Abstract
Recent advances in fluorescent protein technology provide a wide variety of biological imaging applications; however current tools for bio-imaging in the Gram-positive bacterium Staphylococcus aureus has necessitated further developments for fluorescence intensity and for a multicolor palette of fluorescent proteins. To enhance the expression of multicolor fluorescent proteins in clinical S. aureus strains, we developed new fluorescent protein expression vectors, containing the blaZ/sodp promoter consisting of the β-lactamase gene (blaZ) promoter and the ribosome binding site (RBS) of superoxide dismutase gene (sod). We found S. aureus-adapted GFP (GFPsa) driven by the blaZ/sodp promoter was highly expressed in the S. aureus laboratory strain RN4220, but not in the clinical strains, MW2 and N315, harboring the endogenous blaI gene, a repressor of the blaZ gene promoter. We therefore constructed a constitutively induced blaZ/sodp promoter (blaZ/sodp(Con)) by introducing substitution mutations into the BlaI binding motif, and this modification allowed enhanced expression of the multicolor GFP variants (GFPsa, EGFP, mEmerald, Citrine, Cerulean, and BFP) as well as codon-optimized reef coral fluorescent proteins (mCherry and AmCyan) in the S. aureus clinical strains. These new fluorescent probes provide new tools to enhance expression of multicolor fluorescent proteins and facilitate clear visualization of clinical S. aureus strains.
Introduction
Fluorescent proteins are widely used as biological markers that enable visualization of subcellular protein localization, gene expressions, protein-protein interactions, and in vivo monitoring of bacterial infection1–3. Currently available fluorescent proteins are primarily derived from either the green fluorescent protein (GFP) originating in the jellyfish Aequorea victoria 4, 5, or reef coral fluorescent proteins (RCFP) derived from Discosoma sp5–7. The wild type A. victoria GFP exhibits poor fluorescent brightness in Escherichia coli and mammalian cell lines; and many of the wild type GFP have a strong tendency to be expressed as an insoluble protein, showing cytotoxicity in E. coli 8, 9. To date, extensive studies have reported numerous types of A. victoria GFP variants that provide significant improvements in brightness, protein solubility, stability, pH-sensitivity, and yield4, 5, 9. Further, numerous variants of GFP and RCFP with distinct colors have been engineered using a combination of random mutagenesis and directed evolution. This has enabled co-visualization of several proteins in a single cell, selective identification of particular cells in co-culture systems, and detection of protein-protein interactions based on a measurement of fluorescence resonance energy transfer (FRET)1–5.
Staphylococcus aureus is a low-GC Gram-positive bacterium that causes a variety of diseases, e.g., abscess, bullous impetigo, toxic shock syndrome, pneumonia, sepsis, and food poisoning. Multidrug-resistant strains such as methicillin-resistant S. aureus (MRSA) cause severe hospital-acquired infections such as pneumonia and sepsis; and the resistance makes the treatment increasingly difficult. The development of molecular genetic tools including gene deletion, controllable gene expression, and bio-imaging is essential for our understanding of the mechanisms underlying the pathogenesis of S. aureus infections10, 11. Currently, for bio-imaging numerous GFP and RCFP variants are commercially available from many distributors; however, these FPs were not optimized for less common bacterial strains (e.g., clinical strains of S. aureus). Bio-imaging tools based on fluorescent proteins have also been widely used in the studies of S. aureus to visualize subcellular proteins or for biofilm formation as well as to establish a S. aureus infection model using fluorescent proteins, in which optimization of codon usage and replacement of the region surrounding the ribosome binding sequence (RBS) have been reported to enhance the expression of the fluorescent proteins in S. aureus 12–16. However, the current methods often entail several limitations as to color palette and its brightness and therefore necessitated the development of new fluorescent vectors that can efficiently enhance fluorescence intensity and the multicolor palette in S. aureus strains. Many proteins are generally difficult to highly express in heterologous host organisms. In E. coli, extensive research has established many different types of promoters and a great number of engineered laboratory strains for heterologous protein production17. However, a well-established method for the heterologous protein expression is still lacking in S. aureus, much less clinically isolated strains. Previous studies have focused on the β-lactamase gene (blaZ) promoter P blaZ for the gfp expression in S. aureus. Notably, the GFPmut2 was constitutively expressed in the laboratory strain RN422018–20. This study therefore developed fluorescent protein expression vectors with the P blaZ promoter to highly express GFP and RCFP variants in clinically isolated S. aureus.
Here, we describe novel fluorescent protein expression vectors, which were shown to exhibit greater fluorescence intensity in clinical S. aureus strains. To improve the expression of an exogenous fluorescent protein in S. aureus clinical strains, we used the blaZ/sodp(Con) promoter and codon-optimized fluorescent protein genes, and showed that the fluorescence intensity in the series of GFP and RCFP variants were significantly enhanced in the clinical strains. These new tools efficiently expressing fluorescent proteins in clinical S. aureus strains are valuable for understanding the pathogenic mechanism of S. aureus.
Results
Adaptation of the green fluorescent protein to S. aureus
We aimed to develop fluorescent protein expression vectors that can produce sufficient fluorescence intensity to visually identify fluorescing colonies of S. aureus on agar plates with the naked eye. S. aureus strain RN4220 colonies containing pS1GFP in which GFPmut3b was expressed under the control of the β-lactamase gene (blaZ) promoter exhibited faint fluorescence on TSB agar plates (Fig. 1b). When we replaced the 13-bp sequence containing RBS sequence of the blaZ gene with the corresponding region of the superoxide dismutase gene (sod) (Fig. 1a) that has been reported to enhance the expression of fluorescent protein in S. aureus 13, the fluorescent activity of the colonies expressing GFPmut3b was significantly improved on the agar plates (Fig. 1b). Further, the fluorescence intensity increased by an approximately 10-fold using the microplate assay reader (Fig. 1c). The high expression of GFPmut3b was confirmed as a major protein band corresponding to its molecular weight, 26.8 kDa, in SDS-PAGE (Fig. 1d). Although the GFPmut3b is one of the faster folding GFP variants that has been optimized for bacteria and has minimal toxicity21, overexpression of GFPmut3b in S. aureus showed adverse effects such as cell growth inhibition, producing smaller colonies, and frequent co-occurrence of larger colonies exhibiting no fluorescence signal (Fig. S1a). To overcome these adverse effects, GFPmut3b was modified by introducing Cycle3 mutations (F99S/M153T/V163A) that are known to improve the solubility and reduce the toxicity9, 22, yielding S. aureus-adapted GFPmut3b, GFPsa (S65G/S72A/F99S/M153T/V163A). As a result, the mutations antagonized the appearance of non-fluorescent colonies and maintained high levels of the fluorescent intensity (Figs 1b,c and S1b) and GFPsa expression (Fig. 1d) without affecting its excitation and emission wavelengths (Fig. 1e,f). Further, confocal laser scanning microscopic analysis revealed that almost all of S. aureus cells expressed GFPsa protein and were clearly visualized (Fig. 1g).
Figure 1.
