Abstract
Both p150 and p110 isoforms of ADAR1 convert adenosine to inosine in double-stranded RNA (dsRNA). ADAR1p150 suppresses the dsRNA sensing mechanism that activates MDA5-MAVS-IFN signaling in the cytoplasm. In contrast, the biological function of the ADAR1p110 isoform, usually located in the nucleus, remains largely unknown. Here we show that stress-activated phosphorylation of ADAR1p110 by MKK6-p38-MSK MAP kinases promotes its binding to Exportin-5 and export from the nucleus. Once translocated to the cytoplasm, ADAR1p110 suppresses apoptosis of stressed cells by protecting many anti-apoptotic gene transcripts that contain 3′UTR dsRNA structures primarily made from inverted Alu repeats. ADAR1p110 competitively inhibits binding of Staufen1 to the 3′UTR dsRNAs and antagonizes the Staufen1-mediated mRNA decay. Our studies revealed a new stress response mechanism, in which human ADAR1p110 and Staufen1 regulate surveillance of a set of mRNAs required for survival of stressed cells.
Keywords: RNA editing, ADAR1, dsRNA, Alu, stress, apoptosis, MAP kinase, Exportin-5, Staufen1, mRNA decay, SMD
Introduction
One type of RNA editing that converts adenosine residues to inosine (A-to-I RNA editing) is catalyzed by adenosine deaminases acting on RNA (ADARs). Three ADAR gene family members, ADAR1 (ADAR), ADAR2 (ADARB1), and ADAR3 (ADARB2), have been identified in human1,2. The translation machinery reads inosine as guanosine, which could result in recoding and diversification of gene functions3,4. Precursors of certain microRNA (miRNA) also undergo A-to-I RNA editing, which regulates their processing as well as the function of mature miRNAs5,6. Furthermore, A-to-I RNA editing occurs frequently in non-coding regions that contain inversely oriented repeat elements such as Alu and LINE, although its biological significance remains largely unknown7,8.
Two isoforms of ADAR1, a full-length interferon-inducible p150 and a shorter and constitutive p110, are generated through usage of different promoters and alternative splicing9. Two repeats of Z-DNA binding domains (Zα and Zβ), three repeats of dsRNA binding domains (dsRBD1-3), and a separate deaminase domain are present in ADAR1p150, whereas ADAR1p110 has a truncated N-terminus and lacks the Zα domain (Fig. 1a). Although both isoforms are catalytically active enzymes10, their distinct localization within the cell, i.e., p110 detected mainly in the nucleoplasm and nucleoli whereas p150 is mainly found in the cytoplasm, suggests that their target RNAs and biological functions are different. Adar-/- (ADAR1 null) mouse embryos die around E12 due to fetal liver disintegration, defective hematopoiesis, and widespread apoptosis, indicating that ADAR1 is absolutely required for embryonic development11-13. The aberrantly activated dsRNA sensing mechanism, which in turn induces interferon responses, seems to underlie the embryonic lethal Adar-/- mouse phenotype14-16. Long and unedited dsRNAs made from inverted SINE repeats present in 3′UTRs of certain mRNAs have been proposed as endogenous triggers of the dsRNA sensing mechanism. A-to-I editing of those 3′UTR dsRNA, specifically by ADAR1p150, suppresses the dsRNA sensing and consequent activation of interferon responses15,17. This particular ADAR1p150 function has been linked to Aicardi-Goutièrs syndrome, a severe human autoimmune disease14,15, establishing one important function of ADAR1p150.
