Abstract
The role of multiple gonadotropin-releasing hormone receptor (GnRH-R) types in the regulation of gonadotropic and nongonadotropic cells remains speculative. To address this issue, we developed a technology integrating laser-captured microdissection of single digoxigenin-labeled pituitary cells coupled with real-time quantitative PCR to examine the expression profiles of three endogenous GnRH-R types (R1, R2, and R3) in immature and mature males of tilapia Oreochromis niloticus. Here, in addition to gonadotropes (luteinizing and folicle-stimulating hormone, FSH), we show GnRH-Rs are also present in lactotropes, somatotropes, thyrotropes, melanotropes (melanocyte-stimulating hormone, MSH), corticotropes and somatolactin cells. Subpopulations of pituitary cells express single (42.9%), multiple (32.4%) or lack (24.7%) GnRH-Rs. In immature males, the percentage of FSH cells containing combinations of GnRH-Rs was significantly higher (R1+R2: 24%, P < 0.05; R1+R2+R3: 25%, P < 0.01) than in mature males, whereas the percentage showing only R1 and R1 and R3 transcripts (P < 0.05) was higher in mature males. Significantly greater copies of R1 and R3 transcripts were found in MSH cells of immature and mature males, respectively (P < 0.05). GnRH-R transcripts in other pituitary cells (lactotropes, R1 and R2; somatolactin cells/thyrotropes/corticotropes, R1, R2, and R3) were significantly higher in mature males (P < 0.05) but were unaltered in somatotropes and luteinizing hormone cells. Thus, FSH and MSH cells are required for both reproductive states, whereas other pituitary cells are recruited only during testicular maturation. The differential expression of GnRH-Rs in gonadotropic and nongonadotropic cells demonstrates cellular and functional heterogeneity of mechanisms controlling normal sexual development.
Keywords: G protein, in situ hybridization, tilapia
Gonadotropin-releasing hormone (GnRH) is now recognized as a family of 16 multifunctional neuropeptides in vertebrates (1–3). It is well documented that all vertebrate species ranging from fish to humans possess two (hypothalamus, GnRH1; midbrain, GnRH2) or, as in recently derived teleosts, three GnRH types (caudal olfactory bulbs, GnRH3) (1–3). GnRH1, GnRH2, and GnRH3, in addition to stimulating gonadotropes (follicle-stimulating hormone, FSH; luteinizing hormone, LH), are potent regulators of somatotropes [growth hormone (GH) cells], lactotropes [prolactin (PRL) cells], and somatolactin (SL)-containing cells in teleosts (4–9). Because GnRH exerts its actions through binding to GnRH receptors (GnRH-Rs) (10), it is, therefore, conceivable that the three GnRH types have their respective cognate receptors expressed in different pituitary cells. Multiple GnRH-Rs have been cloned (10) and their transcripts identified in LH and GH cells (11), and GnRH-R proteins in LH, GH, and PRL cells (4) of teleosts. However, the distribution of GnRH-R transcripts in other endocrine cell types of the adenohypophysis has never been evaluated. Besides, it is unknown whether transcripts of multiple GnRH-R types are coexpressed in individual pituitary cells. Therefore, there is the need to precisely identify individual cells in the pituitary that express GnRH-Rs to formulate possible roles of GnRHs and their cognate receptors.
We have successfully cloned three GnRH-R types from the brain and pituitary of the tilapia Oreochromis niloticus [GenBank accession nos.: AB111356 (GnRHR1), AB111357 (GnRHR2), and AB158490 (GnRHR3); unpublished data]. Furthermore, we developed a technology integrating laser-captured microdissection (LCM) of single digoxigenin (DIG)-labeled pituitary cells coupled with real-time quantitative RT-PCR (RT-Q-RT-PCR) that allows harvesting identified individual pituitary cells with precision and high preservation of mRNA for analysis. Thus, to increase understanding of the role of GnRH types on the pituitary–gonadal axis, the present study was designed to analyze the effects of sexual maturity on the expression profiles and the functional states of the three GnRH-R types in single gonadotropic and nongonadotropic cells. For this purpose, we used immature and mature males of tilapia because sexual maturity in males is marked by the development of pinkish-red coloration, initiation of nest-building behavior, and increase aggressiveness, and there is evidence in males that shows GnRH1 is important for sexual maturation, whereas GnRH2 and GnRH3 might have roles in reproductive behaviors or nonreproductive functions (12).
Materials and Methods
Experimental procedures in the present study were performed under the guidelines of the Animal Care Committee of Nippon Medical School, Tokyo. Male tilapia O. niloticus, maintained in fresh water at 27 ± 1°C with a natural photo regime (10/14-h light/dark cycle), were used in the present study.
Tissue Preparation and in Situ Hybridization for Pituitary Hormones. Immature [standard length, 5.25 ± 0.24 cm; body weight, 4.77 ± 0.51 g; gonadosomatic index (GSI), 0.032 ± 0.018; n = 5] and mature (standard length, 11.13 ± 0.68 cm; body weight, 45.73 ± 7.02 g; GSI, 1.28 ± 0.39; n = 5) males were anesthetized by immersing in a 0.01% solution of 3-aminobenzonic acid ethyl ester (MS222; Sigma) before they were killed by decapitation. The brains with pituitaries attached were dissected and fixed in 4% buffered paraformaldehyde for 6 h at room temperature, cryoprotected in 20% sucrose, and embedded in Tissue Tek OCT compound (Sakura Finetechnical, Tokyo). Pituitary sections were cut in sagittal planes (6 μm) and mounted onto aminopropyl triethoxy silane-treated slide glass (Matsunami Glass, Tokyo) and stored at -80°C until use.
