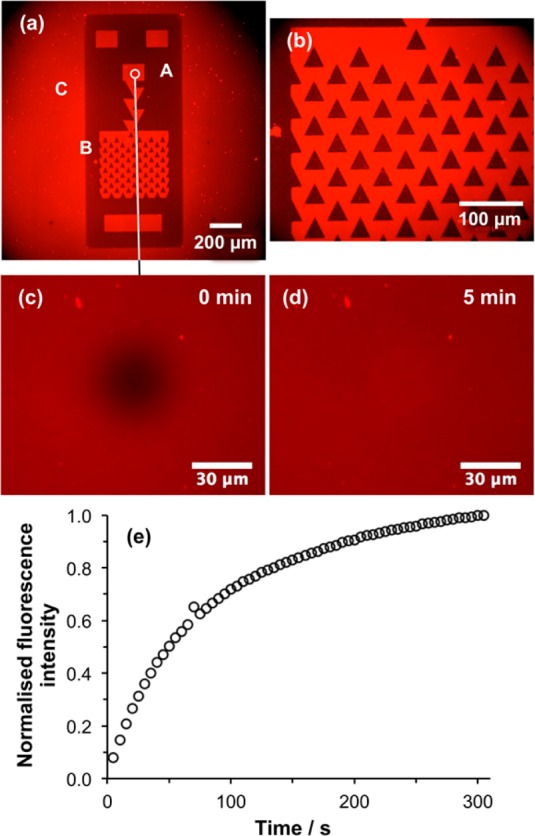
(a,b) Fluorescence micrographs of a trap structure formed by UV exposure of a CMPTS film, ATRP of OEGMEMA and SLB deposition. Areas masked during UV light (for example the region marked A in panel a) support growth of POEGMEMA by ATRP, and thus resist SLB formation, while exposed regions such as B are polymer-free and facilitate SLB formation. The laser spot used during patterning was somewhat larger than the dimensions of the trap structure defined by the photolithography mask, hence removal of halogen also occurs outside the trap structure, enabling formation of an SLB there (e.g., at C). (c) Fluorescence micrograph showing a bleached spot formed in the small circular region identified in panel a. (d) Fluorescence micrograph of the same region acquired after 5 min. (e) Variation in fluorescence intensity in the bleached region as a function of time after exposure.
FRAP measurements were made to test the mobility of lipids in these patterned bilayers. Figure 5c shows a fluorescence micrograph acquired of a bleached spot (the dark, central feature in the image) formed within the small circular region indicated in the upper central portion of Figure 5a. Figure 5d shows a fluorescence micrograph of the same region acquired 5 min later. It is clear that intensity has recovered fully in the bleached spot as a consequence of lipid diffusion in the SLB. The fluorescence intensity in the bleached region is shown as a function of time in Figure 5e. This fluorescence recovery plot was analyzed using the method of Axelrod et al. The Axelrod method is a well-established method for the analysis of diffusion in supported lipid bilayers. It involves fitting the recovery curve to yield a mathematical relationship between fluorescence intensity and time after bleaching, from which the diffusion coefficient and mobile fraction may be calculated.38 Analysis of the data in Figure 5e using this method indicated that the mobile fraction was 98% and the diffusion coefficient was 0.84 μm2 s–1, comparable to values obtained for SLBs formed from the same lipids on glass. These data demonstrate that the carboxylic acid-functionalized surface produced by photochemical modification of the CMPTS film is an excellent substrate for SLB formation.
Electrophoresis
Trap structures were defined by using mask-based photolithography to expose CMPTS films, and the resulting carboxylic acid functionalized regions were enclosed by POEGMEMA by using ATRP to grow brushes from intact Cl in regions that were masked during exposure. After incubation in Atto590-labeled DOTAP-POPC lipid vesicles, the sample was imaged using fluorescence microscopy (Figure 6a). It may be seen that the fluorescence intensity is confined to the traps, and that it is uniformly distributed across their length.
Figure 6.
