Abstract
Pre-BCR signaling and SYK recently were introduced as therapeutic targets for patients with B-cell acute lymphoblastic leukemia (B-ALL), but the importance of this pathway in B-ALL subsets and mechanism of downstream signaling have not fully been elucidated. Here, we provide new detailed insight into the mechanism of pre-BCR signaling in B-ALL. We compared the effects of pharmacologic and genetic disruption of pre-BCR signaling in vitro and in mouse models for B-ALL, demonstrating exquisite dependency of pre-BCR+ B-ALL, but not other B-ALL subsets, on this signaling pathway. We demonstrate that SYK, PI3K/AKT, FOXO1, and MYC are important downstream mediators of pre-BCR signaling in B-ALL. Furthermore, we define a characteristic immune phenotype and gene expression signature of pre-BCR+ ALL to distinguish them from other B-ALL subsets. These data provide comprehensive new insight into pre-BCR signaling in B-ALL and corroborate pre-BCR signaling and SYK as promising new therapeutic targets in pre-BCR+ B-ALL.
Keywords: SYK, B-ALL, pre-BCR
INTRODUCTION
The accumulation of B-ALL blasts results from the disruption of developmental checkpoints that normally regulate the generation of functional mature B-cells.1,2 One of these checkpoints, as pro-B lymphocytes (cytoplasmic Igμ−/surface IgM−) transition to become pre-B lymphocytes (cytoplasmic Igμ+/surface IgM−), involves surface expression of the precursor-B-cell receptor (pre-BCR).3 Expression of a functional pre-BCR supports the survival of pre-B cells, induces their rapid proliferative expansion, and ensures their differentiation into more mature B-cells.4 B-cell progenitors that fail to assemble a functional pre-BCR lack pre-BCR-dependent survival signals and are removed from the pool of developing B-cells.5,6
The structure of the pre-BCR closely resembles that of the BCR on mature B-cells.7 Both contain two immunoglobulin μ (Igμ) heavy chains and the signal-transducing heterodimer of Igα (CD79a) and Igβ (CD79b). However, instead of the immunoglobulin light chain (LC), the Igμ of the pre-BCR pairs with the surrogate light chain (SLC), a heterodimer consisting of the invariant proteins λ5 (CD179b) and VpreB (CD179a). The SLC is not only required for proper assembly and surface expression of the pre-BCR, but also for the distinctive mechanism by which it is activated, referred to as “cell-autonomous signaling”. Highly conserved regions within the Igμ CH1 domain and λ5 facilitate the aggregation of multiple pre-BCR complexes, followed by the engagement of LYN and spleen tyrosine kinase (SYK) and subsequent activation of downstream signaling cascades including PI3K/AKT, ERK and BTK/PLCγ2.8–11 Pre-BCR aggregation also induces its rapid internalization; consequently, active pre-BCR signaling is associated with low pre-BCR surface expression.
Besides its importance for normal B-cell development, several studies suggest a role for the pre-BCR and its downstream signaling components in B-ALL.12–17 In Ph+-ALL, the oncogenic fusion kinase BCR-ABL activates pre-BCR downstream effectors, such as BTK, BACH2 and BCL6.15,18,19 TCF3-PBX1+ B-ALL cells depend on Igα and Igβ expression and are sensitive to the combined BTK-, SRC- and ABL kinase inhibitor dasatinib.20 Similarly, small-molecular inhibitors of SYK were shown to induce apoptosis and reduce tumor cell burden in several xenograft models of B-ALL21,22 and recently Geng et al. identified a subgroup of B-ALL, characterized by the presence of tonic pre-BCR signaling and the sensitivity to small-molecular inhibitors of SYK and BTK.14 Whether inhibition of pre-BCR signaling using, for example, SYK inhibitors is more broadly applicable across B-ALL subsets, as suggested by earlier studies21–23, or restricted to pre-BCR+ subsets of patients based on more recent studies, remains controversial, given these discrepant data.14,20,24 This has major impact on the conceptualization of future clinical trials, which could be restricted to pre-BCR+ patient subsets or not.
Prior studies of pre-BCR signaling in normal and malignant B-cells relied on the use of tyrosine kinase inhibitors or mouse models of B-cell development, two approaches that have some drawbacks. Tyrosine kinase inhibitors harbor off-target effects, and pre-BCR checkpoints in humans and mice differ significantly, which both can introduce variables and complicate data interpretation.25,26 Our comparative approach of gene deletion and pharmacologic inhibition of pre-BCR signaling avoids some of these limitations, and therefore sheds new light onto these issues.
MATERIALS AND METHODS
Cell lines, xenografts and patient samples
Please see Supplemental Methods
Flow Cytometry
The following antibodies were used throughout the study: PE anti-human pre-BCR, PE anti-human λ5, PE anti-human VpreB (BioLegend, San Diego, CA), R-PE anti-human IgM, APC anti-human IgM (SouthernBiotech, Birmingham, AL), APC anti-human SYK (eBioscience, San Diego, CA), PE anti-human CD19 (BD Pharmingen, San Diego, CA), PE anti-human AKT (pS473) (BD Phosflow, San Jose, CA). To detect cell surface antigens 5*105 cells were stained with the respective antibodies diluted in 100 μl FACS-Buffer (0.5% bovine serum albumin (BSA) w/v in RPMI1640) for 30 min at 4°C protected from light. To measure intracellular antigens cells were fixed, permeabilized and stained using the BD Cytofix/Cytoperm™ Fixation/Permeabilization Kit (BD Bioscience, San Jose, CA). For each antibody respective Isotype antibodies were used as controls. After staining cells were washed once and mean fluorescence was assessed on a FACSCalibur (BD, Franklin Lakes, NJ).
Proliferation and Viability
For in vitro efficacy studies of PRT318 and LY294002 2*104 cells were seeded in 100μl in 96 well plates in cell culture medium without phenol red and treated with PRT318 or LY294002 for 72 h, followed by XTT assays (XTT Cell Proliferation Assay, Trevigen, Gaithersburg, MD, USA).
To assess cell proliferation after Igμ-KO, MYC-KO and FOXO1-AAA expression, 2*104 cells were seeded in 100 μl in 96 well plates. At the indicated time points cells were transferred into FACS tubes followed by the addition of 300 μl viability staining solution (see below) containing 10 μl CountBright™ counting beads (LifeTechnologies). Cells were analyzed via flow cytometry and absolute numbers of live cells per well were calculated based on the ratio of acquired beads and total numbers of beads/tube.