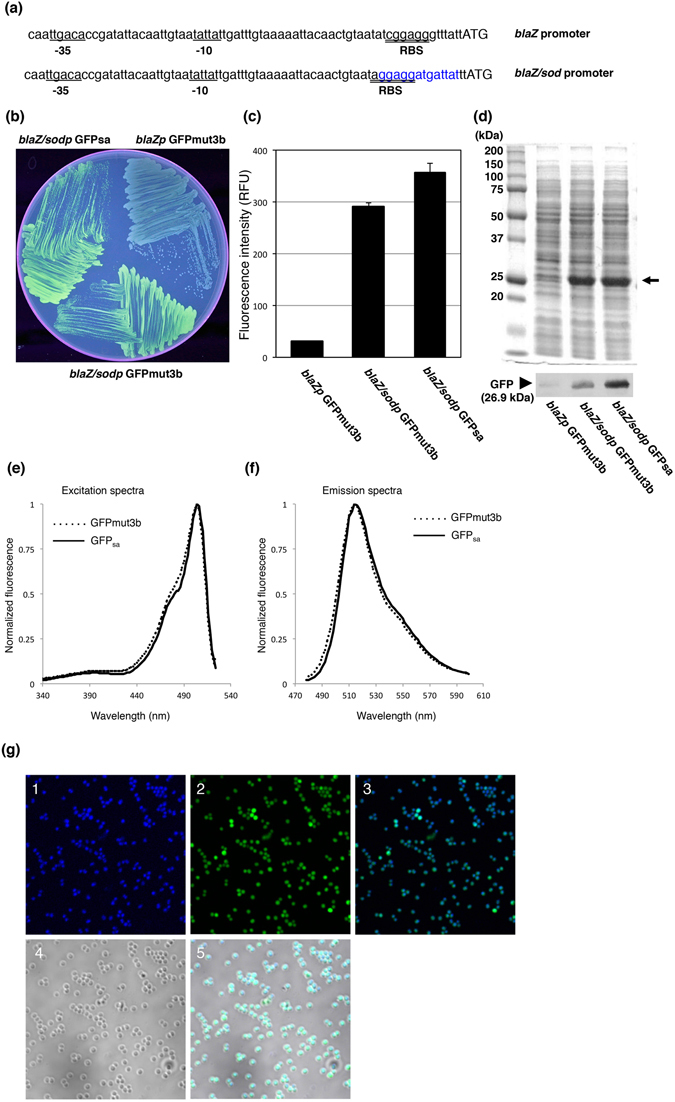
Replacement of RBS sequence and introduction of Cycle3 mutations into GFPmut3. (a) Sequence alignment of the blaZp and its derivative blaZ/sodp promoters. Blue bold face indicates the surrounding sequence containing RBS derived from the sod gene. Underlined sequences indicate −35 and −10 elements of the blaZ promoter. Double underlines indicate the RBS sequence. Initiation codons are shown in uppercase. (b) The fluorescing colonies were photographed under UV excitation. S. aureus RN4220 containing pS1GFP, pFK51, and pFK52 were grown on TSB agar plate containing chloramphenicol. (c) The fluorescent intensities of GFPmut3b, and GFPsa in S. aureus RN4220. Cell homogenate of S. aureus containing pS1GFP, pFKS1 and pFK52 was prepared and the fluorescent intensities at 513 nm were measured with a microplate reader, λex = 490 nm. The data represent mean values ± standard deviation. (d) The expression efficiency of pS1GFP, pFK51, and pFK52 in S. aureus RN4220. SDS-PAGE and Western blot analysis show the relative quantities of the GFP in the whole cell lysates. The Western blot gel was cropped and the full-length image is included in Supplemental Fig. 5(a). (e) Excitation and (f) Emission spectra for GFPmut3b and GFPsa in S. aureus RN4220. Each spectrum was normalized to a maximum value of 1. Excitation spectra were recorded with emission at 540 nm. Emission spectra were recorded with excitation at 460 nm. GFPmut3b and GFPsa were depicted by a solid line and dotted line, respectively. (g) S. aureus RN4220 containing pMK4blaZ/sodpGFPsa (pFK52) was grown on TSB containing chloramphenicol and visualized using an FV1000 confocal scanning laser microscope (Olympus). Panels show (1) DAPI, (2) GFP, (3) overlay of DAPI and GFP images, (4) differential interference contrast (DIC), (5) overlay of DAPI, GFP, and DIC images.
Introducing suppressor mutations into the blaZ gene promoter
S. aureus-adapted GFPsa demonstrated higher expression under the control of the hybrid blaZ/sodp promoter in laboratory strain RN4220. However, in clinical strains, MW2 and N315, with the same vector, the expression of GFPsa was very low, and the fluorescence intensity was poor (Fig. 2b,c,d). BlaZ plasmid is prevalent in clinical strains23, 24, and the blaIRZ gene operon is natively present on a plasmid in both clinical strains, MW2 and N315, but not in RN422025–27. The BlaI and MecI specifically bind to the same dyad symmetry (TACA/TGTA) sequence and repress the blaZ promoter activity (Fig. 2a)28. To minimize the possible negative effect by the endogenous BlaI/MecI on the blaZ/sodp promoter activity, we introduced suppressor mutations into the BlaI/MecI binding motif (TACA/TGTA), yielding a constitutively induced blaZ/sodp(Con) promoter (Fig. 2a). As a consequence, GFPsa expression was significantly elevated, and the resulting fluorescence intensity was highly improved in both N315 and MW2, whereas the fluorescence intensity was slightly reduced in strain RN4220, compared with the parental sequence (Fig. 2b,c,d). Taken together, the data demonstrated the modified blaZ/sodp promoter, (blaZ/sodp(Con)) can effectively enhance the expression of GFPsa in S. aureus clinical strains.
Figure 2.