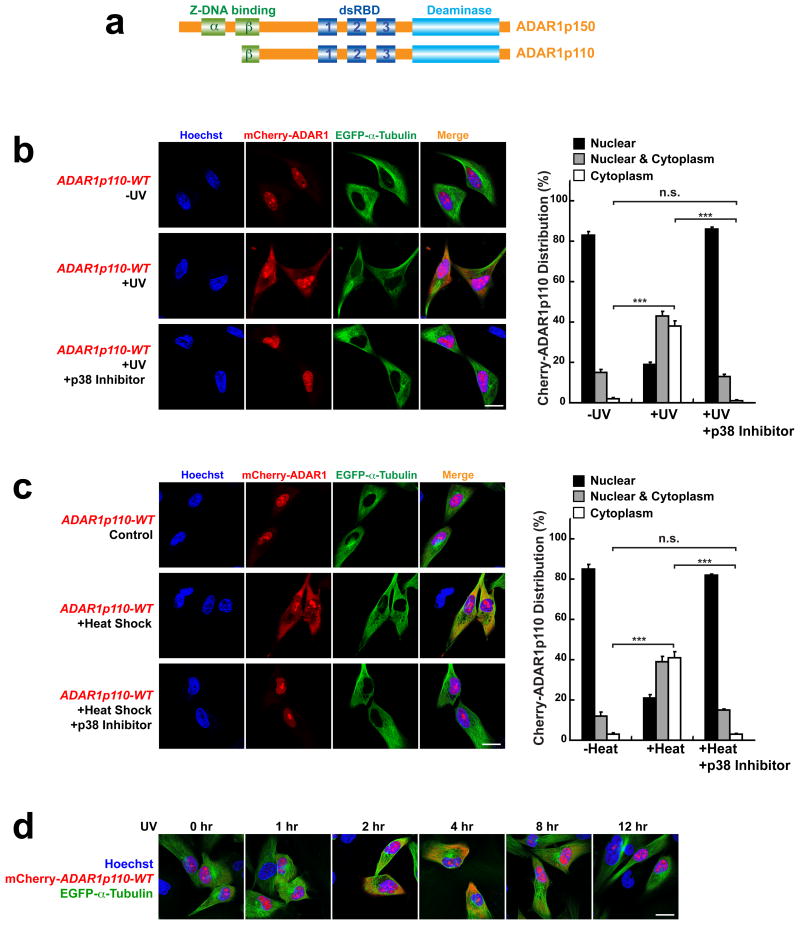
Figure 1. Stress-induced cytoplasmic localization of ADAR1p110.
(a) Schematic domain structure of two ADAR1 isoforms. (b) UV irradiation induced cytoplasmic localization of mCherry-ADAR1p110-WT in A172 cells, which was blocked by p38 inhibitor SB203580. (c) Heat-shock stress induced cytoplasmic localization of mCherry-ADAR1p110-WT in A172 cells, which was blocked by p38 inhibitor SB203580. (b, c) For quantitation (right summary graphs), images of ∼30 cells each from (x02267)7 separate slides (n (x02267) 7) prepared independently ((x02267)180 cells) were examined for mCherry-ADAR1p110 distribution between nucleus and cytoplasm. Data are mean ± S.D.; significant differences were identified by two-tailed Student's t-tests: ***P <0.001, n.s., not significant. Source data are available online in Source Data Set 1. (d) Time-course analysis of mCherry-ADAR1p110-WT localization after UV irradiation. (b-d) Scale bars, 20 μm.
In contrast, the in vivo functions of ADAR1p110, which is usually expressed at much higher levels compared to ADAR1p150, remain largely unknown. In the present study, we identified a new ADAR1p110 function regulated through its phosphorylation by MKK6-p38-MSK1&2 MAP kinases. The MKK6-p38-MSK1&2 signaling pathway is linked to the mechanism that responds to various stresses such as UV irradiation and heat shock18,19. We show that stress induced phosphorylation promotes ADAR1p110 binding to the nuclear exporter protein Exportin-5 (Xpo5), resulting in a dramatic increase in cytoplasmic ADAR1p110. In the cytoplasm, ADAR1p110 protects many anti-apoptotic gene transcripts containing 3′UTR dsRNA structures such as those made from inverted Alu repeats, which are otherwise subjected to Staufen1 mediated mRNA decay (SMD)20-22. We demonstrate that ADAR1p110, independent of its A-to-I editing function, competes with Staufen1 for binding to the 3′UTR dsRNAs. Posttranscriptional mRNA decay mechanisms are often used for regulation of genes that need to respond swiftly to environmental changes23,24. Our studies revealed a new function of ADAR1p110 in regulation of stress-induced SMD and protection of stressed cells from apoptosis.
