The pre-mRNA splicing factor PRPF8 is a crucial component of the U5 snRNP. Using quantitative proteomics, Malinová et al. show that assembly of the U5 snRNP is controlled by the HSP90/R2TP chaperones and that Retinitis pigmentosa–associated mutations in PRPF8 impair PRPF8 quality control and U5 snRNP chaperone-mediated assembly.
Abstract
Splicing is catalyzed by the spliceosome, a complex of five major small nuclear ribonucleoprotein particles (snRNPs). The pre-mRNA splicing factor PRPF8 is a crucial component of the U5 snRNP, and together with EFTUD2 and SNRNP200, it forms a central module of the spliceosome. Using quantitative proteomics, we identified assembly intermediates containing PRPF8, EFTUD2, and SNRNP200 in association with the HSP90/R2TP complex, its ZNHIT2 cofactor, and additional proteins. HSP90 and R2TP bind unassembled U5 proteins in the cytoplasm, stabilize them, and promote the formation of the U5 snRNP. We further found that PRPF8 mutants causing Retinitis pigmentosa assemble less efficiently with the U5 snRNP and bind more strongly to R2TP, with one mutant retained in the cytoplasm in an R2TP-dependent manner. We propose that the HSP90/R2TP chaperone system promotes the assembly of a key module of U5 snRNP while assuring the quality control of PRPF8. The proteomics data further reveal new interactions between R2TP and the tuberous sclerosis complex (TSC), pointing to a potential link between growth signals and the assembly of key cellular machines.
Introduction
Pre-mRNA splicing is a crucial step in eukaryotic gene expression that greatly diversifies the proteome. Splicing is catalyzed by a complex called the spliceosome, which consists of five major small nuclear ribonucleoprotein particles (snRNPs) and numerous non-snRNP proteins. Each snRNP contains a short noncoding uridine-rich RNA (snRNA), a ring of seven Sm or Like-Sm (in the case of U6 RNP) proteins, and 1–12 proteins that are specific for each snRNP (Matera and Wang, 2014).
The assembly of Sm-type snRNPs is a complex process that involves both cytoplasmic and nuclear phases. In metazoans, the nascent snRNAs are rapidly exported to the cytoplasm, where they associate with the survival of motor neurons (SMN) complex. This promotes the assembly of the Sm ring around a specific snRNA sequence (Matera and Wang, 2014; Raimer et al., 2016), and this core snRNP is then reimported to the nucleus, where it first appears in nuclear structures called Cajal bodies (CBs; Sleeman and Lamond, 1999). Here, the final stages of snRNP biogenesis occur, namely snRNA modifications and formation of mature snRNPs, which include the addition of snRNP-specific proteins and assembly of complex snRNPs (Staněk, 2016). Although the assembly of the Sm core by the SMN complex has been studied in great detail, little is known about the biogenesis of snRNP-specific proteins despite their importance for splicing.
Recent structural studies show that the U5 snRNP plays a central role in the spliceosome and particularly the highly conserved U5 protein PRPF8 (Yan et al., 2015; Bertram et al., 2017). PRPF8 adopts a complex 3D structure with a central cavity that forms the catalytic center of the spliceosome. PRPF8 also holds two other U5-specific proteins that perform essential functions during splicing: the helicase SNRNP200 (Brr2 in yeast) and the GTPase EFTUD2 (Snu114 in yeast). SNRNP200 unwinds the U4/U6 duplex to activate the spliceosome, whereas EFTUD2, along with PRPF8, controls SNRNP200 activity (Kuhn et al., 1999; Small et al., 2006; Maeder et al., 2009; Mozaffari-Jovin et al., 2012, 2013, 2014; Nguyen et al., 2013). Studies of the biogenesis of Prp8 in Saccharomyces cerevisiae showed that yeast Prp8 has a dedicated chaperone called Aar2, which forms a cytoplasmic complex with Prp8 and Snu114 (Gottschalk et al., 2001; Boon et al., 2007). This complex then binds U5 snRNA and translocates to the nucleus, where Aar2 is released and replaced by the helicase Brr2 (Boon et al., 2007). The biogenesis of snRNPs differs significantly between S. cerevisiae and metazoans (e.g., S. cerevisiae does not have the SMN protein), and despite the crucial roles of PRPF8, EFTUD2, and SNRNP200 in splicing, the biogenesis of these proteins is still uncharacterized in mammals.
It is noteworthy that investigation of Prp8 maturation in yeast used mutations that cause a hereditary retina dystrophy in humans (Boon et al., 2007). This disease, called Retinitis pigmentosa (RP), affects 1 out of 4,000 people worldwide and causes the gradual damage of rods and cones in the retina (Hartong et al., 2006; Mordes et al., 2006). There are nearly 90 genes that are associated with RP (RetNet Database), and many of them are known to be involved in retinal function, structure, and metabolism. However, the second most common target of RP mutations codes for ubiquitously expressed pre-mRNA splicing factors, including PRPF8 (Růžičková and Staněk, 2016). In PRPF8, all RP mutations cluster within the C-terminal Jab1/MPN (Mpr1/Pad1 N-terminal) domain that binds SNRNP200 (McKie et al., 2001; Kondo et al., 2003; Martínez-Gimeno et al., 2003; Ziviello et al., 2005; Towns et al., 2010). RP mutations in Prp8 have been originally studied in yeast, where different mutants exhibited growth and splicing defects (Boon et al., 2007; Maeder et al., 2009; Mozaffari-Jovin et al., 2013). Yeast and in vitro studies further demonstrated that mutations in the Jab1/MPN domain negatively affect the interaction with Brr2 and Snu114 proteins and alter the regulation of Brr2 helicase activity (Boon et al., 2007; Pena et al., 2007; Maeder et al., 2009; Mozaffari-Jovin et al., 2013, 2014).
In this study, we used RP mutations to study the biogenesis of the human PRPF8 protein, and we analyzed factors involved in the maturation of U5-specific proteins. We identified assembly intermediates containing PRPF8, EFTUD2, and SNRNP200 together with the chaperones HSP90/R2TP and AAR2. Furthermore, we provide evidence that the HSP90/R2TP machinery is involved in the assembly of this key module of U5 snRNP and that it helps discriminate between wild-type (WT) and assembly-defective mutants of PRPF8.
Results
RP mutations affect PRPF8 stability and localization
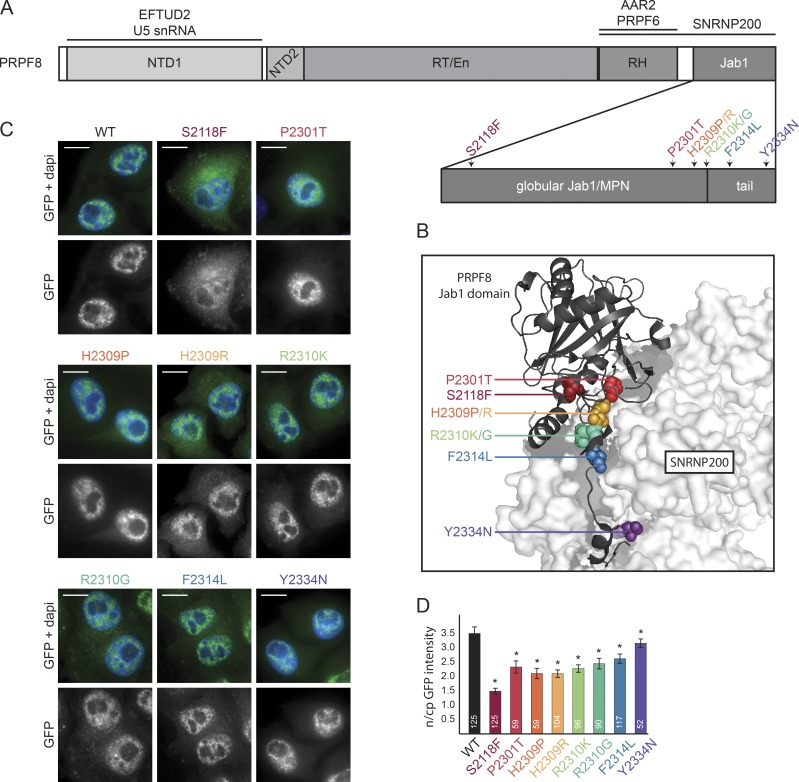
To study how RP-linked mutations influence the function of the PRPF8 protein, we introduced mutations into a bacterial artificial chromosome (BAC) that contained the entire human PRPF8 gene tagged at the protein C terminus with a localization and affinity purification (LAP) tag. This preserves the gene regulatory sequences and allows for protein expression at endogenous levels (Poser et al., 2008). We chose a C-terminal tag because an N-terminal FLAG-tag was previously shown to hinder the nuclear localization of Prp8 in Drosophila melanogaster (Claudius et al., 2014). In addition, PRPF8-WT-LAP fully complemented the depletion of endogenous PRPF8 (see next section), showing that the tag at the C terminus did not conflict with the function of the protein. There are 19 known mutations in the PRPF8 gene that cause RP (Růžičková and Staněk, 2016). We selected mutations that result in single amino acid changes, including the most common substitutions, H2309P, H2309R, and R2310K. We further chose S2118F, which is the first mutation found outside exon 42, and mutations P2301T, R2310G, F2314L, and Y2334N, which are found at different positions within the Jab1/MPN domain (Fig. 1, A and B). Using a two-step scarless mutagenesis method (Poser et al., 2008; Cvačková et al., 2014), individual mutations were introduced into the PRPF8-LAP BAC, and all LAP-tagged proteins were stably expressed in HeLa cells (Fig. 1 C).
Figure 1.
Description of PRPF8 and its RP-linked mutations. (A) A schematic organization of PRPF8 domains. RP-related mutations from this study are indicated by different colors. RH, RNase H-like domain. (B) View of the interaction between the C-terminal Jab1/MPN domain of PRPF8 (cartoon representation) and SNRNP200 (surface representation). The PRPF8 RP mutations are indicated with the same color code as in A. The picture is based on Mozaffari-Jovin et al. (2013); Protein Data Bank accession no. 4KIT. (C) Intracellular localization of PRPF8 variants. Panels are microscopy images of HeLa cells stably expressing the indicated LAP-tagged PRPF8 variants. Green/bottom panels, GFP; Blue, DAPI. Bars, 10 µm. (D) Nucleocytoplasmic (n/cp) localization of PRPF8 RP mutants. The ratio of nuclear and cytoplasmic GFP intensity was quantified using ImageJ. The mean of 52–125 cells (numbers of experiments are indicated in individual columns) together with the SD is shown. *, P < 0.05, as measured with t test comparing the indicated sample and the PRPF8-WT-LAP–expressing cells.
Neither WT nor RP mutants altered cell proliferation (not depicted), and WT and all mutated proteins localized to the cell nucleus. However, most mutated proteins were also partially present in the cytoplasm, with the S2118F mutant exhibiting the strongest cytoplasmic accumulation (Fig. 1, C and D). In addition, the expression of all RP mutants, with the exception of F2314L, was lower than PRPF8-WT-LAP (Fig. S1 A). The decrease in protein level may be caused by reduced expression or faster degradation. Because the promoter and cis-regulatory elements are identical for all PRPF8 variants, we examined the protein degradation rate. We blocked protein synthesis using cycloheximide and measured GFP fluorescence intensity at 15-min intervals over an 8-h period (Fig. S1 B). We observed that the stability of mutant proteins correlated with their expression level. Of the mutants analyzed, F2314L exhibited a degradation rate similar to the WT protein, whereas the other mutants were degraded more rapidly. S2118F and H2309P/R mutants were found to be the least stable.