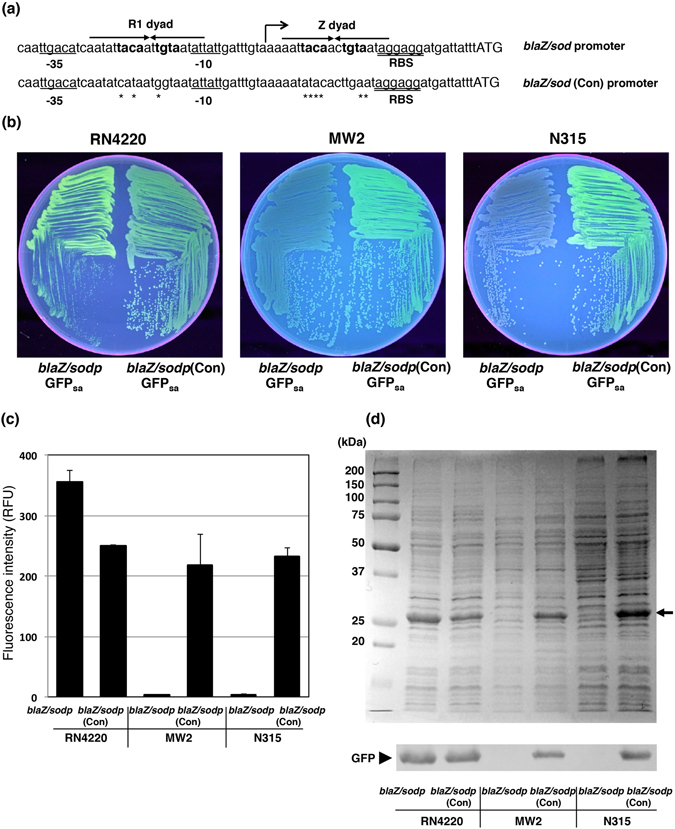
Site-directed mutagenesis into the BlaI/MecI binding sequence. (a) Sequence alignment of the blaZ/sodp and its constitutively induced blaZ/sodp(Con) promoter region. Asterisks indicate the sequence exchanged by site-directed mutagenesis. Bold face indicates the BlaI/MecI binding motif (TACA/TGTA) within the larger palindromes, the R1 dyad and Z dyad are indicated by arrows. Up arrows with the tip to right indicates the transcription initiation site. Underlined sequences indicate −35 and −10 elements. Double underline indicates the RBS sequence. Initiation codons are shown in uppercase. (b) The fluorescing colonies were photographed under UV excitation. S. aureus RN4220, MW2 and N315 containing either pFK52, or pFK54 were grown on TSB agar plate containing chloramphenicol. (c) The comparison of fluorescence intensity among S. aureus strain RN4220, MW2, and N315 containing either pFK52, or pFK54. The fluorescent intensities at 513 nm were measured with a microplate reader, λex = 490 nm. The data represent mean values ± standard deviation. (d) The comparison of GFPsa expression efficiency among S. aureus strain RN4220, MW2, and N315 containing either pFK52, or pFK54. SDS-PAGE and Western blot analysis show the relative quantities of GFPsa in the whole cell lysates. The arrow indicates the position of GFPsa in gel. The Western blot gel was cropped and the full-length image is included in Supplemental Fig. 5(b).
Expression of multicolor GFP variants in clinical strains
A. victoria GFP and its variants contribute to multicolor imaging with spectral profiles ranging in color from blue to yellow, and have been engineered by site-directed mutagenesis29–34. Using GFPsa as the standard, we next constructed these GFP color variants (EGFP, mEmerald, Citrine, Cerulean, and BFP) using amino acid substitutions considering the codon usage patterns in S. aureus. Since several BFP variants were engineered using substitution of the T66H mutation; we constructed three BFP variants, EBFP (F64L/S65T/Y66H/Y145F), 1EMF (F64L/Y66H/V163A), and P4–3E (F64L/Y66H/Y145F/V163A) and evaluated which BFP variants would be more suitable for S. aureus MW2 and N315. Fluorescence assay showed that homogenates of P4-3E and EBFP exhibited somewhat greater fluorescence intensity than 1EMF when S. aureus MW2 were grown in TSB medium (Fig. S2a), while only S. aureus expressing P4-3E was visually identified as fluorescing colonies on agar plates by the naked eye (Fig. S2b). Each strain expressing the multicolor GFP variant exhibited sufficient fluorescence intensity to visually identify fluorescing colonies by the naked eye (Fig. 3a), and SDS-PAGE and Western blot analyses showed that like GFPsa all of these multicolor variants were expressed with high yield in S. aureus MW2 harboring the corresponding vectors with the blaZ/sodp(Con) promoter (Fig. 3b,c). The multicolor GFP variant genes driven by the blaZ/sodp(Con) promoter also exhibited high fluorescence intensity in another clinical strain, N315 (Fig. S3a). Further, fluorescence excitation and emission spectrum analysis showed these strains expressing the multicolor GFP variants reflected the previously published spectrum profiles (Fig. S4). This data indicated that multicolor GFP variants were highly expressed in clinical strains MW2 and N315 with the blaZ/sodp(Con) promoter.
Figure 3.
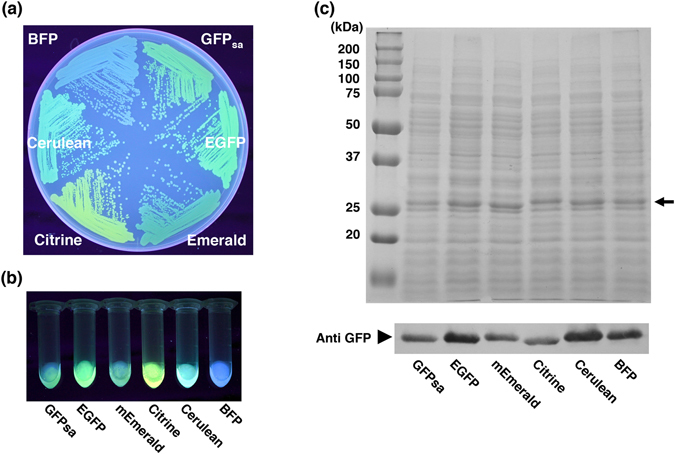
Detection of multicolor GFP variants in the clinical strain, MW2. (a) The fluorescing colonies were photographed using UV excitation. S. aureus strain MW2 expressing multicolor GFP variants (GFPsa, EGFP, mEmerald, Citrine, Cerulean, and BFP) were grown on TSB agar plate containing chloramphenicol. (b) The whole cell lysates of S. aureus MW2 expressing multicolor variants used for SDS-PAGE and Western blot analysis were photographed under UV excitation. (c) The comparison of expression efficiency among multicolor GFP variants in S. aureus strain MW2. SDS-PAGE and Western blot analysis show the relative quantities of the multicolor GFP variants in the whole cell lysates. The arrow indicates the position of GFP variants in gel. The Western blot gel was cropped and the full-length image is included in Supplemental Fig. 5(c).
Codon optimization of AmCyan and mCherry for S. aureus
Like Aequorea GFP, reef coral fluorescent proteins (RCFP) and their variants also contribute to multicolor imaging with spectral profiles ranging in color from cyan to far red1, 5. We constructed the RCFP expression vectors for S. aureus clinical strains, in which the amCyan or mCherry gene was expressed under the control of the hybrid blaZ/sodp(Con) promoter. Unexpectedly, S. aureus expressing the commercially available amCyan or mCherry gene formed faintly fluorescent colonies on agar plates (Fig. 4a), we therefore optimized the codon usage of amCyan and mCherry genes by replacing rare GC- rich codons with AT-rich ones. As a consequence, the codon-optimized AmCyan (AmCyan(S.a)) and mCherry (mCherry(S.a)) exhibited sufficient fluorescence intensity to visually identify fluorescing colonies on agar plates by the naked eye (Fig. 4a), and the corresponding proteins were confirmed to their molecular weights, 25.2 kDa and 26.7 kDa, respectively (Fig. 4b) and detected by Western blot (Fig. 4c). The fluorescence assay shows the codon adapted genes resulted in increased fluorescence intensity of AmCyan and mCherry (Fig. 4d,e). The amCyan and mCherry genes driven by the blaZ/sodp(Con) promoter also exhibited a significant fluorescence intensity in strain N315 (Fig. S3b). Further, fluorescence excitation and emission spectrum analysis showed these strains expressing AmCyan or mCherry reflected previously published spectrum profiles (Fig. S4). Taken together, the expression of AmCyan and mCherry were significantly improved in S. aureus clinical strains.