For DIG in situ hybridization, we cloned and identified partial sequences of SL (GenBank accession no. AB120767; unpublished data) and thyroid-stimulating hormone β subunit (TSHβ, GenBank accession no. AB120769; unpublished data) whereas the sequences of other tilapia pituitary hormones were obtained from GenBank [accession nos.: M27010 (PRL188), M97766 (GH1), AF289173 (FSHβ subunit), AY294016 (LHβ subunit), and AF116240 (proopiomelanocortin, POMC; adenocorticotropin, ACTH; and melanocyte-stimulating hormone, MSH)]. Because GH1 and GH2 encode an identical polypeptide (13) and PRL188 and PRL177 are colocalized in the same pituitary cells in the tilapia (14), we used the sequences of GH1 and PRL188 to synthesize riboprobes for in situ hybridization. Sense and anti-sense riboprobes were synthesized by using the pGEM-T easy transcription vector constructs (Promega), linearized with SpeI or NcoI endonuclease (Nippon Gene) as a template for T7 or SP6 RNA polymerase (Toyobo, Tokyo). The RNA probes were labeled by using DIG RNA-labeling mix (Roche Diagnostics). DIG in situ hybridization was carried out as described (4).
LCM of Pituitary Cells. The hydrated pituitary section was overlaid with a thermoplastic membrane mounted on an optically transparent cap (CapSure Macro LCM Caps, Arcturus, Mountain View, CA). We used a Pix Cell II Laser capture instruments (Arcturus), to microdissect DIG-identified pituitary cells by focal melting of the membrane through laser activation (laser pulse power, 25–65 mW; laser pulse duration, 1.5 ms; laser spot size, 10-μm diameter). Heat-pulled borosilicate glass microcapillary pipette (1.5-mm outer diameter, Harvard Apparatus, Edenbridge, Kent, U.K.; micropipet puller, Type PE-2, Narishige, Tokyo) attached to a micromanipulator (Narishige) was used to remove undesirable tissue around the periphery of the single cells. Then, by using a negative pressure, single-cells were harvested from the LCM cap into the micropipette under visual control and subsequently expelled into a sterile 1.5-ml reaction tube containing 50 μl of the lysis buffer and stored at -80°C until total RNA isolation. For unbiased cell sampling, 8–10 cells were harvested at random (≈2 cells per alternate section) along the rostral-caudal extent of the whole population of each pituitary cell type in each animal (n = 5 per age group). Only those cells positive for each pituitary hormone and free from genomic contamination were used for RT-Q-RT-PCR analysis (n = 5–9 cells per animal; 23–44 cells per pituitary cell type per age group).
GnRH-R Types in Pituitary Cells. The cell harvesting protocol and the conditions for RT-PCR were similar to those described elsewhere (12, 15). Briefly, the harvested single-cell from the pituitary was digested with 1 μg of proteinase K (Gentra Systems, Minneapolis, MN) and 10 units of ribonuclease inhibitor (Eppendorf, Hamburg, Germany) for 1 h at 53°C. The cell lysate was incubated for 1 h at 37°C with 1 unit of ribonuclease-free DNase I (Promega) to eliminate genomic DNA and heat denatured at 95°C for 10 min to separate the mRNA from the DIG-labeled riboprobe. Total RNA was extracted from the cell lysate by using ISOGEN (Nippon Gene) and reverse transcribed to cDNA with 0.1 pmol of random primers (TaKaRa) by using 40 units of SuperScript III reverse transcriptase (Invitrogen).
To confirm the presence of GnRH-Rs and pituitary hormone transcripts, the single cell's cDNA was subjected to RT-PCR using gene-specific primers for GnRH-Rs and pituitary hormones [GenBank accession nos.: AB120767 (SL), AB120769 (TSHβ), M27010 (PRL188), M27011 (PRL177), M97766 (GH1), M97765 (GH2), AF289173 (FSHβ), AY294016 (LHβ), and AF116240 (POMC); Table 2, which is published as supporting information on the PNAS web site] and 1/20th of a single cell's RT cDNA solution. To confirm the sequences, some bands were subcloned and both strands of the DNA were sequenced as described above. Several controls were included for the RT-PCR: buffer without harvested cells and no reverse transcriptase.
Quantitative Analysis of GnRH-Rs in Pituitary Cells. To quantify copies of GnRH-R transcripts in pituitary cells, cDNAs from single cells were subjected to RT-Q-RT-PCR, which was performed in 10-μl reaction volumes consisting of 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 300 nM GnRH-R primers, and 200 nM GnRH-R hybridization probes (GR7 through GR15; Table 2), and 1/20th of a single cell's RT cDNA or absolute standard cDNA by using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The PCR conditions were as described (12, 15). The GnRH-Rs hybridization probes spanned an intron and complemented the sequence on either side of the splice site of the gene. For each animal and pituitary cell type, average copies of transcripts per cell were determined, and these values were combined to give experimental group means. All values are expressed as the mean ± SEM, and statistical comparisons were made between immature and mature males (n = 5 per group) by using nonparametric ANOVA followed by Fisher's probable least-squares difference test. P < 0.01 or P < 0.05 were considered statistically significant.
Results
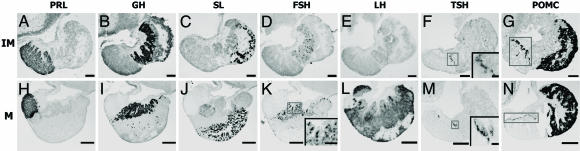
DIG in Situ Hybridization for Pituitary Hormones. In immature and mature males, DIG in situ hybridization for pituitary hormones (Fig. 1) showed cells expressing PRL188 mRNA localized in the rostral pars distalis (RPD, Fig. 1 A and H). GH cells were seen in the dorsal and LH cells in the ventral proximal pars distalis of mature males (PPD; Fig. 1 B, I, and L). The expression of LHβ transcripts was undetectable by in situ hybridization in immature males (Fig. 1E). FSH cells were scattered among GH cell population in the PPD (Fig. 1 D and K). TSH cells were located at the boundary between PPD and the RPD (Fig. 1 F and M). SL cells were located along the edge of the neurohypophysis in the pars intermedia (PI; Fig. 1 C and J). The POMC probe hybridized with both ACTH and MSH cells. ACTH cells were located in the dorsal periphery of the RPD in contact with the neurohypophysis and MSH cells were located in the PI (Fig. 1 G and N).