RP mutations reduce splicing efficiency
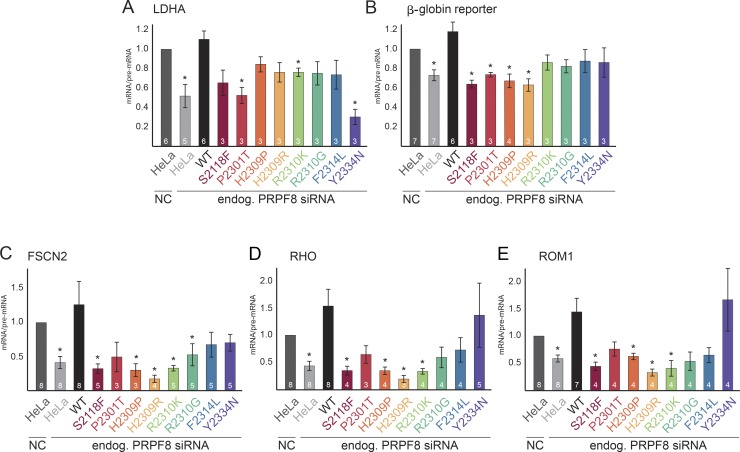
We then tested whether PRPF8 proteins with RP mutations were able to complement the depletion of endogenous PRPF8. To specifically knock down (KD) PRPF8, we used siRNA that was designed over the stop codon and that did not target the PRPF8-LAP fusion protein mRNA (Novotný et al., 2015). We frequently observed an increase in the expression of PRPF8-LAP proteins upon the KD of PRPF8 (Fig. S4 and not depicted), which likely reflects compensation for reduced levels of PRPF8. To assay the functionality of PRPF8-LAP fusion proteins in splicing, we analyzed the splicing efficiency of the endogenous LDHA gene or used an ectopically expressed reporter which was derived from the β-globin gene. Finally, we used three reporters that were derived from retina-specific genes: rhodopsin (RHO), fascin actin-bundling protein 2 (FSCN2), and retinal outer segment membrane protein 1 (ROM1). The KD of PRPF8 in parental HeLa cells significantly reduced splicing efficiency of all tested genes and was rescued by the expression of PRPF8-WT-LAP (Fig. 2, A–E; and Fig. S1, C–F). In contrast, none of the RP mutants was able to completely rescue splicing, but splicing defects varied among the tested genes and mutants. The strongest defects were observed for S2118F and H2309R mutations, which were not able to rescue splicing of four tested genes (except for LDHA). Out of all the tested RP mutations, the Y2334N mutant exhibited notably different behavior. It strongly reduced LDHA splicing, but other splicing reporters were unaffected. Interestingly, we observed diverse effects of PRPF8 KD on splicing efficiency of the tested genes. The β-globin reporter was less sensitive to the reduction of PRPF8 (Figs. 2 B and S1 C), whereas the splicing efficiency of two retina-specific reporters (FSCN2 and RHO) dropped below 50% after PRPF8 KD. Four RP mutants (S2118F, H2309P, H2309R, and R2310K) were not able to rescue the splicing defects of all tested retina-specific reporters (Fig. 2, C–E; and Fig. S1, D–F). In general, splicing of retina-specific reporters was more sensitive to the reduction of PRPF8 levels, and RP mutants rescued retina-specific genes less efficiently than other genes. Collectively, these data show that the tested RP mutations affect the splicing activity of PRPF8.
Figure 2.
PRPF8 RP mutations affect splicing. (A) Splicing efficiencies of PRPF8 mutants. Graphs display the splicing efficiencies of the endogenous (endog.) LDHA expressed as the ratio of mRNA over pre-mRNA and measured by reverse transcription quantitative PCR in cells knocked down for PRPF8 and expressing the indicated LAP-tagged PRPF8 variants. Results have been normalized to the ratio obtained in parental HeLa cells treated with an NC siRNA. (B–E) Graphs displaying the splicing efficiency of β-globin gene reporter (B) and retina-specific gene reporters FSCN2 (C), RHO (D), and ROM1 (E). Legend is the same as in A except that RT-PCR was used instead of reverse transcription quantitative PCR. Error bars represent SEM. Numbers of experiments are indicated in individual columns. *, P < 0.05, as measured with t test comparing the indicated sample and the parental cells treated with the control siRNA.
RP mutations compromise U5 snRNP maturation and formation of the U4/U6.U5 tri-snRNP
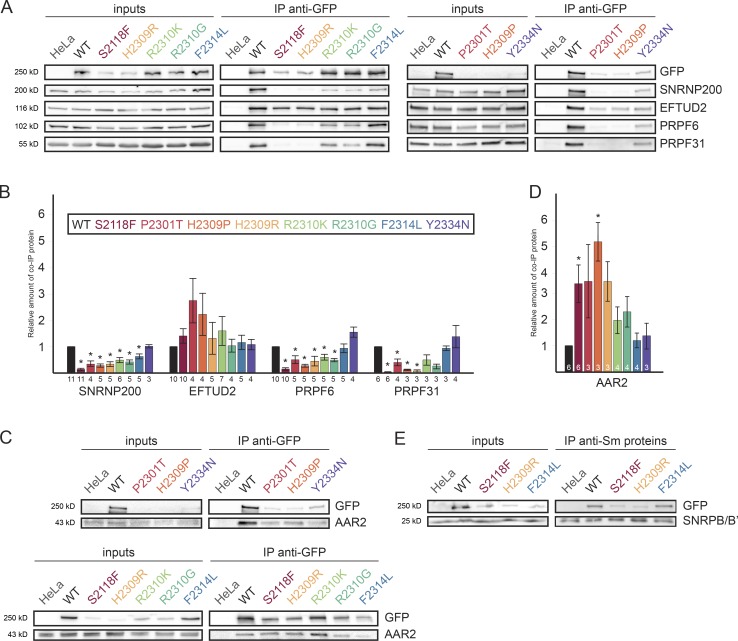
To test whether the splicing defects observed with the PRPF8 mutants were caused by a reduced snRNP assembly, we measured the interaction of RP mutants with several snRNP-specific proteins. PRPF8 RP mutants and WT proteins were immunoprecipitated using anti-GFP antibodies, and the coprecipitated proteins were detected by Western blotting (WB; Fig. 3 A). PRPF8-WT-LAP was properly incorporated into U5 and tri-snRNPs, as documented by the coprecipitation of U5-specific proteins (SNRNP200, EFTUD2, and PRPF6) and of the U4/U6-specific factor PRPF31. To quantitatively evaluate the PRPF8 mutants, the amount of coprecipitated proteins was quantified and normalized to the GFP signal in the pellets (Fig. 3 B). With the exception of Y2334N, which did not display any visible alterations, the mutants exhibited defects in the binding of SNRNP200, which is consistent with the location of the mutations in the SNRNP200 interaction domain Jab1/MPN. This translated into a reduced snRNP formation, demonstrated by significantly lower amounts of PRPF6 and PRPF31 copurified with the mutants as compared with WT (except for the F2314L mutant). These data show that most RP mutations in the PRPF8 gene reduce the formation of snRNPs.
Figure 3.
RP mutations inhibit U5 snRNP assembly. (A) Association of PRPF8 variants with U5 and U4/U6.U5 snRNPs. PRPF8-LAP variants are indicated at the top. IP was done with anti-GFP antibodies. Inputs contained 2% of the materials in the IP, and the antibodies used for WB are indicated at the right. (B) Quantification of the immunoprecipitated material. The graph displays the amount of the protein present in the pellet and normalized, first to the amount of copelleted GFP and then to the same ratio measured in the PRPF8-WT-LAP pellets. (C and D) Association of PRPF8 variants with AAR2. Legend is as in A and B. Error bars represent SEM. The exact number of each experiment is indicated below/in each column. *, P < 0.05, as measured with t tests comparing the indicated sample and WT PRPF8-LAP IPs. (E) Association of three selected PRPF8 variants with snRNPs. WBs were made with extracts of cells expressing the PRPF8-LAP variant indicated at the top (input lanes) and immunoprecipitated with anti-Sm Y12 antibody. PRPF8-LAP proteins were detected with anti-GFP antibody and SNRPB/B’ with the Y12 antibody. Inputs contain 2% of the materials in the IP.
In yeast, Prp8 and Snu114 form an intermediate complex with the chaperone Aar2 before joining U5 snRNA. To test whether this was also the case in human cells, we tested the association of PRPF8 RP mutants with AAR2 and EFTUD2. Both AAR2 and EFTUD2 efficiently copurified with all mutated proteins (Fig. 3, A–D), and two RP mutants even showed a significantly enhanced coprecipitation in comparison with WT. Increased interactions with EFTUD2 and AAR2 correlated with a decreased binding of the mutants to the other U5-specific proteins. These results support the idea that PRPF8, AAR2, and EFTUD2 form an assembly intermediate in human cells and that some of the mutants are trapped at this stage. To test whether this intermediate complex associates with the core U5 snRNP, we measured the interaction of selected PRPF8 RP mutants with Sm proteins, which stably associate with U5 snRNA. We compared two PRPF8 mutations, S2118F and H2309R, which stall its biogenesis, with WT and the F2314L mutant that are both incorporated into U5 and the tri-snRNPs (Fig. 3, A and B). We immunoprecipitated snRNPs via Sm proteins and analyzed coprecipitated PRPF8-LAP fusion proteins (Fig. 3 E). We found that the F2314L mutant copurified with Sm proteins to the same extent as WT. In contrast, the S2118F and H2309R mutants showed only negligible interaction with Sm proteins. Collectively, these data provide strong support for the idea that PRPF8 associates with EFTUD2 and AAR2 early in its assembly pathway. Immunoprecipitation (IP) of Sm proteins further suggests that the PRPF8-AAR2-EFTUD2 is formed before its loading onto the core U5 snRNP and that at least two tested RP mutations (S2118F and H2309R) stall assembly at this stage.
AAR2, ZNHIT2, and the HSP90/R2TP complex associate with the U5-specific proteins EFTUD2, PRPF8, and SNRNP200
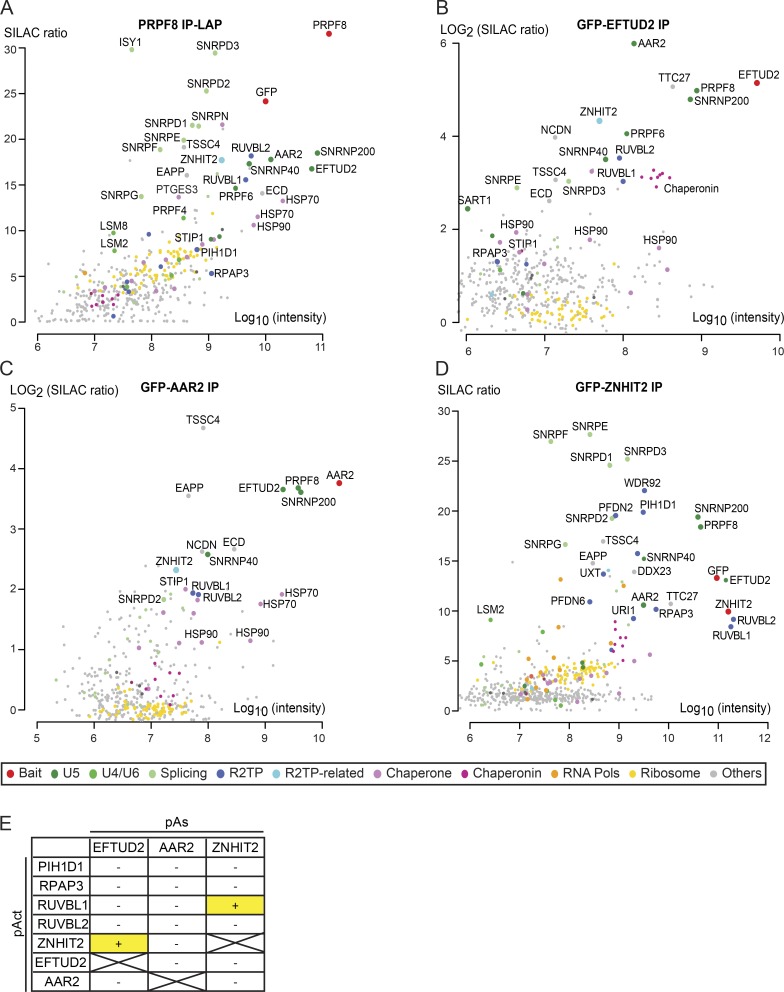
These data indicate that the assembly pathway of PRPF8 appears to be key to understanding the molecular defects that are caused by RP mutations. To define this pathway in more detail, we used an unbiased quantitative proteomic approach based on stable isotope labeling in cell culture (SILAC) in conjunction with anti-GFP IP (Ong et al., 2002; Trinkle-Mulcahy et al., 2008). We first used HeLa cell–expressing PRPF8-LAP (see the first Results section) and GFP-EFTUD2 as bait (Fig. 4, A and B). In the latter cells, GFP-EFTUD2 localized mainly to nuclear speckles, as expected, and was also weakly detected in the cytoplasm (Fig. S2 A). The proteomic analysis of the GFP IPs showed a strong association of PRPF8-LAP with its partners EFTUD2 and SNRNP200, and conversely, GFP-EFTUD2 strongly bound PRPF8 and SNRNP200. Several other components of U5 snRNP were identified with high SILAC ratios in the two IPs, namely SNRNP40, PRPF6, and Sm proteins. Additional tri-snRNP components were also detected (e.g., SART1, USP39, and PRPF4), indicating that the tagged proteins were incorporated into U5 and tri-snRNPs. In agreement with our coimmunoprecipitations (Fig. 3, C and E), AAR2 was also detected with high SILAC ratios in both the PRPF8 and EFTUD2 IPs, indicating that in addition to the mature U5 snRNP, the IPs also detected assembly intermediates. Interestingly, additional chaperones were found, including HSP70 and HSP90, regulators of HSP90 (PTGES3 and STIP1), and subunits of R2TP (RUVBL1 and 2), a known HSP90 cochaperone. Finally, a series of poorly characterized proteins were also enriched in one or both of the IPs: ECD, TTC27, TSSC4, EAPP, NCND, and ZNHIT2 (see Table S1 for the complete list of coprecipitated proteins). None of these proteins is part of the mature U5 snRNP (Makarov et al., 2002), and they thus represent potential U5 snRNP assembly factors.