To assess cell viability cells were stained with 2 μg/ml propidium iodide (PI) and DiOC6 (60 nmol/l) as described previously.27
CRISPR/Cas9 and FOXO1-AAA expression
Guide RNA (gRNA) design and cloning was performed according to the protocol by Ran et al.28 For a more detailed description of the procedure please refer to the supplemental information. For gRNA sequences please refer to Supplemental Table 4.
1099 pcDNA GFP FKHR AAA (FOXO1-AAA) was a gift from William Sellers (Addgene plasmid # 9023).29 Cells were transfected as described above. GFP+ cells were sorted 24 h after electroporation and subjected to functional and molecular testing.
Confocal microscopy
The following dyes and antibodies were used for confocal microscopy: CellTracker™ Green CMFDA (Molecular Probes®, Life Technologies). Anti-human FOXO1 primary antibody (Cell Signaling Technologies), Alexa Fluor® 647-conjugated species-specific secondary antibody (Molecular Probes®, Life Technologies). For a detailed description of the staining protocol please refer to supplementary information.
Gene Expression Profiling and Gene Set Enrichment Analysis (GSEA)
Total RNA was extracted with the RNAqueous kit (Ambion, Austin, TX). After confirmation of RNA quality (RIN > 6.6, median 7.2) using a Bioanalyzer 2100 instrument (Agilent), 300 ng of total RNA was amplified and biotin-labeled through an Eberwine procedure using an Illumina TotalPrep RNA Amplification kit (Ambion) and hybridized to Illumina HT12 version 4 human whole-genome arrays. Data processing was as previously described.30 The B-cell progenitor GEP dataset (GSE45460)31 and St. Jude B-ALL dataset (GSE33315)32 were obtained from the NCBI GEO gene expression database and processed using the Gene Pattern Server provided by the Broad Institute (Cambridge, MA). GSEA was performed using the GSEA software provided by the Broad Institute.33,34
Hierarchical clustering with the Average linkage clustering method was performed with Cluster 3.0 (University of Tokyo, Human Genome Center).
Heatmaps and dendrograms were visualized with Java TreeView (v1.1.6r4).
Statistical Analysis
Statistical analyses were performed with GraphPad Prism v6.
Western Blotting
Please see Supplemental Methods
B-ALL xenograft mouse models
Please see Supplemental Methods
RESULTS
Gene expression profiling identifies a subgroup of B-ALL with features of normal pre-B cell progenitors and active pre-BCR signaling
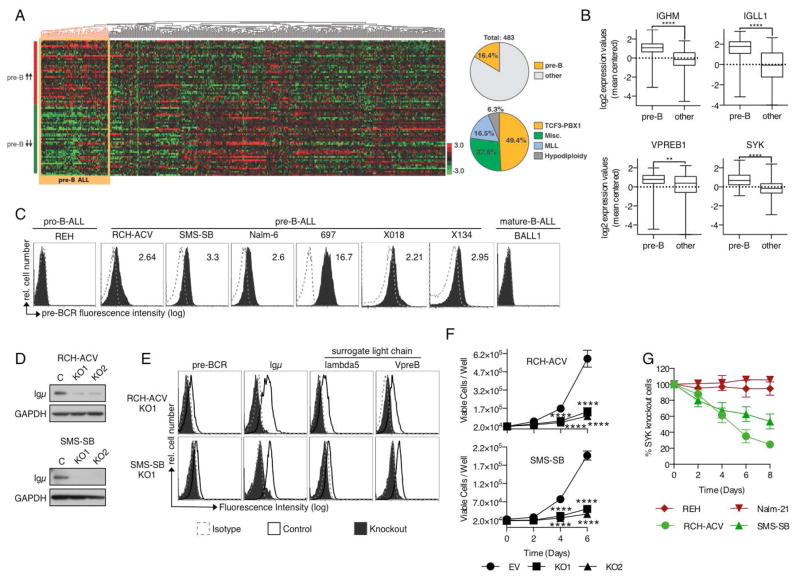
Pre-BCR+ B cell progenitors (CD19+, CD34−, cyto-Igμ+, surface IgM−) have a defined gene expression signature that distinguishes them from early lymphoid progenitors (CD34+, CD19−) and pro-B cells (CD19+, CD34+, cyto-Igμ−).31,35,36 We used publicly available gene expression profiling (GEP) microarray-based data from non-malignant B-cell progenitors31 to create signatures (up and down) of genes distinguishing pre-B from pro-B cells (Supplemental Figure 1A, Supplemental Table 1). We applied these signatures to microarray data of 483 primary B-ALL samples, with various cytogenetic abnormalities, from the St. Jude Children’s Hospital.32 Unsupervised hierarchical clustering of the arrays revealed one group of B-ALL samples with GEP expression similar to normal pre-B cells (Figure 1A), hereafter referred to as pre-B ALL. This cluster comprised 79 (16.4%) cases; half of these (49.4%) harbored t(1;19), followed by cases with non-recurrent cytogenetic abnormalities (27.8%), MLL rearrangements (16.5%) and hypodiploidy (6.3%) (Figure 1A, Supplemental Figure 1B). Pre-B ALL cases conserved several features of their non-malignant counterparts, including high levels of the pre-BCR components IGHM, IGLL1 (λ5) and VPREB1 as well as increased expression of SYK (Figure 1B), all likely indicating active pre-BCR signaling.
Figure 1. Pre-B ALL resembles normal pre-B cells by GEP and depends on constitutive pre-BCR signaling.
(A) Pre-B ALL resembles normal pre-B cell progenitors and exhibits signs of active pre-BCR signaling. Pre-B cell specific genes were used to cluster gene expression arrays from B-ALL patient samples (GSE33315, Zhang J et al. 2012). (B) Increased expression of pre-BCR pathway components in pre-B cell-derived ALL. Graphs depict log2-transformed mean centered gene expression values. ****p<0.001, **p<0.01 Mann-Whitney test. (C) Pre-BCR surface expression in pro-B, pre-B and mature-B-derived ALL cells (dotted line: Isotype; solid: pre-BCR). Numbers indicate mean fluorescent intensity ratios (MFIR). Histograms are representative for 3 independent experiments. (D) Igμ protein expression after targeted knockout of Igμ (KO1 and KO2) in comparison to control cells. (E) Flow cytometric analysis of the pre-BCR and its individual components Igμ, λ5 and VpreB after Igμ-KO. (F) Igμ-KO inhibits proliferation of pre-BCR+ ALL cells. 48h after transfection Igμ− cells were sorted, seeded in equal numbers in 96 well plates (2*104/100μl) and viable cells were counted via flow cytometry at the indicated time points. Displayed are Means ± SD (N=3). ****p<0.001 Two-way ANOVA and Sidak’s test for multiple comparisons (G) Sensitivity of pre-BCR− (red) and pre-BCR+ (green) cells to the knockout of SYK. Cells were transfected with CRISPR/Cas9 vectors specific for the SYK gene, followed by assessment of SYK protein expression via intracellular flow cytometry every other day. After complete loss of SYK expression the fraction of SYK-KO cells in a mixed culture with SYK-WT cells was quantified at the indicated time points via flow cytometry. Results for each cell line represent means ± SD for 3 independent experiments; each experiment was conducted with a distinct SYK-specific gRNA.