Results
Stress induced cytoplasmic localization of ADAR1p110
We reasoned that the cellular location of ADAR1 might be closely linked to its functions. Accordingly, we attempted to identify the mechanism that regulates cellular distribution of ADAR1. We prepared mCherry-fusion constructs of two ADAR1 isoforms and tested them in transiently transfected A172 glioblastoma cells by fluorescence microscopy. We first confirmed that mCherry-ADAR1p110 localized almost exclusively in the nucleoplasm and nucleoli (Fig. 1b, left upper and right panels), whereas mCherry-ADAR1p150 localized mainly in the cytoplasm (Supplementary Fig. 1a, upper panels). We then examined various conditions and found that stress such as UV irradiation (Fig. 1b, left middle and right panels) and heat shock (Fig. 1c, left middle and right panels) increased significantly cytoplasmic localization of ADAR1p110. The UV irradiation-induced cytoplasmic localization of ADAR1p110 was detected by 1 hr and peaked at 4 hrs after UV irradiation. ADAR1p110 returned to the nucleus by 12 hrs post UV irradiation, indicating that its cytoplasmic localization is temporal and reversible (Fig. 1d). In contrast to dramatic effects on ADAR1p110, UV irradiation did not change the cytoplasmic localization of ADAR1p150 (Supplementary Fig. 1a, lower panels). As reported previously25, however, a fraction of the cytoplasmic ADAR1p150 moved to stress granules (Supplementary Fig. 1b, lower panels). The UV irradiation-induced cytoplasmic localization of ADAR1p110 was further confirmed by western blotting analysis of cytoplasmic and nuclear fractions of UV-irradiated A172 cells (Supplementary Fig. 2).
ADAR1p110 is phosphorylated by MKK6-p38-MSK MAP kinases
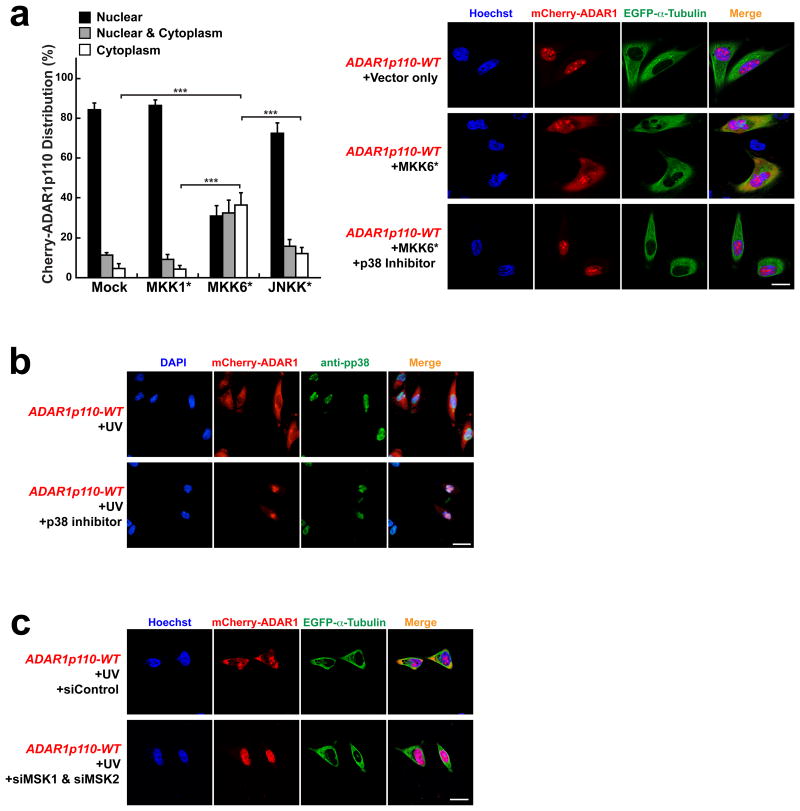
Phosphorylation plays important roles in regulating functions as well as cellular distribution of many proteins26. Furthermore, signaling of MAP kinases, p38 and JNK, is activated in response to stress such as UV irradiation and heat shock18,19. In order to identify the kinase responsible for phosphorylation of ADAR1, we tested various MAP kinase activating pathways by transiently transfecting mCherry-ADAR1p110-WT together with MAP kinase activating kinase expression constructs such as constitutively activated MKK1* carrying S218E and S222E mutations27, MKK6* carrying S207E and T211E mutations28, or JNKK* carrying S257E and T261D mutations29. We found that only