Figure 4.
SILAC quantitative proteomic and yeast two-hybrid analyses of U5 snRNP proteins and assembly factors. (A) Proteomic analyses of the partners of PRPF8-WT-LAP. (B–D) SILAC proteomic analyses of GFP-EFTUD2 (B), GFP-AAR2 (C), or GFP-ZNHIT2 (D). Graphs display SILAC ratios (y axis) as a function of signal abundance (x axis) measured by quantitative proteomic analysis of extracts from HeLa cells expressing the indicated GFP-protein and immunoprecipitated with anti-GFP antibodies. SILAC ratios were calculated from a control IP done with parental HeLa cells. Each dot represents a protein and is color coded according to the classification shown below SILAC panels. The labeled dots were arbitrarily selected to highlight proteins relevant to this study. Significance values are given in Table S1. (E) Yeast two-hybrid analysis of interactions between R2TP, ZNHIT2, EFTUD2, and AAR2. The presence or absence of interactions is indicated by “+” and “−,” respectively, for the pairs of interactions tested.
To characterize in more detail the U5 assembly intermediates, we performed a SILAC IP with GFP-AAR2 as bait (Fig. 4 C and Table S1). The GFP–AAR2 fusion was stably expressed in HeLa cells and was detected in the cytoplasm and, to a lesser extent, in the nucleus (Fig. S2 B). GFP-AAR2 associated not only with PRPF8 and EFTUD2 as expected, but also with SNRNP200, which was surprising because the yeast homologues of AAR2 and SNRNP200 were shown to bind PRPF8 in a mutually exclusive manner (Weber et al., 2011, 2013; Nguyen et al., 2013). Nevertheless, the GFP-AAR2 IP purified only a few other U5 proteins, which is in agreement with the idea that AAR2 binds PRPF8 mostly before its association with U5 snRNA. Interestingly, many chaperones and assembly factors found in the previous IPs were also associated with GFP-AAR2: HSP70, HSP90, STIP1, the R2TP components RUVBL1/2, as well as the putative assembly factors TSSC4, EAPP, NCDN, ECD, and ZNHIT2.
Next, we turned our attention to the R2TP complex. This complex functions as a cochaperone of HSP90, and it comprises four subunits: RPAP3, PIH1D1, and RUVBL1/2 (McKeegan et al., 2007; Boulon et al., 2012). The HSP90/R2TP system has been shown to participate in the assembly of multisubunit complexes including the U4 snRNP (Bizarro et al., 2015), small nucleolar ribonucleoprotein particles (snoRNPs; Boulon et al., 2008; Zhao et al., 2008), nuclear RNA polymerases (Boulon et al., 2010; Forget et al., 2010), and PIKKs-containing complexes (Hořejší et al., 2010, 2014; Kamano et al., 2013). The detection of R2TP subunits in association with PRPF8, EFTUD2, and AAR2 thus suggests that it could participate in the biogenesis of U5 snRNA. We have previously proposed that proteins of the ZNHIT family function as cofactors for the RUVBL1/2 ATPases of the HSP90/R2TP chaperone system (Bizarro et al., 2014; Verheggen et al., 2015). The presence of both ZNHIT2 and R2TP in the previous IPs thus suggested that ZNHIT2 could work as an R2TP cofactor specialized for U5 biogenesis. To test this idea, we investigated the partners of ZNHIT2 by performing a systematic pairwise two-hybrid analysis. This revealed interactions between ZNHIT2 and both RUVBL1 and EFTUD2 (Fig. 4 E). ZNHIT2 could thus potentially bridge R2TP with U5-specific proteins. Next, we stably expressed GFP-ZNHIT2 in HeLa cells (Fig. S2 C) and performed a SILAC proteomic analysis (Fig. 4 D and Table S1). Remarkably, the GFP-ZNHIT2 SILAC IP revealed a strong association of this factor with EFTUD2, PRPF8, and SNRNP200. Association of ZNHIT2 with PRPF8 was confirmed by coimmunoprecipitation (Fig. S2 D). GFP-ZHNIT2 further copurified large amounts of the R2TP complex, especially RUVBL1/2 and a set of prefoldins known to associate with R2TP (e.g., URI1, UXT, and PFDN6; Boulon et al., 2010; Cloutier and Coulombe, 2010). Consistent with a role in PRPF8 biogenesis, the GFP-ZNHIT2 SILAC IP was enriched for AAR2 and other putative assembly factors, including TTC27, TSSC4, EAPP, NCDN, and ECD. Interestingly, GFP-ZNHIT2 also copurified large amounts of Sm proteins, indicating that it remained present until the late stages of U5 snRNP biogenesis.
The HSP90/R2TP machinery binds unassembled cytoplasmic forms of PRPF8 and EFTUD2
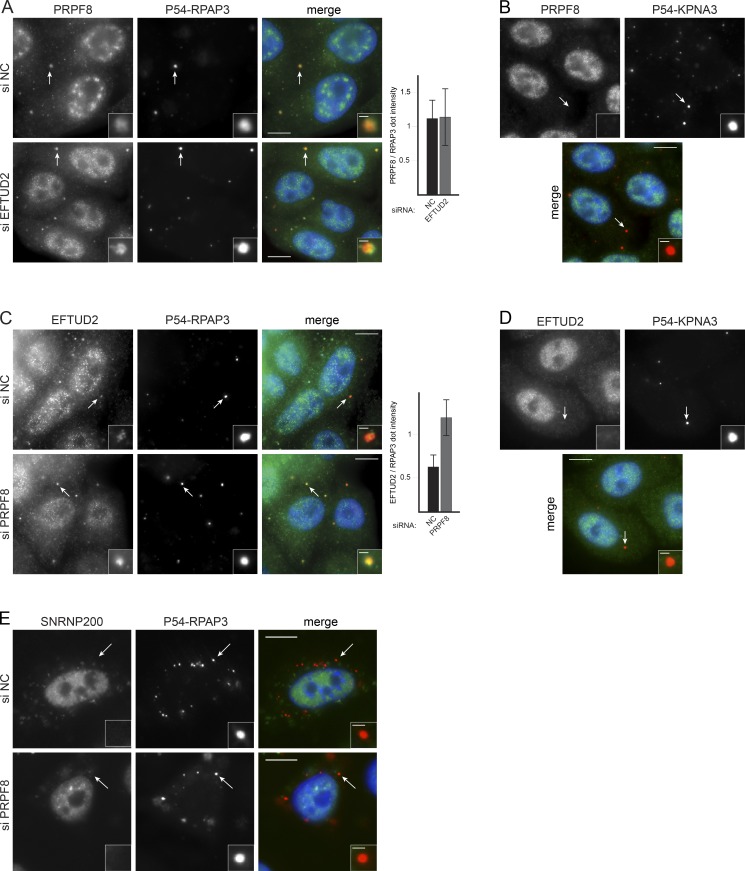
PRPF8 is synthesized in the cytoplasm and must be transported to the nucleus to perform its role in splicing. To determine whether the interactions between R2TP and PRPF8 or EFTUD2 occur at an early cytoplasmic stage, we implemented a corecruitment assay using a fusion of RPAP3 with a fragment of p54, which targets the fusion protein to cytoplasmic P-bodies (Boulon et al., 2010). Using immunofluorescence (IF), we detected PRPF8 and GFP-AAR2 localizing in P-bodies that contained p54-RPAP3 (Figs. 5 A and S3 C) but not in P-bodies labeled by p54-KPNA3, a nuclear transport factor that served as a negative control (NC; Figs. 5 B and S3 C). Interestingly, the interaction between p54-RPAP3 and PRPF8 also occurred when EFTUD2 was depleted, which was further confirmed by regular coimmunoprecipitation (Fig. S3 A). This demonstrated that the interaction between PRPF8 and RPAP3 can take place in the cytoplasm and is independent of EFTUD2.
Figure 5.
R2TP interacts with EFTUD2 and PRPF8 in the cytoplasm. (A and B) Analysis of the interaction between PRPF8 and RPAP3 with a cytoplasmic corecruitment assay. (A) HeLa cells stably expressing a p54-DsRed2-RPAP3 fusion and treated with a siRNA against EFTUD2 or NC siRNA (siNC). Green/left panels, PRPF8 as detected by IF; red/middle panels, p54-DsRed2-RPAP3; blue, DAPI. A quantification of green over red signals in dots is shown on the right. (B) Same as in A, except that cells express p54-DsRed2-KPNA3 as controls. (C and D) Interaction between RPAP3 and EFTUD2 tested with a cytoplasmic corecruitment assay with and without PRPF8 KD. (C) Legend is as in A. (D) control with cells expressing p54-DsRed2-KPNA3. Values are means calculated from >25 dots. Error bars indicate SD. (E) Interaction between SNRNP200 and RPAP3 tested with a cytoplasmic corecruitment assay with and without PRPF8 KD. Legend is as in A. Insets are magnified images of the dots marked by the white arrows. Bars: (main images) 10 µm; (insets) 1 µm.
Next, we tested the interaction of EFTUD2 and SNRNP200 with p54-RPAP3. EFTUD2 was weakly corecruited to p54-RPAP3 P-bodies under normal conditions, and this interaction increased after the KD of PRPF8 (Fig. 5, C and D), a result also confirmed by coimmunoprecipitations (Fig. S3 B). This indicated that EFTUD2 interacts with R2TP in the cytoplasm independently of PRPF8. In contrast, the SNRNP200-GFP fusion failed to be recruited with p54-RPAP3 even after KD of PRPF8, EFTUD2, or AAR2 (Figs. 5 E and S3 D). Furthermore, SNRNP200-GFP was not recruited on p54-ZNHIT2 P-bodies, whereas EFTUD2 and PRPF8 were (Fig. S3 E). The lack of cytoplasmic interaction of SNRNP200 with RPAP3 and ZNHIT2 suggests that SNRNP200 becomes stably associated into these complexes later in the nucleus.
Collectively, our data confirm that interactions of the R2TP complex with PRPF8 and EFTUD2 occur early on in the cytoplasm and are independent of each other. In contrast, recruitment of SNRNP200 occurs at a later step and most likely in the nucleus.