Expression and function of the pre-BCR in pre-B ALL
To study the role of pre-BCR signaling in this subgroup of B-ALL, we assessed pre-BCR expression and function in Igμ+, surface IgM− B-ALL cells. Flow cytometry with the pre-BCR-specific monoclonal antibody HSL237 showed that the pre-BCR was expressed in the majority of pre-B ALL cells (9/11) (Figure 1C, Supplemental Figure 1C, Supplemental Table 2). Western Blot analysis revealed the presence of key constituents of the pre-BCR signaling cascade in all pre-B-derived ALL cells tested (Supplemental Figure 1D).
To assess the role of pre-BCR signaling in pre-BCR+ ALL we subsequently studied the effects of pre-BCR knockout (KO) on proliferation and survival of the pre-BCR+ ALL cells RCH-ACV and SMS-SB. Targeting the VDJ-segment of the expressed IGHM gene with CRISPR/Cas9 gene editing permanently eliminated Igμ protein expression (Figure 1D) and resulted in the loss of pre-BCR and SLC cell surface expression (Figure 1E, Supplemental Figure 1E). Loss of pre-BCR expression significantly reduced cell proliferation and exhibited minor effects on cell viability (Figure 1F, Supplemental Figure 1F), suggesting that pre-BCR+ ALL depends on constitutive signals from the pre-BCR. Accordingly, pre-BCR-dependent cells were also sensitive to CRISPR/Cas9 mediated knockout of SYK (Figure 1G; Supplemental Figure 1G). Although SYK is ubiquitously expressed in B-ALL, the pre-BCR− cells REH (t12;21) and Nalm-21 (t9;22), both were highly resistant to SYK knockout, suggesting that SYK is predominantly involved in the maintenance of pre-BCR+ ALL.
SYK serves as a therapeutic target in pre-BCR+ ALL
To delineate the role of SYK as a therapeutic target for B-ALL, we treated pre-BCR+ and pre-BCR− B-ALL cells with the highly specific SYK inhibitor PRT318 (Figure 2A and Supplemental Figure 2A).38,39 In line with our previous results, pre-BCR+ cells were particularly sensitive to SYK inhibition and IC50 values of PRT318 in these cells corresponded closely to IC50 values of PRT318 in SYK-dependent diffuse large B-cell lymphoma cells (Supplemental Figure 2B).39,40 Similarly, pre-BCR+ ALL cells were also more sensitive to R406 (Fostamatinib), an alternative ATP-competitive inhibitor of SYK (Supplemental Figure 2C – D).
Figure 2. SYK constitutes a potential therapeutic target in pre-BCR+ ALL.
(A) Pre-BCR+ ALL cells (green) are sensitive to the SYK inhibitor PRT318. Pre-BCR+ and pre-BCR− cells were incubated with increasing concentrations of PRT318 for 72h. Effects were analyzed using XTT assays followed by the calculation of IC50 values. Results represent 3 independent experiments. (B) PRT318 reduces leukemia cell burden in a pre-BCR+ (X018) but not in a pre-BCR− (X089) in vivo model of B-ALL. HuCD19+ blast cells in the peripheral blood 7 and 10 days after the start of therapy with PRT318 (30mg/kg BID) or Captisol (5%). ***p<0.005 Two-way ANOVA and Sidak’s test for multiple comparisons (C) Pre-B (cyto-Igμ+/surface IgM−) and pro-B (cyto-Igμ−/surface IgM−) ALL primary patient samples were treated with increasing concentrations of PRT318 for 48h followed by the assessment of cell viability. (D) The SYK inhibitor PRT062607 prolongs survival in an alternative model of pre-BCR+ ALL. ***p<0.001 Mantel-Cox survival analysis. (E) Igμ-KO and control cells were flow sorted followed by the assessment of the phosphorylation level of the indicated proteins by western blot analysis. To assess phosphorylation changes in response to PRT318, SMS-SB cells were pretreated with the indicated concentrations of PRT318 for 2 h. Immunoblots are representative for two independent experiments. (F) Pre-BCR− (REH, RS4;11, Tanoue, BALL-1) and Pre-BCR+ (RCH-ACV, SMS-SB, Nalm-6) cells were treated with 0.5 μM PRT318 for 2 h, followed by the assessment of pAKT and its densitometric quantification in ImageJ. Bar graphs depict the average reduction of pAKT relative to control (± SD). ****p<0.0001 Mann-Whitney test (G) Effects of PRT318 on pAKT levels in pre-BCR+ and pre-BCR− xenograft cells. Cell were incubated with 1 μM PRT318 for 2 h before assessing AKT phosphorylation via flow cytometry. **p<0.001 Mann-Whitney test (H) Cells were treated for 72 h with the indicated concentrations of LY294002 before assessing its effects via XTT proliferation assays. Results depict means ± SD of three independent experiments.
To further investigate the efficacy of pharmacologic SYK inhibition, we studied the effects of PRT318 in a xenograft mouse model of B-ALL. We inoculated NOD/SCID mice with pre-BCR+ (X018) or pre-BCR− (X089) B-ALL cells, and following engraftment (>5% huCD19+ cells in peripheral blood) initiated therapy with PRT318 (30mg/kg BID) or vehicle control (5% Captisol). After 10 days mice were sacrificed and subjected to the assessment of leukemia cell burden. Treatment with PRT318 resulted in a significant reduction of B-ALL blasts in the peripheral blood (PB) and bone marrow (BM) selectively in mice inoculated with pre-BCR+ B-ALL cells (Figure 2B, Supplemental Figure 2E). PRT318 also reduced infiltration of B-ALL cells in extramedullary tissues, including the spleen (SP) and the central nervous system (CNS), only in mice with pre-BCR+ ALL (Supplemental Figure 2E). Similarly, we noted that primary B-ALL cells that were isolated directly from patient samples and that had a pre-B immunophenotype (cyto-Igμ+, surface IgM−) were more sensitive to SYK inhibition with PRT318 than B-ALL cells with a pro-B immunophenotype (cyto-Igμ−, surface IgM−; Figure 2C, Supplemental Table 3). We furthermore tested the activity of the SYK-selective inhibitor PRT06260741 in an alternative model of pre-BCR+ ALL, using pre-BCR+ patient-derived, xenograft-expanded ICN12 cells (Supplemental Figure 2F). We found that PRT062607 significantly prolonged survival of mice inoculated with ICN12 cells (Figure 2D), further corroborating the activity of SYK inhibition in this disease subset.
