IL-22 binding protein inhibits IL-22 signaling, which is important for intestinal homeostasis. Jinnohara et al. report that IL-22 binding protein is strongly expressed by Peyer’s patch dendritic cells and facilitates the M cell uptake of bacterial antigens into Peyer’s patches.
Abstract
Interleukin-22 (IL-22) acts protectively and harmfully on intestinal tissue depending on the situation; therefore, IL-22 signaling needs to be tightly regulated. IL-22 binding protein (IL-22BP) binds IL-22 to inhibit IL-22 signaling. It is expressed in intestinal and lymphoid tissues, although its precise distribution and roles have remained unclear. In this study, we show that IL-22BP is highly expressed by CD11b+CD8α− dendritic cells in the subepithelial dome region of Peyer’s patches (PPs). We found that IL-22BP blocks IL-22 signaling in the follicle-associated epithelium (FAE) covering PPs, indicating that IL-22BP plays a role in regulating the characteristics of the FAE. As expected, FAE of IL-22BP–deficient (Il22ra2−/−) mice exhibited altered properties such as the enhanced expression of mucus and antimicrobial proteins as well as prominent fucosylation, which are normally suppressed in FAE. Additionally, Il22ra2−/− mice exhibited the decreased uptake of bacterial antigens into PPs without affecting M cell function. Our present study thus demonstrates that IL-22BP promotes bacterial uptake into PPs by influencing FAE gene expression and function.
Introduction
The mucosal surface of the intestinal tract is exposed to a variety of foreign antigens, including pathogens for host animals. To avoid the infectious risks of these pathogens, intestinal epithelial cells (IECs) develop several layers of barriers, such as mucus secreted from goblet cells and antimicrobial proteins (AMPs). To efficiently regulate these epithelial barriers, IECs sense the condition of the intestinal lumen, which is largely mediated by intracellular signaling pathways evoked by pattern recognition receptors and cytokine receptors (Nenci et al., 2007; Rubino et al., 2012).
IL-22 is one of the IL-10 family cytokines, and its functions are well characterized in terms of intestinal homeostasis, especially in the regulation of the IEC barrier (Sabat et al., 2014). IL-22 is produced by immune cells, such as innate lymphoid cells and T helper 17 cells, in response to microbial stimulation, whereas the IL-22 receptor (IL-22R; also known as IL-22Ra1) is predominantly expressed on IECs (Rubino et al., 2012; Sabat et al., 2014). When IL-22 binds IL-22R, signal transducer and activator of transcription 3 (STAT3) is phosphorylated, which results in enhancing the expression of AMPs and mucus-associated molecules. These are essential for the protection against pathogens because IL-22–deficient mice become more susceptible to the infection by Citrobacter rodentium and Klebsiella pneumoniae (Aujla et al., 2008; Zheng et al., 2008). In addition, IL-22 signaling induces the fucosylation of IECs, which contributes to the protection against Salmonella enterica serovar Typhimurium (S. Typhimurium) infection (Goto et al., 2014).
IL-22 also plays an important role in the maintenance of intestinal epithelial integrity when intestinal tissues are damaged by various disorders, such as inflammatory bowel disease and graft-versus-host disease (Sugimoto et al., 2008; Pickert et al., 2009; Hanash et al., 2012). In these situations, IL-22 enhances the proliferation and wound healing of IECs, which results in the repair of the IEC barrier. However, it has been reported that IL-22 signaling might be deleterious for epithelial tissues under certain conditions, including Toxoplasma gondii–infectious immunopathology, suggesting that IL-22 signaling needs to be finely regulated to maintain IEC integrity (Muñoz et al., 2009).
IL-22 signaling is negatively regulated by IL-22 binding protein (IL-22BP; also known as IL-22Ra2). IL-22BP shares 34% amino acid sequence homology with the extracellular domain of IL-22Ra1 (Dumoutier et al., 2001). Importantly, IL-22BP possesses a much higher affinity to IL-22 than IL-22Ra1 (20- to 1,000-fold) and prevents the binding of IL-22 to IL-22Ra1 (Wolk et al., 2007). Previous studies have demonstrated that IL-22BP is expressed in mucosal tissues, including the intestinal tract (Huber et al., 2012; Martin et al., 2014). In the colon, DCs constitutively express IL-22BP, which is involved in the suppression of tumor development (Huber et al., 2012). This observation suggests that IL-22BP also acts to maintain intestinal homeostasis. In addition, it has been reported that IL-22BP is highly expressed in lymphoid tissues such as the spleen and mesenteric lymph nodes (MLNs) in rats (Martin et al., 2014), suggesting that IL-22BP may also play an important role in adaptive immune responses, although little is known in this regard.
Peyer’s patches (PPs) are a part of the gut-associated lymphoid tissue in the small intestine and serve as an important inductive site for the generation of antigen-specific intestinal immune responses (Pearson et al., 2012). To evoke these responses, PPs need to take up antigens from the intestinal lumen. The luminal side of PPs is covered with follicle-associated epithelia (FAE), which is distinct from villous epithelia (VE), especially in terms of the presence of M (microfold or membranous) cells. M cells possess high phagocytic and transcytotic capacities, which allow efficient bacterial uptake into PPs (Ohno, 2016). Subsequently, taken-up bacteria are transferred to DCs in the subepithelial dome (SED) of PPs, eventually leading to the production of antigen-specific immunoglobulin A (IgA; Hase et al., 2009).
FAE also displays a more distinct phenotype than VE (Ohno, 2016). The features of FAE help the closer association to FAE and efficient uptake to gut-associated lymphoid tissue thereafter of luminal bacteria. Although we and others have previously identified that the transcription factor Spi-B is essential for M cell development (de Lau et al., 2012; Kanaya et al., 2012; Sato et al., 2013), molecular mechanisms for these FAE characteristics have been poorly understood.
In this study, we show that IL-22BP expression is the highest in PPs among the intestinal tissues in mice, and we identify that IL-22BP–positive cells that predominantly accumulate in PP SED are CD11chighCD11b+CD8α− DCs. In addition, we demonstrate that IL-22BP expressed by SED-DCs is important for facilitating antigen uptake into PPs by negatively regulating IL-22 signaling in FAE. Our study thus identifies IL-22BP as one of the important determinants for FAE characteristics.