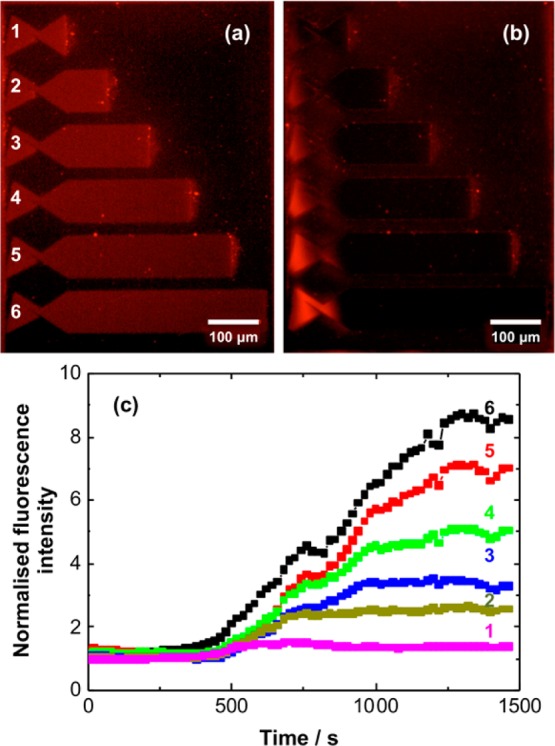
Fluorescence micrographs of trap structures before (a) and after (b) application of a 100 V dc potential for 20 min. (c) Time dependence of the fluorescence intensity measured at the left-hand side of the trap structures in panels a and b.
A 100 V dc potential was applied parallel to the long axes of the trap structures. After 20 min, the distribution of fluorescence intensity was nonuniform. Intensity was found to have accumulated in the “nest” of the trap, at the left-hand side of the structures in Figure 6a,b. This is consistent with movement of lipids opposite to the electric field direction. This movement of lipids was confirmed by measuring the intensity of fluorescence in the nest as a function of time (Figure 6c). Initially the fluorescence intensity changes slowly, but after 400 s, the intensity starts to rise rapidly, indicating the presence of highly mobile lipids in the traps. The increase in fluorescence intensity in the nest is proportional to the length of the trap, so the brightest fluorescence is observed for the longest trap. The longest traps also require the longest time to reach a limiting value, consistent with the fact that lipid transport occurs over longer distances. For the shorter traps, a limiting intensity is reached much more quickly. The proportionality between fluorescence intensity and trap length indicates that the charged fluorescent species are mobile along the lengths of the traps.
Lipid Diffusion in Nanostructures
It is known that rates of diffusion of lipids in SLBs may be reduced when the bilayers are formed into channels narrower than 50 nm.41,42 For example, Tsai et al. used electron beam lithography to fabricate barriers with periods of 125 and 250 nm, and containing gaps that varied from 30–50 nm.42 They found that such structures were useful in capturing the diffusional behavior of membrane lipids. To examine the feasibility of using polymer brushes for studies of lipid diffusion in confined geometries, nanostructures were fabricated by interferometric lithography (IL). A particularly attractive feature of IL for such studies is the fact that patterning occurs simultaneously over a macroscopic region (∼1 cm2 in the apparatus used here). Portions of silicon wafer derivatized with a CMPTS film were placed in the interferometer and exposed to UV light, before growth of polymer brushes by ATRP. In IL, the sample is exposed to an interferogram with a sinusoidal cross-section; hence the resulting patterns exhibit a gradient character because the intensity of illumination varies in a gradient fashion. Control of the exposure conditions and development process (in this case, brush growth) provides control over the dimensions and properties of the resulting structures. The period may be controlled by changing the angle between the sample and mirror in the interferometer, with a theoretical minimum period of λ/2.