The PIH1D1 N-terminal domain coprecipitates several R2TP clients but not U5 proteins
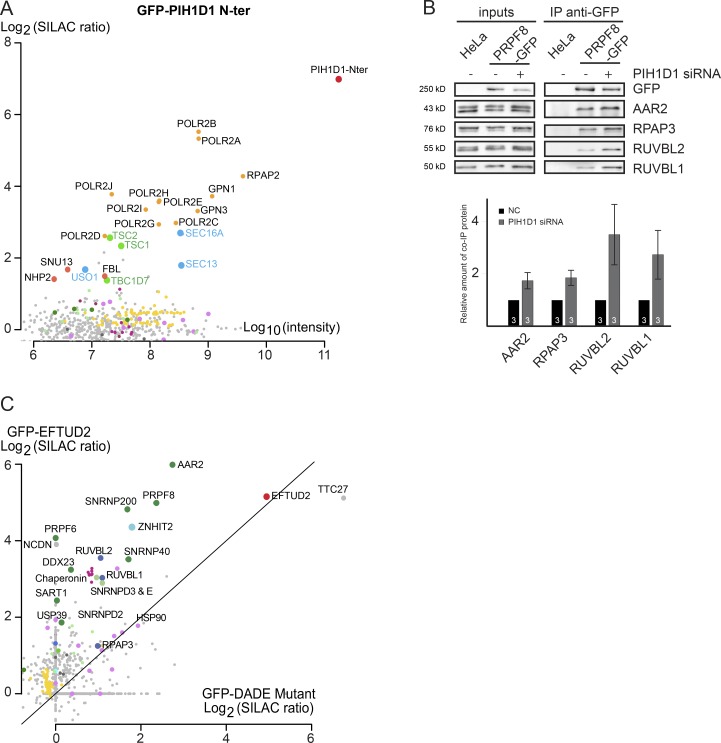
These data indicate that EFTUD2 and PRPF8 make early interactions with R2TP. PIH1D1 is a client-binding subunit of R2TP, and its N-terminal domain binds phosphorylated peptides with a DSDD(D/E) consensus sequence (Hořejší et al., 2014). EFTUD2 possesses a conserved phosphorylated DSDED sequence at its N terminus (Hornbeck et al., 2015), and it was shown to make phosphorylation-dependent interactions with PIH1D1 in vitro (Hořejší et al., 2014). The N-terminal domain of PIH1D1 could thus recruit EFTUD2 to R2TP via an interaction with its DSDED motif. To test whether the PIH1D1 N terminus brings EFTUD2 into the R2TP complex and to determine the role of the DSDED motif, we analyzed the interactomes of (i) the PIH1D1 N-terminal domain and (ii) EFTUD2 with a mutated DSDED motif.
We fused the N-terminal domain of PIH1D1 to GFP, stably expressed it in HeLa cells, and performed a SILAC proteomic analysis (Fig. 6 A). The N terminus of PIH1D1 was not incorporated into R2TP because it did not copurify other subunits of this complex, which is in agreement with previous studies (Hořejší et al., 2014; Pal et al., 2014). Although the PIH1D1 N-terminal domain bound tightly to some known R2TP substrates—two box C/D core proteins (SNU13 and FBL) and factors involved in RNA pol II assembly (polymerase subunits RPAP2, GPN1, and GPN3)—we did not detect any enrichment for EFTUD2 or other U5 proteins. Interestingly, the PIH1D1 N terminus also bound two complexes not previously known to associate with R2TP: a SEC16A complex containing SEC13 and USO1 and also all the three subunits of the tuberous sclerosis complex (TSC; TSC1, TSC2, and TBC1D7), which is a major regulator of mTOR (Dibble and Cantley, 2015). To further test whether PIH1D1 is important for the recruitment of U5 proteins to the R2TP complex, we used siRNA to reduce PIH1D1 expression in the PRPF8-LAP cell line and immunoprecipitated PRPF8-LAP with anti-GFP antibodies (Fig. 6 B). The results showed that the interactions of PRPF8 with R2TP and AAR2 increased in the absence of PIH1D1. This indicates that although PIH1D1 is not required to recruit PRPF8 to the R2TP, it may be important for the release of AAR2 and thus the maturation of PRPF8.
Figure 6.
PIH1D1 is not essential for PRPF8–R2TP interaction. (A) Proteomic analyses of the partners of the N-terminal domain of PIH1D1 tagged with GFP. The graph displays SILAC ratios (y axis) as a function of signal abundance (x axis) measured by a quantitative proteomic of extracts from HeLa cells expressing the GFP-PIH1D1 N-terminal domain and immunoprecipitated with anti-GFP antibody. SILAC ratios are calculated from a control IP done with parental cells. Each dot represents a protein and is color-coded according to the classification shown in Fig. 4. Significance values are given in Table S1. (B) Changes in the partners of PRPF8 upon depletion of PIH1D1. The top image represents WBs made with extracts of cells expressing PRPF8-WT-LAP and treated with PIH1D1 or control siRNA (lanes “+” and “−,” respectively) and then immunoprecipitated with anti-GFP antibody (IP anti-GFP lane). Inputs contain 2% of the materials in the IP, and the antibodies used to probe the WB are indicated at the right. The bottom graph displays the amount of the protein present in the pellet and normalized, first to the amount of copelleted GFP and then to the same ratio measured in the NC siRNA. Values were measured with ImageJ and are the means of three experiments. The exact number of each experiment is indicated in each column. Error bars indicate SEM. (C) Comparison of GFP-EFTUD2 WT and the DADE mutant interactomes as detected by SILAC IPs. Log2 of SILAC ratios of the two proteins are plotted against each other.
The DSDED motif of EFTUD2 is important for the assembly of U5 snRNP
To study the role of the DSDED motif in EFTUD2, we generated a phospho-dead mutant of this motif (S19A; DADE mutant). This mutant was then fused to GFP and stably expressed in HeLa cells. Interestingly, the mutant displayed an altered localization with increased levels in the cytoplasm and lower accumulation in the nucleus (Fig. S2 F). We then performed a quantitative SILAC proteomic analysis of the mutant (Fig. S2 E). Direct comparison with WT GFP-EFTUD2 revealed that the DADE mutant had a dramatically altered interactome (Fig. 6 C). First, it bound more strongly to the poorly characterized factor TTC27. Second, it bound less strongly to a set of factors that included Sm proteins, U5-specific proteins (PRPF8, SNRNP200, and SNRNP40), and proteins found in U5 assembly intermediates (AAR2, RUVBL1/2, ZNHIT2, and Chaperonin). Third, it lost interactions with another set of U5-specific proteins (PRPF6 and DDX23) as well as with U4/U6 and U4/U6.U5 proteins (SART1, USP39, and PRPF4). The proteomic data thus indicated major defects in the formation of snRNP particles and pointed to the essential role of the phosphorylated DSDED motif of EFTUD2 during the biogenesis of U5 and the tri-snRNP.
Collectively, our data show two partially contradictory observations: (i) that the PIH1D1 N-terminal domain is not sufficient to stably bind EFTUD2 and (ii) that the DSDED motif of EFTUD2 is important for U5 maturation. In vitro, the affinity of the PIH1D1 N-terminal domain with the EFTUD2 phosphorylated peptide is ∼10 times weaker than previously published PIH1D1 phosphointeractions (Hořejší et al., 2014; Pal et al., 2014; Smerdon, S., personal communication). This may be insufficient to ensure a stable binding in vivo, and additional factors such as ZNHIT2 may be essential to secure the interaction between EFTUD2 and R2TP. A similar situation has been described for ECD binding to R2TP when two independent interaction sites (one PIH1D1-dependent and one PIH1D1-independent) were together necessary for a stable ECD–R2TP binding (Mir et al., 2015). The phosphorylated DSDED motif may therefore stabilize EFTUD2 binding to R2TP after its initial recruitment and/or may trigger a conformational change important for later assembly steps of U5 snRNP. We can also not formally exclude that the effect of the S→A mutation in the DSDED motif is independent of R2TP.
The R2TP complex is important for the stability of EFTUD2 and U5 snRNP maturation
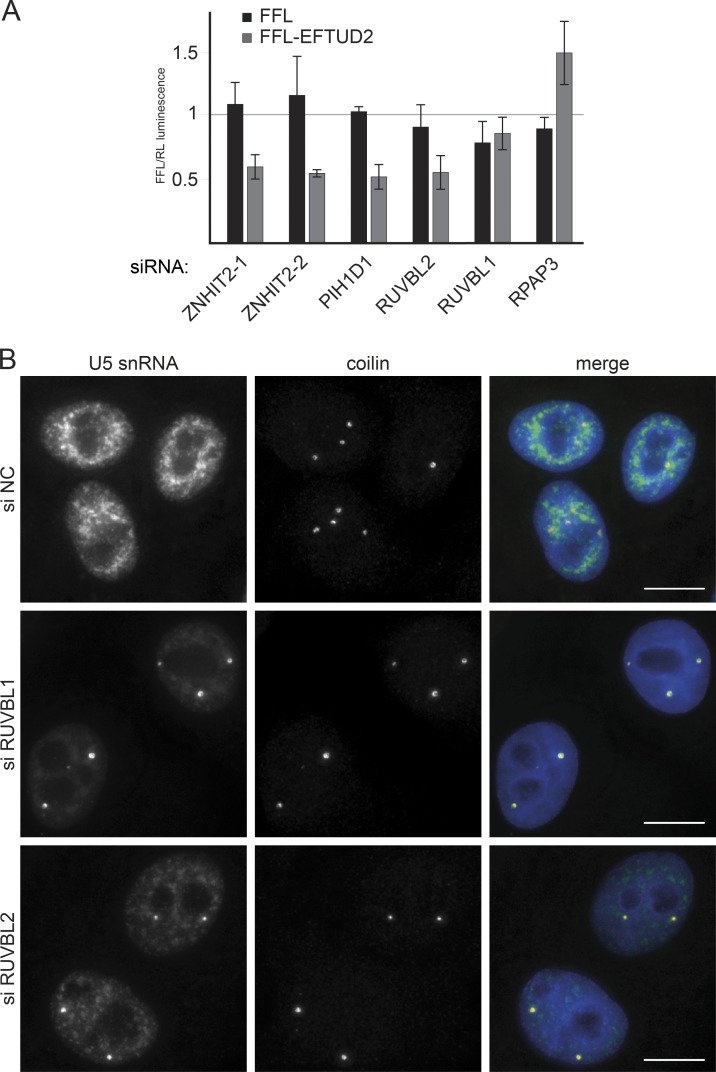
To confirm that R2TP is involved in EFTUD2 maturation, we knocked down its individual components with siRNAs and measured the level of newly synthesized EFTUD2. To this end, we fused EFTUD2 with firefly luciferase (FFL) and transiently transfected the corresponding expression vector in HeLa cells. This transient expression system ensured that the analysis focused on newly synthesized proteins and not on proteins that had been already incorporated into U5 snRNP (Boulon et al., 2008; Bizarro et al., 2014). KD of ZNHIT2, PIH1D1, and to a lesser extent RUVBL2 reduced the levels of FFL-EFTUD2 but not of FFL alone, which was used as a control (Fig. 7 A). This indicated that ZNHIT2 and several subunits of the R2TP complex were important for the stability of the nascent EFTUD2 protein.
Figure 7.
R2TP components are important for EFTUD2 stability and U5 snRNP maturation. (A) Levels of newly synthesized FFL-EFTUD2 after KD of R2TP and ZNHIT2. The graph displays FFL activity measured in extracts of HeLa cells transiently transfected with the indicated plasmid and siRNAs and then normalized first to RL activity (expressed from a cotransfected plasmid) and then to the ratio obtained with control siRNAs. Values are the means of at least three experiments, each done in triplicate. Error bars indicate SEM. (B) U5 snRNA accumulation in CBs after KD of RUVBL1/2. HeLa cells were treated with siRNAs against RUVBL1/2 or an NC siRNA. Green/left panels, U5 snRNA was detected by FISH; red/middle panels, coilin detected by IF; blue, DAPI. Bars, 10 µm.
A previous study has shown that defects in U5 snRNP maturation cause the accumulation of U5 snRNA in CBs (Novotný et al., 2015). To confirm the role of R2TP in U5 biogenesis, we treated HeLa cells with siRNAs targeting RUVBL1 and 2 and determined the intracellular localization of U5 snRNA. Depleted cells displayed a clear accumulation of U5 snRNA in CBs (Fig. 7 B), thus confirming that these key R2TP components are required for the maturation of U5 snRNP.
