Abstract
Tibetans are well adapted to high-altitude hypoxia. Previous genome-wide scans have reported many candidate genes for this adaptation, but only a few have been studied. Here we report on a hypoxia gene (GCH1, GTP-cyclohydrolase I), involved in maintaining nitric oxide synthetase (NOS) function and normal blood pressure, that harbors many potentially adaptive variants in Tibetans. We resequenced an 80.8 kb fragment covering the entire gene region of GCH1 in 50 unrelated Tibetans. Combined with previously published data, we demonstrated many GCH1 variants showing deep divergence between highlander Tibetans and lowlander Han Chinese. Neutrality tests confirmed a signal of positive Darwinian selection on GCH1 in Tibetans. Moreover, association analysis indicated that the Tibetan version of GCH1 was significantly associated with multiple physiological traits in Tibetans, including blood nitric oxide concentration, blood oxygen saturation, and hemoglobin concentration. Taken together, we propose that GCH1 plays a role in the genetic adaptation of Tibetans to high altitude hypoxia.
Keywords: GCH1, Positive selection, Tibetan, Hypoxia adaptation, Nitric oxide, Hemoglobin, Oxygen saturation
INTRODUCTION
Tibetans are a well-known example of successful adaptation to extreme environments at high-altitude. Compared to lowlanders moving to high altitude, Tibetans show greater lung capacity, function, diffusion, and ventilation, as well as lower hemoglobin (Hb) levels, better blood oxygen saturation, low hypoxic pulmonary vasoconstriction, high nitric oxide (NO) concentrations, and lower incidence of reduced birth weight (Beall, 2006; Beall et al., 1997; Erzurum et al., 2007; Wu & Kayser, 2006). These traits have been acquired during a long period of natural selection at high altitude after the ancestors of modern Tibetans permanently settled on the Qinghai-Tibetan Plateau during the early Upper Paleolithic period (Qi et al., 2013).
Various research groups have compared the genetic differences between Tibetan and Han Chinese using genome-wide scans, with two key genes identified (hypoxia-inducible factor 2α, HIF2α, also called EPAS1, and EGLN1) in the hypoxic pathway showing deep between-population divergence (Beall et al., 2010; Bigham et al., 2010; Peng et al., 2011; Simonson et al., 2010; Xu et al., 2011; Yi et al., 2010). For these two genes, Tibetan-specific haplotypes have been found, which are highly enriched in Tibetans (~80%), but rare or absent in other world populations (Lorenzo et al., 2014; Peng et al., 2011, 2017; Xiang et al., 2013). The function of selection on these two genes have been shown to cause blunted physiological responses under high altitude hypoxic conditions (Lorenzo et al., 2014; Peng et al., 2017; Xiang et al., 2013). For example, the Tibetan versions of EPAS1 and EGLN1 protect Tibetans from the over production of red blood cells (polycythemia), a common deleterious physiological response when lowlanders migrate to high altitude areas (Beall et al., 2010; Erzurum et al., 2007; Lorenzo et al., 2014; Peng et al., 2011, 2017; Petousi et al., 2014; Xiang et al., 2013; Xu et al., 2011; Zhang et al., 2010).
Many genes are likely involved in complex traits like high altitude adaptation. In addition to EPAS1 and EGLN1, previous genome-wide studies have identified other candidate genes that might also contribute to genetic adaptation in Tibetans (Beall et al., 2010; Bigham et al., 2010; Peng et al., 2011; Simonson et al., 2010; Xu et al., 2011; Yi et al., 2010). The GTP-cyclohydrolase I (GCH1) gene is a reported candidate that shows relatively deep allelic divergence between Tibetans and Han Chinese, a sign of positive Darwinian selection (Peng et al., 2011). GCH1 is located on human chromosome 14 (14q22.1-q22.2), spanning 60.8 kb with six exons. Furthermore, GCH1 is a rate-limiting enzyme in the de novo synthesis of tetrahydrobiopterin (BH4). It has been reported that under hypoxia, GCH1 can promote cancer growth, and its expression and that of endothelial nitric oxide synthetase (eNOS) is upregulated (Pickert et al., 2013). GCH1 is considered as a major factor in maintaining nitric oxide synthetase (NOS) function and normal blood pressure, and its inhibition can increase blood pressure due to NOS uncoupling, which is found in many cardiovascular diseases such as hypertension and atherosclerosis (Antoniades et al., 2006). Hence, the known functional role of GCH1 also makes it a candidate for high-altitude adaptation in Tibetans.
We resequenced an 80.8 kb fragment covering the entire gene region of GCH1 in 50 unrelated Tibetans. Combined with published data, we found signals of positive selection on GCH1 in Tibetans, with multiple sequence variants showing deep genetic divergence between highlander Tibetans and lowlander Han Chinese. Genetic association analysis detected significant correlation of the GCH1 variants with multiple physiological traits of Tibetans, including blood nitric oxide (NO) concentration, blood oxygen saturation level, and hemoglobin concentration. Hence, GCH1 might play an essential role in high-altitude adaptation in Tibetans.