Figure 7 shows an AFM topographical image of a nanostructured sample formed as described above. The period was selected to be large (1.39 μm) because it was intended to use fluorescence microscopy to characterize the structure. The cross section reveals that the polymer structures have a fwhm of 750 nm. The line section indicates that the polymer-free region, where the CMPTS film was exposed to a maximum in the interferogram, has a width of ∼300 nm. To test the effectiveness of these structures at confining vesicle deposition, fluorescence microscopy was carried out after incubation of the sample with DOTAP:POPC:Atto488-DOPE vesicles. Very narrow bands of fluorescence were observed (Figure 7c). The widths of these features are similar to the diffraction limit for this dye, ∼ 300 nm, consistent with the approximate widths of the polymer-free regions observed in the AFM images. Clearly a precise estimation of the feature sizes is not possible by fluorescence microscopy, but the data provide very good evidence that nanostructured POEGMEMA brushes are effective at localizing vesicles in narrow regions.
Figure 7.
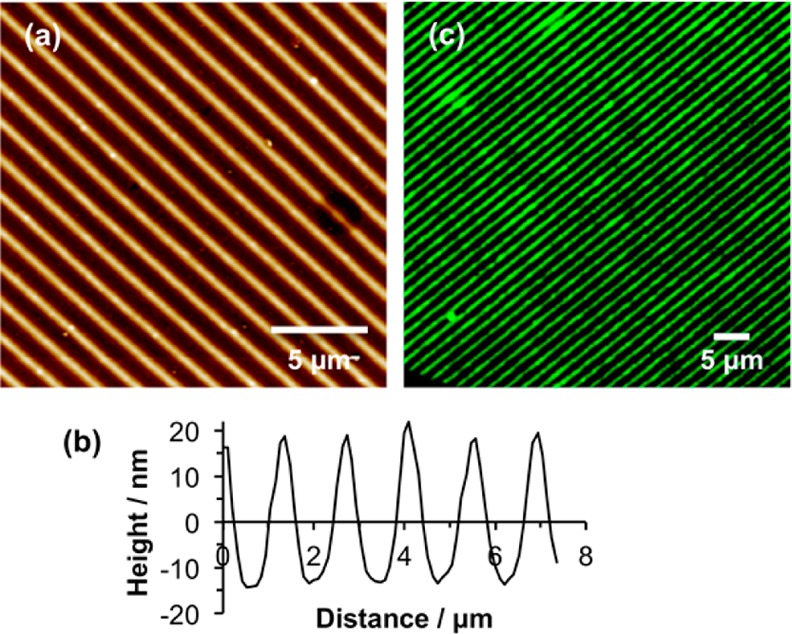
(a) AFM height image of POEGMEMA nanostructures formed by using IL to expose a CMPTS film, followed by ATRP. (b) Line section through the height image in panel a. (c) Fluorescence microscopy image of the sample shown in panel a after immersion in a suspension of Atto488-labeled vesicles.
To determine whether the lipids in these structures remained mobile, DOTAP:POPC:Atto590-DOPE vesicles were deposited into nanolines and ruptured to yield SLBs. The resulting nanostructured bilayers were investigated using FRAP. Figure 8a shows an AFM topographical image of a nanostructured surface prior to SLB formation. The associated line section is shown in Figure 8b. The period of the POEGMEMA nanolines was 926 nm, slightly smaller than the period in Figure 7a, but the width of the polymer-free region was similar (∼300 nm). After deposition of vesicles, lines of lipids could not be resolved because the microscope used for FRAP measurements was fitted with a less powerful objective. After photobleaching, a dark spot was observed (Figure 8c). After 375 s, the fluorescence had recovered in the bleached region, indicating that the lipids were mobile in the nanostructured channels formed between POEGMEMA structures. Analysis of the fluorescence recovery curve (Figure 8e) yielded a diffusion rate of 0.47 μm2 s–1. Although this is smaller than the value measured for trap structures such as the one in Figure 5, it remains within the range normally expected for mobile lipid bilayers supported on glass substrates. Moreover, the mobile fraction was calculated to be 0.96, indicating a fully mobile lipid bilayer. A systematic investigation of the relationship between channel dimensions and diffusional behavior is beyond the scope of the present study. However, the data presented here demonstrate that fabrication of polymer brush structures by IL is a convenient and effective way of producing structures that facilitate uniform confinement of SLBs over macroscopic areas.
Figure 8.