HSP90 stabilizes cytoplasmic unassembled PRPF8
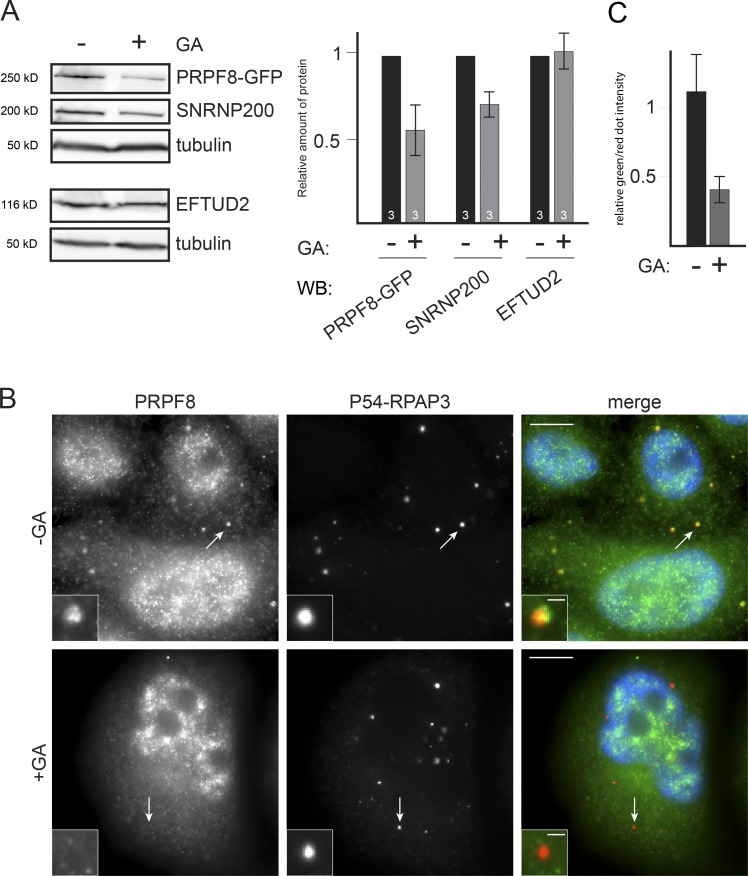
The R2TP complex is a cochaperone for HSP90, and the interactions described in this study suggest that HSP90 could be involved in the biogenesis of PRPF8 and other U5-specific proteins. To test whether PRPF8, EFTUD2, and SNRNP200 were clients of HSP90, we inhibited its activity using geldanamycin (GA). This locks HSP90 in a closed state and usually results in client degradation (Whitesell et al., 1994). HSP90 inhibition reduced the levels of PRPF8 and SNRNP200, but not of EFTUD2 (Fig. 8 A). This suggests that PRPF8 and SNRNP200 are potential client proteins of HSP90.
Figure 8.
HSP90 is required for PRPF8 stability in the cytoplasm. (A) Effect of GA on the levels of U5 proteins. Left images represent WBs made with extracts of HeLa cells expressing WT PRPF8-LAP and treated (+) or untreated (−) with GA. WBs were probed with the antibodies indicated at the right. On the right is a quantification of the signals as measured with ImageJ and expressed as percentages of the untreated controls. Values are means of three experiments. (B) Effect of GA on the levels of RPAP3-bound cytoplasmic PRPF8. Legend as in Fig. 5, except cells were treated with GA. Insets are magnified images of the dots marked by the white arrows. Bars: (main images) 10 µm; (insets) 1 µm. (C) Quantification of the green (PRPF8) and red (p54-RPAP3) signals in 30 dots shown at B. Error bars indicate SEM.
To determine whether HSP90 stabilizes nuclear or cytoplasmic forms of PRPF8, we performed the same corecruitment assay as in Fig. 5 A but in the presence of GA. Interestingly, although GA treatment had no obvious effect on the nuclear pool of PRPF8 (Fig. 8 B and unpublished data), it reduced the amount of PRPF8 present in p54-RPAP3 P-bodies (Fig. 8, B and C). These data suggest that HSP90 activity is required to stabilize cytoplasmic forms of PRPF8.
The R2TP complex associates with PRPF8 assembly-deficient mutants and sequesters them in the cytoplasm
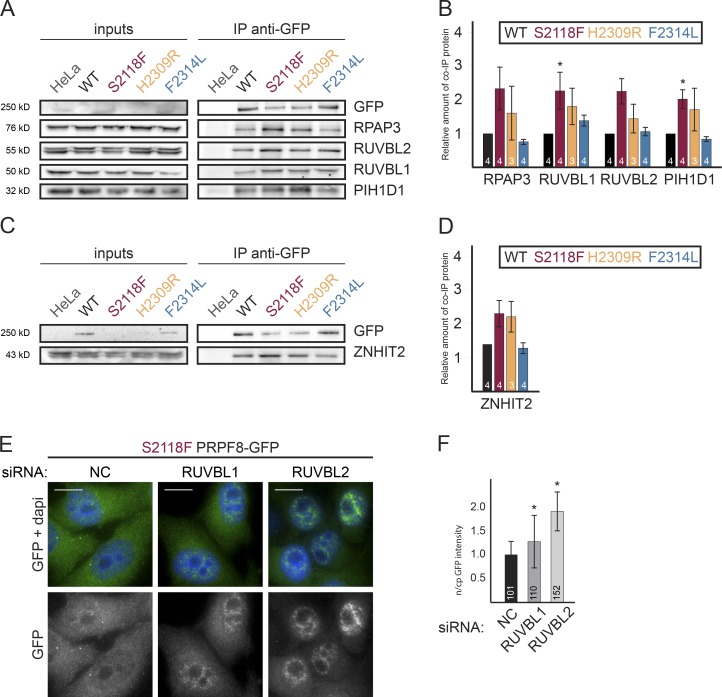
The aforementioned data indicate that HSP90 and its R2TP cochaperone are involved in the biogenesis of the U5-specific proteins PRPF8 and EFTUD2. At the same time, we have shown that RP mutations stall PRPF8 maturation and impair its interaction with SNRNP200 and U5 snRNA (Fig. 3). To test whether R2TP associates with the stalled RP mutants, we used PRPF8-LAP cell lines. We focused on mutations S2118F and H2309R, which exhibited the strongest assembly phenotype. As controls, we used WT PRPF8 and the F2314L mutant, both of which incorporate into the tri-snRNP. S2118F and H2309R mutations both increased the association of PRPF8 with R2TP (Fig. 9, A and B) and also with ZNHIT2 (Fig. 9, C and D). These results show that R2TP binds more strongly to PRPF8 assembly-deficient mutants.
Figure 9.
The R2TP complex binds more strongly to RP PRPF8 assembly-deficient mutants and is required for their cytoplasmic retention. (A) WB made with extracts of cells expressing the indicated PRPF8-LAP variants (input lane) and immunoprecipitated with an anti-GFP antibody. Inputs contain 2% of the materials in the IP, and the antibodies used to probe the WB are indicated at the right. (B) The graph displays the amount of the protein present in the pellet and normalized first to the amount of copelleted GFP and then to the same ratio measured in the NC siRNA. Values were measured with ImageJ and are the means of 3–4 experiments (indicated in each bar). (C and D) Increased binding of ZNHIT2 to PRPF8 RP mutants. Legend is as in A and B. Error bars indicate SEM. (E and F) Relocalization of PRPF8-S2118F-LAP upon RUVBL1/2 KDs. (E) HeLa cells stably expressing PRPF8-S2118F-LAP fusion protein and treated with siRNAs against RUVBL1/2 or a control siRNA. Green/bottom panels, PRPF8-LAP–S2118F; blue, DAPI. Bars, 10 µm. (F) The ratio of nuclear and cytoplasmic (n/cp) GFP signal intensity was quantified using ImageJ software. The means of the measurement of 101–152 cells along with the SD is shown. The exact number of each experiment is indicated in each column. *, P < 0.05, as measured with a t test comparing the indicated sample and the control siRNA.
PRPF8 contains a putative nuclear localization signal at its N terminus. However, RP mutants are not efficiently imported to the nucleus, resulting in a significant fraction remaining in the cytoplasm where the interaction with R2TP occurs (Fig. 1, C and D; and Fig. 5 A). To test whether R2TP is involved in the cytoplasmic accumulation of PRPF8 mutants, we depleted RUVBL1 or 2 in the S2118F-LAP cell line that exhibited the strongest accumulation in the cytoplasm (Fig. 1 D). Both KDs resulted in the relocalization of the S2118F-LAP protein to the nucleus (Fig. 9, E and F). These findings suggest that the HSP90/R2TP chaperone complex retains assembly-defective PRPF8 mutants in the cytoplasm and thus performs an early discrimination of WT versus mutant PRPF8 proteins.
Discussion
PRPF8 RP mutants are defective in snRNP assembly and splicing
RP is a progressive degenerative disease that can be caused by mutations in genes coding for snRNP-specific proteins. In this study, we studied eight point mutations in PRPF8. None of the mutants were able to rescue the splicing deficiency caused by the depletion of the endogenous PRPF8 protein (Figs. 2 and S1). For six out of the eight analyzed mutations, this could be explained by an inhibition of tri-snRNP formation (Fig. 3, A and B). However, this is not a general case because the Y2334N and F2314L mutations in PRPF8 do not block snRNP biogenesis while affecting splicing, similar to two RP mutations in the SNRNP200 gene (Cvačková et al., 2014). The F2314L mutation weakens association with the essential helicase SNRNP200, which could explain the splicing defects. The Y2334 mutant binds SNRNP200 as efficiently as WT. This mutation is in the C-terminal tail that inhibits SNRNP200 helicase activity (Mozaffari-Jovin et al., 2013), and it may thus affect the proper regulation of SNRNP200 during spliceosome activation. Based on these findings, we conclude that most mutations in PRPF8 exhibit a phenotype similar to RP mutations in other snRNP genes, which often reduce snRNP assembly and, consequently, splicing (Gonzalez-Santos et al., 2008; Huranová et al., 2009; Tanackovic et al., 2011; Linder et al., 2014).
AAR2 identifies a PRPF8 assembly intermediate containing EFTUD2 and SNRNP200
Considering the essential function of PRPF8 and U5 snRNP during splicing, surprisingly little is known about their biogenesis in metazoans. In yeast, Prp8 first interacts with Aar2 and Snu114, and this complex then joins the core U5 snRNP, followed by the replacement of Aar2 with the helicase Brr2 (Gottschalk et al., 2001; Boon et al., 2007). In the human pathway characterized here, PRPF8, AAR2, and EFTUD2 also make an intermediate assembly complex. GFP-AAR2 IP copurified large amounts of PRPF8, EFTUD2, and SNRNP200 but only small amounts of Sm proteins or other U5-specific proteins (Fig. 4 C). In addition, RP mutants stalled in the intermediate assembly complexes with AAR2 do not associate with Sm proteins (Fig. 3, C–E). These findings collectively suggest that AAR2-containing intermediates are loosely or not associated with U5 snRNA.
ZNHIT2 and the HSP90/R2TP chaperone system are involved in the assembly of U5-specific proteins
Our SILAC IPs identified a set of proteins that are not present in the mature U5 snRNP (TTC27, TSSC4, EAPP, NCDN, and ECD). These factors might thus represent additional U5 assembly factors as proposed for the ECD homologue in flies (Claudius et al., 2014). Most importantly, these IPs identified the HSP90/R2TP complex and its cofactor ZNHIT2 as key U5 snRNP assembly factors (Fig. 4, A and B). This chaperone machinery was previously found to be involved in the snoRNP assembly as well as in the assembly of U4-specific proteins (Bizarro et al., 2014, 2015). In this study, we show that the U5-specific proteins PRPF8 and EFTUD2 interact in vivo with HSP90 and R2TP and that these interactions arise early in the cytoplasm. Functional experiments demonstrate that both HSP90 and R2TP are required for the biogenesis of U5 snRNP. First, inhibiting HSP90 leads to decreased levels of PRPF8 and SNRNP200. Second, KD of R2TP components reduces the amount of newly synthesized EFTUD2 and also leads to the accumulation of U5 in CB, a signature of assembly defects (Novotný et al., 2015).
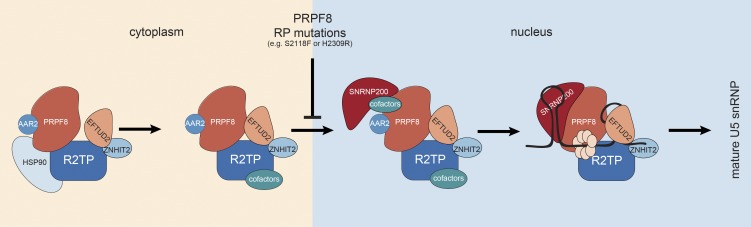
Based on the available data, we propose the following model (Fig. 10). First, the HSP90/R2TP chaperone independently recruits PRPF8 and EFTUD2 and promotes the assembly of a cytoplasmic complex containing these proteins in association with AAR2, ZNHIT2, and other factors. SNRNP200 does not interact with this complex and would be recruited later in the nucleus. Because U5 snRNA accumulates in CBs when PRPF8 is absent (Novotný et al., 2015), it is likely that the preassembly complex interacts with U5 snRNA in CBs, where final maturation steps would take place. Recycling of U5 snRNP and reformation of the U4/U6.U5 tri-snRNP have also been suggested to occur in CBs (Staněk et al., 2008). In yeast, this process also requires Aar2 (Gottschalk et al., 2001). In future studies, it will be interesting to determine whether the R2TP complex is also involved in snRNP recycling.