Results and discussion
IL-22BP is highly expressed by DCs in PPs
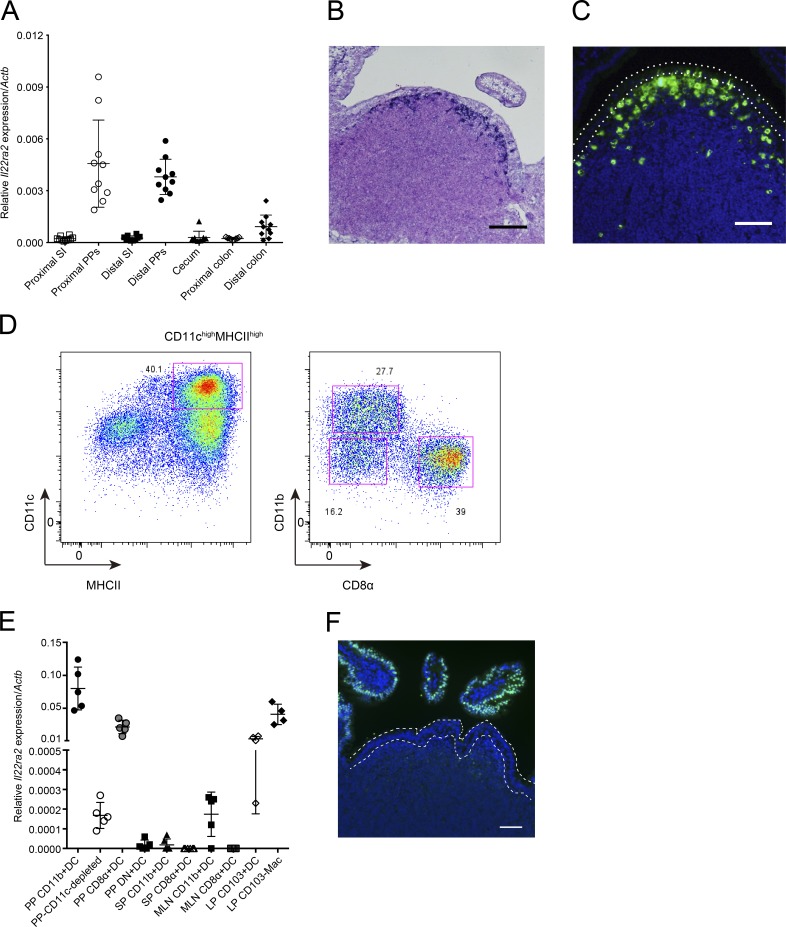
To understand the role of IL-22BP in the intestinal homeostasis, including mucosal immune responses, we first examined the expression of IL-22BP throughout the intestinal tract and found much higher expression of Il22ra2 mRNA in PPs than in other parts of the intestine in mice (Fig. 1 A). To verify as well as examine the localization of Il22ra2 expression morphologically, we performed in situ hybridization. We identified the accumulation of Il22ra2-positive cells in the SED of PPs (Fig. 1 B). We also confirmed the existence of IL-22BP protein-expressing cells in the PP SED by immunostaining (Fig. 1 C). We next determined the cellular identity of IL-22BP expression in the SED region. Previous studies have demonstrated that a main source of IL-22BP is DCs (Huber et al., 2012; Martin et al., 2014). In addition, DCs are known to abundantly accumulate in the SEDs of PPs (Iwasaki and Kelsall, 2000). We therefore speculated that DCs could be responsible for IL-22BP expression in PPs as well. We isolated CD11chighMHCIIhigh DCs from PPs (Fig. 1 D). PP DCs can be subdivided into CD11b+CD8α−, CD11b−CD8α+, and CD11b−CD8α− (double negative; DN) subsets (Fig. 1 D) localizing in the SED, the interfollicular region, and both the SED and interfollicular region, respectively (Iwasaki and Kelsall, 2000). We next assessed the expression of Il22ra2 in these three subsets. Consistent with the distribution of IL-22BP protein and the localization of DC subpopulations in PPs, CD11b+CD8α− DCs exhibited the highest expression of Il22ra2 among them (Fig. 1 E). DN DCs exhibited modest expression of Il22ra2, which probably derived from DN DCs localizing in the SED. However, the CD11b−CD8α+ DCs and the CD11c− (CD11c+ cell-depleted) population only showed low levels of Il22ra2 expression. These results suggest that CD11b+CD8α− DCs are a major source of IL-22BP in PPs.
Figure 1.
IL-22BP is expressed by DCs located on SED of PPs. (A) Relative Il22ra2 mRNA expression of intestinal tissue, small intestine (SI), colon, cecum, and PPs (normalized with Actb; n = 10). Data are pooled from two independent experiments. (B) In situ hybridization analysis of PPs with a digoxigenin-labeled specific RNA probe for Il22ra2 mRNA. Nuclei were counterstained with Nuclear Fast Red. (C) Immunohistochemistry of PP tissues with anti–IL-22BP antibody. Green colors show IL-22BP–positive cells, and blue colors show nuclei. Dotted lines indicate FAE. Data are representative of three independent experiments. (D) CD11c-enriched cells were stained with antibodies to isolate DCs. CD3−B220−CD45+ PP cells were selected and analyzed by the expression of CD11c and MHCII. CD11chighMHCIIhigh cells were isolated as DCs, and CD11b+CD8α−, CD11b−CD8α+, and CD11b−CD8α− (DN) DCs were sorted from this DC population. Data are representative of four independent experiments. (E) Relative Il22ra2 mRNA expression of sorted DC and macrophage (Mac) populations from PPs (n = 5), MLNs (n = 5), spleens (SPs; n = 5), and LPs (n = 4). Data were normalized with Actb. Data are pooled from four independent experiments. Means ± SD are shown. (F) Recombinant IL-22 was intravenously injected to a WT mouse. After 15 min, PPs were collected and immunostained with anti-pSTAT3 antibody. Green colors show pSTAT3, and blue colors show nuclei. Dotted lines indicate FAE. Data are representative of at least four independent experiments. Bars: (B) 100 µm; (C and F) 50 µm.
We also examined the similar DC subsets from the MLN and spleen for the expression of Il22ra2 (Fig. S1 A); in contrast to PPs, the expression of Il22ra2 was fairly low in CD11b+CD8α− DCs even though it was higher than in CD11b−CD8α+ DCs (Fig. 1 E), as described by a previous study (Martin et al., 2014).
We next evaluated the expression of Il22ra2 mRNA by lamina propria (LP) CD103+ DCs and CD103−macrophages, which were identified to express Il22ra2 by a previous study (Martin et al., 2014) and by the ImmGen database (Immunological Genome Project, 2017). We isolated these cell populations (Fig. S1 B) and found that CD103+ DCs slightly and CD103−macrophages modestly express Il22ra2 (Fig. 1 E). We next carefully observed the intestinal sections to identify IL-22BP protein-expressing cells in the LP. Immunostaining of IL-22BP protein demonstrated that IL-22BP protein-expressing cells also accumulate in the SED of colonic patches and isolated lymphoid follicles as well as that of PPs, but not in the LP of either the small intestine or the colon (Fig. S1 C). These data suggest that the microenvironment established in the SED region might be required for the expression of IL-22BP protein.
IL-22BP blocks IL-22 signaling on the FAE
The preferential expression of IL-22BP in SED DCs suggests that IL-22 signaling may be suppressed in the FAE compared with the VE. To assess this, we examined the activation of IL-22 signaling in FAE in mice treated with recombinant IL-22 protein. Binding of IL-22 to IL-22R phosphorylates STAT3, which results in translocation of phosphorylated STAT3 (pSTAT3) into nucleus. As expected, we observed the nuclear translocation of pSTAT3 in the VE of WT mice upon IL-22 administration. In contrast, nuclear pSTAT3 was almost absent in the FAE of the same mice, suggesting that IL-22BP expressed in SED preferentially blocks IL-22 signaling in the FAE (Fig. 1 F).