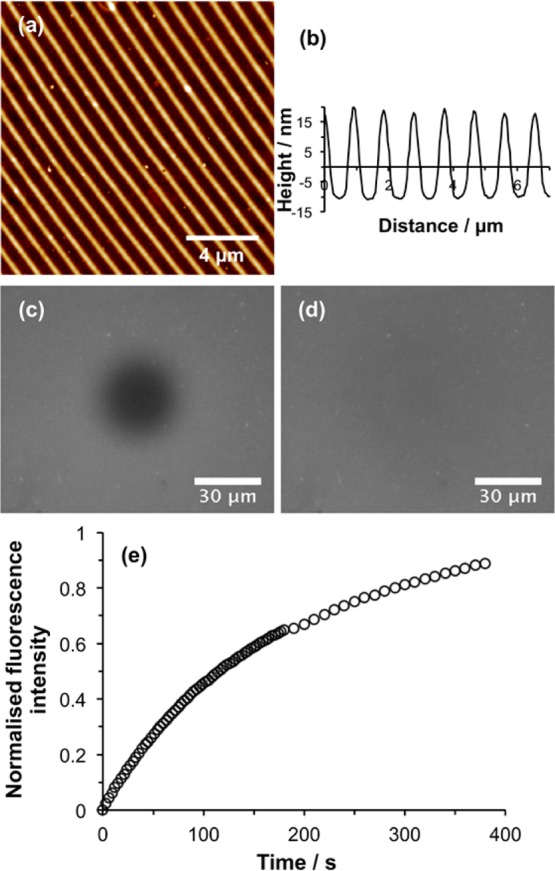
(a) AFM height image of POEGMEMA nanostructures formed by using IL to expose a CMPTS film, followed by ATRP. (b) Line section through the height image in panel a. (c) Fluorescence microscopy image of the sample shown in panel a after deposition of an SLB and photobleaching. (d) Fluorescence microscopy image acquired 375 s after photobleaching. (e) Recovery in fluorescence as a function of time after photobleaching.
Conclusions
Photolysis of C–Cl bonds in CMPTS films leads to the formation of carboxylic acid-functionalized surfaces. Mobile SLBs are formed on these surfaces. Unmodified regions of the CMPTS film retain Cl, which is an initiator for ATRP. POEGMEMA brushes may be grown to high thicknesses from these surfaces. The brushes resist the deposition of proteins, vesicles, and lipid bilayers. The combination of ATRP and photopatterning thus provides a very effective method to form patterned POEGMEMA brushes. These structures in turn provide a highly effective means to organize the formation of supported lipid bilayers. The spatial confinement of the bilayers is precise, and they exhibit similar mobilities to those observed for the same lipids on glass surfaces. For both micrometer-scale and nanometer-scale structures, the mobile fraction is close to unity. While the mobility is slightly reduced in nanostructured channels, probably as a consequence of the lateral confinement, it is still significant. This combination of patterning approaches and surface-initiated polymerization used here seems to be a promising approach for the formation of spatially organized supported lipid bilayers.
Acknowledgments
The authors thank Dr. P. Chapman for helpful contributions to the development of the strategies described here, and Dr J. Roth for designing the traps that were used to generate the data in Figure 6. The authors are grateful to EPSRC (Programme Grant EP/I012060/1) for Financial Support. CNH gratefully acknowledges financial support from the Biotechnology and Biological Sciences Research Council (BBSRC UK), award number BB/M000265/1. CNH was also supported by Advanced Award 338895 from the European Research Council. This work was also supported as part of the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC 0001035. PARC’s role was to provide partial support for CNH.
Author Present Address
⊥ Department of Physics, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL UK.
The authors declare no competing financial interest.