MKK6* significantly increased cytoplasmic mCherry-ADAR1p110-WT localization (Fig. 2a). MKK6 phosphorylates and activates its target p38 kinase28, and, as expected, the cytoplasmic localization of mCherry-ADAR1p110 co-transfected with MKK6* was completely blocked by a p38-specific inhibitor SB20358030 (Fig. 2a, right lower panels). The inhibitor also blocked the stress-induced cytoplasmic localization of mCherry-ADAR1p110-WT (Fig. 1b, c, left lower panels) as well as endogenous ADAR1p110 (Supplementary Fig. 2), confirming that phosphorylation and consequent cytoplasmic localization of ADAR1p110 is indeed regulated by activation of the MKK6-p38 signaling pathway. The activated p38 kinase translocates to the nucleus (Fig. 2b) and phosphorylates its nuclear targets including its downstream kinases such as MAPKAPK-2-5, MNK1 & MNK2, and MSK1 & MSK218,19,31. Accordingly, we next tested the possibility that ADAR1p110 may be directly phosphorylated by these p38 downstream kinases by knocking down them individually or combinatorially. We found that only simultaneous knockdown of MSK1 and MSK2 abolished the UV-induced cytoplasmic localization of ADAR1p110, suggesting that these nuclear p38 target kinases most likely phosphorylate ADARp110 (Fig. 2c).
Figure 2. Cytoplasmic localization of ADAR1p110 is regulated by p38 MAP kinase signaling.
(a) Various MAP kinase activating kinase expression constructs were co-transfected with mCherry-ADAR1p110-WT into A172 cells (left summary graph). Only MKK6* (constitutively active MKK6) significantly increased cytoplasmic localization of ADAR1p110 in UV un-irradiated A172 cells. Images of ∼30 cells each from (x02267)7 separate slides (n (x02267) 7) prepared independently (∼200 cells) were examined for mCherry-ADAR1p110 distribution between nucleus and cytoplasm. Data are mean ± S.D.; significant differences were identified by two-tailed Student's t-tests: ***P <0.001. Source data are available online in Source Data Set 1. The cytoplasmic localization was completely blocked by SB203580 (p38 inhibitor). (b) Phosphorylated p38 (pp38) localizes exclusively in the nucleus in UV irradiated A172 cells. The p38 inhibitor, although not affecting the nuclear localization of pp38, blocks the cytoplasmic localization of mCherry-ADAR1p110. (c) UV-irradiation induced cytoplasmic localization of mCherry-ADAR1p110-WT was blocked by simultaneous knockdown of MSK1 and MSK2. (a-c) Scale bars, 20 μm.