Fig. 1.
Photomicrographs of DIG-labeled endocrine cells of the pituitary. Immature (IM) (A–G) and mature (M) (H–N) males are shown. (A and H) PRL cells. (B and I) GH cells. (C and J) SL cells. (D and K) FSH cells. (E and L) LH cells. (F and M) TSH cells. (G and N) POMC (MSH, ACTH); ACTH cells are shown in rectangles. (Scale bars: 70 μm in A–G; 200 μm in H–N; 20 μm in F and M Insets; and 70 μm in K Inset.) Cells shown in F, K, and M Insets are from within the rectangles.
GnRH-R Types in Pituitary Cells. There was no genomic DNA contamination in the harvested single-cells of the pituitary (Figs. 2 and 3). RT-PCR showed that 70–85% of DIG-labeled pituitary cells had pituitary hormone transcripts (Fig. 3). The amplicon sizes of PRL188, PRL177, GH1, GH2, SL, FSHβ, LHβ, TSHβ, and POMC and their sequences were identical with tilapia pituitary hormones (Fig. 3; see Materials and Methods for GenBank accession numbers). Because PRL188 and PRL177 and GH1 and GH2 transcripts were colocalized (Fig. 3), these are described as single PRL and GH molecules throughout the text. Nested PCR was necessary to observe R1, R2, and R3 in pituitary cells. The amplicon sizes of R1 (371 bp), R2 (322 bp), and R3 (277 bp) and their sequences were identical with tilapia GnRH-Rs (see Materials and Methods for GenBank accession numbers). RT-PCR revealed that 40–60% of all pituitary cell types had GnRH-Rs (Fig. 3).
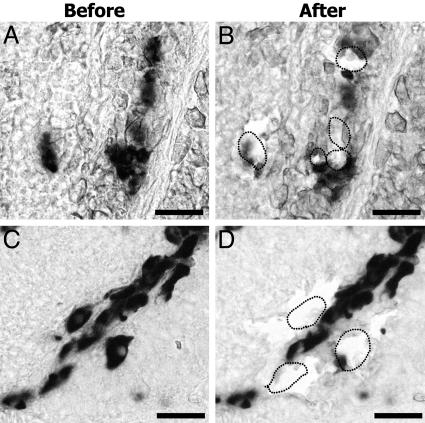
Fig. 2.
DIG-labeled FSH (A and B) and GH (C and D) cells before and after LCM. (Scale bar, 20 μm.)
Fig. 3.
Composite gel showing expression of amplicons of pituitary hormones and GnRH-Rs (R1, R2, and R3) in representative PRL cells (lanes 1–3), GH cells (lanes 4–6), SL cells (lanes 7–9), FSH cells (lanes 10–12), LH cells (lanes 13–15), TSH cells (lanes 16–18), MSH cells (lanes 19–21), and ACTH cells (lanes 22–24) taken from mature males. BF, buffer control; RT-, without reverse transcriptase; PC, whole pituitary cDNA as positive control for PCR; M, marker, DNA 100-bp size ladder. Note that the expressions of PRL188 and PRL177 and GH1 and GH2 transcripts are colocalized in PRL and GH cells, respectively. The POMC primer recognizes both MSH and ACTH transcripts. The sizes of the bands, in base pairs, are given in the right margin.
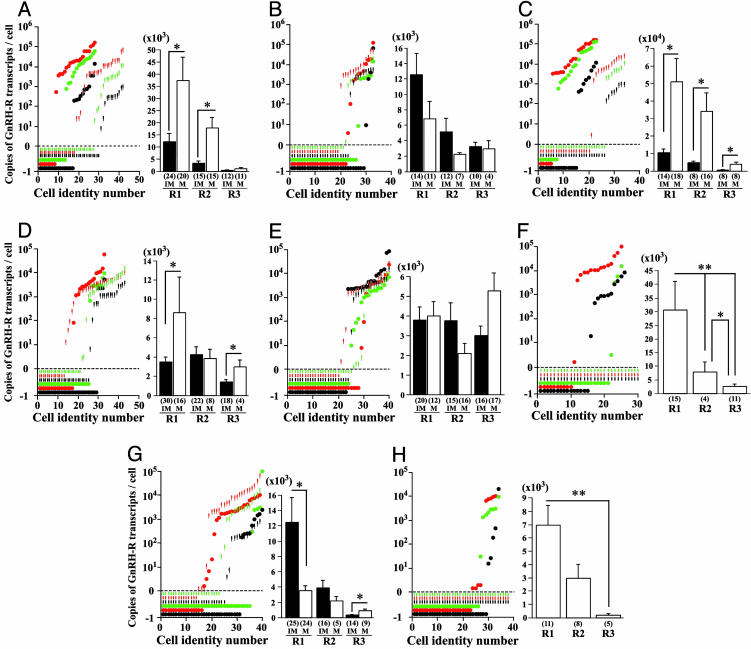
Quantitative Analysis of GnRH-Rs in Pituitary Cells. RT-Q-RT-PCR showed that 58–88% of pituitary cells in mature males expressed single or multiple GnRH-R transcripts (Table 1). In addition, a large variation in copies of R1, R2, and R3 transcripts were observed between individual cells (Fig. 4 A–H Left). R1 and R2 transcripts were predominant in the two reproductive states (Fig. 4 A–H Right). Absolute copies of GnRH-R transcripts in TSH and ACTH cells were below detectable levels in immature males (Fig. 4 F and H).