References
- Stryer L.Biochemistry, 4th ed.; W. H. Freeman: New York, 1995. [Google Scholar]
- Grzybowski B. A.; Huck W. T. S. The nanotechnology of life-inspired systems. Nat. Nanotechnol. 2016, 11, 585–592. 10.1038/nnano.2016.116. [DOI] [PubMed] [Google Scholar]
- Deng N.-N.; Yelleswarapu M.; Zheng L.; Huck W. T. S. Microfluidic Assembly of Monodisperse Vesosomes as Artificial Cell Models. J. Am. Chem. Soc. 2017, 139, 587–590. 10.1021/jacs.6b10977. [DOI] [PubMed] [Google Scholar]
- Citra M. J.; Axelsen P. H. Determination of molecular order in supported lipid membranes by internal reflection Fourier transform infrared spectroscopy. Biophys. J. 1996, 71, 1796–1805. 10.1016/S0006-3495(96)79380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C. A.; Glasmästar K.; Zhdanov V. P.; Kasemo B. Formation of Supported Membranes from Vesicles. Phys. Rev. Lett. 2000, 84, 5443–5446. 10.1103/PhysRevLett.84.5443. [DOI] [PubMed] [Google Scholar]
- Tamm L. K.; McConnell H. M. Supported phospholipid bilayers. Biophys. J. 1985, 47, 105–113. 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R. P.; Bérat R.; Brisson A. R. Formation of Solid-Supported Lipid Bilayers: An Integrated View. Langmuir 2006, 22, 3497–3505. 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]
- Castellana E. T.; Cremer P. S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006, 61, 429–444. 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee P. N.; Lin Y.-S.; Dunphy D. R.; Muñiz A. J.; Butler K. S.; Humphrey K. R.; Lokke A. J.; Agola J. O.; Chou S. S.; Chen I. M.; Wharton W.; Townson J. L.; Willman C. L.; Brinker C. J. Mesoporous Silica Nanoparticle-Supported Lipid Bilayers (Protocells) for Active Targeting and Delivery to Individual Leukemia Cells. ACS Nano 2016, 10, 8325–8345. 10.1021/acsnano.6b02819. [DOI] [PubMed] [Google Scholar]
- Santonicola M. G.; Memesa M.; Meszynska A.; Ma Y.; Vancso G. J. Surface-grafted zwitterionic polymers as platforms for functional supported phospholipid membranes. Soft Matter 2012, 8, 1556–1562. 10.1039/C1SM06709E. [DOI] [Google Scholar]
- Kowal J.; Wu D.; Mikhalevich V.; Palivan C. G.; Meier W. Hybrid Polymer–Lipid Films as Platforms for Directed Membrane Protein Insertion. Langmuir 2015, 31, 4868–4877. 10.1021/acs.langmuir.5b00388. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Inal S.; Hsia C.-Y.; Ferro M.; Ferro M.; Daniel S.; Owens R. M. Supported Lipid Bilayer Assembly on PEDOT:PSS Films and Transistors. Adv. Funct. Mater. 2016, 26, 7304–7313. 10.1002/adfm.201602123. [DOI] [Google Scholar]
- Beltramo P. J.; Van Hooghten R.; Vermant J. Millimeter-area, free standing, phospholipid bilayers. Soft Matter 2016, 12, 4324–4331. 10.1039/C6SM00250A. [DOI] [PubMed] [Google Scholar]
- Cartron M. L.; Olsen J. D.; Sener M.; Jackson P. J.; Brindley A. A.; Qian P.; Dickman M. J.; Leggett G. J.; Schulten K.; Neil Hunter C. Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides. Biochim. Biophys. Acta, Bioenerg. 2014, 1837, 1769–1780. 10.1016/j.bbabio.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Cartron M. L.; Mullin N.; Qian P.; Leggett G. J.; Hunter C. N.; Hobbs J. K. Direct Imaging of Protein Organization in an Intact Bacterial Organelle Using High-Resolution Atomic Force Microscopy. ACS Nano 2017, 11, 126–133. 