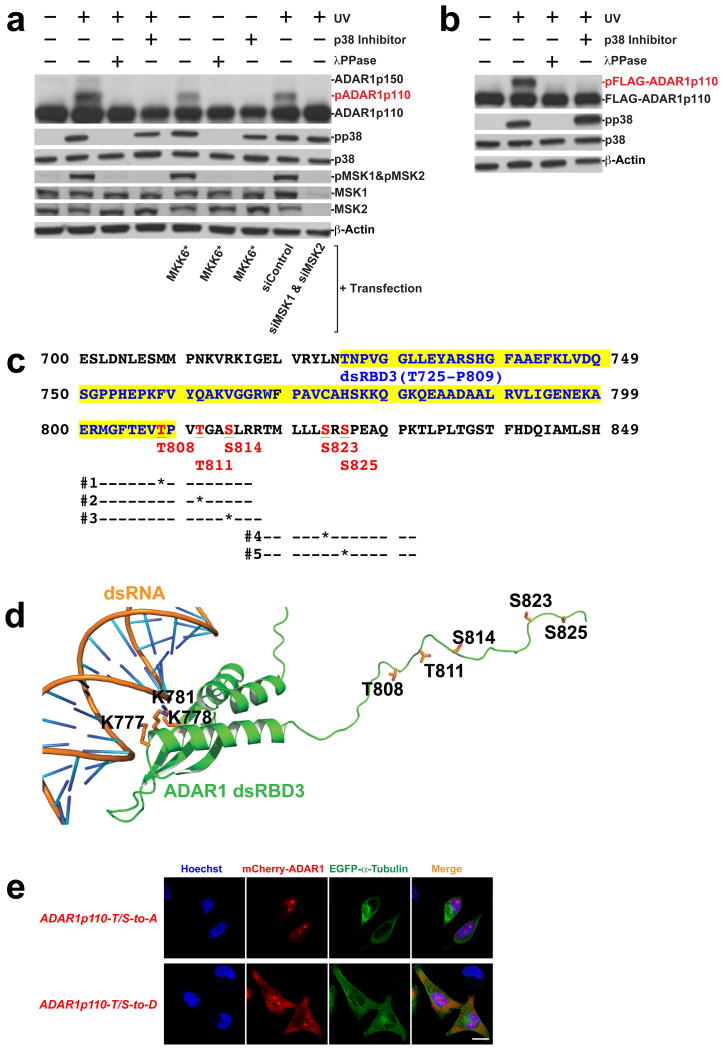
Finally, Phos-tag western blotting analysis revealed a slower migrating ADAR1p110 band, which was detected only in UV-irradiated or MKK6* transfected cells but abolished by addition of the p38 inhibitor or by knockdown of MSK1 and MSK2 (Fig. 3a). Treatment of the cell extract with a Lambda phosphatase abolished the slow migrating ADAR1p110 band (marked as pADAR1p110), indicating that phosphorylation is responsible for this band shift (Fig. 3a). We confirmed that not only endogenous ADAR1p110 but also exogenous FLAG-tagged ADAR1p110 is phosphorylated in a UV irradiation and p38 dependent manner (Fig. 3b). Mass spectrometry of the FLAG-ADAR1p110 phosphorylated band revealed significant phosphorylation of two threonine and three serine (T808, T811, S814, S823, and S825) in the region connecting dsRBD3 and catalytic domains (Fig. 3c, d). The effects of phosphorylation of these five sites on the cytoplasmic localization of ADAR1p110 were examined by using mCherry-fusion constructs of two isogenic ADAR1p110 mutants, T/S-to-A phosphorylation inhibitory mutant (five threonine and serine residues mutated to alanine) and T/S-to-D phosphomimetic mutant (five threonine and serine residues mutated to aspartate). We found that the T/S-to-A mutant, like the wildtype, localized almost exclusively in the nucleoplasm and nucleoli. In contrast, significantly increased cytoplasmic localization of the T/S-to-D mutant, co-localized with EGFP-α-Tubulin, was detected (Fig. 3e), confirming a critical role played by phosphorylation of these sites in the cytoplasmic localization of ADAR1p110. We also investigated the effect of single site T/S-to-D mutation at select sites and found that mutation of even a single site had some effect on cytoplasmic localization, but was not as effective as mutation of the five sites (data not shown), indicating the importance of the multiple site phosphorylation and its cumulative effects. The almost exclusively cytoplasmic localization of ADAR1p150-WT was not affected by either T/S-to-A or T/S-to-D mutation (Supplementary Fig. 1c).
Figure 3. Phosphorylation of ADAR1p110 at five sites.