Figure 4.
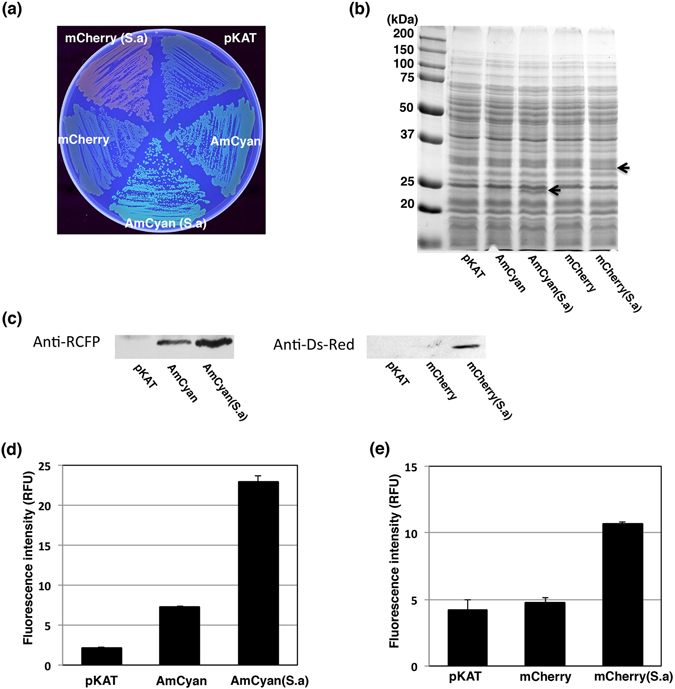
Codon usage optimization of the amCyan and mCherry genes. (a) The fluorescing colonies were photographed under UV excitation. S. aureus strain MW2 containing pKAT (control), pFK62 (AmCyan), pFK64 (AmCyan(S.a)), pFK63 (mCherry), and pFK65 (mCherry(S.a)) were grown on TSB agar plates containing chloramphenicol. (b) SDS-PAGE analysis showed the relative quantities of AmCyan and mCherry in the whole cell lysates. Arrows indicate the position of AmCyan and mCherry in gel. (c) Western blot analysis of AmCyan or mCherry in the whole cell lysates. The Western blot gels were cropped and the full-length image are included in Supplemental Fig. 5(d). The comparison of fluorescent intensities of pKAT (control), pFK62 (AmCyan), and pFK64 (AmCyan(S.a)) in S. aureus MW2. The fluorescent intensities at 489 nm were measured with a microplate reader, λex = 458 nm. The data represent mean values ± standard deviation. (e) The comparison of fluorescent intensities of pKAT (control), pFK63 (mCherry), and pFK65 (mChaerry(S.a)) in S. aureus MW2. The fluorescent intensities at 610 nm were measured with a microplate reader, λex = 586 nm. The data represent mean values ± standard deviation.
Identification of S. aureus cells expressing a specific fluorescent protein in co-culture systems
We next aimed to discriminate S. aureus cells expressing a specific fluorescent protein from the co-existence of bacteria expressing different fluorescent proteins in a co-culture experiment. Confocal laser scanning microscopic analysis revealed that each S. aureus clinical strain expressing different fluorescent proteins was selectively detected in the co-culture system in which Cerulean, Citrine, and mCherry(S.a) were differently expressed in the strain N315, TY3435, and MW2, respectively (Fig. 5a). Further, S. aureus N315 cells expressing mCheery(S.a) were clearly distinguished from E. coli DH5α cells expressing EGFP (Fig. 5b). These results demonstrated that our fluorescent protein expression vectors have the potential to sensitively detect particular S. aureus cells in co-culture with different strains or other bacteria.
Figure 5.
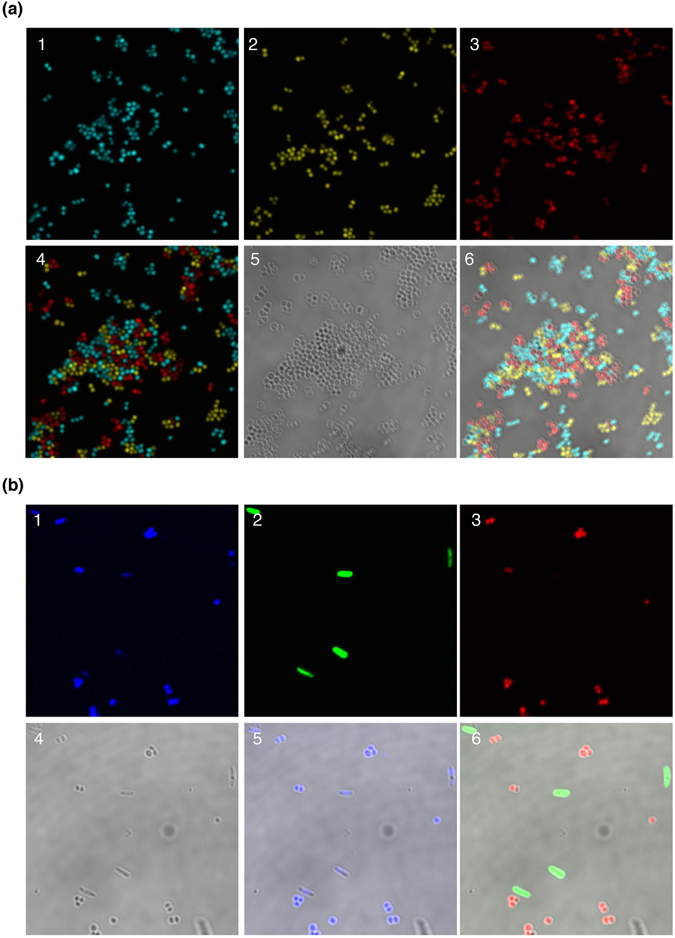
Confocal laser microscopic analysis in co-culture systems. (a) Co-culture of three clinically isolated S. aureus strains. S. aureus strain N315 containing pFK60 (Cerulean), strain TY34 containing pFK56 (Citrine), and strain MW2 containing pFK65 (mCherry(S.a)) were co-cultured in BHI broth. Panels show (1) Cerulean (N315), (2) Citrine (TY34), (3) mCherry(S.a) (MW2), (4) overlay of Cerulean, Citrine, and mCherry images, (5) DIC, (6) overlay of Cerulean, Citrine, mCherry, and DIC images. (b) Co-culture of S. aureus with E. coli. S. aureus N315 containing pFK65 (mCherry(S.a)) and E. coli DH5α containing pFK55 (EGFP) were co-cultured in BHI broth. Panels show (1) DAPI, (2) EGFP, (3) mCherry(S.a), (4) DIC, (5) overlay of DAPI and DIC images, and (6) overlay of EGFP, mCherry(S.a), and DIC images.