MATERIALS AND METHODS
DNA sample collection and resequencing of GCH1 gene fragment
The 50 unrelated Tibetan samples were obtained from previous research (Peng et al., 2011). We resequenced an 80.8 kb fragment covering the gene region of GCH1 and its flanking sequences (10 kb up-and down-stream of GCH1). For association analysis, we collected blood samples and extracted DNA from 226 unrelated adult Tibetans, whose physiological data were also collected with written informed consent. The protocol of this study was evaluated and approved by the Internal Review Board of Kunming Institute of China, Chinese Academy of Sciences.
Detection of selection on GCH1 in Tibetans
The initially identified sequence variants were filtered by removing single nucleotide polymorphisms (SNPs) showing a significant deviation from the Hardy-Weinberg Equilibrium (HWE < 0.000 1), and those with an excessive missing genotype rate (MGR > 0.05). Four methods were used for selection testing, including two allele-frequency-based and two haplotype-based tests. Locus specific FST was calculated between 83 Tibetans and three reference populations (103 Han Chinese, CHB; 99 Europeans, CEU; and 108 Africans, YRI) following Weir & Cockerham (1984). The Tajima's D-test was also performed following the published procedure (Tajima, 1989).
The two haplotype-based tests included the iHS and XP-EHH tests (Sabeti et al., 2002). The iHS score was calculated for each site using selscan (Szpiech & Hernandez, 2014) based on the phased haplotypes, and only those allelic loci whose ancestral alleles were known with certainty were included in the analysis (Voight et al., 2006). The XP-EHH (Sabeti et al., 2007) analysis was used to detect the extended haplotypes due to positive selection. Han Chinese were used as the reference population in the XP-EHH test. We computed XP-EHH scores using selscan (Szpiech & Hernandez, 2014) based on phased haplotypes of Tibetans and Han Chinese. The XP-EHH score of each SNP was standardized by the mean XP-EHH and the standard deviation (SD) over the genome.
Functional prediction of GCH1 candidate SNPs
Functional enrichment analyses of the candidate variants were performed using the Combined Annotation Dependent Depletion (CADD) database (<uritalic>http://krishna.gs.washington.edu/download/CADD/v1.3/1000G_phase3_inclAnno.tsv.gz</uritalic>), which incorporates data from ENCODE (<uritalic>http://genome.ucsc.edu/ENCODE/</uritalic>) and NIH Roadmap Epigenomics using ChromHMM (<uritalic>https://sites.google.com/site/anshulkundaje/projects/epigenomeroadmap#TOC-Core-Integrative-chromatin-state-maps-127-Epigenomes-</uritalic>) (<xref ref-type="bibr" rid="b7-ZoolRes-38-3-155">Ernst & Kellis, 2012</xref>).
Evolutionary constraint is an indication of functional importance. We used Genome Evolutionary Rate Profiling (GERP) to evaluate how conserved a test SNP was compared with SNP-containing sequences from different species (<uritalic>http://mendel.stanford.edu/SidowLab/downloads/gerp/</uritalic>). The GERP++ method was used to calculate site-specific RS scores (<xref ref-type="bibr" rid="b6-ZoolRes-38-3-155">Davydov et al., 2010</xref>). A positive GERP++ score indicates evolutionary constraint, and the greater the score, the greater the level of evolutionary constraint inferred to be acting on that site.
The H3K4Me1 value indicates the maximum ENCODE H3K4 methylation level (maximum value observed across 16 ENCODE cell lines at a given position), suggestive of an enhancer or other regulatory activities. The H3K4Me3 value indicates the maximum ENCODE H3K4 trimethylation level, and is an indication of a promoter. The DNase P value indicates evidence for open chromatin. The transcription factor binding site (TFBS) is indicated by the number of different overlapping ChIP transcription factor binding sites. In addition, the splice site results indicate if the tested variant is within an ACCEPTOR or a DONOR (Supplementary Table S1).
Measurements of physiological traits
Physiological data and blood samples were collected from 226 unrelated Tibetans permanently residing in Bange county (n=135, 37.41±3.8 years old) at an elevation of 4 700 m and Lhasa city (n=91, 35.33±6.8 years old) at an elevation of 3 600 m. Written informed consent from all participants was obtained. For physiological parameters, we collected hemoglobin (Hb) concentration, arterial oxygen saturation (SaO2) level, and blood nitric oxide concentration, representing key adaptive physiological traits in Tibetans (Wu & Kayser, 2006).
The Hb level in blood was measured using a HemoCue Hb 201+ analyzer (Angelholm, Sweden). The SaO2 level was measured using fingertip blood with a hand-held pulse oximeter (Nellcor NPB-40, CA, USA). Blood NO was measured using a nitric oxide analyzer (Sievers Model-280, GE Analytical Instruments; Boulder, CO, USA).