(a) Detection of phosphorylated ADAR1p110 by Phos-tag PAGE and western blotting. A172 cell extracts were prepared following UV irradiation, transient transfection with the MKK6* expression construct in the absence or presence of p38 inhibitor SB203580, or simultaneous knockdown of MSK1 and MSK2. Some samples were treated with λ phosphatase prior to SDS PAGE. (b) Phos-tag SDS PAGE analysis of FLAG-ADAR1p110 recombinant proteins. Cell extracts were prepared from A172 cells transiently transfected with the FLAG-ADAR1p110 expression construct with or without UV irradiation. (a, b) Source data are available online in Source Data Set 2. (c) Phosphorylation sites identified by LC-MS/MS are shown. DsRBD3 region (T725-P809) is highlighted in yellow. Phosphorylation sites identified are highlighted in red and underlined. Each bar indicates peptides identified by LC-MS/MS. (d) Model of human ADAR1 dsRBD3 and the downstream region in complex with dsRNA. The region of ADAR1 spanning residues 715-799 belongs to the standard dsRBD, whereas the region downstream of this domain where T808, T811, S814, S823, S825 are located is disordered. The lysines K777, K778 and K781 coordinating the dsRNA are indicated. (e) Localization of ADAR1p110 phosphorylation inhibitory (T/S-to-A) and phosphorylation mimetic (T/S-to-D) mutants. Scale bar, 20 μm.
We also examined in vitro the effects of mutation on the A-to-I editing activity using ADAR1p110 recombinant proteins made in baculovirus infected Sf9 insect cells32. We found no difference in the A-to-I RNA editing activity among T/S-to-A, T/S-to-D, and wildtype ADAR1 (Supplementary Fig. 3a). We previously reported that ADAR1p110 forms a complex with Dicer and promotes processing of pre-miRNAs to mature miRNAs during mouse embryo development33,34. Accordingly, we also examined processing of pre-let-7a to mature let-7a in vitro using Dicer-ADAR1p110 complexes34. We found again no difference in the pre-miRNA cleavage activity among Dicer-ADAR1p110 complexes consisting of wildtype, T/S-to-D, and T/S-to-A mutant (Supplementary Fig. 3b).
Finally, we examined global miRNA expression profile in UV irradiated and ADAR1 knocked down cells by high throughput sequencing of small RNAs. We anticipated that ADAR1p110 newly translocated to the cytoplasm might be involved in expression of a special set of miRNAs, perhaps required for stress response35,36. Surprisingly, knockdown of ADAR1 caused very little change in global expression of miRNAs in UV irradiated cells (data not shown). Furthermore, we found very little formation of the Dicer-ADAR1p110 complex in UV-irradiated A172 cells (data not shown). This is possibly due to the presence of abundant TRBP and thus Dicer-TRBP complexes in A172 cells used for the present studies, in contrast to the low TRBP expression in E11-12 mouse embryos34. It seems that ADAR1p110 is exported to the cytoplasm for reasons other than its RNAi promoting function34.
Phosphorylation of ADAR1p110 promotes its binding to nuclear exporter protein Exportin-5
Although ADAR1p110 usually localizes in the nucleus, nuclear export of ADAR1p110 mediated by Xpo5-RanGTP has been reported37. Furthermore, nuclear import of ADAR1p110 is regulated by Transportin-1 (TRN1). Cellular distribution of ADAR1p110 seems to be regulated by a balance between Xpo5 mediated export and TRN1 mediated import37. We already showed that ADAR1p110 is phosphorylated in the nucleus by the nuclear targets of pp38, MSK1 and MSK2 (Fig. 2c, Fig. 3a). Furthermore, UV irradiation-induced cytoplasmic localization of wildtype ADAR1p110 (Supplementary Fig. 4a) and also cytoplasmic localization of ADAR1p110-T/S-to-D mutant (Supplementary Fig. 4b) were completely blocked upon knocking down of Xpo5, indicating that the nuclear export of both phosphorylated ADAR1-WT and ADAR1p110-T/S-to-D mutant depends on Xpo5. Finally, the almost exclusive nuclear localization of ADAR1p110-T/S-to-A mutant was abolished upon knocking down of TRN1, indicating that the nuclear import of the T/S-to-A mutant still depends on TRN1 (Supplementary Fig. 4c). Taken together, these results suggest that phosphorylation of ADAR1p110, which occurs in the nucleus, most likely affects the Xpo5 mediated nuclear export mechanism rather than the TRN1 mediated nuclear import mechanism.