Discussion
In this study, we developed new fluorescent protein vectors to express a bright fluorescence intensity enough to visually identify fluorescent colonies of S. aureus macroscopically on agar plates. Practically available high-level expression of fluorescent protein has not been achieved in previous studies in which fluorescent proteins were detected only with fluorescence microscopy10–14. Our findings provide new applications to enhance the expression of multicolor GFP variants and would allow us to sensitively detect particular S. aureus cells in bacterial populations and in animal infection models.
To enhance fluorescent brightness, we replaced the sequence from the Shine-Dalgarno (SD) sequence to the start codon with sod that was previously reported with the sod RBS to lead to the highest fluorescence intensity with the sarA promoter13. Consequentially, we not only modified the SD sequence of blaZ gene to an optimal SD sequence (AGGAGG), but also altered the distance from the SD sequence to the start codon. The distance between the RBS and the start codon is also known to affect the efficiency of translation initiation36. Our results support the concept that the improvement of the sequence from the SD sequence to the start codon sequence significantly enhances the expression of the GFP variant, although the exact mechanism responsible for this effect remains unclear (Fig. 1b). However, recent studies suggest the interaction among the RBS, the initiation codon, the 5′-coding region in translation initiation, and RNA secondary structure at the 5′ terminus affects the protein expression in bacteria37, 38. Therefore, the enhancement of GFP production may be accounted for by the decrease of free fold energy of the 5′ end of mRNA transcripts.
At the beginning of this study, we evaluated GFPsa expression vectors with the blaZ/sodp promoter in a laboratory strain, RN4220. This strain is easily genetically manipulated; however, it carries a number of genetic mutations that may affect the virulence of the strain20, 27. Like RN4220, laboratory strains may not be suitable for the evaluation of pathogenesis, because laboratory strains often lack important pathophysiological characters39. Hence, clinical strains should be used to properly evaluate the pathogenesis and virulence of S. aureus. S. aureus harboring the bla gene locus appeared in the 1940s after the introduction of penicillin; and at present, most clinical isolates carry the blaIRZ gene on plasmids or on the chromosome23, 40, 41. Because the GFPsa expression driven by the blaZ/sodp promoter was not detectable in S. aureus clinical strains, MW2 and N315 (Fig. 2), we hypothesized the endogenous BlaI may inhibit the transcription of the blaZ/sodp promoter in trans; and we therefore generated the constitutively induced blaZ/sodp(Con) promoter to overcome the limitation of the clinical strains. The blaZ/sodp(Con) promoter can be adapted to enable expression of not only fluorescent proteins but also various exogenous proteins (toxins) from clinical S. aureus strains.
Codon optimization frequently plays a key role in exogenous protein expression37, 42. The GC contents of human codon-optimized amCyan and mCherry are 46.8% and 62.5%, respectively, showing strong preferences for G + C at the third codon position is distinct from the codon usage patterns in S. aureus with 33.5% GC content. Exchanging the 92 nucleotides of amCyan gene and 76 nucleotides of mCherry gene, respectively, both fluorescence intensities were significantly improved overcoming the codon usage bias in S. aureus. However, SDS-PAGE analysis showed the level of RCFPs production did not reach the level of GFP variants (Figs 3c and 4b). These results suggest the possibility that further investigation with these vectors could improve the expression level of RCFPs by further adapting of codon usage or decreasing the free folding energy of the initial 5′-coding region.
In summary, we have developed new multicolor fluorescent protein vectors that efficiently enhance fluorescence intensity in S. aureus clinical strains where the greater fluorescence intensity of multicolor fluorescent proteins may facilitate clear visualization of S. aureus clinical strains. Ultimately, these findings may help in better understanding the pathogenic mechanisms of S. aureus.
Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are in Table 1. S. aureus and E. coli were cultured at 37 °C with shaking at 140 rpm in test tube (25 mm × 150 mm) containing 3 ml of trypticase soy broth (TSB) (Becton, Dickinson and Company) or 3 ml of lysogeny broth (5 g yeast extract, 10 g polypeptone, 10 g NaCl per liter; pH 7.2), respectively. Ampicillin (Amp, 100 µg/ml) and chloramphenicol (Cp, 10 µg/ml) were added to the medium if necessary. The plasmids encoding AmCyan or mCherry were purchased from Clontech, TaKaRa Bio Inc., Japan.
Table 1.
Bacterial strains and plasmids used in this study.
| Bacterial strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F−, φ80dlacZΔM15, Δ(lacZYA−argF)U169, deoR, recA1, endA1, hsdR17(rk−, mk+), phoA, supE44, λ−, thi-1, gyrA96, relA1 | TaKaRa |
| S. aureus | ||
| RN4220 | NCTC8325-4, r- m+ | 20 |
| N315 | hospital-aquired MRSA | 25 |
| MW2 | community-aquired MRSA | 26 |
| TY34 | clinical isolate MRSA from patient with impetigo | 35 |
| Plasmids | ||
| pMK4 | Shuttle vector between E. coli and S. aureus, Amr in E. coli, Cmr in S. aureus | 46 |
| pS1GFP | pMK4 containing GFPmut3b gene fused to the blaZp promoter | 19 |
| pND50 | Shuttle vector between E. coli and S. aureus, Cmr | 44 |
| pKAT | pND50 derivative containing the lacZ(α) gene from pUC19, Cmr | 43 |
| pAmCyan | Plasmid encoding AmCyan gene, Ampr | TaKaRa Clontech |
| pmCherry | Plasmid encoding mCherry gene, Ampr | TaKaRa Clontech |
| pFK51 | pMK4 containing GFPmut3b gene fused to the blaZ/sodp promoter | This study |
| pFK52 | pMK4 containing GFPsa gene fused to the blaZ/sodp promoter | This study |
| pFK53 | pMK4 containing GFPmut3b gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK54 | pMK4 containing GFPsa gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK55 | pMK4 containing EGFP gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK56 | pMK4 containing Citrine gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK57 | pMK4 containing EBFP gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK58 | pMK4 containing BFP(P4-3E) gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK59 | pMK4 containing BFP(1EMF) gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK60 | pMK4 containing Cerulean gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK61 | pMK4 containing mEmerald gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK62 | pKAT containing AmCyan gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK63 | pKAT containing mCherry gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK64 | pKAT containing codon-optimized AmCyan(S.a) gene fused to the blaZ/sodp(Con) promoter | This study |
| pFK65 | pKAT containing codon-optimized mCherry(S.a) gene fused to the blaZ/sodp(Con) promoter | This study |
Improvement of GFP expression vector using site-directed mutagenesis
To increase expression of fluorescent proteins in S. aureus clinical strains, we improved a GFP expression vector based on pS1GFP carrying GFPmut3b gene (a kind gift from Prof. M. Krönke)19. The hybrid blaZ/sodp promoter was constructed by replacing the sequence containing the RBS in the blaZ gene in the sod gene with blaZPR and GFP-F primers using the inverse PCR method with KOD Plus Neo DNA polymerase (TOYOBO, Japan). The PCR product was phosphorylated with T4 Polynucleotide Kinase (TaKaRa Bio Inc, Japan), and circularized by self-ligation with Ligation high Ver.2 (TOYOBO, Japan); and then the circular DNA was transformed into E. coli DH5α. Plasmid DNA was extracted from the transformed E. coli DH5α using FastGeneTM Plasmid Mini Kit (Nippon Genetics Co., Ltd. Japan) and the resultant plasmid was verified using ABI 3130 DNA sequencer (Applied Biosystems). To express the fluorescent protein in S. aureus clinical strains, the substitution of the repressor BlaI/MecI binding motif (TACA/TGTA) was performed with the blaZ-mutF and blaZ-mutR primers using inverse PCR as above, yielding the constitutively induced blaZ/sodp(Con) promoter.