SNP genotyping and association analysis
We selected nine tag SNPs, covering the entire gene region of GCH1 (Table 1). Genotyping was conducted by SNaPshot on an ABI 3130 sequencer (Applied Bio-Systems, Forster City, CA, USA). Genetic association analysis was executed using PLINK 1.07 (Purcell et al., 2007). We used an additive model in the association analysis as all candidate SNPs were non-coding and likely influenced the level of gene expression. Permutations (100 000 times for each test) were performed for statistical assessment and correction for multiple tests.
1.
Association of nine GCH1 variants with three physiological traits in Tibetans
| Traits | SNP_ID | Male (n=91) | Female (n=135) | All (n=226) | ||||
| Beta | EMP' | Beta | EMP' | Beta | EMP'' | R2(%) | ||
| Hb, hemoglobin concentration; NO, blood nitric oxide concentration; SaO2, blood oxygen saturation level. EMP', P value after multiple test corrections; EMP'', P value after multiple test corrections with sex as the covariant. | ||||||||
| Hb | rs7148266 | -4.33 | 0.48 | -0.53 | 0.78 | -1.51 | 0.78 | 0.05 |
| rs17128004 | -3.02 | 0.65 | -0.49 | 0.86 | -1.10 | 0.86 | 0.01 | |
| rs4411417 | -4.33 | 0.48 | -1.27 | 0.86 | -1.85 | 0.69 | 0.23 | |
| rs2183082 | 1.17 | 0.86 | -2.10 | 0.57 | -0.65 | 0.86 | 0.08 | |
| rs10220344 | 1.17 | 0.86 | -2.10 | 0.57 | -0.65 | 0.86 | 0.08 | |
| rs146540091 | -62.95 | 0.05 | -17.31 | 0.25 | -35.15 | 0.02 | 1.65 | |
| rs117863726 | -62.95 | 0.05 | -17.61 | 0.47 | -36.27 | 0.02 | 2.39 | |
| rs10136972 | 12.97 | 0.03 | 0.21 | 1.00 | 4.28 | 0.28 | 1.29 | |
| rs112700866 | 22.04 | 0.05 | -0.78 | 1.00 | 10.25 | 0.16 | 0.67 | |
| NO | rs7148266 | -15.97 | 0.04 | -10.82 | 0.04 | -12.96 | 2.32E-03 | 3.05 |
| rs17128004 | -17.23 | 0.03 | -10.77 | 0.04 | -13.42 | 2.32E-03 | 3.08 | |
| rs4411417 | -15.97 | 0.04 | -11.67 | 0.04 | -13.63 | 2.75E-03 | 3.76 | |
| rs2183082 | -6.35 | 0.54 | -6.05 | 0.17 | -6.32 | 0.08 | 1.28 | |
| rs10220344 | -6.35 | 0.54 | -6.05 | 0.17 | -6.32 | 0.08 | 1.28 | |
| rs146540091 | -162.70 | 3.53E-03 | 13.14 | 0.78 | -41.65 | 0.07 | 3.36 | |
| rs117863726 | -162.70 | 3.53E-03 | 13.07 | 0.65 | -41.05 | 0.07 | 1.40 | |
| rs10136972 | -5.71 | 0.65 | -14.60 | 0.01 | -11.35 | 0.02 | 1.88 | |
| rs112700866 | -10.98 | 0.35 | 4.97 | 0.73 | -2.78 | 0.86 | 0.03 | |
| SaO2 | rs7148266 | -0.84 | 0.38 | 0.55 | 0.78 | 0.05 | 0.86 | 0.01 |
| rs17128004 | -1.08 | 0.33 | -0.05 | 1.00 | -0.44 | 0.48 | 0.07 | |
| rs4411417 | -0.84 | 0.38 | 0.57 | 0.65 | 0.03 | 1.00 | 2.50E-03 | |
| rs2183082 | -0.50 | 0.57 | -0.38 | 0.78 | -0.44 | 0.52 | 0.03 | |
| rs10220344 | -0.50 | 0.57 | -0.38 | 0.78 | -0.44 | 0.52 | 0.03 | |
| rs146540091 | 12.53 | 0.05 | -1.61 | 0.78 | 3.07 | 0.54 | 3.22 | |
| rs117863726 | 12.53 | 0.05 | 6.66 | 0.14 | 8.53 | 0.02 | 2.47 | |
| rs10136972 | -1.86 | 0.27 | -1.84 | 0.11 | -1.84 | 0.05 | 0.75 | |
| rs112700866 | -1.68 | 0.67 | -3.46 | 0.17 | -2.76 | 0.16 | 0.75 | |
RESULTS
Resequencing of GCH1 in Tibetans and tests of selection
Previous DNA array-based genome-wide studies have only covered a limited number of GCH1 sequence variants (Peng et al., 2011). To obtain complete sequence data of GCH1, we first resequenced an 80.8 kb fragment covering the entire gene region of GCH1(60.8 kb) as well as the flanking sequences (10 kb from upstream and 10 kb from downstream regions). In total, we sequenced 50 unrelated Tibetan individuals, as reported previously (Peng et al., 2011). In addition, we obtained the GCH1 sequences of 33 Tibetans from recently published whole genome sequencing data (Lu et al., 2016), for a final sample size of 83.