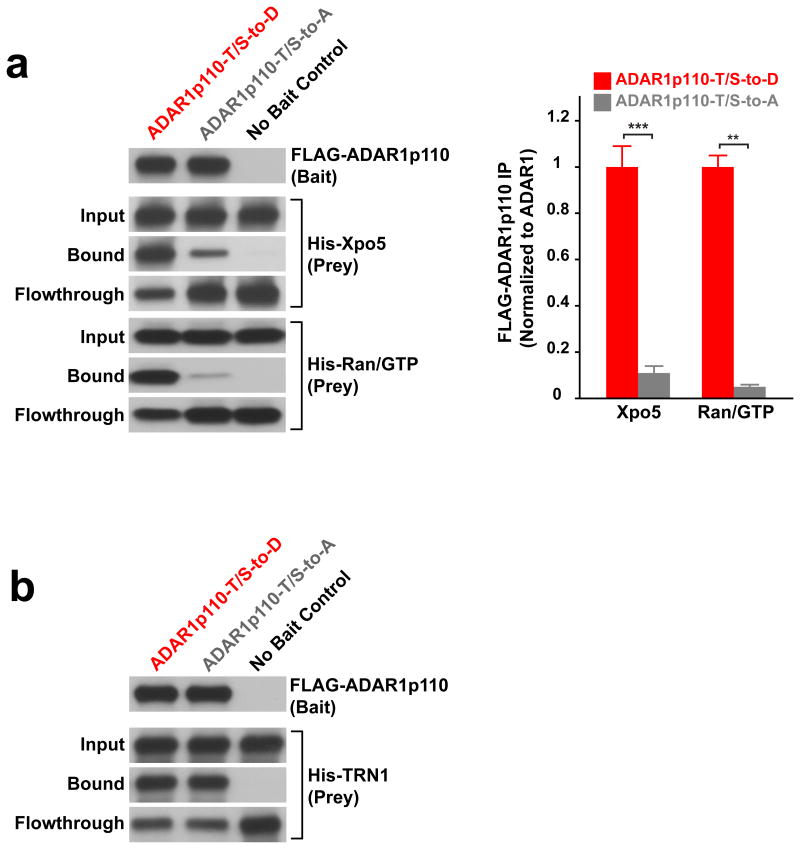
Accordingly, we next investigated the effects of ADAR1p110 phosphorylation on binding to Xpo5 by an in vitro binding assay as described previously37,38. We found that the amount of ADAR1p110-T/S-to-D bound to the Xpo5-RanGTP complex indeed increased significantly (∼10-20 fold) as compared to that of ADAR1p110-T/S-to-A (Fig. 4a). As expected, no difference between ADAR1p110-T/S-to-A and -T/S-to-D mutants was detected for their binding to TRN1 (Fig. 4b). We conclude that phosphorylation of ADAR1p10 regulated by MKK6-p38-MSK1&MSK2 MAP kinases promotes its binding to the Xpo5-RanGTP complex and consequently its nuclear export to cytoplasm.
Figure 4. Phosphorylation of ADAR1p110 promotes its binding to Xpo5-RanGTP.