IL-22BP deficiency gains the activation of IL-22 signaling in the FAE
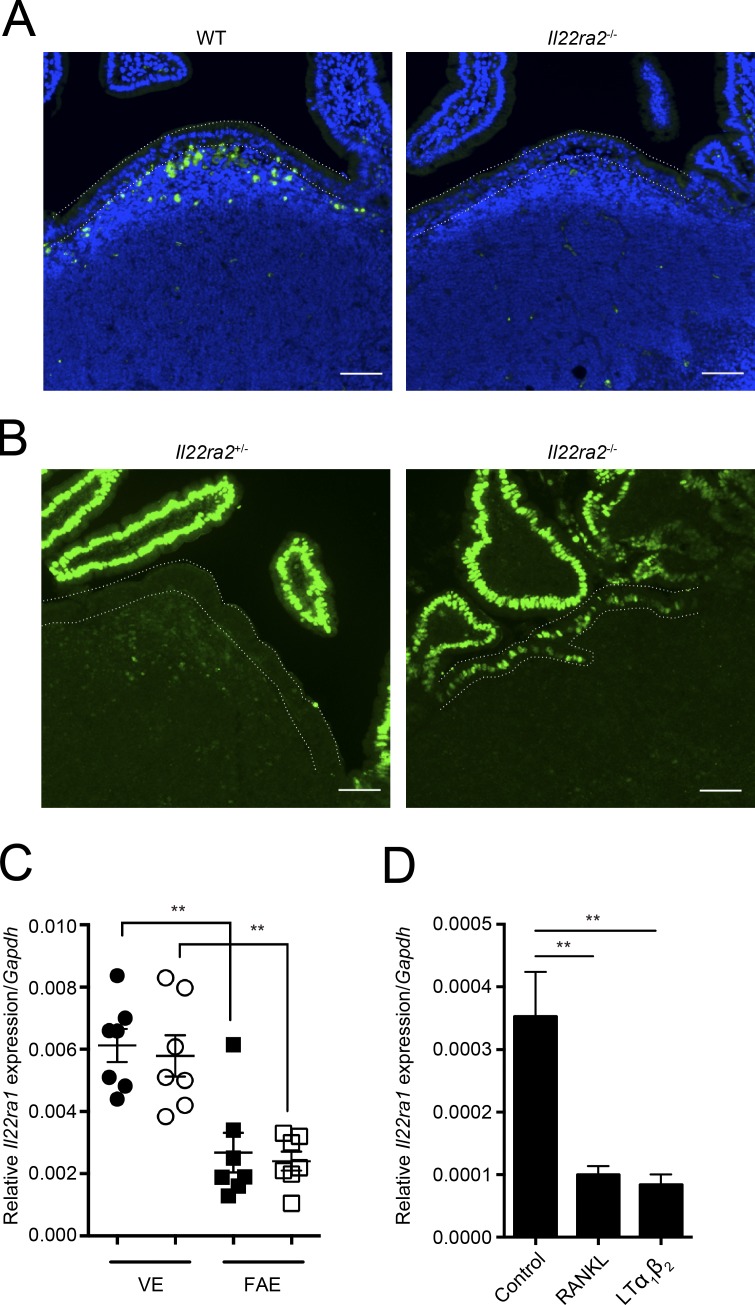
To further evaluate the role of IL-22BP in vivo, we generated mice lacking the Il22ra2 gene (Il22ra2−/−; Fig. S2). In these mice, cells expressing IL-22BP protein were absent from the SED of PPs (Fig. 2 A). We next verified whether IL-22 signaling was restored in the FAE of Il22ra2−/− mice. Both Il22ra2−/− and heterozygous Il22ra2+/− were injected with IL-22 and then nuclear pSTAT3 was examined. Similar to WT mice, IL-22 signaling was almost totally blocked in the FAE of Il22ra2+/− mice. In contrast, the nuclear pSTAT3 was clearly detected in the FAE of Il22ra2−/− mice, suggesting that IL-22BP deficiency enhanced the ability of FAE cells to respond to IL-22 (Fig. 2 B).
Figure 2.
IL-22BP deficiency releases IL-22 signaling on the FAE. (A) PP tissues from WT and Il22ra2−/− mice were immunostained with anti–IL-22BP antibody. Green colors show IL-22BP–positive cells, and blue show nuclei. (B) Both Il22ra2+/− and Il22ra2−/− mice were intravenously administered with recombinant IL-22 protein. After 15 min, PP tissues were collected. Tissue sections prepared from these mice were immunostained with anti-pSTAT3 antibody. Green colors show pSTAT3, and blue show nuclei. Dotted lines indicate FAE. Bars, 40 µm. Data are pooled from at least three mice of each genotype. (C) Relative Il22ra1 mRNA expression of FAE and VE from both Il22ra2+/− and Il22ra2−/− mice. Circles and squares show VE and FAE, respectively. Solid symbols indicate expression profiles of Il22ra2+/− mice, and open ones indicate those of Il22ra2−/− mice. Data were normalized with Gapdh (n = 7). Data are representative of three independent experiments. (D) Relative Il22ra1 mRNA expression of intestinal organoids stimulated with RANKL and LT α1β2 for 24 h (n = 4). Data are representative of two independent experiments. Means ± SEM. are shown. One-way ANOVAs with Bonferroni’s (C) or Dunnett’s (D) post hoc test were used for statistical analyses. **, P < 0.01.
We also noticed that the signal of pSTAT3 in FAE was weaker than those of VE in Il22ra2−/− mice. We therefore compared the expression of Il22ra1 between FAE and VE and found that the expression of Il22ra1 was significantly lower in FAE than in VE (Fig. 2 C). Because the FAE is closely associated with the SED, we postulated that a factor or factors derived from the SED could suppress the expression of Il22ra1 in the FAE. To examine the possibility, we generated intestinal organoids (Sato et al., 2009) and stimulated them with receptor activator of NF-κB ligand (RANKL) and lymphotoxin (LT) α1β2, which are expressed by stromal cells and immune cells of the SED, respectively (Rumbo et al., 2004; Taylor et al., 2007). Both RANKL and LT α1β2 stimulation decreased the expression of Il22ra1 in organoids (Fig. 2 D). This suggests that the expression of Il22ra1 was suppressed by RANKL–RANK and LT β receptor signaling pathways in the FAE. Collectively, IL-22 signaling in FAE is blocked under physiological conditions because of a combination of IL-22BP expressed in the SED and suppressed expression of Il22ra1 in the FAE.
IL-22BP deficiency causes excessive expression of IL-22–responsive molecules in FAE
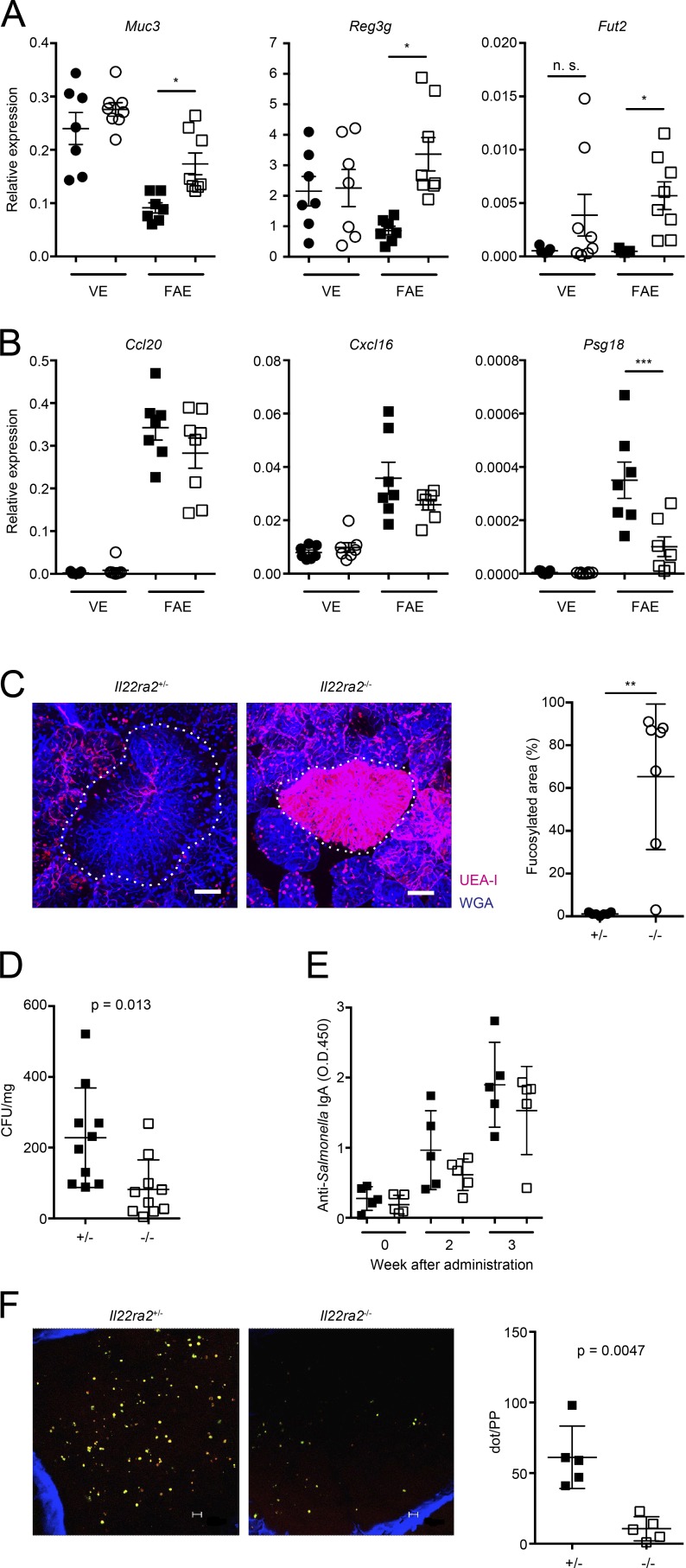
Given that IL-22BP deficiency causes an excessive activation of IL-22 signaling, Il22ra2−/− mice might exhibit abnormalities in FAE properties. To evaluate the possibility, we examined the expression of IL-22–responsive genes in FAE. IL-22 signaling is known to target the genes associated with mucus, AMPs, and fucosylation. As expected, Muc3, Reg3g, and Fut2 were significantly increased in FAE of Il22ra2−/− mice (Fig. 3 A). We also investigated the expression of FAE-associated chemokines such as Ccl20 and Cxcl16; however, expression of these genes was comparable between Il22ra2−/− and Il22ra2+/− mice. We also found that Psg18, another FAE-associated gene, was significantly decreased in Il22ra2−/− mice (Fig. 3 B). In addition, IL-22 signaling is known to activate the genes involved in epithelial cell proliferation and wound healing, implying that the renewal of FAE might be accelerated in Il22ra2−/− mice. We therefore injected 5-ethynyl-2’-deoxyuridine (EdU) into Il22ra2−/− and Il22ra2+/− mice and traced the EdU-labeled cells to observe the zone of proliferation within the FAE. However, there was no apparent difference in the labeled area of FAE after 48 h injection (Fig. S2 C). Collectively, these observations demonstrate that IL-22BP deficiency causes the augmented activation of IL-22–responsive genes in FAE and also influences expression of some FAE-associated genes but does not alter the distribution of proliferating epithelial cells within the FAE.