We identified a total of 384 <italic>GCH1</italic> sequence variants (SNPs) in the 83 unrelated Tibetans, among which 245 were shared between Tibetans and the three lowland reference populations from the 1000 Genomes Project (<uritalic>http://www.1000genomes.org</uritalic>) (103 Han Chinese, CHB; 99 Europeans, CEU and 108 Africans, YRI). The remaining 139 SNPs were rare in Tibetans ( < 1.0%), and therefore not included in our analysis. The 245 SNPs were all located in the non-coding regions of <italic>GCH1</italic>.
To detect whether GCH1 was under selection in Tibetans, we conducted four different tests of selection, including two allele-frequency-based tests (FST and Tajima's D) and two haplotype-based tests (iHS and XP-EHH). We identified 49 GCH1 SNPs with large between-population (Tibetan vs. Han) divergence (FST > 0.2), much larger than the genome average (FST=0.03). These high-FST variants also showed high iHS and XP-EHH values (iHS > 0.2 and/or XP-EHH > 0.2) (Figure 1). They were aggregated in a relatively short region (7.5 kb) covering intron-1 and intron-2 of GCH1. These results suggest a clear signal of positive Darwinian selection on GCH1 in Tibetans (Supplementary Table S1)
Figure 1.
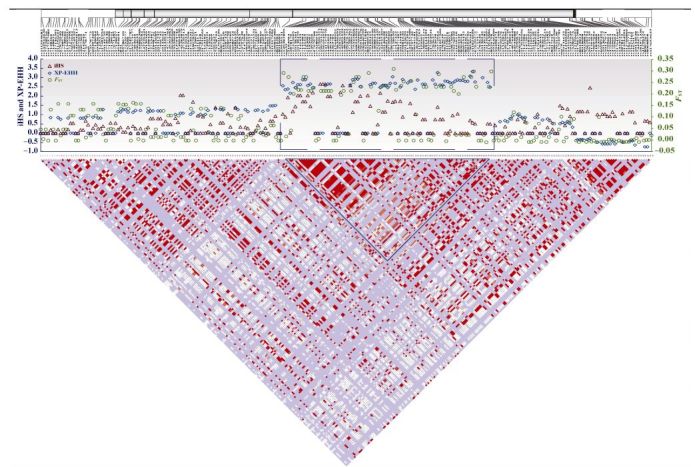
Genetic divergence of 245 GCH1 variants between Tibetans and Han Chinese (FST, iHS, and XP-EHH)
Functional prediction and genetic association analysis
To determine the potential function of the SNPs with selection signals, we performed functional predictions using evolutionary constraint (GERP), transcription factor binding sites (TFBS), splicing motif, H3K4Me1/H3K4Me3 sites, and DNase-I hypersensitive sites. Results showed that there might be multiple functional sites, reflected by the consistent signals of different functional predictions (Supplementary Table S1).
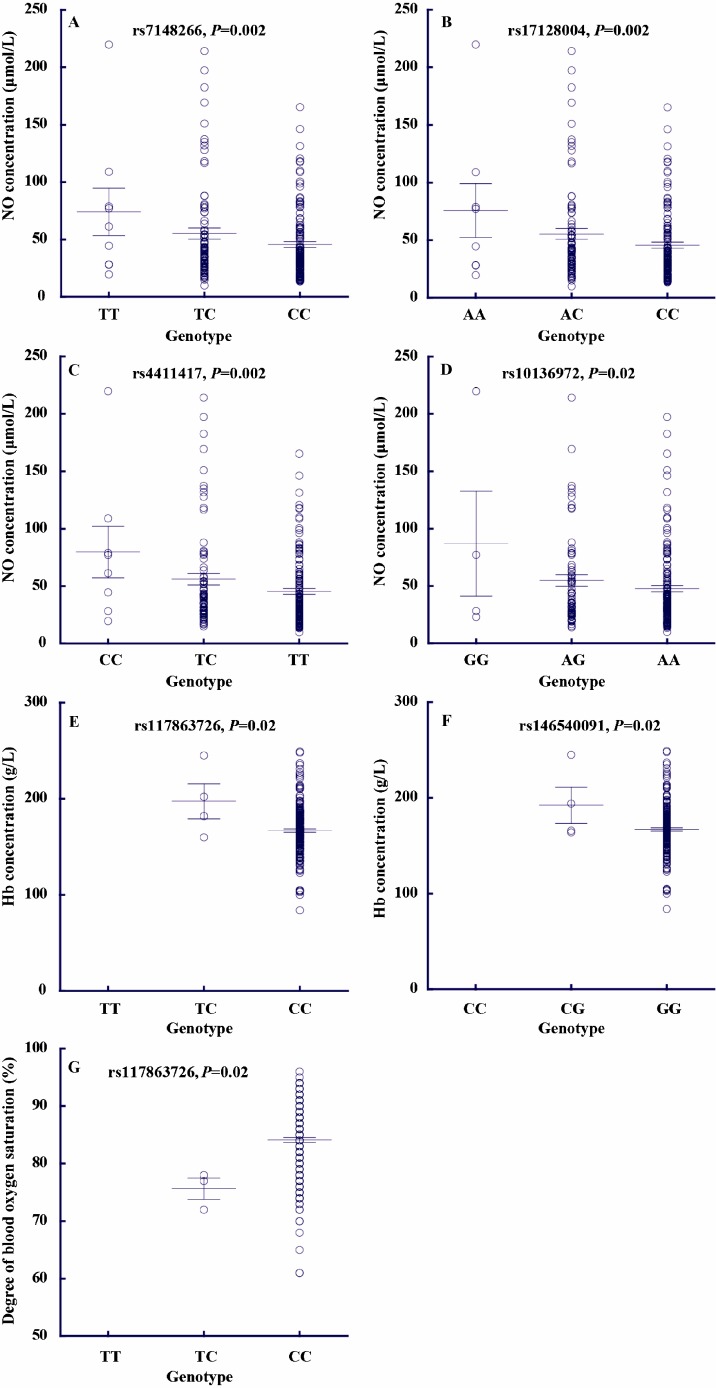
To test whether these candidate GCH1 variants contribute to the adaptive physiological traits of Tibetans, we measured blood NO concentrations, SaO2 levels, and Hb concentrations in 226 unrelated adult Tibetans (refer to methods for details). For genotyping, we selected nine tag SNPs to represent the entire gene region of GCH1. Notably, four SNPs (rs7148266, rs17128004, rs4411417, and rs10136972) showed significant association with blood NO levels, with the adaptive alleles exhibiting decreased NO. Each SNP accounted for 1.88%-3.76% of NO variance (Figure 2). Similar results were observed when males and females were analyzed separately (Table 1). Additionally, two SNPs (rs146540091 and rs137863726) showed association with hemoglobin concentration, with the adaptive alleles having lower Hb levels, consistent with previous results for EPAS1 and EGLN1 (Peng et al., 2011, 2017; Xiang et al., 2013). Another SNP (rs137863726) was associated with blood oxygen saturation, with the adaptive allele exhibiting higher SaO2 levels.