Table 1. Percentage of pituitary cells with single or multiple GnRH-R transcripts.
| PRL | GH | SL | FSH | LH | TSH | MSH | ACTH | ||
|---|---|---|---|---|---|---|---|---|---|
| R1 | IM | 20.8 ± 4.1 | 11.4 ± 7.0 | 20.5 ± 7.2 | 16.1 ± 8.4** | 20.0 ± 5.0 | 0.0 | 23.2 ± 6.2 | 0.0 |
| M | 18.7 ± 6.4 | 23.8 ± 7.5 | 9.0 ± 5.6 | 38.1 ± 10.5 | 12.5 ± 4.0 | 46.0 ± 11.7 | 35.0 ± 4.7 | 31.9 ± 8.1 | |
| R2 | IM | 13.9 ± 4.5 | 9.1 ± 3.7 | 6.2 ± 3.8 | 2.2 ± 2.2 | 7.5 ± 5.0 | 0.0 | 2.9 ± 2.9 | 0.0 |
| M | 6.7 ± 4.1 | 18.1 ± 7.4 | 0.0 | 15.7 ± 7.0 | 15.0 ± 4.7 | 0.0 | 2.5 ± 2.5 | 9.0 ± 5.9 | |
| R3 | IM | 2.2 ± 2.2 | 6.7 ± 6.7 | 8.6 ± 5.7 | 13.6 ± 5.4 | 12.5 ± 4.0 | 0.0 | 10.7 ± 5.3 | 0.0 |
| M | 3.3 ± 3.3 | 6.7 ± 4.1 | 5.0 ± 5.0 | 9.0 ± 3.7 | 12.5 ± 0.0 | 3.3 ± 3.3 | 10.0 ± 4.7 | 11.4 ± 5.3 | |
| R1+R2 | IM | 11.9 ± 4.0 | 12.9 ± 6.2 | 11.4 ± 11.4** | 24.7 ± 9.5* | 12.5 ± 4.0 | 0.0 | 22.5 ± 6.1 | 0.0 |
| M | 24.0 ± 8.4 | 2.9 ± 2.9 | 42.0 ± 10.1 | 6.2 ± 3.8 | 2.5 ± 2.5 | 0.0 | 12.5 ± 5.6 | 11.9 ± 8.4 | |
| R1+R3 | IM | 8.9 ± 4.2 | 8.6 ± 3.5 | 6.2 ± 3.8 | 2.5 ± 2.5 | 10.0 ± 2.5 | 0.0 | 10.0 ± 2.5 | 0.0 |
| M | 3.3 ± 3.3 | 3.3 ± 3.3 | 5.0 ± 5.0 | 0.0 | 5.0 ± 5.0 | 19.3 ± 9.0 | 12.5 ± 5.6 | 0.0 | |
| R2+R3 | IM | 2.2 ± 2.2 | 6.2 ± 3.8 | 2.9 ± 2.9 | 2.2 ± 2.2 | 10.0 ± 7.3 | 0.0 | 2.5 ± 2.5 | 0.0 |
| M | 3.3 ± 3.3 | 0.0 | 5.0 ± 5.0 | 0.0 | 15.0 ± 9.2 | 0.0 | 0.0 | 2.9 ± 2.9 | |
| R1+R2+R3 | IM | 16.9 ± 7.4 | 9.1 ± 3.7 | 6.2 ± 3.8 | 25.3 ± 4.7** | 7.5 ± 5.0 | 0.0 | 12.9 ± 4.0 | 0.0 |
| M | 24.7 ± 12.8 | 3.3 ± 3.3 | 22.0 ± 10.2 | 3.3 ± 3.3 | 10.0 ± 2.5 | 19.3 ± 6.4 | 0.0 | 0.0 | |
| R-positive | IM | 77.0 ± 6.5 | 63.8 ± 7.5 | 61.9 ± 5.4 | 88.9 ± 6.1 | 80.0 ± 3.1 | 0.0 | 84.6 ± 4.6 | 0.0 |
| M | 84.0 ± 11.7 | 58.1 ± 11.9 | 88.0 ± 8.0 | 72.4 ± 6.3 | 72.5 ± 7.3 | 88.0 ± 8.0 | 72.5 ± 7.3 | 67.1 ± 7.7 | |
| R-negative
|
IM | 23.0 ± 6.5 | 36.2 ± 7.5 | 38.1 ± 5.4 | 11.1 ± 6.1* | 20.0 ± 3.1 | 0.0 | 15.4 ± 4.6 | 0.0 |
| M | 16.0 ± 11.7 | 41.9 ± 11.9 | 12.0 ± 8.0 | 27.6 ± 6.3 | 27.5 ± 7.3 | 12.0 ± 8.0 | 27.5 ± 7.3 | 32.9 ± 7.7 |
R-positive and R-negative represent the total percentage of cells with and without GnRH-Rs, respectively. Statistical analysis on an animal basis (n = 5 per age group) show significant differences in the percentage of SL and FSH cells expressing GnRH-Rs in immature and mature males (bolded values). *, P<0.05; **, P<0.01, Fisher's probable least-squares difference test.
Fig. 4.