Figure 3.
Excessive expression of IL-22 signaling in the FAE of Il22ra2−/− mice decreased the uptake of bacterial antigens into PPs. (A and B) Relative expressions of IL-22–responsive genes (A) and those of FAE-associated genes (B) on the FAE and VE from both Il22ra2+/− and Il22ra2−/− mice are shown. Circles and squares indicate VE and FAE, respectively. Solid symbols indicate the expression profile of Il22ra2+/− mice, and open ones indicate those of Il22ra2−/− mice. Data were normalized with Gapdh (n = 7). *, P < 0.05; ***, P < 0.001. Data are representative of three independent experiments. (C) PP tissues from both Il22ra2+/− and Il22ra2−/− mice were fixed and stained with fluorescent-labeled lectins. Red colors show UEA-I, and blue show WGA. Dotted lines indicate FAE. Data are pooled from three mice of each genotype. Fucosylated area of FAE was determined by MetaMorph software (n = 7). **, P < 0.01. (D) Both Il22ra2+/− and Il22ra2−/− mice were fed with S. Typhimurium (χ3306), and 24 h after the infection, PPs were collected. Then, PPs were homogenized and plated on Luria broth plates to calculate the number of S. Typhimurium taken up into PPs (n = 10 of each genotype). Data are representative of three independent experiments. (E) Both Il22ra2+/− and Il22ra2−/− mice were gavaged with 5 × 109 CFUs of ΔaroA S. Typhimurium (UF20). After 2 and 3 wk, feces were collected, and S. Typhimurium–specific IgA was analyzed by ELISA (n = 5 of each genotype). Data are representative of four independent experiments. (F) The presence of Alcaligenes spp. was visually analyzed by wholemount fluorescent in situ hybridization on the interior of PPs. Small dots indicate Alcaligenes spp. The left graph shows the dot count of Alcaligenes spp. (n = 5). Data are representative of two independent experiments. Bars: (B) 40 µm; (F) 10 µm. Means ± SD are shown. One-way ANOVA with Bonferroni’s post hoc test (A and B) and Student’s t test (C, D, and F) were used for statistical analysis.
The internalization of pathogenic and commensal bacteria into PPs is hampered in Il22ra2−/− mice
The enhanced expression of IL-22–responsive genes could result in lowering the accessibility of bacteria for uptake into the PPs of Il22ra2−/− mice. Muc3 is one of the mucus proteins and is distributed in the cell surface (Linden et al., 2008). A previous study demonstrated that Muc3 is correlated with decreased binding of enteropathogenic Escherichia coli (Mack et al., 2003), indicating that the up-regulation of Muc3 might impair the adhesion of bacteria. Fut2 is responsible for the fucosylation of IECs, which contributes to protection against S. Typhimurium infection (Goto et al., 2014). We therefore examined the fucosylation of the FAE of Il22ra2−/− mice by wholemount staining of PPs with fluorescent-labeled Ulex europaeus agglutinin-1 (UEA-I), which recognizes α (1,2)-fucose. As expected, Il22ra2−/− mice exhibited far more prominent fucosylation in FAE compared with Il22ra2+/− mice (Fig. 3 C).
We hypothesized that the enhanced expression of Muc3 and Fut2 in FAE impairs the internalization of bacteria into PPs. Orally administered S. Typhimurium is preferentially internalized into PPs via M cell–mediated transport (Hase et al., 2009; Kanaya et al., 2012). We therefore evaluated whether the FAE of Il22ra2−/− mice exhibit the resistance to the internalization of S. Typhimurium into PPs. Both Il22ra2−/− and Il22ra2+/− mice were gavaged with S. Typhimurium, and the number of S. Typhimurium translocated in PPs was determined. Consistently, the internalization of S. Typhimurium into PPs was significantly decreased in Il22ra2−/− mice compared with Il22ra2+/− mice (Fig. 3 D). We also examined the amount of S. Typhimurium–specific fecal IgA after the oral gavage with attenuated S. Typhimurium and found that S. Typhimurium–specific IgA response tended to decrease in Il22ra2−/− mice (not significant; Fig. 3 E). We also assessed the possibility that bacteria other than S. Typhimurium are also less internalized into PPs in Il22ra2−/− mice. To examine the possibility, we evaluated the internalization of one of the commensal gram-negative bacteria, Alcaligenes spp., which can colonize inside PPs (Obata et al., 2010; Sato et al., 2013). As expected, the internalization of Alcaligenes spp. into PPs was drastically decreased in Il22ra2−/− mice (Fig. 3 F). These observations indicate that enhanced fucosylation and mucin production in FAE of Il22ra2−/− mice impairs both pathogenic and commensal bacterial internalization into PPs. The mechanisms of protection against bacteria by fucosylation have not been elucidated; however, the bacteria that can use fucose for their proliferation might inhibit the uptake of S. Typhimurium or Alcaligenes spp. It is thought that the production of mucus and AMPs is normally suppressed in FAE to facilitate bacterial antigen uptake into PPs (Kraehenbuhl and Neutra, 2003). Our data support and further this notion by showing that the local action of IL-22BP can account for these properties of the FAE.