Figure 2.
Genetic association of seven GCH1 variants with three physiological traits (blood NO concentration, blood oxygen saturation level, and hemoglobin concentration)
For the four SNPs (rs7148266, rs17128004, rs4411417, and rs10136972) showing significant association with blood NO levels, functional prediction analysis indicated that they were located in the GCH1 intron regions with peak signals for H3K4Me1, H3K4Me3, DNase-I, and TFBS. For example, the H3K4Me1 peak values for rs10136972 and rs4411417 were 8.2 and 5.9, respectively, indicating their potential role in gene expression regulation of GCH1.
DISCUSSION
Hypoxia serves as a key stress in high altitude environments. For lowlanders, prolonged exposure to high altitude hypoxia can cause chronic mountain sickness, reflected by the over production of red blood cells (polycythemia) as well as other deleterious physiological changes (Hackett & Roach, 2001; Macinnis et al., 2010). Tibetans are genetically adapted to high altitude hypoxia, and exhibit blunted physiological responses, e.g., relatively low hemoglobin levels (Lorenzo et al., 2014; Peng et al., 2011, 2017; Xiang et al., 2013). Key hypoxic pathway genes EPAS1 and EGLN1 were reported to be responsible for these blunted physiological responses (Lorenzo et al., 2014; Peng et al., 2017; Xiang et al., 2013). However, while the Tibetan version of these two genes provide protection against polycythemia, they do not explain all physiological changes in Tibetans, suggesting there might be other genes involved given the complexity of high altitude adaptation.
Based on resequencing and population analysis, we confirmed a signal of selection on GCH1 in Tibetans. We identified more than 40 variants showing deep allelic divergence between highlander Tibetans and lowlander Han Chinese, with some having potential functional effects based on prediction using ENCODE. GCH1 is a rate-limiting enzyme, acting as a crucial factor for maintaining normal NO synthetase function and blood pressure. Inhibition of GCH1 activity is related to several cardiovascular diseases, with GCH1 found to prevent hypoxia-induced pulmonary hypertension (Khoo et al., 2005). Hence, the function of selection on GCH1in Tibetans is expected to help maintain proper cardiovascular function at high altitude.