10.1021/acsnano.6b05647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond A. L.; Mossman K. D.; Starr T.; Dustin M. L.; Groves J. T. T Cell Receptor Microcluster Transport through Molecular Mazes Reveals Mechanism of Translocation. Biophys. J. 2008, 94, 3286–3292. 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman N. C.; Nye J. A.; Groves J. T. Cluster size regulates protein sorting in the immunological synapse. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 12729–12734. 10.1073/pnas.0902621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond A. L.; Groves J. T. Interrogating the T cell synapse with patterned surfaces and photoactivated proteins. Curr. Opin. Immunol. 2007, 19, 722–727. 10.1016/j.coi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Stelzle M.; Miehlich R.; Sackmann E. Two-dimensional microelectrophoresis in supported lipid bilayers. Biophys. J. 1992, 63, 1346–1354. 10.1016/S0006-3495(92)81712-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshina-Ishii C.; Boxer S. G. Controlling Two-Dimensional Tethered Vesicle Motion Using an Electric Field: Interplay of Electrophoresis and Electro-Osmosis. Langmuir 2006, 22, 2384–2391. 10.1021/la0526277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham M. R.; Bramble J. P.; McMillan D. G. G.; Krzeminski L.; Han X.; Johnson B. R. G.; Bushby R. J.; Olmsted P. D.; Jeuken L. J. C.; Marritt S. J.; Butt J. N.; Evans S. D. Concentrating Membrane Proteins Using Asymmetric Traps and AC Electric Fields. J. Am. Chem. Soc. 2011, 133, 6521–6524. 10.1021/ja2007615. [DOI] [PubMed] [Google Scholar]
- Cheetham M. R.; Bramble J. P.; McMillan D. G. G.; Bushby R. J.; Olmsted P. D.; Jeuken L. J. C.; Evans S. D. Manipulation and sorting of membrane proteins using patterned diffusion-aided ratchets with AC fields in supported lipid bilayers. Soft Matter 2012, 8, 5459–5465. 10.1039/c2sm25473e. [DOI] [Google Scholar]
- Han X. J.; Achalkumar A. S.; Bushby R. J.; Evans S. D. A Cholesterol-Based Tether for Creating Photopatterned Lipid Membrane Arrays on both a Silica and Gold Surface. Chem. - Eur. J. 2009, 15, 6363–6370. 10.1002/chem.200900404. [DOI] [PubMed] [Google Scholar]
- Roth J. S.; Zhang Y.; Bao P.; Cheetham M. R.; Han X.; Evans S. D. Optimization of Brownian ratchets for the manipulation of charged components within supported lipid bilayers. Appl. Phys. Lett. 2015, 106, 183703. 10.1063/1.4919801. [DOI] [Google Scholar]
- Sun S.; Montague M.; Critchley K.; Chen M.-S.; Dressick W. J.; Evans S. D.; Leggett G. J. Fabrication of Biological Nanostructures by Scanning Near-field Photolithography of Chloromethylphenylsiloxane Monolayers. Nano Lett. 2006, 6, 29–33. 10.1021/nl051804l. [DOI] [PubMed] [Google Scholar]
- Patten T. E.; Matyjaszewski K. Atom Transfer Radical Polymerization and the Synthesis of Polymeric Materials. Adv. Mater. 1998, 10, 901–915. . [DOI] [Google Scholar]
- Matyjaszewski K.; Xia J. Atom Transfer Radical Polymerization. Chem. Rev. 2001, 101, 2921–2990. 10.1021/cr940534g. [DOI] [PubMed] [Google Scholar]
- Ma H. W.; Hyun J. H.; Stiller P.; Chilkoti A. Non-Fouling” Oligo(ethylene glycol)- Functionalized Polymer Brushes Synthesized by Surface-Initiated Atom Transfer Radical Polymerization. Adv. Mater. 2004, 16, 338. 10.1002/adma.200305830. [DOI] [Google Scholar]
- Ma H.; Wells M. Jr.; Beebe T. P.; Chilkoti A. Surface initiated polymerization of nonfouling polymer brushes of oligoethylene glycol methacrylate on gold. Adv. Funct. Mater. 2006, 16, 640–648. 10.1002/adfm.200500426. [DOI] [Google Scholar]