Distribution of GnRH-R transcripts in individual pituitary cells. (Left) Graphs showing the distribution of R1 (red), R2 (green), and R3 (black) transcripts in individual cells expressing PRL (immature, 28 cells; mature, 43 cells) (A), GH (immature 33 cells; mature, 33 cells) (B), SL (immature 34 cells; mature, 23 cells) (C), FSH (immature, 44 cells; mature, 33 cells) (D), LH (immature, 40 cells; mature, 40 cells) (E), TSH (immature, 30 cells; mature, 26 cells) (F), MSH (immature, 39 cells; mature, 40 cells) (G), and ACTH (immature, 38 cells; mature, 34 cells) (H) taken from immature (short bars) and mature (circles) males. The x axis represents cell identity numbers and the y axis represents copies of GnRH-R transcripts per cell. The short bars and circles below zero are undetectable levels of GnRH-Rs in positively identified pituitary cells. (Right) Histograms showing the average copies of R1, R2, and R3 transcripts per cell deduced from the total number of cells expressing PRL (A), GH (B), SL (C), FSH (D), LH (E), TSH (F), MSH (G), and ACTH (H) taken from immature (IM, filled bars) and mature males (M, open bars). Statistical comparisons are per cell basis using nonparametric ANOVA followed by Fisher's probable least-squares difference test. *, P < 0.05; **, P < 0.01. Cell numbers are given in parentheses.
GH Family (PRL/SL/GH). PRL cells. The total percentage of PRL cells in immature (77%, 33 of 43 cells) and mature (84%, 24 of 28 cells) males expressing single or multiple GnRH-Rs were statistically nonsignificant between the two reproductive states (n = 5 per age group; Table 1).
The absolute copies of R1 (mature, 37,478.3 ± 9,965.8 vs. immature, 12,201.3 ± 3,299.9 copies per cell) and R2 transcripts (mature, 17,779.2 ± 4,436.2 vs. immature, 3,275.0 ± 882.0 copies per cell, P < 0.05) were significantly higher in PRL cells in mature males (Fig. 4A).
GH cells. The total percentage of GH cells in immature (64%, 21 of 33 cells) and mature males (58%, 19 of 33 cells) expressing single or multiple GnRH-Rs were statistically nonsignificant between the two reproductive states (n = 5 per age group; Table 1). The R2+R3 combination was absent in GH cells in mature males (Table 1).
Absolute copies of R1, R2 and R3 transcripts in GH cells were nonsignificant between the two reproductive states (Fig. 4B). SL cells. The total percentage of SL cells in immature (62%, 21 of 34 cells) and mature males (88%, 20 of 23 cells) expressing single or multiple GnRH-Rs in the two reproductive states were statistically nonsignificant (n = 5 per age group; Table 1). A significantly higher percentage of SL cells in mature males had R1+R2 (42.0 ± 10.1% vs. 11.4 ± 11.4%; P < 0.01, n = 5 per age group) but lacked R2 subtype (Table 1).
Absolute copies of R1 (mature, 51,048.1 ± 13,240.3 vs. immature, 10,469.0 ± 2,148.7 copies per cell), R2 (mature, 34,210.6 ± 10,625.0 vs. immature, 4,739.8 ± 1,007.4 copies per cell) and R3 transcripts (mature, 3,657.5 ± 1,403.0 vs. immature, 524.1 ± 112.1 copies per cell, P < 0.05) were significantly higher in SL cells in mature males (Fig. 4C).
Glycoproteins (FSH/LH/TSH). FSH cells. The total percentage of FSH cells expressing single or multiple GnRH-Rs were statistically nonsignificant in immature (89%, 39 of 44 cells) and mature (72%, 24 of 33 cells; Table 1) males. A significantly higher percentage of FSH cells in mature males had R1 (38.1 ± 10.5% vs. 16.1 ± 8.4%; P < 0.01; n = 5 per age group). A significantly higher percentage of FSH cells in immature males had R1+R2 (24.7 ± 9.5% vs. 6.2 ± 3.8%, P < 0.05) and R1+R2+R3 transcripts (25.3 ± 4.7% vs. 3.3 ± 3.3%, P < 0.01; n = 5 per age group) (Table 1).
Absolute copies of R1 (mature, 8,561.5 ± 3,600.2 vs. immature, 3,476.5 ± 472.7 copies per cell) and R3 (mature, 2,965.8 ± 707.6 vs. immature, 1,411.8 ± 221.9 copies per cell, P < 0.05) were significantly higher in FSH cells in mature males (Fig. 4D). LH cells. The total percentage of LH cells expressing single or multiple GnRH-Rs were statistically nonsignificant between immature (80%, 32 of 40 cells) and mature males (73%, 29 of 40 cells; Table 1).
Absolute copies of single and multiple transcripts of GnRH-Rs were statistically nonsignificant between the two reproductive states (Fig. 4E).
TSH cells. TSH cells were devoid of GnRH-R transcripts in immature (0%, 0 of 30 cells) but not in mature (89%, 23 of 26 cells) males (Table 1). R1 type was dominant in TSH cells in mature males (R1, 46.0 ± 11.7% vs. R3, 3.3 ± 3.3%). However, mature males lacked TSH cells with R2, R1+R2 and R2+R3 transcripts (Table 1).
Absolute copies of R1 (mature, 30,687.9 ± 10,408.4 copies per cell) were significantly higher than R2 (mature, 7,930.1 ± 3,663.5 copies per cell) and R3 (mature, 2,568.3 ± 850.8 copies per cell; P < 0.05) transcripts in TSH cells in mature males (Fig. 4F).
POMC Family (MSH/ACTH). MSH cells. The total percentage of MSH cells in immature (85%, 33 of 39 cells) and mature (73%, 29 of 40) males expressing single or multiple transcripts of GnRH-Rs were statistically nonsignificant (Table 1). MSH cells lacked R2+R3 and R1+R2+R3 combination in mature males (Table 1).
Immature males had significantly higher absolute copies of R1 transcripts (immature, 12,394.8 ± 3,152.6 vs. mature, 3,477.7 ± 663.4 copies per cell, P < 0.01) but lower copies of R3 transcripts (immature, 320.7 ± 62.3 vs. mature, 863.9 ± 259.1 copies per cell, P < 0.05) compared to mature males (Fig. 4G).