IL-22BP deficiency does not influence M cell differentiation and function
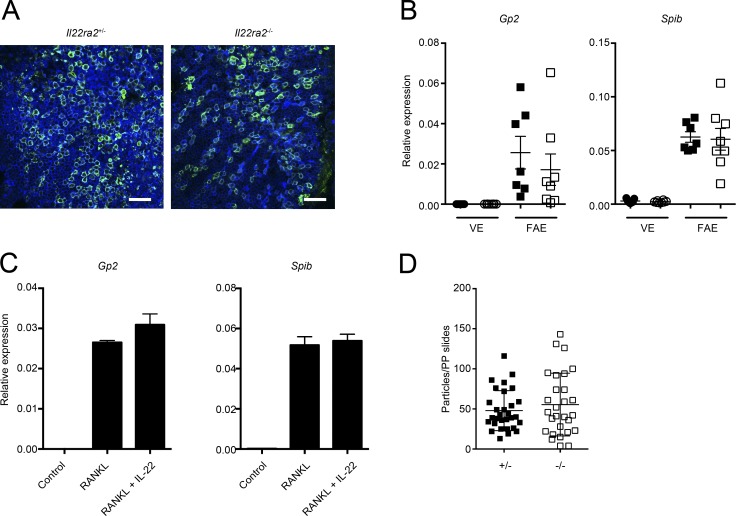
It could be possible that IL-22BP deficiency directly affects M cells, which leads to the reduction in the internalization of S. Typhimurium into PPs. To assess this possibility, we examined the differentiation and function of M cells in Il22ra2−/− mice. Wholemount immunostaining of FAE did not show an apparent difference in the expression level of the M cell–specific marker GP2 (Hase et al., 2009) nor the number of GP2-positive M cells between Il22ra2−/− and Il22ra2+/− mice (Fig. 4 A). Consistently, the mRNA expression of Spib, a transcription factor essential for M cell differentiation (Kanaya et al., 2012), was not significantly different in Il22ra2+/− and Il22ra2−/− mice (Fig. 4 B). To further assess the potential effects of IL-22 signaling on M cell differentiation, we used an in vitro M cell differentiation model generated from intestinal organoids. M cells can be induced in intestinal organoids by the addition of RANKL (de Lau et al., 2012; Rios et al., 2016). We stimulated intestinal organoids with RANKL or RANKL together with IL-22 and evaluated M cell differentiation. Both Gp2 and Spib were prominently increased in organoids upon RANKL stimulation and were not suppressed by the addition of IL-22, suggesting that IL-22 signaling does not impair M cell differentiation (Fig. 4 C). In addition, we evaluated the capacity for antigen uptake of M cells in Il22ra2−/− mice by using fluorescent nanoparticles as a model particulate antigen. We gavaged Il22ra2−/− and Il22ra2+/− mice with fluorescent nanoparticles and quantified the number of particles by counting them in fixed PP sections. There were no significant differences in the number of nanoparticles taken up into PPs between Il22ra2−/− and Il22ra2+/− mice (Fig. 4 D). Collectively, these results indicate that IL-22BP deficiency does not impair M cell differentiation nor M cell–mediated antigen uptake and that the decreased bacterial uptake into PPs in Il22ra2−/− mice is not caused by functional or developmental defects in M cells.
Figure 4.
IL-22BP deficiency does not affect the differentiation and function of M cells. (A) The FAE isolated from both Il22ra2+/− and Il22ra2−/− mice were immunostained with anti-GP2 (green), and samples were counterstained with fluorescent-labeled phalloidin (blue). Bar, 40 µm. Data are pooled from three mice of each genotype. (B) Relative mRNA expression of M cell–marker genes on the FAE and VE from both Il22ra2+/− and Il22ra2−/− mice are shown. Circles and squares indicate VE and FAE, respectively. Solid symbols indicate the expression profile of Il22ra2+/− mice, and open ones indicate those of Il22ra2−/− mice. Data were normalized with Gapdh (n = 7). Data are representative of three independent experiments. (C) Intestinal organoids were stimulated with GST-RANKL or GST-RANKL together with recombinant IL-22 protein for 3 d. Then, expressions of M cell marker genes were analyzed. Data were normalized with Gapdh (n = 4). Data are representative of two independent experiments. (D) Both Il22ra2+/− and Il22ra2−/− mice were orally gavaged with fluorescent-labeled nanoparticles (0.2-µm diameter). After 4 h, particles taken up into PPs were counted. Results show the number of nanoparticles in PPs per section. Ten sections from distal PPs from each three mice are shown (n = 3). Data are pooled by two independent experiments. Means ± SD are shown.
Bacterial colonization does not trigger the expression of IL-22BP
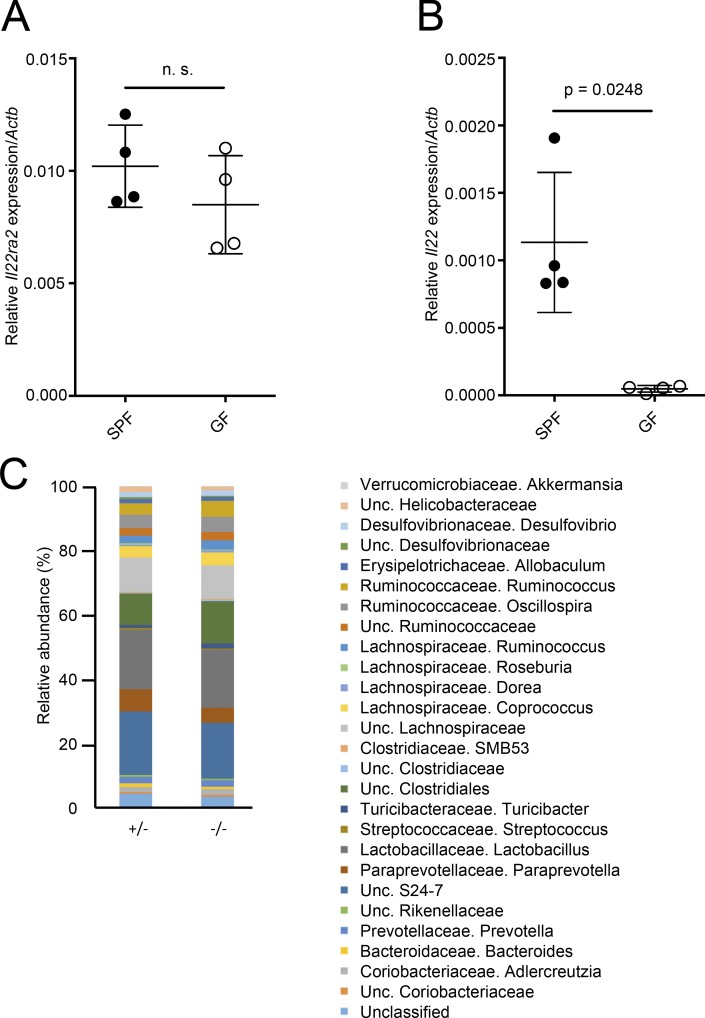
Because bacterial stimulation triggers the expression of IL-22 (Schreiber et al., 2015), we wondered whether the expression of IL-22BP could also be influenced by bacterial colonization. To examine the possibility, we compared the expression of IL-22BP mRNA between germ-free (GF) and specific pathogen–free (SPF) mice. In contrast to the significantly higher expression of Il22 in SPF mice compared with GF mice, the expression of Il22ra2 was comparable between GF and SPF mice (Fig. 5, A and B), suggesting that microbial stimulation does not trigger the expression of IL-22BP by SED DCs. Conversely, we assessed whether IL-22BP deficiency could affect microbiota composition; however, there was no significant difference in microbiota composition of feces between Il22ra2−/− and Il22ra2+/− mice (Fig. 5 C).
Figure 5.
Bacterial colonization does not affect the expression of IL-22BP. (A) Relative Il22ra2 mRNA expression of PPs from SPF and GF C57BL/6N mice are shown (normalized with Actb). (B) Relative Il22 mRNA expression of PPs from SPF and GF C57BL/6N mice are shown (normalized with Actb). n = 4. Means ± SD are shown. (C) Microbiota compositions of Il22ra2+/− and Il22ra2−/− mice are shown. n = 5. Data are representative of two independent experiments. Student’s t test was used for statistical analysis.