Hypoxic pulmonary vasoconstriction and pulmonary vascular structural remodeling are dominant pathophysiological characteristics of hypoxic pulmonary hypertension (Galiè et al., 2016; McLaughlin & McGoon, 2006; Schermuly et al., 2011). When lowlanders move to high altitudes, pulmonary hypertension usually occurs within a few weeks (Wilkins et al., 2015; Wu & Kayser, 2006); however, Tibetans rarely develop this condition. We showed that GCH1 SNPs were associated with NO levels in the blood. GCH1 is involved in the synthesis of tetrahydrobiopterin (BH4), a vital regulator of eNOS, the endothelial-form enzyme producing NO, an important molecule for vasodilation, which is considered the main reason for the superior blood flow and pulmonary pressure in Tibetans (Pickert et al., 2013). In hph-1 mice, deficiency of BH4 causes hypoxia-induced pulmonary hypertension even under normoxic conditions (Khoo et al., 2005). The overexpression of GCH1 in mice could prevent hypoxia-induced pulmonary hypertension due to the augmentation of BH4 (Khoo et al., 2005). Hence, it is possible that GCH1 regulates pulmonary vasoconstriction responses in Tibetans by influencing NO production in the blood. We observed four GCH1 variants showing significant association with blood NO levels. As these variants are located in the GCH1 intron regions with peak enhancer and/or promoter activity signals, they are likely involved in the regulation of GCH1 expression and eventually affect blood NO production, which needs further investigation. We also observed associations of GCH1 SNPs with oxygen saturation and hemoglobin; however, the underlying molecular mechanisms are yet to be studied.
In summary, we demonstrated that GCH1 has been under positive selection in Tibetans. We identified many variants with deep allelic divergence between Tibetans and lowlanders. The association and known function results suggest the potential involvement of GCH1 in the regulation of multiple physiological traits in Tibetans.
ACKNOWLEDGEMENTS
We are grateful to all the volunteers participated in this study.
Funding Statement
This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13010000), the National Natural Science Foundation of China (91631306 to BS, 31671329 to XQ, 31460287 to Ou., 31501013 to HZ and 31360032 to CC), the National 973 program (2012CB518202 to TW), the State Key Laboratory of Genetic Resources and Evolution (GREKF15-05, GREKF16-04), and the Zhufeng Scholar Program of Tibetan University
REFERENCES
- 1. Antoniades C, Shirodaria C, Warrick N, Cai SJ, de Bono J, Lee J, Leeson P, Neubauer S, Ratnatunga C, Pillai R, Refsum H, Channon KM 5-methyltetrahydrofolate rapidly improves endothelial function and decreases superoxide production in human vessels:effects on vascular tetrahydrobiopterin availability and endothelial nitric oxide synthase coupling. Circulation, 2006. 114 (11): 1193- 1201. [DOI] [PubMed] [Google Scholar]
- 2. Beall CM, Strohl KP, Blangero J, Williams-Blangero S, Almasy LA, Decker MJ, Worthman CM, Goldstein MC, Vargas E, Villena M, Soria R, Alarcon AM, Gonzales C Ventilation and hypoxic ventilatory response of Tibetan and Aymara high altitude natives. American Journal of Physical Anthropology, 1997. 104 (4): 427- 447. [DOI] [PubMed] [Google Scholar]
- 3. Beall CM Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integrative and Comparative Biology, 2006. 46 (1): 18- 24. [DOI] [PubMed] [Google Scholar]
- 4. Beall CM, Cavalleri GL, Deng LB, Elston RC, Gao Y, Knight J, Li CH, Li JC, Liang Y, McCormack M, Montgomery HE, Pan H, Robbins PA, Shianna KV, Tam SC, Tsering N, Veeramah KR, Wang W, Wangdui P, Weale ME, Xu YM, Xu Z, Yang L, Zaman MJ, Zeng CQ, Zhang L, Zhang XL, Zhaxi P, Zheng YT Natural selection on EPAS1(HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences of the United States of America, 2010. 107 (25): 11459- 11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bigham A, Bauchet M, Pinto D, Mao XY, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, Herráez DL, Brutsaert T, Parra EJ, Moore LG, Shriver MD Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genetics, 2010. 6 (9): e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Computational Biology, 2010. 6 (12): e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ernst J, Kellis M ChromHMM:automating chromatin-state discovery and characterization. Nature Methods, 2012. 