Figure 10.
Model for the assembly of U5-specific proteins. The HSP90/R2TP chaperone recruits PRPF8 and EFTUD2 rapidly after their synthesis and promotes the formation of a cytoplasmic assembly intermediate complex containing AAR2, ZNHIT2, and possibly other factors. This complex translocates to the nucleus and binds SNRNP200. AAR2 is released upon binding of U5 snRNA, and maturation is complete after the release of ZNHIT2 and R2TP proteins.
Client selection and function of the R2TP complex
We show here that R2TP makes independent interactions with EFTUD2 and PRPF8, thereby bringing these proteins together and providing a simple way to promote complex assembly. It is, however, less clear how these U5 proteins are recruited to R2TP. It was previously shown that the N-terminal domain of PIH1D1 binds phosphorylated DSDD(D/E) peptides, and it was proposed that this could be a determinant for R2TP client selection (Hořejší et al., 2014). We did not detect interaction between PIH1D1 N-terminal and U5 proteins, including EFTUD2, even though this protein carries a phosphorylated DSDED motif and binds PIH1D1 in a phospho-dependent manner in vitro (Hořejší et al., 2014). Nevertheless, the N-terminal domain of PIH1D1 recruits some R2TP clients, e.g., subunits of RNA pol II and box C/D snoRNPs (Fig. 6 A). Interestingly, these factors lack known phosphorylated DSDD(D/E) motifs and may thus interact with PIH1D1 using other interaction surfaces.
Mutation of the phosphoserine in the EFTUD2 DSDED motif affects the composition of U5 snRNP, indicating that it is functionally relevant. However, the binding affinity of the EFTUD2 phosphopeptide may be too weak to ensure a stable in vivo binding to the PIH1D1 N-terminal domain, and additional bridging factors such as ZNHIT2 may be essential to secure this interaction. ZNHIT2 belongs to the Zinc-Finger histidine triad motif (HIT) protein family, which contains six members in humans. These proteins have been recently proposed to function as cofactors for the RUVBL1/2 ATPases, and they may target them to specific substrates (Verheggen et al., 2015). Our two-hybrid analysis demonstrates that ZNHIT2 forms a bridging interaction between EFTUD2 and RUVBL1 (Fig. 4 E) and is in agreement with this hypothesis, which suggests that ZNHIT2 targets RUVBL1/2 toward EFTUD2.
The mechanism of action of R2TP is still unclear. In the case of box C/D snoRNPs, we proposed that R2TP recruits snoRNP subunits and then transfers RUVBL1/2 from R2TP to these proteins, possibly in an ATP- and ZNHIT-dependent manner (Bizarro et al., 2014). Indeed, ATP dissociates RUVBL1/2 from R2TP, whereas it promotes binding to snoRNP proteins and to ZNHIT6, an essential C/D assembly factor (McKeegan et al., 2007). A similar model could be proposed in the case of U5 snRNP (Fig. 10) because our proteomic data indicate a much tighter binding of U5 proteins to ZNHIT2 and RUVBL1/2 than to the other R2TP components. In the assembly intermediates, the role of the RUVBL proteins could be to maintain together U5 subunits and/or to dissociate assembly factors at late assembly stages, as proposed for snoRNPs (Machado-Pinilla et al., 2012; Bizarro et al., 2014).
The HSP90/R2TP machinery performs a quality control of PRPF8
Depletion of U5 proteins and PRPF8 RP mutation both increase the association of PRPF8 and EFTUD2 with R2TP. Furthermore, assembly-defective PRPF8 RP mutants accumulate in the cytoplasm, and one such mutant relocates to the nucleus upon KD of RUVBL1 or 2 (Fig. 9, E and F). These data suggest a model in which incomplete assembly intermediates remain associated with R2TP in the cytoplasm, and only the properly formed complex translocates to the nucleus. The R2TP machinery would thus provide a quality control to ensure that only correctly folded proteins and assembled complexes are imported to the nucleus, as proposed previously for RNA pol II (Boulon et al., 2010).
Interestingly, the pulldown of the N-terminal domain of PIH1D1 revealed a strong interaction with the TSC complex. This complex is a tumor suppressor that integrates extracellular signals and energy availability to regulate mTORC1 (Islam and Roach, 2015). The TSC complex could be another R2TP client but could also regulate it. This raises the fascinating possibility that R2TP could link energy and cell growth signals to RNP biogenesis.
Materials and methods
Cell culture, siRNA, and plasmid transfection
HeLa cells were cultured at 37°C and 5% CO2 in DMEM containing 4.5 g glucose per liter (Sigma-Aldrich) supplemented with 10% fetal calf serum (Biochrom AG) and 1% penicillin/streptomycin (Gibco).
For KDs, siRNAs were introduced into cells with oligofectamine transfection reagent (Invitrogen) or JetPRIME (Polyplus) according to the manufacturer’s protocol, and cells were then grown for 48 to 96 h. KD efficiencies were assayed by WB (Fig. S4). The siRNA against PRPF8 (5′-CCUGUAUGCCUGACCGUUUTT-3′) was designed against the sequence around the STOP codon and did not target the PRPF8-LAP fusion used in the BAC cell lines (Novotný et al., 2015). Additional KDs were performed using the following siRNAs: PRPF8 (5′-CCUGUAUGCCUGACCGUUUTT-3′), PIH1D1 (5′-UUUCCUUAGCCGCCUCUCCTT-3′), mixture of siRNA PIH1D1-1 (5′-GAAUGGAAAUGUAGUCUUATT-3′) and PIH1D1-2 (5′-GAGAAGAGGCUGCUGGCUUTT-3′), RUVBL1 (5′-CCUUGAAGCUGAAGAGUAUTT-3′), RUVBL2 (5′-AGGAAGAAGAUGUGGAGAUTT-3′), RPAP3-1 (5′-CAACAGAAGGAGAGCGAAATT-3′), RPAP3-2 (5′-GGACUAUCUUUGAACAUAA-3′), EFTUD2 (5′-CUAUACUGACAUCCUCUUCTT-3′), AAR2 (5′-UUGUCUCCCAAGACAACUUTT-3′), SNRNP200 (5′-GUGAUUCAGAUUGAGUCCUTT-3′), ZNHIT2-1 (5′-CGGUAAAGAUGAUCUCGCUTT-3′), and ZNHIT2-2 (5′-CUGGUCAUUAAUAAAGCUGTT-3′). As a control, we used scrambled siRNA (5′-GCUAAAUACACUAGGCUACGTT-3′) or NC #5 siRNA (Thermo Fisher Scientific).
Plasmids were transfected into cells using X-tremeGENE HP (Roche) according to manufacturer’s protocol, and cells were analyzed 24 h after transfection. For splicing experiments, we used a β-globin reporter derived from the E3 vector containing the β-globin gene (Darzacq et al., 2006), a gift from Y. Shav-Tal (Bar-Ilan University, Tel Aviv, Israel). We deleted the second β-globin intron (nucleotides 496–1,536). These changes were confirmed by sequencing. Retina-specific reporters, RHO, ROM1, and FSCN2 (Yuan et al., 2005; Mordes et al., 2007) were provided by J. Wu (Northwestern University, Chicago, IL).
For IP and SILAC IP experiments, GFP-tagged proteins were stably expressed in HeLa cells using the Flp-In system, and stable clones were maintained in DMEM with 0.04 µg/ml hygromycin B. HeLa clones expressing GFP-AAR2, GFP-EFTUD2, GFP-ZNHIT2, and GFP-PIH1D1 N-terminal (aa 1–180) were used for SILAC IP. For corecruitment assays, we used HeLa cells that stably expressed p54-DsRed-RPAP3. For HSP90 inhibition, cells were treated with 2 µM GA for 16 h.
Plasmids
DNA cloning was performed with the Gateway system (Invitrogen). The N-terminal fragment of PIH1D1 was generated by PCR amplification (nucleotides 1–540) and cloned in pcDNA5-GFP using Gateway technology. GFP-EFTUD2-DADE was generated by oligonucleotide-directed mutagenesis using the QuikChange II kit (Agilent Technologies) followed by Gateway cloning.
BAC recombineering
The BAC clone containing the PRPF8 gene tagged with the LAP tag (containing GFP) was provided by I. Poser (Poser et al., 2008). All RP-related mutations investigated in this study were prepared by Red/ET recombineering using the Counter-Selection BAC Modification kit (Gene Bridges). The selection marker cassette (Rpsl-amp) was PCR amplified using PRPF8 specific primers carrying 50 nt of homology surrounding the sequence to be modified and inserted into the target sequence by DNA recombination. In the second step, the cassette was replaced with a modified version of the target sequence containing the appropriate point mutation using recombination. The synthesis of individual mutations was verified by sequencing. BAC DNA was transfected into HeLa cells using X-tremeGENE HP (Roche) according to the manufacturer’s protocol. Stable cell lines were selected with G418 (Gibco) and sorted by a cell sorter.
Antibodies
We used the following primary antibodies for IF, WB, or IP: rabbit anti-PRPF8 (Santa Cruz Biotechnology, Inc.; Thermo Fisher Scientific; Bethyl Laboratories, Inc.), rabbit anti-SNRNP200 (Sigma-Aldrich), rabbit anti-C20orf4 35–50 (AAR2 splicing factor homologue; Sigma-Aldrich), rabbit anti-PRPF6 (Santa Cruz Biotechnology, Inc.), rabbit anti-EFTUD2 (Thermo Fisher Scientific; Bethyl Laboratories, Inc.), mouse anti-GFP (Santa Cruz Biotechnology, Inc.), rabbit anti-GFP (Torrey Pines Biolabs), mouse anti-RUVBL1 (Abcam), rabbit anti-RUVBL1 (Proteintech), rabbit anti-RUVBL2 (GeneTex; Proteintech), mouse anti-PIH1D1 (Abcam), rabbit anti-RPAP3 (Abcam), rabbit anti-ZNHIT2 (Abcam), rabbit anti-GAPDH (Abcam), anti-coilin (5P10) mouse antibody (provided by M. Carmo-Fonseca, Institute of Molecular Medicine, Lisbon, Portugal; Almeida et al., 1998), goat anti-GFP (provided by D. Drechsler, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany), rabbit anti-EFTUD2 and anti-PRPF31 (provided by R. Lührmann, Max Planck Institute, Göttingen, Germany), and mouse anti-tubulin (provided by P. Dráber; Institute of Molecular Genetics, Prague, Czech Republic). The anti-Sm antibody Y12 was produced from a hybridoma cell line (a gift from K. Neugebauer, Yale University, New Haven, CT) at the Antibody Facility of the Institute of Molecular Genetics, Czech Academy of Sciences. For RPAP3 IP, the anti-RPAP3 antibody was produced from the hybridoma cell line 19B11.
For WB, we used secondary antibodies conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc.). For IF, we used secondary anti–rabbit or anti–mouse antibodies coupled to FITC (Sigma-Aldrich) or DyLight488 (Jackson ImmunoResearch Laboratories, Inc.).
IF and light microscopy
Cells grown on coverslips were washed with PBS and fixed with 4% (weight/volume) paraformaldehyde/Pipes for 10–20 min at RT. Permeabilization was done in 0.1–0.2% Triton X-100 in PBS for 5–10 min at RT. For IF, coverslips were incubated with primary and secondary antibodies. Coverslips were mounted with Fluoromount G (Southern Biotech) containing DAPI for DNA staining or with Vectashield (Vector Laboratories). SnRNAs were visualized by FISH as previously described (Novotný et al., 2015).