Concluding remarks
In this study, we identified that IL-22BP is highly expressed by PP CD11b+CD8α− DCs and that IL-22BP blocks IL-22 signaling in FAE. We also found that Il22ra2−/− mice exhibited enhanced fucosylation as well as increased production of AMPs and the Muc3 mucin, which resulted in impaired bacterial translocation into PPs. Collectively, our findings suggest that the extent of bacterial uptake into PPs can be regulated both at the level of the FAE and at that of M cells and provide novel information with respect to the microenvironment and molecular mechanisms required for FAE characteristics.
Materials and methods
Mice
BALB/c, C57BL/6J, and C57BL/6N mice were purchased from CLEA Japan, Inc. All mice were maintained under SPF or GF conditions in Institute of Physical and Chemical Research and Yokohama City University animal facilities until use in experiments at 8–12 wk old. All animal experiments were approved by the Animal Research Committees of the Institute of Physical and Chemical Research Yokohama Research Institute and of Yokohama City University.
Generation of Il22ra2−/− mice
Il22ra2 genomic sequences were amplified from a bacterial artificial chromosome (clone 353K11; Thermo Fisher Scientific) and inserted into the targeting vector with restriction enzymes (SacII–NotI) or the Gateway Recombination System (Thermo Fisher Scientific; Fig. S2 A). The targeting vector was transfected into embryonic stem cells to obtain the clone with homologous recombination. The successful targeting was confirmed by Southern blotting analysis (Fig. S2 B). Mice with the floxed Il22ra2 allele were crossed with CAG-Cre transgenic mice to delete exon 3 of Il22ra2. Il22ra2+/− and Il22ra2−/− mice were cohoused for at least 4 wk, and then mice were used for each experiment.
Cell isolation
PPs, the spleen, and MLNs were minced gently and then digested with 1 mg/ml collagenase (Wako Pure Chemical Industries) and 40 µg/ml DNaseI (Roche) containing RPMI 1640 (Sigma-Aldrich) supplemented with 2% fetal bovine serum for 30 min at 37°C twice. Digested tissues were homogenized with a syringe with an 18G needle and passed through a 70-µm mesh to obtain the single cell suspension. LP cells were prepared according to a previous study (Moro et al., 2015).
Cell sorting
CD11c-enriched cells were obtained by autoMACS with CD11c-microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. CD11c-enriched cell suspensions were stained with antibodies, PerCP/Cy5.5–anti-CD3 (145-2C11), PerCP/Cy5.5–anti-B220 (RA3-6B2), APC-eFluor780–anti-CD45 (30F-11), PE-Cy7 or PE–anti-CD11c (N418), Brilliant violet 421–anti-MHCII (M5/114.15.2), PE-Cy7 or APC–anti-CD11b (M1/70), PE–anti-CD8α (53-6.7), and FITC–anti-F4/80 (BM8; BioLegend) for separation of DC or macrophage subsets. Dead cells were removed by Fixable Viability Dye eFluor 506 (eBioscience). Finally, DCs were sorted by FACSAria (BD). The flow cytometry data were analyzed by FlowJo software (Tree Star).
Isolation of FAE and VE
PPs were dissected from the small intestine and were soaked in HBSS containing 30 mM EDTA for 15 min. After the incubation, FAE and VE were isolated by manipulation with needles under a stereomicroscope. Isolated FAE were used for wholemount immunostaining or expression analyses.
Intestinal organoids culture
Proximal small intestine was incubated with 2 mM EDTA/PBS for 30 min on ice. Then, intestinal crypts were isolated by vigorous pipetting and embedded in Matrigel (Corning). Crypts were cultured in Advanced DMEM/F12 (Thermo Fisher Scientific) containing 10 ng/ml EGF (PeproTech), R-spondin1 conditioning medium (provided by C.J. Kuo; Ootani et al., 2009), 100 ng/ml Noggin (PeproTech), B27 supplement (Thermo Fisher Scientific), N2 supplement (Thermo Fisher Scientific), and 1 mM N-acetylcysteine (Sigma-Aldrich). Recombinant glutathione S-transferase–mouse RANKL fusion protein (GST-RANKL; 500 ng/ml; Knoop et al., 2009) and LT α1β2 (1 µg/ml; R&D Systems) were used for the stimulation of organoids. Recombinant mouse IL-22 (100 ng/ml; PeproTech) was added into RANKL-stimulated organoids to evaluate the effect of IL-22 signaling on M cell differentiation.
Expression analyses
Total RNA from each sample was extracted with the RNeasy mini kit (QIAGEN) and was reverse transcribed with ReverTra Ace (Toyobo) to obtain cDNA. Gene expression was analyzed by real-time quantitative PCR with SYBR Premix Ex Taq (Takara Bio Inc.) and specific primers (Table S1). The expression of target genes was assessed by a comparative cycle threshold method using expression of Gapdh or Actb.
In situ hybridization
PP tissues were fixed for 24 h with 4% paraformaldehyde and were embedded into paraffin. The cDNA fragments for RNA probes of mouse Il22ra2 were amplified by PCR and subcloned into pcDNA3 plasmid vector. The primer sequences are described in Table S1. A digoxigenin-labeled antisense RNA probe was prepared by in vitro transcription with SP6 RNA polymerase (Roche), with pcDNA3 including subcloned sequences as templates. All procedures of in situ hybridization were performed by a VENTANA DISCOVERY system (Roche) following the manufacturer’s instructions.
Immunohistochemistry
PP tissues were fixed with zinc formalin fixative (Polysciences) and embedded into paraffin. Paraffin sections of PPs were deparaffinized and treated with 0.3% H2O2 in PBS for 20 min at room temperature to quench an endogenous peroxidase activity and then sections were boiled with citrate buffer, pH 6.0, for 20 min to antigen retrieval of both IL-22BP and pSTAT3. The sections were incubated with 0.5% blocking buffer (PerkinElmer) in PBS for 30 min at room temperature and then with primary antibodies overnight at 4°C. The binding of anti–IL-22BP (AF1087; R&D Systems) and anti–phospho-STAT3 (Tyr705; Cell Signaling Technology) was followed by biotinylated secondary antibodies and visualized by a Tyramide Signal Amplification system (PerkinElmer). The sections were analyzed with an SP5 confocal laser microscope (Leica Microsystems).
Detection of EdU-incorporating cells in the FAE
EdU (5 mg/kg of body weight) was intraperitoneally injected to mice. After 48 h, EdU was detected using the Click-iT EdU Alexa Fluor 488 imaging kit (Thermo Fisher Scientific) according to manufacturer’s protocol.
Wholemount staining
To observe GP2, isolated FAE were fixed with 4% paraformaldehyde for 1 h on ice and then incubated with primary antibodies overnight at 4°C. After the overnight incubation, specimens were incubated with fluorescent-labeled secondary antibodies for 1 h at room temperature. To examine the fucosylation of epithelial cells, PPs were fixed with periodate lysine paraformaldehyde solution and stained with 20 µg/ml Rhodamine–UEA-I (Vector Laboratories) and 10 µg/ml Alexa Fluor 633–wheat-germ agglutinin (WGA; Thermo Fisher Scientific). The specimens were analyzed with an SP5 confocal laser microscope.
Intravenous injection of recombinant IL-22 protein
Recombinant mouse IL-22 (3 µg/mouse; PeproTech) was intravenously injected to mice. After 15 min, mice were sacrificed and PPs were collected. The PPs were fixed and embedded in paraffin blocks for sectioning.
Quantification of fluorescent nanoparticles taken up into PPs
Fluorescent-labeled nanoparticles (0.2-µm diameter; Molecular Probes) were administered to fasted mice. After 4 h, mice were sacrificed and distal PPs were collected. Then, PP tissues were embedded into optimum cutting temperature compound (Sakura). Five-µm-thick sections were prepared from embedded PPs, and particles taken up into PPs were counted.