9 (3): 215- 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erzurum SC, Ghosh S, Janocha AJ, Xu W, Bauer S, Bryan NS, Tejero J, Hemann C, Hille R, Stuehr DJ, Feelisch M, Beall CM Higher blood flow and circulating NO products offset high-altitude hypoxia among Tibetans. Proceedings of the National Academy of Sciences of the United States of America, 2007. 104 (45): 17593- 17598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galiè N, Humbert M, Vachiery JL, Gibbs S, Lang I, Torbicki A, Simonneau G, Peacock A, Noordegraaf AV, Beghetti M, Ghofrani A, Sanchez MAG, Hansmann G, Klepetko W, Lancellotti P, Matucci M, McDonagh T, Pierard LA, Trindade PT, Zompatori M, Hoeper M, Aboyans V, Vaz Carneiro A, Achenbach S, Agewall S, Allanore Y, Asteggiano R, Badano LP, Barberà JA, Bouvaist H, Bueno H, Byrne RA, Carerj S, Castro G, Erol C, Falk V, FunckBrentano C, Gorenflo M, Granton J, Iung B, Kiely DG, Kirchhof P, Kjellstrom B, Landmesser U, Lekakis J, Lionis C, Lip GY, Orfanos SE, Park MH, Piepoli MF, Ponikowski P, Revel MP, Rigau D, Rosenkranz S, Völler H, Zamorano JL 2015 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension:the joint task force for the diagnosis and treatment of pulmonary hypertension of the european society of cardiology (ESC) and the european respiratory society (ERS):endorsed by:association for european paediatric and congenital cardiology (AEPC), international society for heart and lung transplantation (ISHLT). European Heart Journal, 2016. 37 (1): 67- 119. [DOI] [PubMed] [Google Scholar]
- 10. Hackett PH, Roach RC High-altitude illness. The New England Journal of Medicine, 2001. 345 (2): 107- 114. [DOI] [PubMed] [Google Scholar]
- 11. Khoo JP, Zhao L, Alp NJ, Bendall JK, Nicoli T, Rockett K, Wilkins MR, Channon KM Pivotal role for endothelial tetrahydrobiopterin in pulmonary hypertension. Circulation, 2005. 111 (16): 2126- 2133. [DOI] [PubMed] [Google Scholar]
- 12. Lorenzo FR, Huff C, Myllymäki M, Olenchock B, Swierczek S, Tashi T, Gordeuk V, Wuren T, Ri-Li G, McClain DA, Khan TM, Koul PA, Guchhait P, Salama ME, Xing JC, Semenza GL, Liberzon E, Wilson A, Simonson TS, Jorde LB, Kaelin WG, J r., Koivunen P, Prchal JT A genetic mechanism for Tibetan high-altitude adaptation. Nature Genetics, 2014. 46 (9): 951- 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu DS, Lou HY, Yuan K, Wang XJ, Wang YC, Zhang C, Lu Y, Yang X, Deng L, Zhou Y, Feng QD, Hu Y, Ding QL, Yang YJ, Li SL, Jin L, Guan YQ, Su B, Kang LL, Xu SH Ancestral origins and genetic history of tibetan highlanders. The American Journal of Human Genetics, 2016. 99 (3): 580- 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macinnis MJ, Koehle MS, Rupert JL Evidence for a genetic basis for altitude illness:2010 update. High Altitude Medicine & Biology, 2010. 11 (4): 349- 368. [DOI] [PubMed] [Google Scholar]
- 15. McLaughlin VV, McGoon MD Pulmonary arterial hypertension. Circulation, 2006. 114 (13): 1417- 1431. [DOI] [PubMed] [Google Scholar]
- 16. Peng Y, Yang ZH, Zhang H, Cui CY, Qi XB, Luo XJ, Tao X, Wu TY, Ouzhuluobu, Basang, Ciwangsangbu, Danzengduojie, Chen H, Shi H, Su B Genetic variations in tibetan populations and high-altitude adaptation at the himalayas. Molecular Biology and Evolution, 2011. 28 (2): 1075- 1081. [DOI] [PubMed] [Google Scholar]
- 17. Peng Y, Cui CY, He YX, Ouzhuluobu, Zhang H, Yang DY, Zhang Q, Bianbazhuoma, Yang LX, He YB, Xiang K, Zhang XM, Bhandari S, Shi P, Yangla, Dejiquzong, Baimakangzhuo, Duojizhuoma, Pan YY, Cirenyangji, Baimayangji, Gonggalanzi, Bai CJ, Bianba, Basang, Ciwangsangbu, Xu SH, Chen H, Liu SM, Wu TY, Qi XB, Su B Down-regulation of EPAS1 transcription and genetic adaptation of tibetans to high-altitude hypoxia. Molecular Biology and Evolution, 2017. 34 (4): 818- 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petousi N, Croft QPP, Cavalleri GL, Cheng HY, Formenti F, Ishida K, Lunn D, McCormack M, Shianna KV, Talbot NP, Ratcliffe PJ, Robbins PA Tibetans living at sea level have a hyporesponsive hypoxia-inducible factor system and blunted physiological responses to hypoxia. Journal of Applied Physiology, 2014. 116 (7): 893- 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pickert G, Lim HY, Weigert A, Häussler A, Myrczek T, Waldner M, Labocha S, Ferreirós N, Geisslinger G, Lötsch J, Becker C, Brüne B, Tegeder I Inhibition of GTP cyclohydrolase attenuates tumor growth by reducing angiogenesis and M2-like polarization of tumor associated macrophages. International Journal of Cancer, 2013. 132 (3): 591- 604. [DOI] [PubMed] [Google Scholar]