- Ma H.; Textor M.; Clark R. L.; Chilkoti A. Monitoring kinetics of surface initiated atom transfer radical polymerization by quartz crystal microbalance with dissipation. Biointerphases 2006, 1, 35–39. 10.1116/1.2190697. [DOI] [PubMed] [Google Scholar]
- Ma H.; Li D.; Sheng X.; Zhao B.; Chilkoti A. Protein resistant polymer brushes on silicon oxide by surface initiated atom transfer radical polymerization. Langmuir 2006, 22, 3751–3756. 10.1021/la052796r. [DOI] [PubMed] [Google Scholar]
- Feng W.; Zhu S.; Ishihara K.; Brash J. L. Adsorption of Fibrinogen and Lysozyme on Silicon Grafted with Poly(2-methacryloyloxyethyl Phosphorylcholine) via Surface-Initiated Atom Transfer Radical Polymerization. Langmuir 2005, 21, 5980–5987. 10.1021/la050277i. [DOI] [PubMed] [Google Scholar]
- Feng W.; Brash J. L.; Zhu S. Non-biofouling materials prepared by atom transfer radical polymerization grafting of 2-methacryloloxyethyl phosphorylcholine: Separate effects of graft density and chain length on protein repulsion. Biomaterials 2006, 27, 847–855. 10.1016/j.biomaterials.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Chao T.; Chen S.; Jiang S. Superlow Fouling Sulfobetaine and Carboxybetaine Polymers on Glass Slides. Langmuir 2006, 22, 10072–10077. 10.1021/la062175d. [DOI] [PubMed] [Google Scholar]
- Alswieleh A. M.; Cheng N.; Canton I.; Ustbas B.; Xue X.; Ladmiral V.; Xia S.; Ducker R. E.; El Zubir O.; Cartron M. L.; Hunter C. N.; Leggett G. J.; Armes S. P. Zwitterionic Poly(amino acid methacrylate) Brushes. J. Am. Chem. Soc. 2014, 136, 9404–9413. 10.1021/ja503400r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey R.; Lavanant L.; Paripovic D.; Schüwer N.; Sugnaux C.; Tugulu S.; Klok H.-A. Polymer Brushes via Surface-Initiated Controlled Radical Polymerization: Synthesis, Characterization, Properties, and Applications. Chem. Rev. 2009, 109, 5437–5527. 10.1021/cr900045a. [DOI] [PubMed] [Google Scholar]
- Hucknall A.; Rangarajan S.; Chilkoti A. In Pursuit of Zero: Polymer Brushes that Resist the Adsorption of Proteins. Adv. Mater. 2009, 21, 2441–2446. 10.1002/adma.200900383. [DOI] [Google Scholar]
- Axelrod D.; Koppel D. E.; Schlessinger J.; Elson E.; Webb W. W. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976, 16, 1055–1069. 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alang Ahmad S. A.; Leggett G. J.; Hucknall A.; Chilkoti A. Micro- and Nanostructured Poly[oligo(ethylene glycol)methacrylate] Brushes Grown From Photopatterned Halogen Initiators by Atom Transfer Radical Polymerization. Biointerphases 2011, 6, 8–15. 10.1116/1.3553579. [DOI] [PubMed] [Google Scholar]
- Hucknall A.; Kim D. H.; Rangarajan S.; Hill R. T.; Reichert W. M.; Chilkoti A. Simple Fabrication of Antibody Microarrays on Nonfouling Polymer Brushes with Femtomolar Sensitivity for Protein Analytes in Serum and Blood. Adv. Mater. 2009, 21, 1968–1971. 10.1002/adma.200803125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath G. R.; Roth J.; Connell S. D.; Evans S. D. Diffusion in Low-Dimensional Lipid Membranes. Nano Lett. 2014, 14, 5984–5988. 10.1021/nl503024v. [DOI] [PubMed] [Google Scholar]
- Tsai J.; Sun E.; Gao Y.; Hone J. C.; Kam L. C. Non-Brownian Diffusion of Membrane Molecules in Nanopatterned Supported Lipid Bilayers. Nano Lett. 2008, 8, 425–430. 10.1021/nl072304q. [DOI] [PMC free article] [PubMed] [Google Scholar]