Constitutive pre-BCR signaling selectively modulates PI3K pathway activity
Studies of the pre-BCR in normal B-cell development demonstrated that activation of the BTK/BLNK/PLCγ2, PI3K/AKT, and ERK pathways are essential for transmitting pre-BCR-derived downstream signals.10,11,42 To understand the molecular mechanisms underlying the dependence of pre-BCR+ ALL on the pre-BCR and SYK, we examined the activity of these signaling molecules after Igμ-KO or treatment with PRT318. Both approaches consistently demonstrated a marked reduction in AKT phosphorylation, but only minor reductions in ERK and BTK phosphorylation (Figure 2E; Supplemental Figure 2G), pointing towards a central role of PI3K in pre-BCR-signaling in B-ALL. To determine whether the effects of SYK inhibition on PI3K signaling were specific for pre-BCR+ ALL, we treated pre-BCR− and pre-BCR+ B-ALL cells with increasing concentrations of PRT318 and assessed changes in AKT phosphorylation (Figure 2F; Supplemental Figure 2H). In accordance with our results from the functional assays, pre-BCR+ ALL exhibited an almost complete loss of AKT phosphorylation in response to PRT318, but there was only minor pAKT modulation by PRT318 in the other groups of B-ALL. We similarly found higher pAKT levels in pre-BCR+ xenograft cells (Supplemental Figure 2I), which were reduced selectively by PRT318 (Figure 2G), indicating that constitutive activation of AKT by the pre-BCR and SYK characteristically occur in pre-BCR+ ALL. The importance of PI3K was further supported by the finding that PRT318-sensitive cells were significantly more sensitive to the pan-PI3K inhibitor LY294002 than were PRT318-resistant cells (Figure 2H).
Inhibition of pre-BCR signaling activates FOXO1 transcriptional activity
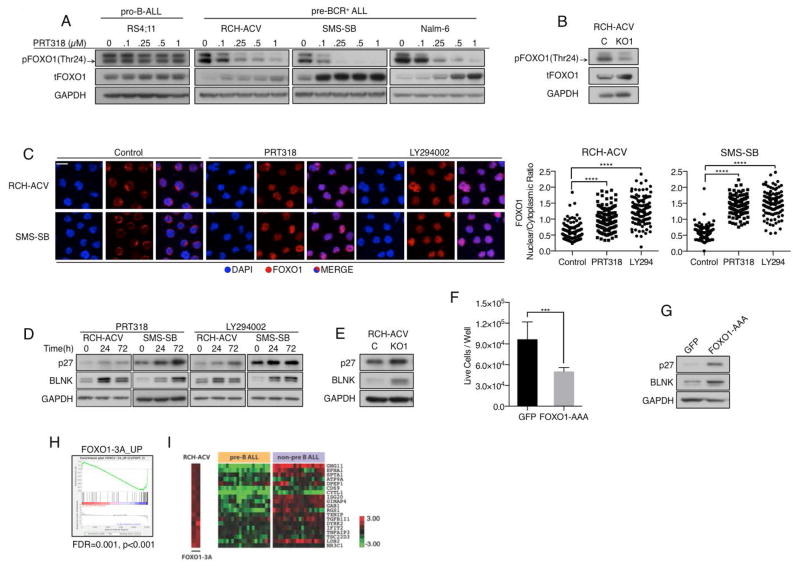
Effects on FOXO transcription factors (TF) are important downstream consequences of PI3K/AKT signaling in various tissues. During B-cell development, inactivation of FOXO TFs through AKT-mediated phosphorylation has been shown to be essential for delaying immunoglobulin gene rearrangement and facilitating the expansion of pre-BCR+ B-cell progenitors.11 Moreover, inactivation of FOXO family members was shown to be involved in the development of a variety of malignancies, including leukemia.43–45 To determine whether inactivation of FOXO TFs is also involved in the maintenance of pre-BCR+ ALL, we first analyzed expression levels of FOXO family members (FOXO1, FOXO3a, FOXO4, FOXO6) in RNA microarray data from the Immunological Genome Project46 at various stages of B-cell development (Supplemental Figure 3A). FOXO1 predominates in B-lineage-restricted progenitor cells (Fraction A to Fraction E), and its expression levels inversely correlate with the degree of cell proliferation at each developmental stage (Fraction A/C: high proliferation, low FOXO1; Fraction BC/D/E: no proliferation, high FOXO1). In accordance with these data, our immunoblot analyses of pre-BCR+ ALL cells demonstrated expression of FOXO1 and its phosphorylation at residues responsible for nuclear export and cytoplasmic degradation (Supplemental Figure 3B). Treatment with increasing concentrations of PRT318, or Igμ-KO, resulted in reduced FOXO1 phosphorylation and increased total protein levels of FOXO1 selectively in pre-BCR+ ALL cells (Figure 3A–B and Supplemental Figure 3C). This was accompanied by the translocation of FOXO1 from the cytoplasm to the nucleus (Figure 3C), suggesting an increase in FOXO1 transcriptional activity in response to the inhibition of pre-BCR signaling. Accordingly, we also observed increased protein levels of established FOXO1 transcriptional targets, such as BLNK and p27 after treatment with PRT318 or Igμ-KO (Figure 3D–E). We subsequently transfected RCH-ACV cells with a constitutively active form of FOXO1 (FOXO1-AAA). Similar to the effects of Igμ-KO and PRT318, FOXO1-AAA-expressing cells proliferated less than control cells and displayed up-regulation of p27 and BLNK (Figure 3F–G). To evaluate the role of FOXO1 inactivation in B-ALL patient samples, we compared gene expression profiles of RCH-ACV cells expressing either FOXO1-AAA or the respective empty vector control. Using a minimum absolute fold-change threshold of ≥ 2, we identified genes that were induced in response to FOXO1-AAA (Supplemental Table 5). GSEA revealed that this set of genes was significantly enriched, in the expected direction, in the spectrum of genes differentially expressed between the pre-B ALL and non-pre-B ALL samples of the St. Jude GEP dataset (Figure 3H–I, Supplemental Table 6). This confirms that constitutive pre-BCR signaling and FOXO1 inactivation contribute significantly to the distinct phenotype of pre-B ALL.
Figure 3. Pre-BCR signaling promotes B-ALL proliferation through inactivation of FOXO1.