Construction of the multicolor GFP variants using amino acid substitution
Amino acid substitution was performed using inverse PCR with pMK4blaZ/sodp(Con)GFPmut3b (pFK53) as the initial template with the KOD Plus Neo DNA polymerase (TOYOBO, Japan). The primers described in Table 2 were used to construct the GFP variants (Table 3). The resulting plasmids were confirmed using DNA sequencing.
Table 2.
Oligonucleotide primers.
| Name | primer sequence (5' to 3') | Purpose |
|---|---|---|
| blaZPR | AAATAATCATCCTCCTATTACAGTTGTAA | replacement of RBS sequence |
| GFP-F | ATGAGTAAAGGAGAAGAACTTTTCAC | |
| blaZPmut-F | TTTGTAAAAATATACACTTGAATAGGAGGATGAT | destruction of BlaI motif |
| blaZPmut-R | TCAATAATATTACCATTATGATATTGATG | |
| F64LS65T-R | TTGAACACCATATGTTAAAGTAGTAACAAG | construction of Emerald and EGFP |
| F64LY66H-R | TTGAACACCATGAGATAAAGTAGTAACAAG | construction of BFP(P4-3E) and BFP(1EMF) |
| F64LS65TY66H-R | TTGAACACCATGTGTTAATGTAGTAACAAG | construction of EBFP |
| F64LS65TY66W-R | TTGAACACCCCATGTTAAAGTAGTAACAAGTGTTG | construction of Cerulean |
| S65GV68LQ69M-R | CATTAAACCATAACCAAATGTAGTAACAAG | construction of Citrine |
| S72-F | TGTTTTTCAAGATATCCAGATCATATG | construction of EGFP and BFP |
| S72A-F | TGTTTTGCAAGATATCCAGATCATATG | construction of Cerulean, Emerald, and Citrine |
| F99S-F | TTCAAAGATGACGGTAACTACAAGAC | construction of GFPsa |
| F99S-R | AGATATAGTTCTTTCCTGTACATAACC | construction of GFPsa |
| Y145F-R | GATATATACATTATGTGAGTTAAAGTTATATTC | construction of EBFP, P4-3E(BFP) |
| Y145AN146IH148D-R | AATATATACATTGTCTGAGATAGCGTTATATTCCAA | construction of Cerulean |
| N149KM153T-R | CTTTTGTTTGTCTGCTGTAATATATACTTTATGTGAG | construction of Emerald |
| I152-R | AATATATACATTATGTGAATTATAGTTATATTCC | construction of GFPsa and BFP(1EMF) |
| M153-F | ATGGCAGACAAACAAAAGAATGGAATC | construction of EBFP, P4-3E(BFP) |
| M153TV163A-F | ACAGCAGACAAACAAAAGAATGGAATCAAAGCTAAC | construction of GFPsa and Cerulean |
| V163A-F | ATGGCAGACAAACAAAAGAATGGAATCAAAGCTAAC | construction of 1EMF(BFP) |
| I167T-F | AATGGAATTAAAGTTAACTTCAAAACAAGACAC | construction of Emerald |
| T203Y-F | TATCAATCTGCATTATCAAAAGATCCAAAC | construction of Citrine |
| T203Y-R | TGACAAATAATGATTGTCTGGTAAAAGAAC | construction of Citrine |
| A206K-F | ACACAATCTAAATTATCAAAAGATCCAAACG | monomerization |
| gapRF | AGAGAGGATCCTTAAATAGTTAGTTG | amplification of gapR promoter |
| gapRGFPR | GAAAAGTTCTTCTCCTTTACTCATTACTACCTCCTCCTTATATTTATA | amplification of gapR promoter fused to GFP |
| gapRGFPF | TATAAATATAAGGAGGAGGTAGTAATGAGTAAAGGAGAAGAACTTTTC | amplification of gfp gene fused to gapR promoter |
| GFPRB | GTCTAGATCTTTATTTGTATAGTTCATC | amplification of gfp gene |
| blaZPF | ACAAAAGCTTACTATGCTCATTATTAA | amplification of blaZ gene promoter |
| blaZPR-Cyan | AACTTGTTTGAAAGAGCCATAAATAATCATCCTCCTATTA | amplification of blaZ gene promoter fused to AmCyan |
| blaZP-CyanF | TAATAGGAGGATGATTATTTATGGCTCTTTCAAACAAGTT | amplification of amCyan gene fused to blaZ promoter |
| blaZPR-mCherry | TCCTCGCCCTTGCTCACCATAAATAATCATCCTTCCTATTA | amplification of blaZ gene promoter fused to mCherry |
| blaZP-mCherryF | TAATAGGAGGATGATTATTTATGGTGAGCAAGGGCGAGGA | amplification of mCherry gene fused to blaZ promoter |
| pUC-RH | AATGGAAGCTTCCGGCGCTCAGTTGG | amplification of amCyan and mCherry |
| Cyan1F | CATATGAAGGTACACAAACATCAACTTTTAAAGTTACAATGGCAAACGGTGGTCCACTTGCATTCTCATT | codon optimization for amCyan |
| Cyan1R | GTTTACCACTACCTTCACCTTTAACTGTAAAATAATGACCGTTAACACAACCATCCATATGATATGT | |
| Cyan2F | ATGCCAGATTATTTTAAACAAGCATTTCCTGATGGTATGTCATATGAACGTACTTTTACA | codon optimization for amCyan |
| Cyan2R | ACTTGTAGGATATGCAGTAAAACAACGATTACCATACATAAAAACTGTTGATAGAATAC | |
| Cyan3F | GAACATAAATCAACATTTCATGGAGTTAACTTTCC | codon optimization for amCyan |
| Cyan3R | AAAACAGTTACCTTTAAGACTTATTTCCCAACTTGC | |
| Cyan4F | CAAGGAGGTGGTAATTATAGATGTCAATTTCATACTTCTTATAAGACA | codon optimization for amCyan |
| Cyan4R | TAACATTAAAAATGCTGTAACATCACCCTTCAATATTCCATCACAAACAGTC | |
| Cyan5F | AAGGTGGTAATAGTGTTCAATTAACAGAACATGCTGTTGCACATATAACATCTGTTGTTCC | codon optimization for amCyan |
| Cyan5R | TATCTAAATCTGTTCTTGCAATACGATGTTCAACTGCATGGTTTGGTGGCATTGTAACTGGTTT | |
| Cherry1F | AGTTAATGGTCATGAATTCGAAATCGAGGGCGAGGGCGAGGGTCGTCCATATGAGGGCACACAAACAGC | codon optimization for mCherry |
| Cherry1R | GAACCTTCCATATGAACTTTAAAACGCATGAACTCCTTGATGATTGCCATGTTATCCTCCTCACCCTTAC | |
| Cherry2F | TATGGTTCAAAAGCATATGTTAAGCATCCAGCAGACATCCCAGACTATTTGAAGTTGTCATTCCCAGAGG | codon optimization for mCherry |
| Cherry2R | CATAAATTGAGGTGATAAGATGTCCCATGCGAATGGCAATGGACCACCCTTTGTCACCTTCAACTTTGC | |
| Cherry3F | TATTTATAAAGTTAAGTTGCGTGGTACAAACTTCCCATCAGACGGCCCAGTAATGCAGAAGAAGACAATG | codon optimization for mCherry |
| Cherry3R | AATTCACCATCTTGCAATGATGAGTCTTGTGTCACTGTAACCACACCGCCGTCCTCGAAGTTCATCACAC |
Table 3.