Quantification of S. Typhimurium taken up into PPs
Mice were orally administered with 109 CFUs of S. Typhimurium (χ3306; provided by H. Matsui, Kitasato University, Tokyo, Japan; Gulig et al., 1998). After 24 h, PPs were dissected and incubated for 30 min at 20°C in sterile PBS containing 100 µg/ml gentamicin. Pooled PP tissue was weighed and homogenized in sterile PBS. The homogenates were serially diluted in sterile PBS and plated on a Luria broth agar plate containing 25 µg/ml nalidixic acid.
Evaluation of S. Typhimurium–specific IgA in feces
Mice were orally administered ΔaroA S. Typhimurium (UF20; 5 × 109 CFU/mouse; provided by H. Matsui; Gulig and Doyle, 1993). 2 and 3 wk after the infection, feces were collected and suspended in sterile PBS (100 µl to 10 mg feces), homogenized, and centrifuged at 12,000 g for 10 min, and then supernatant was collected. The supernatants were analyzed by the mouse IgA ELISA kit (Bethyl Laboratories) to measure the amount of S. Typhimurium–specific IgA. In this study, 96-well plates were coated with lysates prepared from S. Typhimurium to capture S. Typhimurium–specific IgA in supernatant.
Wholemount fluorescence in situ hybridization analysis
To detect Alcaligenes, oligonucleotide probes ALBO, which detects the 699–716 position in the 23S ribosomal RNA of Alcaligenes spp., and BPA were purchased from Thermo Fisher Scientific. Isolated tissue segments were fixed in 4% paraformaldehyde for 16 h at 4°C and washed with PBS. Tissues were hybridized for 16 h in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl, 45% formamide, 0.1% SDS, and 10 µg/ml DNA probe) at 60°C. After washing twice in washing buffer (0.45 M NaCl, 20 mM Tris-HCl, 45% formamide, and 0.01% SDS) for 10 min at 60°C, tissue segments were flushed with PBS. To detect the epithelial layer, the specimen was stained with the Alexa Fluor 633–labeled WGA (10 µg/ml; Thermo Fisher Scientific) for 1 h. After being washed with PBS, the tissue samples were mounted and examined using an SP5 confocal laser microscope, and the fluorescent dots in each dome were counted.
Microbiota analysis by 16S ribosomal RNA sequencing
Feces were collected and stored at −80°C until analysis. Fecal DNA extraction was performed according to previous studies but with minor modifications (Kim et al., 2013; Kato et al., 2014). Feces (10 mg) were suspended with sterile sticks in 300 µl of TE10. The fecal suspension was incubated with 15 mg/ml lysozyme (Wako Pure Chemical Industries) at 37°C for 1 h. A purified achromopeptidase (Wako Pure Chemical Industries) was added to be the final concentration at 2,000 U/ml and then was incubated at 37°C for 30 min. The suspension was added at 1% (weight/volume) sodium dodecyl sulfate and 1 mg/ml proteinase K (Merck) and incubated at 55°C for 1 h. After centrifugation, the bacterial DNA was purified using phenol/chloroform/isoamyl alcohol (25:24:1) solution. The DNA was precipitated by adding ethanol and sodium acetate. RNase treatment and polyethylene glycol precipitation was performed. The V4 variable region (515F–806R) was sequenced on an Illumina Miseq following the method described previously by Kozich et al. (2013). Each reaction mixture contained 15 pmol of each primer, 0.2 mM deoxyribonucleoside triphosphates, 5 µl of 10× Ex Taq HS buffer, 1.25 U Ex Taq HS polymerase (Takara Bio Inc.), 50 ng extracted DNA, and sterilized water to reach a final volume of 50 µl. PCR conditions were as follows: 95°C for 2 min and then 25 cycles of 95°C for 20 s, 55°C for 15 s, and 72°C for 5 min, followed by 72°C for 10 min. The PCR product was purified by AMPure XP (Beckman Coulter) and quantified using a Quant-iT PicoGreen dsDNA Assay kit (Thermo Fisher Scientific). Mixed samples were prepared by pooling approximately equal amounts of PCR amplicons from each sample. The pooled library was analyzed with a High Sensitivity DNA kit on a 2100 Bioanalyzer (Agilent Technologies). Real-time PCR for quantification was performed on a pooled library using a KAPA Library Quantification kit (Illumina) following the manufacturer’s instructions. Based on the quantification, the sample library was denatured and diluted. A sample library with 20% denatured PhiX spike-in was sequenced by Miseq using a 500 Cycles kit (Illumina). We obtained 2 × 250–bp paired end reads. Taxonomic assignments and estimation of relative abundance of sequencing data were performed using the analysis pipeline of the QIIME software package (Caporaso et al., 2010). Chimera checking was performed using UCHIME (Edgar et al., 2011). An operational taxonomic unit (OTU) was defined at 97% similarity. OTUs indicating relative abundance of <1% were filtered to remove noise. The OTU was assigned a taxonomy based on a comparison with the Greengenes database using RDPclassifier (Wang et al., 2007; McDonald et al., 2012).
Statistical analyses
Student’s t test and one-way ANOVA followed by the Bonferroni’s post hoc test were used. P-values of <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows an analysis of IL-22BP expression in intestine and lymphoid tissues. Fig. S2 shows the generation of IL-22BP–deficient mice. Table S1 lists the primer sequences used for this study.
Supplementary Material
Acknowledgments
We thank the members of laboratory for Intestinal Ecosystem for technical support and experimental assistance. We also thank Dr. Calvin Kuo (Stanford University, Stanford, CA) for providing the R-spondin1–producing cell line and Dr. Hidenori Matsui for S. Typhimurium.
This work was supported in part by Junior Research Associates grant from Institute of Physical and Chemical Research (to T. Jinnohara), Japan Society for the Promotion of Science KAKENHI (26460584 to T. Kanaya, 25293114 and 26116709 to K. Hase, 26293111 to J. Kunisawa, 23229004 to H. Kiyono, and 24249029 and 16H05207 to H. Ohno), Ministry of Education, Culture, Sports, Science and Technology KAKENHI (15H01165 to T. Kanaya), Japan Agency for Medical Research and Development–Core Research for Evolutional Science and Technology (15652274 to H. Ohno), Core Research for Evolutional Science and Technology from the Japan Science and Technology Agency (to H. Kiyono), The Uehara Memorial Foundation (to T. Kanaya), The Naito Foundation Natural Science Scholarship (K. Hase), and the Terumo Foundation for Life Sciences and Arts (to J. Kunisawa).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AMP
- antimicrobial protein
- DN
- double negative
- EdU
- 5-ethynyl-2’-deoxyuridine
- FAE
- follicle-associated epithelium
- GF
- germ free
- IEC
- intestinal epithelial cell
- IL-22BP
- IL-22 binding protein
- IL-22R
- IL-22 receptor
- LP
- lamina propria
- LT
- lymphotoxin
- MLN
- mesenteric lymph node
- PP
- Peyer’s patch
- RANKL
- receptor activator of NF-κB ligand
- SED
- subepithelial dome
- SPF
- specific pathogen free
- VE
- villous epithelium
- WGA
- wheat-germ agglutinin
References
- Aujla S.J., Chan Y.R., Zheng M., Fei M., Askew D.J., Pociask D.A., Reinhart T.A., McAllister F., Edeal J., Gaus K., et al. 2008. IL-22 mediates mucosal host defense against gram-negative bacterial pneumonia. Nat. Med. 14:275–281. 10.1038/nm1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Kujala P., Schneeberger K., Middendorp S., Li V.S.W., Barker N., Martens A., Hofhuis F., DeKoter R.P., Peters P.J., et al. 2012. Peyer’s patch M cells derived from Lgr5+ stem cells require SpiB and are induced by RankL in cultured “miniguts”. Mol. Cell. Biol. 32:3639–3647. 10.1128/MCB.00434-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoutier L., Lejeune D., Colau D., and Renauld J.C.. 2001. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J. Immunol. 166:7090–7095. 10.4049/jimmunol.166.12.7090 [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., and Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Obata T., Kunisawa J., Sato S., Ivanov I.I., Lamichhane A., Takeyama N., Kamioka M., Sakamoto M., Matsuki T., et al. 2014. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 345:1254009 10.1126/science.1254009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P.A., and Doyle T.J.. 1993. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect. Immun. 61:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P.A., Doyle T.J., Hughes J.A., and Matsui H.. 1998. Analysis of host cells associated with the Spv-mediated increased intracellular growth rate of Salmonella typhimurium in mice. Infect. Immun. 66:2471–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash A.M., Dudakov J.A., Hua G., O’Connor M.H., Young L.F., Singer N.V., West M.L., Jenq R.R., Holland A.M., Kappel L.W., et al. 2012. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 37:339–350. 10.1016/j.immuni.2012.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K., Kawano K., Nochi T., Pontes G.S., Fukuda S., Ebisawa M., Kadokura K., Tobe T., Fujimura Y., Kawano S., et al. 2009. Uptake through glycoprotein 2 of FimH+ bacteria by M cells initiates mucosal immune response. Nature. 462:226–230. 10.1038/nature08529 [DOI] [PubMed] [Google Scholar]
- Huber S., Gagliani N., Zenewicz L.A., Huber F.J., Bosurgi L., Hu B., Hedl M., Zhang W., O’Connor W. Jr., Murphy A.J., et al. 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 491:259–263. 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immunological Genome Project 2017. ImmGen Database. Available at: http://www.immgen.org/databrowser/index.html (accessed May 2017).