- 20. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC PLINK:a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 2007. 81 (3): 559- 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qi XB, Cui CY, Peng Y, Zhang XM, Yang ZH, Zhong H, Zhang H, Xiang K, Cao XY, Wang Y, Ouzhuluobu, Basang, Ciwangsangbu, Bianba, Gonggalanzi, Wu TY, Chen H, Shi H, Su B Genetic evidence of paleolithic colonization and neolithic expansion of modern humans on the tibetan plateau. Molecular Biology and Evolution, 2013. 30 (8): 1761- 1778. [DOI] [PubMed] [Google Scholar]
- 22. Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES Detecting recent positive selection in the human genome from haplotype structure. Nature, 2002. 419 (6909): 832- 837. [DOI] [PubMed] [Google Scholar]
- 23. Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie XH, Byrne EH, McCarroll SA, Gaudet R, Schaffner SF, Lander ES, The International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature, 2007. 449 (7164): 913- 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schermuly RT, Ghofrani HA, Wilkins MR, Grimminger F Mechanisms of disease:pulmonary arterial hypertension. Nature Reviews Cardiology, 2011. 8 (8): 443- 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simonson TS, Yang YZ, Huff CD, Yun HX, Qin G, Witherspoon DJ, Bai ZZ, Lorenzo FR, Xing JC, Jorde LB, Prchal JT, Ge RL Genetic evidence for high-altitude adaptation in Tibet. Science, 2010. 329 (5987): 72- 75. [DOI] [PubMed] [Google Scholar]
- 26. Szpiech ZA, Hernandez RD Selscan:an efficient multithreaded program to perform EHH-based scans for positive selection. Molecular Biology and Evolution, 2014. 31 (10): 2824- 2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tajima F Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 1989. 123 (3): 585- 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voight BF, Kudaravalli S, Wen XQ, Pritchard JK A map of recent positive selection in the human genome. PLoS Biology, 2006. 4 (3): e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weir BS, Cockerham CC Estimating F-statistics for the analysis of population structure. Evolution, 1984. 38 (6): 1358- 1370. [DOI] [PubMed] [Google Scholar]
- 30. Wilkins MR, Ghofrani HA, Weissmann N, Aldashev A, Zhao L Pathophysiology and treatment of high-altitude pulmonary vascular disease. Circulation, 2015. 131 (6): 582- 590. [DOI] [PubMed] [Google Scholar]
- 31. Wu TY, Kayser B High altitude adaptation in Tibetans. High Altitude Medicine & Biology, 2006. 7 (3): 193- 208. [DOI] [PubMed] [Google Scholar]
- 32. Xiang K, Ouzhuluobu, Peng Y, Yang ZH, Zhang XM, Cui CY, Zhang H, Li M, Zhang YF, Bianba, Gonggalanzi, Basang, Ciwangsangbu, Wu TY, Chen H, Shi H, Qi XB, Su B Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Molecular Biology and Evolution, 2013. 30 (8): 1889- 1898. [DOI] [PubMed] [Google Scholar]
- 33. Xu SH, Li SL, Yang YJ, Tan JZ, Lou HY, Jin WF, Yang L, Pan XD, Wang JC, Shen YP, Wu BL, Wang HY, Jin L A genome-wide search for signals of high-altitude adaptation in Tibetans. Molecular Biology and Evolution, 2011. 28 (2): 1003- 1011. [DOI] [PubMed] [Google Scholar]
- 34. Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng HC, Liu T, He WM, Li K, Luo RB, Nie XF, Wu HL, Zhao MR, Cao HZ, Zou J, Shan Y, Li SZ, Yang Q, As an, Ni PX, Tian G, Xu JM, Liu X, Jiang T, Wu RH, Zhou GY, Tang MF, Qin JJ, Wang T, Feng SJ, Li GH, Huasang, Luosang JB, Wang W, Chen F, Wang YD, Zheng XG, Li Z, Bianba Z, Yang G, Wang XP, Tang SH, Gao GY, Chen Y, Luo Z, Gusang L, Cao Z, Zhang QH, Ouyang WH, Ren XL, Liang HQ, Zheng HS, Huang YB, Li JX, Bolund L, Kristiansen K, Li YR, Zhang Y, Zhang XQ, Li RQ, Li SG, Yang HM, Nielsen R, Wang J, Wang J Sequencing of 50 human exomes reveals adaptation to high altitude. Science, 2010. 329 (5987): 75- 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L, Chen WZ, Liu YJ, Hu X, Zhou K, Chen L, Peng S, Zhu H, Zou HL, Bai J, Wang ZB Feasibility of magnetic resonance imaging-guided high intensity focused ultrasound therapy for ablating uterine fibroids in patients with bowel lies anterior to uterus. European Journal of Radiology, 2010. 73 (2): 396- 403. [DOI] [PubMed] [Google Scholar]