GFP variants used in this study.
| GFP variant | Mutations relative to wtGFP | Reference |
|---|---|---|
| GFPmut3b | S65G, S72A | 21 |
| GFPsa | S65G, S72A, F99S, M153T, V163A | This study |
| EGFP | F64L, S65T | 4 |
| mEmerald | F64L, S65T, S72A, N149K, M153T, I167T, A206K | 4 |
| Citrine | S65G, S72A, V68L, Q69M, T203Y | 30 |
| Cerulean | F64L, S65T, Y66W, S72A, Y145A, N146I, H148D, M153T, V163A | 31 |
| 1EMF(BFP) | F64L, Y66H, V163A | 32 |
| EBFP | F64L, S65T, Y66H, Y145F | 33 |
| P4-3E(BFP) | F64L, Y66H, Y145F, V163A | 34 |
Construction of codon-optimized RCFP expression vectors
The amCyan and mCherry genes were combined with the hybrid blaZ/sodp(Con) promoter using overlap extension PCR and cloned into the pKAT vector43. Plasmid pKAT derived from pND5044 is an E. coli-S. aureus shuttle vector containing the replication origins of pUB110 (S. aureus) and the pUC19 (E. coli) lacZ(α) gene from pUC19. This enables a simple blue-white screening for clones in E. coli, and enables the cat gene conferring resistance to chloramphenicol in both E. coli and S. aureus. All restriction enzyme sites in the multiple cloning site (MCS) located in the lacZ(α) gene can be used for cloning into pKAT. In the first PCR, the hybrid blaZ/sodp(Con) promoter region, amCyan gene, and mCherry gene were amplified from pFK53, pAmCyan (Clontech, TaKaRa Bio Inc., Japan), and pmCherry (Clontech, TaKaRa Bio Inc., Japan) with the following primer sets (AmCyan: blaZp-F and blaZPR-Cyan and blaZP-CyanF and pUC-RH; mCherry: blaZp-F and blaZPR-Cherry and blaZP-mCherryF and pUC-RH), respectively. The second PCR was performed with the mixture of two PCR fragments as the template using the primer set (blaZp-F and pUC-RH). The resulting PCR products were digested with HindIII and cloned into the same site in pKAT. Codon optimization was then repeatedly performed using inverse PCR as mentioned above with the primers described in Table 2 that were designed to optimize the codon usage of amCyan and mCherry gene using the Kazusa Codon Usage Database (http://www.kazusa.or.jp/codon/). The DNA sequences of codon-optimized amCyan and mCherry genes have been submitted to the GenBank and are available under accession numbers LC088723 (amCyan) and LC88724 (mCherry).
Transformation of S. aureus and quantification of fluorescence intensity
In brief, S. aureus was individually transformed using electroporation as described previously45. Each plasmid was first transformed into S. aureus RN422020 and selected as chloramphenicol-resistant colonies, then the resulting modified plasmids were isolated and electroporated into S. aureus MW2 and N315. S. aureus was grown as described in the growth conditions. Bacterial cells from overnight cultures were washed with phosphate-buffered saline (PBS) and re-suspended to an optical density at 660 nm of 0.2. The cell suspension was then dispensed into triplicate wells (100 µl/well) of a U-bottom 96 well cell culture plate (Greiner Bio-One). The fluorescence intensity was measured using a Varioskan Flash Multimode Reader (ThermoFisher Scientific Inc.) with two independent samples.
Quantification of fluorescent proteins using SDS-PAGE and Western blot analysis
Fluorescent proteins were detected from whole cell lysates. S. aureus was cultured overnight as described in the growth conditions. The pre-cultured cells were adjusted to an optical density at 660 nm of 0.02 with TSB with chloramphenicol, and 3 ml of the culture were transferred into test tube (25 mm × 150 mm) and then incubated at 37 °C with shaking at 140 rpm for 16 h. The bacterial cells from 1 ml cultures were harvested by centrifugation, and then the whole cell lysates were prepared as follows: cells were re-suspended in 200 μl CS buffer (100 mM Tris-HCI, 150 mM NaCl, 100 mM EDTA, pH7.5) containing 1 μg of lysostaphin (Wako Pure Chemical Industries, Co., Ltd, Japan) and incubated at 37 °C for 30 min. Ten microliters of cell lysates were separated on SDS–PAGE and stained with Coomassie Brilliant Blue (CBB), and the fluorescent proteins were detected using Western blot with the following antibodies: Anti-GFP-HRP-Direct T (MBL Co., Ltd.), Living colors® Anti-RCFP Polyclonal Pan Antibody (Clontech Laboratories) and DsRed Polyclonal Antibody (Clontech Laboratories) as the primary antibody, and HRP-conjugated goat antibodies against rabbit IgG (MP Biomedicals, LLC-Cappel Products) as the secondary antibody. Immuno-detection of protein was performed on Pierce® Western Blot Substrate (Thermo Fisher Scientific Inc.) with X-ray film.
Confocal laser scanning microscopic analysis
S. aureus strains and E. coli were cultured in test tube (25 mm × 150 mm) containing 3 ml of Brain Heart Infusion (BHI) broth with chloramphenicol or ampicillin at 37 °C with shaking at 140 rpm overnight. The pre-cultured cells were mixed and diluted 1:000 into 3 ml fresh BHI, and then incubated at 37 °C with shaking at 140 rpm for 6 h. Bacterial cells were harvested by centrifugation, washed twice with PBS buffer, fixed with 3% parafomaldehyde in PBS, and washed twice with PBS. Bacterial cells were counterstained with 0.1 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI) if necessary. The cell suspensions were spotted onto glass slides and air-dried at room temperature. The coverslips were mounted using VECTASHIELD H-1000 (Vector Laboratories, Inc. Burlingame, CA). All confocal images were recorded using an confocal laser scanning microscopy (Olympus, Fluoview FV1000, Japan).