(A) Pro-B and pre-BCR+ cells were incubated with increasing concentrations of PRT318 for 2 h followed by the assessment of pFOXO1 and tFOXO1. Black arrow indicates pFOXO1. (B) Igμ− RCH-ACV cells stained for pFOXO1 and tFOXO1 via immunoblot. (C) RCH-ACV and SMS-SB cells were serum starved for 12 h followed by 6 h incubation with either 1 μM PRT318 or 20 μM LY294002. After fixation and permeabilization cells were stained with DAPI (blue) and anti-FOXO1 antibody (red). Nuclear and cytoplasmic FOXO1 staining intensity was assessed for single cells and plotted as FOXO1 nuclear/cytoplasmic ratio. Each dot represents a single cell. 120 cells from 3 independent experiments (40 cells/experiment) were analyzed ****p<0.0001. Kruskal-Wallis- and Dunn’s multiple comparison test (D) Cells were incubated with 1 μM PRT318 or 20 μM LY294002 for the indicated time points followed by assessment of FOXO1 transcriptional targets p27 and BLNK via western blot. (E) Igμ-KO and control cells were incubated for 48 h under normal growth conditions followed by the analysis of p27 and BLNK protein levels. (F) RCH-ACV cells were transfected with an expression vector containing GFP-tagged constitutively active FOXO1 (FOXO1-AAA) or a GFP containing control vector. 24 h after transfection GFP+ cells were sorted, seeded at equal numbers (2*10e4/100μL) and incubated for 72 h under normal growth conditions before assessing the number of live cells/well via flow cytometry. ***p<0.001 Unpaired t test with Welch’s correction. (G) After sorting GFP+ cells were incubated for 24h before analyzing p27 and BLNK protein levels. (D), (E) and (G) represent 2 (F) 3 independent experiments. (H–I) FOXO1 target genes are suppressed in pre-B ALL cases from the St. Jude GEP dataset. (I) Relative abundance of FOXO1 target genes in RCH-ACV cells after FOXO1-3A expression and in pre-B ALL and non-pre B ALL cases of the St. Jude GEP dataset.
The pre-BCR regulates MYC in a FOXO1-dependent manner
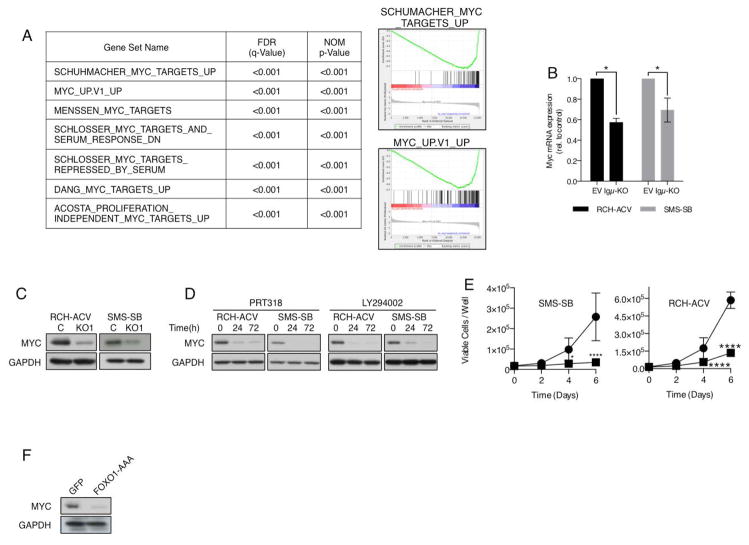
To identify additional targets of the pre-BCR in B-ALL we determined global gene expression changes in RCH-ACV and SMS-SB in response to Igμ-KO (Supplemental Table 7). Using gene set enrichment analysis we probed the list of genes regulated by the pre-BCR for the enrichment of gene sets provided by the Molecular Signatures Database (MSigDB v4.0). Among the most significantly enriched (FDR<0.001) were gene sets associated with the suppression of the transcriptional activity of MYC (Figure 4A). In view of its importance for cell proliferation, survival and metabolism and its frequent deregulation in other B-cell malignancies, the potential involvement of MYC in pre-BCR+ ALL was of particularly interest to us. In agreement with the GSEA, Igμ-KO significantly suppressed MYC mRNA and protein expression levels in RCH-ACV and SMS-SB (Figure 4B–C). Similarly, PRT318 and LY294002 significantly reduced MYC protein levels, indicating that the pre-BCR regulates MYC in a SYK- and PI3K-dependent manner (Figure 4D). Further emphasizing the importance of MYC in transducing signals from the pre-BCR, RCH-ACV and SMS-SB cells were highly sensitive to knockout of MYC (Figure 4E and Supplemental Figure 4A).
Figure 4. The pre-BCR regulates MYC activity in a SYK and FOXO1-dependet manner.
(A) Gene sets associated with the downmodulation of MYC activity are highly enriched in the list of genes regulated by the pre-BCR. (B–C) Reduction of MYC mRNA and protein levels in Igμ− RCH-ACV and SMS-SB cells 48 h after flow sorting or (D) after treatment with 1 μM PRT318 or 20 μM LY294002 for the indicated time points. *p<0.05 Two-tailed Mann-Whitney test (E) Knockout of MYC blocks cell proliferation in RCH-ACV and SMS-SB. MYC-KO cells were sorted and proliferation was assessed as described in Figure 1F. Results indicate means ± SD of three independent experiments, each conducted with a distinct MYC-specific gRNA. *p<0.05 ****p<0.0001 Two-way ANOVA and Sidak’s test for multiple comparisons. (F) RCH-ACV cells were transfected with FOXO1-AAA or GFP (control) and 24 h after sorting MYC was detected via western blot.
Finally, we addressed whether FOXO1 could also be involved in the suppression of MYC in pre-BCR+ ALL. This hypothesis was in part suggested by the nature of differentially expressed genes in RCH-ACV cells in response to FOXO1-AAA expression, which, according to GSEA, also bore evidence of reduced MYC activity (Supplemental Figure 4B). Furthermore, Gan et al. reported that FOXOs are capable of modulating MYC mRNA expression through the induction of Mxi-1 and mir-145.47 Expression of FOXO1-AAA in RCH-ACV cells caused a substantial reduction of MYC protein levels (Figure 4F), indicating that pre-BCR signaling indeed modulates MYC expression in a FOXO1-dependent manner.