- Iwasaki A., and Kelsall B.L.. 2000. Localization of distinct Peyer’s patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3α, MIP-3β, and secondary lymphoid organ chemokine. J. Exp. Med. 191:1381–1394. 10.1084/jem.191.8.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya T., Hase K., Takahashi D., Fukuda S., Hoshino K., Sasaki I., Hemmi H., Knoop K.A., Kumar N., Sato M., et al. 2012. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat. Immunol. 13:729–736. 10.1038/ni.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Fukuda S., Fujiwara A., Suda W., Hattori M., Kikuchi J., and Ohno H.. 2014. Multiple omics uncovers host–gut microbial mutualism during prebiotic fructooligosaccharide supplementation. DNA Res. 21:469–480. 10.1093/dnares/dsu013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-W., Suda W., Kim S., Oshima K., Fukuda S., Ohno H., Morita H., and Hattori M.. 2013. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 20:241–253. 10.1093/dnares/dst006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop K.A., Kumar N., Butler B.R., Sakthivel S.K., Taylor R.T., Nochi T., Akiba H., Yagita H., Kiyono H., and Williams I.R.. 2009. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J. Immunol. 183:5738–5747. 10.4049/jimmunol.0901563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., and Schloss P.D.. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79:5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraehenbuhl J.-P., and Neutra M.R.. 2003. Epithelial M cells: Differentiation and function. Annu. Rev. Cell Dev. Biol. 16:301–332. 10.1146/annurev.cellbio.16.1.301 [DOI] [PubMed] [Google Scholar]
- Linden S.K., Sutton P., Karlsson N.G., Korolik V., and McGuckin M.A.. 2008. Mucins in the mucosal barrier to infection. Mucosal Immunol. 1:183–197. 10.1038/mi.2008.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D.R., Ahrne S., Hyde L., Wei S., and Hollingsworth M.A.. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 52:827–833. 10.1136/gut.52.6.827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.C., Bériou G., Heslan M., Chauvin C., Utriainen L., Aumeunier A., Scott C.L., Mowat A., Cerovic V., Houston S.A., et al. 2014. Interleukin-22 binding protein (IL-22BP) is constitutively expressed by a subset of conventional dendritic cells and is strongly induced by retinoic acid. Mucosal Immunol. 7:101–113. 10.1038/mi.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., and Hugenholtz P.. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K., Ealey K.N., Kabata H., and Koyasu S.. 2015. Isolation and analysis of group 2 innate lymphoid cells in mice. Nat. Protoc. 10:792–806. 10.1038/nprot.2015.047 [DOI] [PubMed] [Google Scholar]
- Muñoz M., Heimesaat M.M., Danker K., Struck D., Lohmann U., Plickert R., Bereswill S., Fischer A., Dunay I.R., Wolk K., et al. 2009. Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J. Exp. Med. 206:3047–3059. 10.1084/jem.20090900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenci A., Becker C., Wullaert A., Gareus R., van Loo G., Danese S., Huth M., Nikolaev A., Neufert C., Madison B., et al. 2007. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 446:557–561. 10.1038/nature05698 [DOI] [PubMed] [Google Scholar]
- Obata T., Goto Y., Kunisawa J., Sato S., Sakamoto M., Setoyama H., Matsuki T., Nonaka K., Shibata N., Gohda M., et al. 2010. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl. Acad. Sci. USA. 107:7419–7424. 10.1073/pnas.1001061107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H. 2016. Intestinal M cells. J. Biochem. 159:151–160. 10.1093/jb/mvv121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I.L., Capecchi M.R., and Kuo C.J.. 2009. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15:701–706. 10.1038/nm.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C., Uhlig H.H., and Powrie F.. 2012. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 33:289–296. 10.1016/j.it.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H.-A., Hirth S., Weigmann B., Wirtz S., et al. 2009. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206:1465–1472. 10.1084/jem.20082683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios D., Wood M.B., Li J., Chassaing B., Gewirtz A.T., and Williams I.R.. 2016. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 9:907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino S.J., Geddes K., and Girardin S.E.. 2012. Innate IL-17 and IL-22 responses to enteric bacterial pathogens. Trends Immunol. 33:112–118. 10.1016/j.it.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Rumbo M., Sierro F., Debard N., Kraehenbuhl J.-P., and Finke D.. 2004. Lymphotoxin β receptor signaling induces the chemokine CCL20 in intestinal epithelium. Gastroenterology. 127:213–223. 10.1053/j.gastro.2004.04.018 [DOI] [PubMed] [Google Scholar]
- Sabat R., Ouyang W., and Wolk K.. 2014. Therapeutic opportunities of the IL-22–IL-22R1 system. Nat. Rev. Drug Discov. 13:21–38. 10.1038/nrd4176 [DOI] [PubMed] [Google Scholar]
- Sato S., Kaneto S., Shibata N., Takahashi Y., Okura H., Yuki Y., Kunisawa J., and Kiyono H.. 2013. Transcription factor Spi-B–dependent and –independent pathways for the development of Peyer’s patch M cells. Mucosal Immunol. 6:838–846. 10.1038/mi.2012.122 [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schreiber F., Arasteh J.M., and Lawley T.D.. 2015. Pathogen resistance mediated by IL-22 signaling at the epithelial–microbiota interface. J. Mol. Biol. 427:3676–3682. 10.1016/j.jmb.2015.10.013 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A.K., Blumberg R.S., Xavier R.J., and Mizoguchi A.. 2008. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118:534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.T., Patel S.R., Lin E., Butler B.R., Lake J.G., Newberry R.D., and Williams I.R.. 2007. Lymphotoxin-independent expression of TNF-related activation-induced cytokine by stromal cells in cryptopatches, isolated lymphoid follicles, and Peyer’s patches. J. Immunol. 178:5659–5667. 10.4049/jimmunol.178.9.5659 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., and Cole J.R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk K., Witte E., Hoffmann U., Doecke W.-D., Endesfelder S., Asadullah K., Sterry W., Volk H.-D., Wittig B.M., and Sabat R.. 2007. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: A potential systemic role of IL-22 in Crohn’s disease. J. Immunol. 178:5973–5981. 10.4049/jimmunol.178.9.5973 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., and Ouyang W.. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.