Images were collected on a DeltaVision microscope system (Applied Precision Ltd.) coupled with a microscope (IX70; Olympus) equipped with a 63× 1.42 NA oil immersion objective and a CoolSNAP HQ2 camera (Photometrics; Princeton Instruments) and the acquisition software SoftWoRx (Applied Precision Ltd.). Stacks of 20 z sections with 200-nm z steps were taken per sample at 37°C and subjected to mathematical deconvolution using SoftWoRx software. Maximal projections of deconvolved pictures were done by SoftWoRx and are presented. Nucleocytoplasmic distribution was analyzed using ImageJ software (National Institutes of Health). Between 52 and 125 cells were analyzed per mutant, and the mean and SEM are shown in the graphs. Alternatively, for corecruitment assays, cells were observed using a wide-field fluorescence microscope with a 63× 1.4 NA oil objective (DMRA; Leica Microsystems; at RT). Images were acquired with a CoolSNAP HQ2 camera. Red/green quantification was performed on red and green images concomitantly using Metamorph (Molecular Devices).
Time-lapse microscopy
For live-cell measurements, cells were cultured on glass-bottomed Petri dishes (MatTek Corporation). Experiments were performed on a DeltaVision microscope system equipped with a chamber for live-cell imaging controlling temperature and CO2 concentration. Other specifications were identical as in the previous section. Cycloheximide was added to the medium (final concentration 30 µg/ml), and GFP fluorescence was recorded at 15-min intervals over an 8-h period. Three experiments, each containing 8–10 cells per cell line, were analyzed, and means are shown in the graphs.
RT-PCR and quantitative PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. When required, RNAs were treated with TURBO DNase (Thermo Fisher Scientific). cDNAs were synthesized using SuperScript III Reverse Transcription (Invitrogen) and random hexamers. Taq polymerase (Thermo Fisher Scientific) was used for PCR reactions.
PCR products were separated on 3% agarose gel, and the intensity of individual bands was determined using ImageJ software. Quantitative PCR was performed using SYBR green on a LightCycler 480 System (Roche). The reaction volume of 5 µl contained 2 µl of template cDNA (diluted to 1:10) and 500 nM of each primer. Denaturation at 95°C for 7 min was followed by 40 cycles for 20 s at 95°C, 20 s at 61°C, and 35 s at 72°C. Splicing efficiency (ratio mRNA/pre-mRNA) was calculated as 2(Ct[pre-mRNA] − Ct[mRNA]). The following primers were used for RT-PCR and quantitative PCR: β-globin forward, 5′-CAAGGTGAACGTGGA-3′; β-globin reverse, 5′-GGACAGATCCCCAAAGGACT-3′; RHO forward, 5′-CGGAGGTCAACAACGAGTCT-3′; RHO reverse, 5′-TCTCTGCCTTCTGTGTGGTG-3′; FSCN2 forward, 5′-TGCCAACACCATGTTTGAGA-3′; FSCN2 reverse, 5′-GCTTGAGGGTGAACTCTTCG-3′; ROM1 forward, 5′-CAACCCAACCAAAACCTCTG-3′; ROM1 reverse, 5′-ATAGCCCTGGGTCTCTCCTC-3′; pre-LDHA forward, 5′-CCTTTCAACTCTCTTTTGGCAACC-3′; pre-LDHA reverse, 5′-AATCTTATTCTGGGGGGTCTGTTC-3′; mLDHA forward, 5′-AGAACACCAAAGATTGTCTCTGGC-3′; and mLDHA reverse, 5′-TTTCCCCCATTAGGTAACGG-3′.
IP and WB
Two alternative IP techniques were used. For the experiments summarized in Figs. 3, 6, and 9, 2 × 107 cells were washed twice with ice-cold PBS, scraped from the dish, and centrifuged at 1,000 g for 10 min. Harvested cells were resuspended in NET-2 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Nonidet P-40) supplemented with protease inhibitor cocktail (EMD Millipore or Roche) and then pulse sonicated on ice (30 pulses; 0.5 s for each pulse at 40% of maximum energy). The cell lysate was centrifuged at 14,000 g, and the supernatant was immunoprecipitated overnight at 4°C with 7.5 µg of goat anti-GFP antibodies prebound to 30 µl of protein G sepharose beads (GE Healthcare). For the IP control, a lysate from parental HeLa cells was used. After five washings with NET-2 buffer, the immunoprecipitated proteins were resuspended in 30 µl of 2× sample buffer.
For the experiments in Figs. S2 and S3, cells were lysed in HNTG buffer (20 mM Hepes, pH 7.9, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM MgCl2, 1 mM EGTA, and protease inhibitors) for 20 min at 4°C. Cell debris were removed by centrifugation (10 min at 9,000 g). Protein G sepharose was preincubated with anti-RPAP3 antibody for 2 h (Fig. S3, A and B). For control IPs, empty beads were preincubated with fetal bovine serum and then incubated with the same lysate used for the anti-RPAP3 IPs. For GFP-fusion proteins (Fig. S2 D), GFP-TRAP beads (ChromoTek) were used, and a control was done with HeLa cells that did not express the GFP protein. Beads were incubated with extracts for 1.5 h at 4°C and then were washed five times with HNTG before resuspending in 2× Laemmli buffer.
Subsequently, proteins were separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane (Protran). Membranes were blocked with 5% nonfat milk (weight/volume) in PBST (0.05% Tween-20 in PBS) and incubated with the appropriate primary antibodies diluted in 1% nonfat milk (weight/volume) in PBST followed by incubation with secondary antibodies conjugated with horseradish peroxidase. Enzymatic activity was detected using the SuperSignal West Pico/Femto Chemiluminescent Substrate (Thermo Fisher Scientific) or ECL kit (Roche). Significance of all experiments was assayed by a two-tailed unpaired t test.
SILAC IP and proteomic analysis
SILAC experiments were performed as previously described (Boulon et al., 2010). H9 HeLa cells were grown for 15 d in each isotopically labeled medium to ensure complete incorporation of isotopic amino acids (CIL/Eurisotop). Six 15-cm diameter plates were used per SILAC condition. Cells were rinsed with PBS, trypsinized, and cryogrinded in SILAC buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitor cocktail). Extracts were incubated for 20 min at 4°C and clarified by centrifugation for 10 min at 20,000 g. For all IP experiments, extracts were precleared by incubation with protein G sepharose beads for 1 h at 4°C. The control was extracted from the light condition prepared from HeLa cells that did not express the GFP fusion. Each extract was incubated with 50 µl of GFP-TRAP beads for 75 min at 4°C, washed five times with SILAC buffer, and beads from the different isotopic conditions were finally pooled. Bound proteins were eluted by adding 1% SDS to the beads and boiling for 10 min. Proteomic analysis was performed as previously described (Bizarro et al., 2014).
Yeast two-hybrid assay
Plasmids pACT2 and pAS2DD were introduced into haploid S. cerevisiae strains (CG1945 and Y187, respectively). Strains were crossed, and diploids were selected on Leu− Trp− selective media and then plated on triple selective media (Leu− Trp− His−). Growth was assessed visually after 3 d at 30°C. The strength of interactions was evaluated by comparing the number of clones growing on Leu− Trp− (selection of diploids) and Leu− Trp− His− plates (selection for interaction).
Luciferase assays
H9 Hela cells were grown on six-well plates and cotransfected with plasmids expressing a FLAG-tagged FFL in fusion with EFTUD2 and with a plasmid-coding Renilla luciferase (RL) as control. After 48 h, cells were extracted in 150 µl of passive lysis buffer and incubated at 4°C for 15 min. RL and FFL activities were measured on 96-well plates using 8 µl of cell extract with 50 µl of each solution of the dual-luciferase assay kit (Promega). Values obtained for FFL were normalized to RL values. Experiments were done in triplicate.
Online supplemental material
Fig. S1 shows the expression of PRPF8 RP mutants, their stability compared with WT, and their effect on splicing. Fig. S2 shows localization of GFP-tagged proteins used in this study and the results of SILAC IP with the GFP-EFTUD2 DADE mutant. Fig. S3 shows interactions of U5-specific proteins with RPAP3 and ZNHIT2 by coimmunoprecipitation and corecruitment assays. Fig. S4 shows the efficiency of siRNAs used in this study. Table S1 is available as an Excel file and is a complete list of proteins coprecipitated in the various SILAC IP experiments.
Supplementary Material
Acknowledgments
We thank J. Wu, Y. Shav-Tal, I. Poser, D. Drechsler, R. Lührmann, K. Neugebauer, M. Carmo-Fonseca, and P. Dráber for the reagents. We thank J. Manning for English proofreading.
This work was supported by grants from the Czech Academy of Sciences (RVO68378050), the Czech Science Foundation (P301/12/P425 to Z. Cvačková, 15-00790S to D. Staněk, and 14-34264S to Z. Hořejší), the National Sustainability Program I (LO14119 to D. Staněk), and the Charles University Grant Agency (460413 to A. Malinová). E. Bertrand and C. Verheggen were supported by grants from Agence Nationale de la Recherche (ANR-11-BSV8-01503) and from the Ligue Nationale Contre le Cancer (équipe labelisée).
The authors declare no competing financial interests.
Author contributions: A. Malinová performed all experiments shown in Figs. 1, 2 (A and B), 3, 6 B, 7 B, 8 A, and 9; C. Abéza and C. Verheggen performed all experiments shown in Figs. 4, 5, 6 A, and 7 A and prepared final figures; F. Vandermoere analyzed proteomics data; Z. Cvačková and A. Malinová performed experiments shown in Fig. 2 (C–E) and both participated in figure preparation and manuscript writing; A. Malinová, Z. Cvačková, and D. Matějů prepared the RP mutants of PRPF8 in BAC and stable cell lines; Z. Hořejší provided reagents and protocols for IP of the R2TP complex and participated in manuscript writing; D. Staněk, E. Bertrand, and C. Verheggen supervised the project, designed and analyzed experiments, and wrote the manuscript.