ACTH cells. ACTH cells were devoid of GnRH-R transcripts in immature (0%, 0 of 38 cells) but not in mature (67%, 23 of 34 cells) males (Table 1). ACTH cells in mature males lacked R1+R3 and R1+R2+R3 combinations (Table 1).
Absolute copies of R1 were significantly higher than R3 transcripts (mature, 6,961.3 ± 1,440.3 vs. immature, 177.5 ± 108.0 copies per cell, P < 0.01) in ACTH cells in mature males (Fig. 4H).
Discussion
Localization. Localization using DIG in situ hybridization showed pituitary cells in the immature and mature male tilapia segregated into three distinct zones: rostral pars distalis (PRL and ACTH cells), proximal pars distalis (GH, LH, FSH, and TSH cells), and pars intermedia (SL and MSH cells). This perfect degree of correspondence between the distributions of transcripts and proteins confirms previous studies in tilapia (4, 16).
We have cloned and obtained full-length sequences for three GnRH-Rs in the tilapia O. niloticus, designated here as tilapia R1, R2, and R3 (unpublished data). We used single-cell RT-PCR to show that, in addition to FSH and LH cells (4), multiple GnRH-Rs are also present in PRL-, GH-, SL-, TSH-, ACTH-, and MSH-producing cells. We are aware that false positives and false negatives may occur from failure of cell harvesting and/or RT-PCR procedures. Therefore, the control measures that we undertook included the use of negative and positive PCR controls (see Materials and Methods). No products were detected in the buffer without harvested cells or without reverse transcriptase, and there was no genomic DNA contamination in the harvested single cells. Furthermore, the amplicon sizes and their sequences were identical with tilapia GnRH-Rs. Together, these results demonstrate the specificity of this procedure. Thus, the amplicons are authentic, and we are confident that the present results provide well controlled evidence for the presence of three GnRH-R transcripts in native pituitary cells.
Cellular and Functional Heterogeneity of GnRH-Rs. The present study provides evidence that subpopulations of pituitary cells express single (42.9%), multiple (32.4%), or lack (24.7%) GnRH-R transcripts and that the frequency with which we detected GnRH-R transcripts varied significantly across the two male reproductive states.
GH Family. Members of the GH family (PRL/SL/GH) have been grouped together because of their structural similarities (17). Our demonstration of multiple GnRH-R transcripts in PRL/SL/GH cells and the presence of GnRH-R protein 1B in PRL cells and R-III in GH cells supports the role of GnRH in the regulation of the GH family (4, 8, 11). In addition, several lines of evidence suggest that GnRH is a secretagogue of PRL/SL/ GH. First, GnRH binding sites have been detected in PRL/SL/GH cells in teleosts (18, 19). Furthermore, GnRH stimulates PRL (6), SL (7), and GH release (8, 20) in teleosts.
In teleosts, PRL is primarily an osmoregulatory hormone (21) and SL is an environmental adaptation hormone (22). However, there is substantial evidence supporting the role of PRL and SL in steroidogenesis and gonadal maturation (23–25). Furthermore, PRL-binding sites and PRL receptors have been detected in the testis of tilapia Oreochromis mossambicus (26). Here, the significant increase in copies of R1 and R2 transcripts in PRL and SL cells in mature males may be involved in the mechanism of activation of PRL and SL release during gonadal maturation, which is supported by activation of PRL and SL genes (25). The absence of any difference in GnRH-R transcripts in GH cells between the two reproductive states suggests a role for GH during early and late testicular development. In contrast, in females, greater levels of GH receptor mRNA have been reported in immature oocytes in the tilapia (27) and in salmonids (28, 29). Whether these differences between males and females are also paralleled by sexually dimorphic GH secretion during development, as in mammals (30), which could initiate the difference in the timing of sex differentiation between tilapia females (28 days) and males (50 days) (see ref. 16) remains to be investigated.
Glycoprotein Family. Members of the glycoprotein family include FSH, LH, and TSH. To be active, FSHβ, LHβ, and TSHβ need to heterodimerize with the α-glycoprotein subunit (31). GnRH-R proteins (IA and IB) have been reported only in LH cells because of the difficulty to distinguish them from FSH cells immunocytochemically (due to lack of specific antibodies) (4). The present study demonstrates multiple GnRH-Rs distinctly in FSH and LH cells, which suggests that the synthesis and release of FSH and LH hormones can be regulated by multiple GnRH ligands. The expression of multiple GnRH-Rs in TSH cells has not been described previously.
The percentage of FSH cells with R1+R2 and R1+R2+R3 combination was significantly higher in immature males, a reproductive stage during which FSH hormone predominates (32). These results support the hypothesis that, in immature males, multiple GnRH-R combinations modulate FSH for spermatogenesis and gonadal steroidogenesis. Furthermore, in mature males, significantly high percentage of FSH cells with R1 and greater GnRH-R transcripts (R1, R3) suggest that FSH hormone levels remain elevated in mature males to maintain spermatogenesis (33).
The percentage of LH cells with GnRH-Rs and the copies of GnRH-R transcripts in LH cells were similar in the two reproductive states. This finding suggests that both LH and FSH hormones could have physiological roles in spermatogenesis and spermiation in male tilapia (34), in contrast to salmonids, in which FSH hormone is important for early gametogenesis and LH is important only for gonadal maturation (32, 33).
Although there is no evidence for GnRH in the regulation of TSH cells, the localization of thyroid hormone receptors in GnRH neurons (35) and the regulation of GnRH gene expression by thyroid hormones (36) demonstrate a relationship between GnRH and TSH cells. In immature males, the differentiation of TSH cells in the absence of GnRH-Rs suggests that GnRH might be important for release rather than synthesis of TSH hormone. On the other hand, in mature males, the activation of GnRH-Rs in TSH cells could have some role in gonadal maturation because thyroid hormones and TSH cells have been implicated in reproductive processes (37).