DISCUSSION
In mature B-cell malignancies the use of microarray-based gene expression profiling (GEP) has contributed significantly to a better understanding of disease biology. In diffuse large B-cell lymphoma (DLBCL) the comparison of GEPs from normal and malignant B-cells resulted in the identification of two DLBCL subtypes, activated B cell-like (ABC) and germinal center B cell-like (GCB) DLBCL, characterized by a distinct cell of origin and differing survival mechanisms.48 Using a similar approach we identified a subgroup of B-ALL with a gene expression profile reminiscent of normal pre-BCR+ pre-B cells (pre-B ALL). Pre-B ALL cells were highly sensitive to the knockout of Igμ and SYK, thereby suggesting that pre-BCR+ ALL cells remain dependent on constitutive pre-BCR signaling despite their malignant transformation. The presence of TCF3-PBX1+, MLL-rearranged and hypodiploid patient samples in the cluster of pre-B ALL samples and the complete absence of ETV6-RUNX1+ and BCR-ABL+ cases indicates that the pre-B phenotype is associated with certain groups of cytogenetic abnormalities. However, whereas almost all TCF3-PBX1+ cases (98%) belong to the group of pre-B ALL only a fraction of MLL-rearranged (45%) and hypodiploid (26%) cases exhibit the pre-B phenotype, suggesting that additional factors shape disease biology in the latter two subtypes. Based on frequency, sample composition and the underlying molecular characteristics the group of pre-B ALL closely resembles the group of B-ALL with tonic pre-BCR signaling, identified by Geng et al. through RPPA.14 Indeed, in our analysis we also observe increased expression of the pre-BCR pathway components Igμ, λ5, VpreB and SYK in pre-B ALL samples.
The knockout of Igμ revealed that proliferation of pre-B ALL cells requires constitutive signals from the pre-BCR. This signaling activates PI3K in a SYK-dependent manner, which results in the inactivation of the tumor suppressor FOXO1 and deregulation of MYC. Importantly, a small-molecule inhibitor of SYK potentially suitable for clinical use (PRT318) abrogated constitutive pre-BCR signaling and its downstream effects in vitro and exhibited promising effects in two in vivo models of pre-BCR+ ALL, thereby providing a rationale for testing the clinical activity of SYK inhibitors in pre-BCR+ B-ALL.
In this respect our data challenge two recent reports in which SYK kinase inhibitors were tested in a variety of B-ALL patient samples and xenograft models of B-ALL, whose authors concluded that SYK is a valuable therapeutic target in all types of B-ALL, irrespective of immunophenotype, cytogenetic abnormalities (including Ph+ cases) and pre-BCR status.21,22 In contrast, our data suggest that SYK is only essential in the subgroup of pre-BCR+ B-ALL cases. We did not observe responses to SYK-KO or PRT318 in pre-BCR− cells in vitro or in vivo. This correlated with a selective effect of SYK inhibition on AKT phosphorylation in pre-BCR+ cases. Part of these discrepancies could be related to of off-target effects of the compounds tested in these previous studies. R406, for instance, inhibits a spectrum of other tyrosine kinases, besides SYK,49,50 including pathways that support B-ALL cell growth or survival (JAK2, PDGFRB and FLT3).1,51,52
Our findings corroborate those of Wossning and colleagues, who demonstrated that the transforming capacity of SYK in B-ALL depends on concurrent expression of the pre-BCR and MYC.53 In regards to Ph+-ALL, several publications indicate that the BCR-ABL fusion kinase is directly involved in the suppression of the pre-BCR and pre-BCR downstream targets, such as SYK and Igβ, in order to maintain its malignant phenotype.18,13,16 Consistently, all BCR-ABL+ B-ALL cells tested by us were pre-BCR− and highly resistant to SYK-KO or SYK inhibition.
We also studied the role of BTK in the pre-BCR signaling cascade. Despite recent reports on the efficacy of the BTK inhibitor ibrutinib in pre-BCR+ ALL, both Igμ-KO and PRT318 did not interfere with BTK signaling activity, suggesting that BTK is dispensable for the pro-proliferating effect of the pre-BCR in B-ALL.24,14 Accordingly, the reported effective concentrations of ibrutinib in pre-BCR+ ALL are considerably higher than in established BTK-dependent B-cell malignancies, indicating that ibrutinib might exert its effects in pre-BCR+ ALL through the inhibition of alternative targets.24,54,55
A translational challenge arising from our data is how best to identify patients with pre-BCR-dependent B-ALL, likely to be sensitive to SYK inhibition, in the clinical setting. Since B-ALL cells must be arrested at the pre-B cell stage (cytoplasmic Igμ+, surface IgM−) in order to express a functional pre-BCR, this immunophenotype should be a minimum requirement, and is already assessed routinely in clinical laboratories. By selecting primary patient samples based on these immune-phenotype criteria we distinguished PRT318-sensitive pre-B-ALL cases from PRT318-resistant pro-B-ALL cases. We also implicated the potential of a gene expression signature to identify suitable cases, but this would also require a method suitable for the clinical laboratory and further validation.
Previous reports suggested that pre-BCR signaling is primarily involved in the maintenance of B-ALL harboring t(1;19), characterized by the expression of the E2A-PBX1 fusion protein14,20,24; indeed, 98% of E2A-PBX1+ cases exhibit the pre-B ALL GEP phenotype, and this group is therefore enriched for pre-BCR+ B-ALL cases.56 However, within the cluster of pre-B ALL cases identified in the St. Jude GEP data, only 50% harbor the E2A-PBX1 fusion protein. Therefore, the t(1;19) does not appear to be a reliable indicator of pre-BCR-dependent ALL cases.
Besides the pre-BCR itself, our data also highlight the importance of the downstream signaling molecules SYK, PI3K, FOXO1 and MYC. As indicated by the highly congruent GEP signature, the addiction of pre-BCR-dependent ALL to this signaling axis is most likely derived from its cell of origin, the pre-B lymphocyte (cytoplasmic Igμ+, surface IgM−). It was previously established that mice deficient for key components of this pathway (including SYK, the catalytic PI3K subunits p110α and p110δ, and MYC) share a common phenotype with respect to B-cell development, characterized by a maturation arrest at the pre-B-cell stage and the absence of mature B cells.57–61 Our data suggest that pre-B ALL cells remain dependent on this signaling axis, despite malignant transformation, and ensure its activation through signals directly from the pre-BCR and SYK.
In summary, we demonstrate that pre-BCR expression defines a subgroup of B-ALL that is exquisitely dependent on SYK and define regulatory downstream signaling and transcriptional events. Results from pharmacologic inhibition suggest that SYK is a valid therapeutic target in this subset of B-ALL patients.
Supplementary Material
Acknowledgments
This work was supported a Cancer Prevention and Research Institute of Texas (CPRIT) grant (to J. A. B.), a Leukemia & Lymphoma Society Scholar Award in Clinical Research (to J. A. B.), the Else-Kröner Forschungskolleg (F.S.), German Research Foundation, SFB 1074 (K.-M.D. and L.H.M.), ‘Förderkreis für Tumor- und Leukämiekranke Kinder Ulm’, and in part by the MD Anderson Cancer Center Support Grant CA016672. STR DNA fingerprinting was done by the Cancer Center Support Grant-funded Characterized Cell Line core, NCI # CA016672. The authors are grateful for access to the data assembled by the ImmGen Consortium.