Electronic supplementary material
Acknowledgements
The authors thank Prof. Martin. Krönke (University of Cologne) for the gift of plasmid pS1GFP; Dr. Junzo Hisatsune for his assistance with the confocal laser scanning microscope; and Dr. Wakako Ikeda-Ohtsubo for critical reading of the manuscript. This study was supported by Grant-in-Aid for Young Scientists (B) Grant Number JP25861744 and Grant-in-Aid for Scientific Research (C) Grant Number JP25460533 from the Japan Society for the promotion of Science (JSPS). A confocal laser scanning microscopy was performed at the Analysis Center of Life Science, Natural Science Center for Basic Research and Development, Hiroshima University.
Author Contributions
F.K. conceived and designed the experiments, performed the experiments, and wrote the main manuscript. M.K. constructed the multicolor-GFP variants. M.S. conceived the experiments and critically revised the manuscript. All authors reviewed approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02930-7
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chudakov, D. M., Matz, M. V., Lukyanov, S., Lukyanov, K. A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 90, 1103–1163 Review, 10.1152/physrev.00038.2009 (2010). [DOI] [PubMed]
- 2.Miyawaki, A., Niino, Y. Molecular spies for bioimaging— fluorescent protein-based probes. Mol. Cell. 58, 632–43 Review, 10.1016/j.molcel.2015.03.002 (2015). [DOI] [PubMed]
- 3.Giepmans, B. N., Adams, S. R., Ellisman, M. H., Tsien, R. Y. The fluorescent toolbox for assessing protein location and function. Science. 312, 217–24 Review, 10.1126/science.1124618 (2006). [DOI] [PubMed]
- 4.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Shaner, N. C, Patterson, G. H., Davidson, M. W. Advances in fluorescent protein technology. J Cell Sci. 120, 4247–60 Review, 10.1242/jcs.005801 (2007). [DOI] [PubMed]
- 6.Dove SG, Hoegh-Guldberg O, Ranganathan S. Major colour patterns of reef-building corals are due to a family of GFP-like proteins. Coral Reefs. 2001;19:197–204. doi: 10.1007/PL00006956. [DOI] [Google Scholar]
- 7.Labas YA, et al. Diversity and evolution of the green fluorescent protein family. Proc Natl Acad Sci. USA. 2002;99:4256–4261. doi: 10.1073/pnas.062552299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heim R, Prasher D, Tsien R. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–9. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 10.Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79:2218–24. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prax, M., Lee, C. Y., Bertram, R. An update on the molecular genetics toolbox for staphylococci. Microbiology. 159, 421–35 Review, 10.1099/mic.0.061705-0 (2013). [DOI] [PMC free article] [PubMed]
- 12.Brzoska AJ, Firth N. Two-Plasmid Vector System for Independently Controlled Expression of Green and Red Fluorescent Fusion Proteins in Staphylococcus aureus. Appl. Environ. Microbiol. 2013;79:3133–6. doi: 10.1128/AEM.00144-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malone CL, et al. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira PM, Veiga H, Jorge AM, Pinho MG. Fluorescent reporters for studies of cellular localization of proteins in Staphylococcus aureus. Appl Environ Microbiol. 2011;76:4346–53. doi: 10.1128/AEM.00359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paprotka K, Giese B, Fraunholz MJ. Codon-improved fluorescent proteins in investigation of Staphylococcus aureus host pathogen interactions. J Microbiol Methods. 2010;83:82–6. doi: 10.1016/j.mimet.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Yu W, Götz F. Cell wall antibiotics provoke accumulation of anchored mCherry in the cross wall of Staphylococcus aureus. PLoS One. 2012;7:e30076. doi: 10.1371/journal.pone.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannig G, Makrides SC. Strategies for the production of recombinant protein in Escherichia coli. Protein J. 2013;32:419–25. doi: 10.1007/s10930-013-9502-5. [DOI] [PubMed] [Google Scholar]
- 18.Charpentier E, et al. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnaith A, et al. Staphylococcus aureus Subvert Autophagy for Induction of Caspase-independent Host Cell Death. J. Biol. Chem. 2007;282:2695–706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- 20.Kreiswirth BN, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 21.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39:12025–32. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 23.Sidhu MS, et al. Frequency of disinfectant resistance genes and genetic linkage with beta-lactamase transposon Tn552 among clinical staphylococci. Antimicrob Agents Chemother. 2002;46:2797–803. doi: 10.1128/AAC.46.9.2797-2803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franklin DL. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 2003;111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda M, et al. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 26.Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–27. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 27.Nair D, et al. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J Bacteriol. 2011;193:2332–5. doi: 10.1128/JB.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Castellanos R, et al. On the transcriptional regulation of methicillin resistance: MecI repressor in complex with its operator. J Biol Chem. 2004;279:17888–96. doi: 10.1074/jbc.M313123200. [DOI] [PubMed] [Google Scholar]
- 29.Richard ND, Michael WD. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griesbeck O, et al. Reducing the Environmental Sensitivity of Yellow Fluorescent Protein. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 31.Markwardt ML, et al. An Improved Cerulean Fluorescent Protein with Enhanced Brightness and Reduced Reversible Photoswitching. PLoS ONE. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palm GJ, et al. The structural basis for spectral variations in green fluorescent protein. Nat Struct Biol. 1997;4:361–5. doi: 10.1038/nsb0597-361. [DOI] [PubMed] [Google Scholar]
- 33.Yang TT, et al. Improved fluorescence and dual color detection with enhanced blue and green variants of the green fluorescent protein. J. Biol. Chem. 1998;273:8212–8216. doi: 10.1074/jbc.273.14.8212. [DOI] [PubMed] [Google Scholar]
- 34.Heim R, Tsien R. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996;6:178–182. doi: 10.1016/S0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 35.Kato F, et al. Regulatory mechanism for exfoliative toxin production in Staphylococcus aureus. Infect Immun. 2011;79:1660–70. doi: 10.1128/IAI.00872-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–9. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 38.Amaral FE, et al. Rational manipulation of mRNA folding free energy allows rheostat control of pneumolysin production by Streptococcus pneumoniae. PLoS One. 2015;10:e0119823. doi: 10.1371/journal.pone.0119823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fux CA, Shirtliff M, Stoodley P, Costerton JW. Can laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol. 2005;13:58–63. doi: 10.1016/j.tim.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Kirby WMM. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science. 1944;99:452–3. doi: 10.1126/science.99.2579.452. [DOI] [PubMed] [Google Scholar]
- 41.Hisatsune J, et al. Emergence of Staphylococcus aureus carrying multiple drug resistance genes on a plasmid encoding exfoliative toxin B. Antimicrob Agents Chemother. 2013;57:6131–40. doi: 10.1128/AAC.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boël G, et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016;529:358–63. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato, F. Ph.D. Thesis, University of Tohoku, Sendai, Japan. NDLC:UT51-2004-P732 (2004).
- 44.Yamagishi J, et al. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1996;40:1157–1163. doi: 10.1128/aac.40.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Löfblom J, et al. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J Appl Microbiol. 2007;102:736–47. doi: 10.1111/j.1365-2672.2006.03127.x. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