POMC Family. Our demonstration of multiple GnRH-R transcripts in ACTH and MSH cells suggests the role of GnRH in the regulation of the POMC-derived family of peptides from ACTH cells (16K fragment, ACTH, γ-lipotropin and β-endorphin) and MSH cells (α-MSH, β-MSH, γ-lipotropin, β-lipotropin and β-endorphin) (38, 39). There are leads in the literature that suggest GnRH1 stimulates ACTH and β-endorphin (40). Therefore, the significantly high R1, R2, and R3 transcripts in ACTH cells in mature males suggests their role during testicular maturation, which may include territoriality and aggressiveness because ACTH induced cortisol has been implicated in social dominance in cichlid fish (41). The role of α-MSH in steroidogenesis and timing of puberty and β-endorphin in reproductive behavior in rats are well documented (42, 43). Therefore, the greater copies of R1 and R3 transcripts in MSH cells in immature and mature males, respectively, suggests differential use of GnRH-Rs, probably to regulate different products of the POMC family of peptides during early and late stages of testicular development.
Summary. Given that pituitary cells are clustered, the present single-cell approach is technically more reliable than in situ hybridization to observe changes in absolute copies of transcripts in individual cells because it avoids the problem of overlap of silver grains from neighboring cells and demonstrates the presence of “silent pituitary cells” with undetectable levels of GnRH-R transcripts during the two reproductive states. Furthermore, this approach allows correlation of multiple gene expression profiles, which would greatly facilitate our understanding of the complex interactions that exist within individual pituitary cells. The presence of GnRH-Rs in MSH, ACTH, and TSH cells are noteworthy, and suggests their role in reproduction and/or novel roles for GnRH molecules in nonreproductive functions during gonadal development.
Supplementary Material
Acknowledgments
We thank Dr. R. Kiyama for providing the laser capture facilities and for valuable discussions. This study was supported in part by Ministry of Education, Culture, Sports, Science and Technology of Japan Grants-in-Aid 14580777 (to I.S.P.) and 4370025 (to Y.S.).
Author contributions: I.S.P. designed research; I.S.P. and S.O. performed research; I.S.P. and Y.S. contributed new reagents/analytic tools; I.S.P., S.O., and Y.S. analyzed data; and I.S.P. and S.O. wrote the paper.
Abbreviations: GnRH, gonadotropin-releasing hormone; GnRH-R, GnRH receptor; FSH, follicle-stimulating hormone; LH, luteinizing hormone; GH, growth hormone; PRL, prolactin; SL, somatolactin; DIG, digoxigenin; RT-Q-RT-PCR, real-time quantitative RT-PCR; LCM, laser-capture microdissection; TSH, thyroid-stimulating hormone; POMC, proopiomelanocortin; ACTH, adenocorticotropin; MSH, melanocyte-stimulating hormone.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AB111356 (GnRH-R1), AB111357 (GnRH-R2), and AB158490 (GnRH-R3)].
References
- 1.King, J. A. & Millar, R. P. (1992) Trends Endocrinol. Metab. 3, 339-346. [DOI] [PubMed] [Google Scholar]
- 2.Sherwood, N. M., Von Schalburg, K. & Lescheid, D. W. (1997) in GnRH Neurons: Gene to Behavior, eds. Parhar, I. S. & Sakuma, Y. (Brain Shuppan, Tokyo), pp. 3-25.
- 3.Parhar, I. S. (2002) Prog. Brain Res. 141, 3-17. [DOI] [PubMed] [Google Scholar]
- 4.Parhar, I. S., Soga, T., Sakuma, Y. & Millar, R. P. (2002) J. Neuroendocrinol. 14, 657-665. [DOI] [PubMed] [Google Scholar]
- 5.Parhar, I. S. & Iwata, M. (1994) Histochemistry 102, 195-203. [DOI] [PubMed] [Google Scholar]
- 6.Weber, G. M., Powell, J. F., Park, M., Fischer, W. H., Craig, A. G., Rivier, J. E., Nanakorn, U., Parhar, I. S., Ngamvongchon, S., Grau, E. G. & Sherwood, N. M. (1997) J. Endocrinol. 155, 121-132. [DOI] [PubMed] [Google Scholar]
- 7.Kakizawa, S., Kaneko, T. & Hirano, T. (1997) Gen. Comp. Endocrinol. 105, 71-78. [DOI] [PubMed] [Google Scholar]
- 8.Melamed, P., Rosenfeld, H., Elizur, A. & Yaron, Z. (1998) Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 119, 325-338. [DOI] [PubMed] [Google Scholar]
- 9.Peter, R. E. & Chang, J. P. (1999) in Neural Regulation in the Vertebrate Endocrine System: Neuroendocrine Regulation, eds. Rao, P. D. & Peter, R. E. (Kluwer Academic/Plenum, New York), pp. 55-67.
- 10.Millar, R. P., Lu, Z. L., Pawson, A. J., Flanagan, C. A., Morgan, K. & Maudsley, S. R. (2004) Endocr. Rev. 25, 235-275. [DOI] [PubMed] [Google Scholar]
- 11.Illing, N., Troskie, B. E., Nahorniak, C. S., Hapgood, J. P., Peter, R. E. & Millar, R. P. (1999) Proc. Natl. Acad. Sci. USA 96, 2526-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parhar, I. S., Ogawa, S., Hamada, T. & Sakuma, Y. (2003) Endocrinology 144, 3297-3300. [DOI] [PubMed] [Google Scholar]
- 13.Ber, R. & Daniel, V. (1993) Gene 125, 143-150. [DOI] [PubMed] [Google Scholar]
- 14.Specker, J. L., Kishida, M., Huang, L., King, D. S., Nagahama, Y., Ueda, H. & Anderson, T. R. (1993) Gen. Comp. Endocrinol. 89, 28-38. [DOI] [PubMed] [Google Scholar]
- 15.Parhar, I. S., Ogawa, S. & Sakuma, Y. (2004) Endocrinology 145, 3613-3618. [DOI] [PubMed] [Google Scholar]
- 16.Parhar, I. S. (1997) in GnRH Neurons: Gene to Behavior, eds. Parhar, I. S. & Sakuma, Y. (Brain Shuppan, Tokyo), pp. 99-122.