Footnotes
This work was presented, in part, in oral sessions at the 2013 annual meeting of the American Society of Hematology (ASH) in New Orleans, and at the 2014 annual meeting of the American Society of Hematology (ASH) in San Francisco.
Disclosure of conflicts of interest
J. A. B. received research funding from Portola Pharmaceuticals, G. P. C. is employee and shareholder of Portola Pharmaceuticals.
Supplementary information is available at Leukemia’s website.
References
- 1.Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic Alterations Activating Kinase and Cytokine Receptor Signaling in High-Risk Acute Lymphoblastic Leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 3.Herzog Sebastian, Reth Michael, Jumaa Hassan. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 4.Mårtensson I-L, Almqvist N, Grimsholm O, Bernardi AI. The pre-B cell receptor checkpoint. FEBS Lett. 2010;584:2572–2579. doi: 10.1016/j.febslet.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 5.Kitamura D, Roes J, Kühn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 6.Kitamura D, Kudo A, Schaal S, Müller W, Melchers F, Rajewsky K. A critical role of λ5 protein in B cell development. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 7.Bankovich AJ, Raunser S, Juo ZS, Walz T, Davis MM, Garcia KC. Structural Insight into Pre-B Cell Receptor Function. Science. 2007;316:291–294. doi: 10.1126/science.1139412. [DOI] [PubMed] [Google Scholar]
- 8.Übelhart Rudolf, Bach Martina P, Eschbach Cathrin, Wossning Thomas, Reth Michael, Jumaa Hassan. N-linked glycosylation selectively regulates autonomous precursor BCR function. Nat Immunol. 2010;11:759–765. doi: 10.1038/ni.1903. [DOI] [PubMed] [Google Scholar]
- 9.Ohnishi Kazuo, Melchers Fritz. The nonimmunoglobulin portion of λ5 mediates cell-autonomous pre-B cell receptor signaling. Nat Immunol. 2003;4:849–856. doi: 10.1038/ni959. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda T, Sanjo H, Pagès G, Kawano Y, Karasuyama H, Pouysségur J, et al. Erk Kinases Link Pre-B Cell Receptor Signaling to Transcriptional Events Required for Early B Cell Expansion. Immunity. 2008;28:499–508. doi: 10.1016/j.immuni.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Herzog S, Hug E, Meixlsperger S, Paik J-H, DePinho RA, Reth M, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. 2008;9:623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 12.Eswaran J, Sinclair P, Heidenreich O, Irving J, Russell LJ, Hall A, et al. The pre-B-cell receptor checkpoint in acute lymphoblastic leukaemia. Leukemia. 2015;29:1623–1631. doi: 10.1038/leu.2015.113. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L, et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 2015 doi: 10.1038/nature14231. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S, et al. Self-Enforcing Feedback Activation between BCL6 and Pre-B Cell Receptor Signaling Defines a Distinct Subtype of Acute Lymphoblastic Leukemia. Cancer Cell. 2015;27:409–425. doi: 10.1016/j.ccell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trageser D, Iacobucci I, Nahar R, Duy C, von Levetzow G, Klemm L, et al. Pre–B cell receptor–mediated cell cycle arrest in Philadelphia chromosome–positive acute lymphoblastic leukemia requires IKAROS function. J Exp Med. 2009;206:1739–1753. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumaa H, Bossaller L, Portugal K, Storch B, Lotz M, Flemming A, et al. Deficiency of the adaptor SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature. 2003;423:452–456. doi: 10.1038/nature01608. [DOI] [PubMed] [Google Scholar]
- 18.Feldhahn N, Klein F, Mooster JL, Hadweh P, Sprangers M, Wartenberg M, et al. Mimicry of a constitutively active pre–B cell receptor in acute lymphoblastic leukemia cells. J Exp Med. 2005;201:1837–1852. doi: 10.1084/jem.20042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19:1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bicocca VT, Chang BH, Masouleh BK, Muschen M, Loriaux MM, Druker BJ, et al. Crosstalk between ROR1 and the Pre-B Cell Receptor Promotes Survival of t(1;19) Acute Lymphoblastic Leukemia. Cancer Cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perova T, Grandal I, Nutter LMJ, Papp E, Matei IR, Beyene J, et al. Therapeutic Potential of Spleen Tyrosine Kinase Inhibition for Treating High-Risk Precursor B Cell Acute Lymphoblastic Leukemia. Sci Transl Med. 2014;6:236ra62–236ra62. doi: 10.1126/scitranslmed.3008661. [DOI] [PubMed] [Google Scholar]
- 22.Uckun FM, Qazi S, Cely I, Sahin K, Shahidzadeh A, Ozercan I, et al. Nanoscale liposomal formulation of a SYK P-site inhibitor against B-precursor leukemia. Blood. 2013;121:4348–4354. doi: 10.1182/blood-2012-11-470633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uckun FM, Ek RO, Jan S-T, Chen C-L, Qazi S. Targeting SYK kinase-dependent anti-apoptotic resistance pathway in B-lineage acute lymphoblastic leukaemia (ALL) cells with a potent SYK inhibitory pentapeptide mimic. Br J Haematol. 2010;149:508–517. doi: 10.1111/j.1365-2141.2010.08106.x. [DOI] [PubMed] [Google Scholar]
- 24.van der Veer A, van der Velden VHJ, Willemse ME, Hoogeveen PG, Petricoin EF, Beverloo HB, et al. Interference with pre-B-cell receptor signaling offers a therapeutic option for TCF3-rearranged childhood acute lymphoblastic leukemia. Blood Cancer J. 2014;4:e181. doi: 10.1038/bcj.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol. 2014;14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Espeli M, Rossi B, Mancini SJC, Roche P, Gauthier L, Schiff C. Initiation of pre-B cell receptor signaling: Common and distinctive features in human and mouse. Semin Immunol. 2006;18:56–66. doi: 10.1016/j.smim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Hoellenriegel J, Coffey GP, Sinha U, Pandey A, Sivina M, Ferrajoli A, et al. Selective, novel spleen tyrosine kinase (Syk) inhibitors suppress chronic lymphocytic leukemia B-cell activation and migration. Leukemia. 2012;26:1576–1583. doi: 10.1038/leu.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ann Ran F, Hsu Patrick D, Wright Jason, Agarwala Vineeta, Scott David A, Zhang Feng. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura N, Ramaswamy S, Vazquez F, Signoretti S, Loda M, Sellers WR. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol Cell Biol. 2000;20:8969–8982. doi: 10.1128/mcb.20.23.8969-8982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma W, Wang M, Wang Z-Q, Sun L, Graber D, Matthews J, et al. Effect of long-term storage in TRIzol on microarray-based gene expression profiling. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2010;19:2445–2452. doi: 10.1158/1055-9965.EPI-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S-T, Xiao Y, Muench MO, Xiao J, Fomin ME, Wiencke JK, et al. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 2012;40:11339–11351. doi: 10.1093/nar/gks957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 35.Hystad ME, Myklebust JH, Bø TH, Sivertsen EA, Rian E, Forfang L, et al. Characterization of Early Stages of Human B Cell Development by Gene Expression Profiling. J Immunol. 2007;179:3662–3671. doi: 10.4049/jimmunol.179.6.3662. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann R, Lottaz C, Kühne T, Rolink A, Melchers F. Neutrality, Compensation, and Negative Selection during Evolution of B-Cell Development Transcriptomes. Mol Biol Evol. 2007;24:2610–2618. doi: 10.1093/molbev/msm198. [DOI] [PubMed] [Google Scholar]
- 37.Tsuganezawa K, Kiyokawa N, Matsuo Y, Kitamura F, Toyama-Sorimachi N, Kuida K, et al. Flow Cytometric Diagnosis of the Cell Lineage and Developmental Stage of Acute Lymphoblastic Leukemia by Novel Monoclonal Antibodies Specific to Human Pre–B-Cell Receptor. Blood. 1998;92:4317–4324. [PubMed] [Google Scholar]
- 38.Reilly MP, Sinha U, André P, Taylor SM, Pak Y, DeGuzman FR, et al. PRT-060318, a novel Syk inhibitor, prevents heparin-induced thrombocytopenia and thrombosis in a transgenic mouse model. Blood. 2011;117:2241–2246. doi: 10.1182/blood-2010-03-274969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng S, Coffey G, Zhang XH, Shaknovich R, Song Z, Lu P, et al. SYK inhibition and response prediction in diffuse large B-cell lymphoma. Blood. 2011;118:6342–6352. doi: 10.1182/blood-2011-02-333773. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Monti S, Juszczynski P, Ouyang J, Chapuy B, Neuberg D, et al. SYK Inhibition Modulates Distinct PI3K/AKT- Dependent Survival Pathways and Cholesterol Biosynthesis in Diffuse Large B Cell Lymphomas. Cancer Cell. 2013;23:826–838. doi: 10.1016/j.ccr.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffey G, DeGuzman F, Inagaki M, Pak Y, Delaney SM, Ives D, et al. Specific Inhibition of Spleen Tyrosine Kinase Suppresses Leukocyte Immune Function and Inflammation in Animal Models of Rheumatoid Arthritis. J Pharmacol Exp Ther. 2012;340:350–359. doi: 10.1124/jpet.111.188441. [DOI] [PubMed] [Google Scholar]
- 42.Guo B, Kato RM, Garcia-Lloret M, Wahl MI, Rawlings DJ. Engagement of the Human Pre-B Cell Receptor Generates a Lipid Raft–Dependent Calcium Signaling Complex. Immunity. 2000;13:243–253. doi: 10.1016/s1074-7613(00)00024-8. [DOI] [PubMed] [Google Scholar]
- 43.Dong X-Y, Chen C, Sun X, Guo P, Vessella RL, Wang R-X, et al. FOXO1A Is a Candidate for the 13q14 Tumor Suppressor Gene Inhibiting Androgen Receptor Signaling in Prostate Cancer. Cancer Res. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- 44.Xie L, Ushmorov A, Leithäuser F, Guan H, Steidl C, Färbinger J, et al. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood. 2012;119:3503–3511. doi: 10.1182/blood-2011-09-381905. [DOI] [PubMed] [Google Scholar]
- 45.Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, et al. AKT/FOXO Signaling Enforces Reversible Differentiation Blockade in Myeloid Leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heng TSP, Painter MW Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 47.Gan Boyi, Lim Carol, Chu Gerald, Hua Sujun, Ding Zhihu, Collins Michael, et al. FoxOs Enforce a Progression Checkpoint to Constrain mTORC1-Activated Renal Tumorigenesis. Cancer Cell. 2010;18:472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 49.Braselmann S, Taylor V, Zhao H, Wang S, Sylvain C, Baluom M, et al. R406, an Orally Available Spleen Tyrosine Kinase Inhibitor Blocks Fc Receptor Signaling and Reduces Immune Complex-Mediated Inflammation. J Pharmacol Exp Ther. 2006;319:998–1008. doi: 10.1124/jpet.106.109058. [DOI] [PubMed] [Google Scholar]
- 50.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 51.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang Y-L, Pei D, et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wossning T, Herzog S, Kohler F, Meixlsperger S, Kulathu Y, Mittler G, et al. Deregulated Syk inhibits differentiation and induces growth factor-independent proliferation of pre-B cells. J Exp Med. 2006;203:2829–2840. doi: 10.1084/jem.20060967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herman SEM, Gordon AL, Hertlein E, Ramanunni A, Zhang X, Jaglowski S, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117:6287–6296. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ponader S, Chen S-S, Buggy JJ, Balakrishnan K, Gandhi V, Wierda WG, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–1189. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunger SP. Chromosomal translocations involving the E2A gene in acute lymphoblastic leukemia: clinical features and molecular pathogenesis. Blood. 1996;87:1211–1224. [PubMed] [Google Scholar]
- 57.Habib T, Park H, Tsang M, de Alborán IM, Nicks A, Wilson L, et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol. 2007;179:717–731. doi: 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki H, Terauchi Y, Fujiwara M, Aizawa S, Yazaki Y, Kadowaki T, et al. Xid-Like Immunodeficiency in Mice with Disruption of the p85α Subunit of Phosphoinositide 3-Kinase. Science. 1999;283:390–392. doi: 10.1126/science.283.5400.390. [DOI] [PubMed] [Google Scholar]
- 59.Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood. 2008;111:1497–1503. doi: 10.1182/blood-2007-08-109769. [DOI] [PubMed] [Google Scholar]
- 60.Ramadani F, Bolland DJ, Garcon F, Emery JL, Vanhaesebroeck B, Corcoran AE, et al. The PI3K Isoforms p110α and p110δ Are Essential for Pre–B Cell Receptor Signaling and B Cell Development. Sci Signal. 2010;3:ra60–ra60. doi: 10.1126/scisignal.2001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner M, Joseph Mee P, Costello PS, Williams O, Price AA, Duddy LP, et al. Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature. 1995;378:298–302. doi: 10.1038/378298a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.