Footnotes
Abbreviations used:
- BAC
- bacterial artificial chromosome
- CB
- Cajal body
- FFL
- firefly luciferase
- GA
- geldanamycin
- HIT
- histidine triad motif
- IF
- immunofluorescence
- IP
- immunoprecipitation
- KD
- knock down
- LAP
- localization and affinity purification
- MPN
- Mpr1/Pad1 N-terminal
- NC
- negative control
- RL
- Renilla luciferase
- RP
- Retinitis pigmentosa
- SILAC
- stable isotope labeling in cell culture
- SMN
- survival of motor neurons
- snoRNP
- small nucleolar ribonucleoprotein particles
- snRNA
- short noncoding uridine-rich RNA
- snRNP
- small nuclear ribonucleoprotein particle
- TSC
- tuberous sclerosis complex
- WB
- Western blotting
- WT
- wild type
References
- Almeida F., Saffrich R., Ansorge W., and Carmo-Fonseca M.. 1998. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 142:899–912. 10.1083/jcb.142.4.899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K., Agafonov D.E., Liu W.-T., Dybkov O., Will C.L., Hartmuth K., Urlaub H., Kastner B., Stark H., and Lührmann R.. 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 542:318–323. 10.1038/nature21079 [DOI] [PubMed] [Google Scholar]
- Bizarro J., Charron C., Boulon S., Westman B., Pradet-Balade B., Vandermoere F., Chagot M.E., Hallais M., Ahmad Y., Leonhardt H., et al. . 2014. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 207:463–480. 10.1083/jcb.201404160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizarro J., Dodré M., Huttin A., Charpentier B., Schlotter F., Branlant C., Verheggen C., Massenet S., and Bertrand E.. 2015. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 43:8973–8989. 10.1093/nar/gkv809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K.L., Grainger R.J., Ehsani P., Barrass J.D., Auchynnikava T., Inglehearn C.F., and Beggs J.D.. 2007. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat. Struct. Mol. Biol. 14:1077–1083. 10.1038/nsmb1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jády B.E., Rothé B., Pescia C., Robert M.C., Kiss T., et al. . 2008. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180:579–595. 10.1083/jcb.200708110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M.C., Ahmad Y., Neel H., et al. . 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell. 39:912–924. 10.1016/j.molcel.2010.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Bertrand E., and Pradet-Balade B.. 2012. HSP90 and the R2TP co-chaperone complex: Building multi-protein machineries essential for cell growth and gene expression. RNA Biol. 9:148–154. 10.4161/rna.18494 [DOI] [PubMed] [Google Scholar]
- Claudius A.K., Romani P., Lamkemeyer T., Jindra M., and Uhlířová M.. 2014. Unexpected role of the steroid-deficiency protein ecdysoneless in pre-mRNA splicing. PLoS Genet. 10:e1004287 10.1371/journal.pgen.1004287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P., and Coulombe B.. 2010. New insights into the biogenesis of nuclear RNA polymerases? Biochem. Cell Biol. 88:211–221. 10.1139/O09-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvačková Z., Matějů D., and Staněk D.. 2014. Retinitis pigmentosa mutations of SNRNP200 enhance cryptic splice-site recognition. Hum. Mutat. 35:308–317. 10.1002/humu.22481 [DOI] [PubMed] [Google Scholar]
- Darzacq X., Kittur N., Roy S., Shav-Tal Y., Singer R.H., and Meier U.T.. 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 173:207–218. 10.1083/jcb.200601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C.C., and Cantley L.C.. 2015. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25:545–555. 10.1016/j.tcb.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget D., Lacombe A.A., Cloutier P., Al-Khoury R., Bouchard A., Lavallée-Adam M., Faubert D., Jeronimo C., Blanchette M., and Coulombe B.. 2010. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteomics. 9:2827–2839. 10.1074/mcp.M110.003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Santos J.M., Cao H., Duan R.C., and Hu J.. 2008. Mutation in the splicing factor Hprp3p linked to retinitis pigmentosa impairs interactions within the U4/U6 snRNP complex. Hum. Mol. Genet. 17:225–239. 10.1093/hmg/ddm300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Kastner B., Lührmann R., and Fabrizio P.. 2001. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA. 7:1554–1565. [PMC free article] [PubMed] [Google Scholar]
- Hartong D.T., Berson E.L., and Dryja T.P.. 2006. Retinitis pigmentosa. Lancet. 368:1795–1809. 10.1016/S0140-6736(06)69740-7 [DOI] [PubMed] [Google Scholar]
- Hořejší Z., Takai H., Adelman C.A., Collis S.J., Flynn H., Maslen S., Skehel J.M., de Lange T., and Boulton S.J.. 2010. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell. 39:839–850. 10.1016/j.molcel.2010.08.037 [DOI] [PubMed] [Google Scholar]
- Hořejší Z., Stach L., Flower T.G., Joshi D., Flynn H., Skehel J.M., O’Reilly N.J., Ogrodowicz R.W., Smerdon S.J., and Boulton S.J.. 2014. Phosphorylation-dependent PIH1D1 interactions define substrate specificity of the R2TP cochaperone complex. Cell Reports. 7:19–26. 10.1016/j.celrep.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., and Skrzypek E.. 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43(D1):D512–D520. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huranová M., Hnilicová J., Fleischer B., Cvačková Z., and Staněk D.. 2009. A mutation linked to retinitis pigmentosa in HPRP31 causes protein instability and impairs its interactions with spliceosomal snRNPs. Hum. Mol. Genet. 18:2014–2023. 10.1093/hmg/ddp125 [DOI] [PubMed] [Google Scholar]
- Islam M.P., and Roach E.S.. 2015. Tuberous sclerosis complex. Handb. Clin. Neurol. 132:97–109. 10.1016/B978-0-444-62702-5.00006-8 [DOI] [PubMed] [Google Scholar]
- Kamano Y., Saeki M., Egusa H., Kakihara Y., Houry W.A., Yatani H., and Kamisaki Y.. 2013. PIH1D1 interacts with mTOR complex 1 and enhances ribosome RNA transcription. FEBS Lett. 587:3303–3308. 10.1016/j.febslet.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Kondo H., Tahira T., Mizota A., Adachi-Usami E., Oshima K., and Hayashi K.. 2003. Diagnosis of autosomal dominant retinitis pigmentosa by linkage-based exclusion screening with multiple locus-specific microsatellite markers. Invest. Ophthalmol. Vis. Sci. 44:1275–1281. 10.1167/iovs.02-0895 [DOI] [PubMed] [Google Scholar]
- Kuhn A.N., Li Z., and Brow D.A.. 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell. 3:65–75. 10.1016/S1097-2765(00)80175-6 [DOI] [PubMed] [Google Scholar]
- Linder B., Hirmer A., Gal A., Rüther K., Bolz H.J., Winkler C., Laggerbauer B., and Fischer U.. 2014. Identification of a PRPF4 loss-of-function variant that abrogates U4/U6.U5 tri-snRNP integration and is associated with retinitis pigmentosa. PLoS One. 9:e111754 10.1371/journal.pone.0111754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Pinilla R., Liger D., Leulliot N., and Meier U.T.. 2012. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA. 18:1833–1845. 10.1261/rna.034942.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder C., Kutach A.K., and Guthrie C.. 2009. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat. Struct. Mol. Biol. 16:42–48. 10.1038/nsmb.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov E.M., Makarova O.V., Urlaub H., Gentzel M., Will C.L., Wilm M., and Lührmann R.. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 298:2205–2208. 10.1126/science.1077783 [DOI] [PubMed] [Google Scholar]
- Martínez-Gimeno M., Gamundi M.J., Hernan I., Maseras M., Millá E., Ayuso C., García-Sandoval B., Beneyto M., Vilela C., Baiget M., et al. . 2003. Mutations in the pre-mRNA splicing-factor genes PRPF3, PRPF8, and PRPF31 in Spanish families with autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 44:2171–2177. 10.1167/iovs.02-0871 [DOI] [PubMed] [Google Scholar]
- Matera A.G., and Wang Z.. 2014. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15:108–121. 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan K.S., Debieux C.M., Boulon S., Bertrand E., and Watkins N.J.. 2007. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 27:6782–6793. 10.1128/MCB.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie A.B., McHale J.C., Keen T.J., Tarttelin E.E., Goliath R., van Lith-Verhoeven J.J., Greenberg J., Ramesar R.S., Hoyng C.B., Cremers F.P., et al. . 2001. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum. Mol. Genet. 10:1555–1562. 10.1093/hmg/10.15.1555 [DOI] [PubMed] [Google Scholar]
- Mir R.A., Bele A., Mirza S., Srivastava S., Olou A.A., Ammons S.A., Kim J.H., Gurumurthy C.B., Qiu F., Band H., and Band V.. 2015. A novel interaction of ecdysoneless (ECD) protein with R2TP complex component RUVBL1 is required for the functional role of ECD in cell cycle progression. Mol. Cell. Biol. 36:886–899. 10.1128/MCB.00594-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D., Luo X., Kar A., Kuo D., Xu L., Fushimi K., Yu G., Sternberg P. Jr., and Wu J.Y.. 2006. Pre-mRNA splicing and retinitis pigmentosa. Mol. Vis. 12:1259–1271. [PMC free article] [PubMed] [Google Scholar]
- Mordes D., Yuan L., Xu L., Kawada M., Molday R.S., and Wu J.Y.. 2007. Identification of photoreceptor genes affected by PRPF31 mutations associated with autosomal dominant retinitis pigmentosa. Neurobiol. Dis. 26:291–300. 10.1016/j.nbd.2006.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S., Santos K.F., Hsiao H.H., Will C.L., Urlaub H., Wahl M.C., and Lührmann R.. 2012. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 26:2422–2434. 10.1101/gad.200949.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S., Wandersleben T., Santos K.F., Will C.L., Lührmann R., and Wahl M.C.. 2013. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science. 341:80–84. 10.1126/science.1237515 [DOI] [PubMed] [Google Scholar]
- Mozaffari-Jovin S., Wandersleben T., Santos K.F., Will C.L., Lührmann R., and Wahl M.C.. 2014. Novel regulatory principles of the spliceosomal Brr2 RNA helicase and links to retinal disease in humans. RNA Biol. 11:298–312. 10.4161/rna.28353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.H.D., Li J., Galej W.P., Oshikane H., Newman A.J., and Nagai K.. 2013. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure. 21:910–919. 10.1016/j.str.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotný I., Malinová A., Stejskalová E., Matějů D., Klimešová K., Roithová A., Švéda M., Knejzlík Z., and Staněk D.. 2015. SART3-dependent accumulation of incomplete spliceosomal snRNPs in Cajal bodies. Cell Reports. 10:429–440. 10.1016/j.celrep.2014.12.030 [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., and Mann M.. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1:376–386. 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Pal M., Morgan M., Phelps S.E.L., Roe S.M., Parry-Morris S., Downs J.A., Polier S., Pearl L.H., and Prodromou C.. 2014. Structural basis for phosphorylation-dependent recruitment of Tel2 to Hsp90 by Pih1. Structure. 22:805–818. 10.1016/j.str.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena V., Liu S., Bujnicki J.M., Lührmann R., and Wahl M.C.. 2007. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol. Cell. 25:615–624. 10.1016/j.molcel.2007.01.023 [DOI] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J.R., Hériché J.K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W., et al. . 2008. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 5:409–415. 10.1038/nmeth.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimer A.C., Gray K.M., and Matera A.G.. 2016. SMN - A chaperone for nuclear RNP social occasions? RNA Biol. 20:1–11. 10.1080/15476286.2016.1236168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžičková Š., and Staněk D.. 2016. Mutations in spliceosomal proteins and retina degeneration. RNA Biol. 14:1–9. 10.1080/15476286.2016.1191735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E., and Lamond A.I.. 1999. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 9:1065–1074. 10.1016/S0960-9822(99)80475-8 [DOI] [PubMed] [Google Scholar]
- Small E.C., Leggett S.R., Winans A.A., and Staley J.P.. 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 23:389–399. 10.1016/j.molcel.2006.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staněk D. 2016. Cajal body and snRNPs - friends with benefits. RNA Biol. 14:1–9. 10.1080/15476286.2016.1231359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staněk D., Pridalová-Hnilicová J., Novotný I., Huranová M., Blazíková M., Wen X., Sapra A.K., and Neugebauer K.M.. 2008. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol. Biol. Cell. 19:2534–2543. 10.1091/mbc.E07-12-1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G., Ransijn A., Thibault P., Abou Elela S., Klinck R., Berson E.L., Chabot B., and Rivolta C.. 2011. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 20:2116–2130. 10.1093/hmg/ddr094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towns K.V., Kipioti A., Long V., McKibbin M., Maubaret C., Vaclavik V., Ehsani P., Springell K., Kamal M., Ramesar R.S., et al. . 2010. Prognosis for splicing factor PRPF8 retinitis pigmentosa, novel mutations and correlation between human and yeast phenotypes. Hum. Mutat. 31:E1361–E1376. 10.1002/humu.21236 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Boulon S., Lam Y.W., Urcia R., Boisvert F.M., Vandermoere F., Morrice N.A., Swift S., Rothbauer U., Leonhardt H., and Lamond A.. 2008. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183:223–239. 10.1083/jcb.200805092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Pradet-Balade B., and Bertrand E.. 2015. SnoRNPs, ZNHIT proteins and the R2TP pathway. Oncotarget. 6:41399–41400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G., Cristão V.F., de L Alves F., Santos K.F., Holton N., Rappsilber J., Beggs J.D., and Wahl M.C.. 2011. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev. 25:1601–1612. 10.1101/gad.635911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G., Cristão V.F., Santos K.F., Jovin S.M., Heroven A.C., Holton N., Lührmann R., Beggs J.D., and Wahl M.C.. 2013. Structural basis for dual roles of Aar2p in U5 snRNP assembly. Genes Dev. 27:525–540. 10.1101/gad.213207.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., Mimnaugh E.G., De Costa B., Myers C.E., and Neckers L.M.. 1994. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA. 91:8324–8328. 10.1073/pnas.91.18.8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Hang J., Wan R., Huang M., Wong C.C.L., and Shi Y.. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 349:1182–1191. 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- Yuan L., Kawada M., Havlioglu N., Tang H., and Wu J.Y.. 2005. Mutations in PRPF31 inhibit pre-mRNA splicing of rhodopsin gene and cause apoptosis of retinal cells. J. Neurosci. 25:748–757. 10.1523/JNEUROSCI.2399-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R.L., Boone C., Hughes T.R., et al. . 2008. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180:563–578. 10.1083/jcb.200709061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviello C., Simonelli F., Testa F., Anastasi M., Marzoli S.B., Falsini B., Ghiglione D., Macaluso C., Manitto M.P., Garrè C., et al. . 2005. Molecular genetics of autosomal dominant retinitis pigmentosa (ADRP): a comprehensive study of 43 Italian families. J. Med. Genet. 42:e47 10.1136/jmg.2005.031682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.