- 17.Ono, M., Takayama, Y., Rand-Weaver, M., Sakata, S., Yasunaga, T., Noso, T. & Kawauchi, H. (1990) Proc. Natl. Acad. Sci. USA 87, 4330-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook, H., Berkenbosch, J. W., Fernhout, M. J., Yu, K. L., Peter, R. E., Chang, J. P. & Rivier, J. E. (1991) Regul. Pept. 36, 369-378. [DOI] [PubMed] [Google Scholar]
- 19.Stefano, A. V., Vissio, P. G., Paz, D. A., Somoza, G. M., Maggese, M. C. & Barrantes, G. E. (1999) Gen. Comp. Endocrinol. 116, 133-139. [DOI] [PubMed] [Google Scholar]
- 20.Marchant, T. A., Chang, J. P., Nahorniak, C. S. & Peter, R. E. (1989) Endocrinology 124, 2509-2518. [DOI] [PubMed] [Google Scholar]
- 21.Hirano, T. (1986) Prog. Clin. Biol. Res. 205, 53-74. [PubMed] [Google Scholar]
- 22.Kaneko, T. (1996) Int. Rev. Cytol. 169, 1-24. [DOI] [PubMed] [Google Scholar]
- 23.Planas, J. V., Swanson, P., Rand-Weaver, M. & Dickhoff, W. W. (1992) Gen. Comp. Endocrinol. 87, 1-5. [DOI] [PubMed] [Google Scholar]
- 24.Rand-Weaver, M., Swanson, P., Kawauchi, H. & Dickhoff, W. W. (1992) J. Endocrinol. 133, 393-403. [DOI] [PubMed] [Google Scholar]
- 25.Bhandari, R. K., Taniyama, S., Kitahashi, T., Ando, H., Yamauchi, K., Zohar, Y., Ueda, H. & Urano, A. (2003) Gen. Comp. Endocrinol. 130, 55-63. [DOI] [PubMed] [Google Scholar]
- 26.Sandra, O., Le Rouzic, P., Cauty, C., Edery, M. & Prunet, P. (2000) J. Mol. Endocrinol. 24, 215-224. [DOI] [PubMed] [Google Scholar]
- 27.Kajimura, S., Kawaguchi, N., Kaneko, T., Kawazoe, I., Hirano, T., Visitacion, N., Grau, E. G. & Aida, K. (2004) J. Endocrinol. 181, 65-76. [DOI] [PubMed] [Google Scholar]
- 28.Le Gac, F., Blaise, O., Fostier, A., Le Bail, P-Y., Loir, M., Mourot, B. & Weil, C. (1993) Fish Physiol. Biochem. 11, 219-232. [DOI] [PubMed] [Google Scholar]
- 29.Gomez, J. M., Mourot, B., Fostier, A. & Le Gac, F. (1999) J. Reprod. Fertil. 115, 275-285. [DOI] [PubMed] [Google Scholar]
- 30.Eden, S. (1979) Endocrinology 105, 555-560. [DOI] [PubMed] [Google Scholar]
- 31.Pierce, J. G. & Parsons, T. F. (1981) Annu. Rev. Biochem. 50, 465-495. [DOI] [PubMed] [Google Scholar]
- 32.Planas, J.V. & Swanson, P. (1995) Biol. Reprod. 52, 697-704. [DOI] [PubMed] [Google Scholar]
- 33.Gen, K., Okuzawa, K., Senthilkumaran, B., Tanaka, H., Moriyama, S. & Kagawa, H. (2000) Biol. Reprod. 63, 308-319. [DOI] [PubMed] [Google Scholar]
- 34.Rosenfeld, H., Levavi-Sivan, B., Melamed, P., Yaron, Z. & Elizur, A. (1997) Fish Physiol. Biochem. 17, 85-92. [Google Scholar]
- 35.Jansen, H. T., Lubbers, L. S., Macchia, E., DeGroot, L. J. & Lehman, M. N. (1997) Endocrinology 138, 5039-5047. [DOI] [PubMed] [Google Scholar]
- 36.Parhar, I. S., Soga, T. & Sakuma, Y. (2000) Endocrinology 141, 1618-1626. [DOI] [PubMed] [Google Scholar]
- 37.Young, G. & Ball, J. N. (1983) Gen. Comp. Endocrinol. 51, 24-38. [DOI] [PubMed] [Google Scholar]
- 38.Dores, R. M. (1990) Prog. Clin. Biol. Res. 342, 22-27. [PubMed] [Google Scholar]
- 39.Kawauchi, H., Kawazoe, I., Adachi, Y., Buckley, D. I. & Ramachandran, J. (1984) Gen. Comp. Endocrinol. 53, 37-48. [DOI] [PubMed] [Google Scholar]
- 40.Gambacciani, M., Yen, S. S. & Rasmussen, D. D. (1988) Life Sci. 43, 755-760. [DOI] [PubMed] [Google Scholar]
- 41.Fox, H. E., White, S. A., Kao, M. H. & Fernald, R. D. (1997) J. Neurosci. 17, 6463-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durando, P. E. & Celis, M. E. (1998) Peptides 19, 667-675. [DOI] [PubMed] [Google Scholar]
- 43.Sirinathsinghji, D. J. (1986) Brain Res. 375, 49-56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.