(a) Binding of the phosphomimetic and phosphorylation inhibitory mutants of FLAG-ADAR1p110 to the Xpo5-RanGTP complex was tested in vitro on the M2 anti-FLAG MAb beads. An equimolar ratio of His-Xpo5 and His-RanGTP prey proteins to their bait proteins (FLAG-ADAR1p110) was used. Judging from the amount of unbound His-Xpo5 proteins detected in flow-through fractions, it seems that ADAR1p110-T/S-to-D binds to the Xpo5-RanGTP complex with ∼70% efficiency, whereas binding of ADAR1p110-T/S-to-A is at most 10% efficiency. Data are mean ± S.E.M.(n = 3, technical replicates); significant differences were identified by two-tailed Student's t-tests: **P < 0.01, ***P < 0.001. Source data are available online in Source Data Set 1. (b) Binding of the phosphomimetic and phosphorylation inhibitory mutants of FLAG-ADAR1p110 to TRN1 was tested in vitro on the M2 anti-FLAG MAb beads. (a, b) Source data for western blotting images are available online in Source Data Set 2.
What is the function of ADAR1p110 exported to the cytoplasm in response to stress?
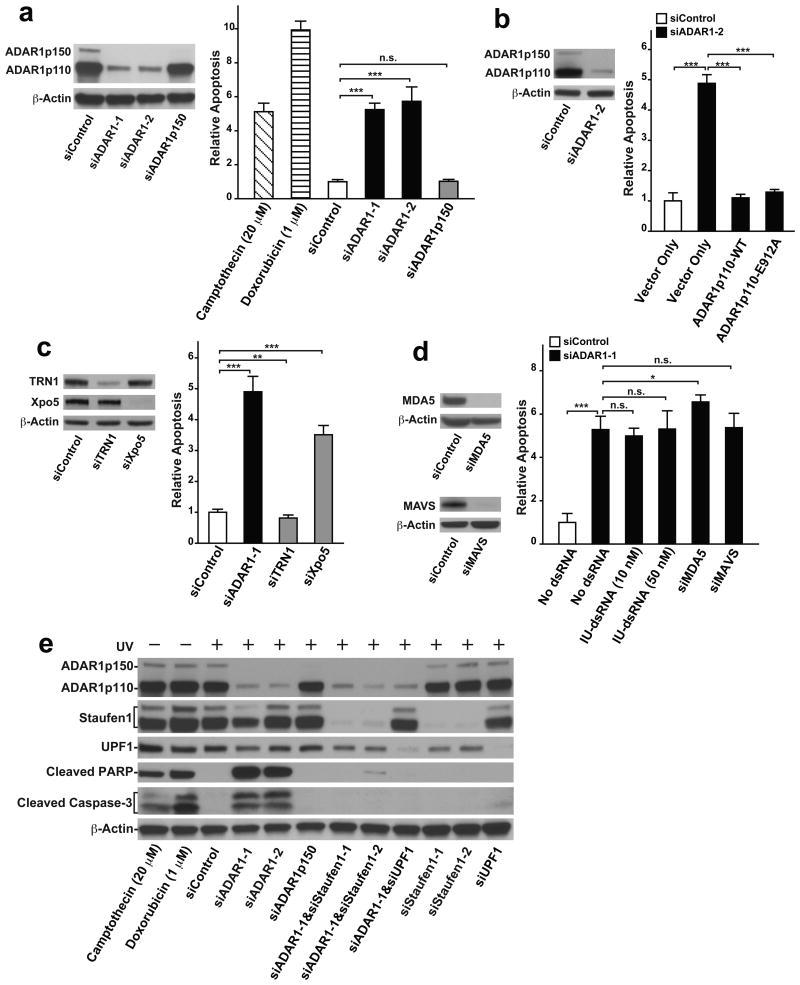
The stress-activated p38 MAPK signaling pathways are known to be involved in regulation of death or survival of stressed cells39,40. We, therefore, examined the possibility that cytoplasmic ADAR1p110 plays a role in the regulation of apoptosis. We found that knocking down ADAR1 indeed induced apoptosis in UV irradiated A172 cells at a significantly higher rate (∼5-6 fold) as compared to the control (Fig. 5a, Supplementary Fig. 5a), revealing a protective role of ADAR1 against induction of apoptosis in cells under stress. Interestingly, ADAR1p110 is the isoform specifically responsible for this anti-apoptotic function, as specific knockdown of ADAR1p150 did not induce apoptosis in UV irradiated cells (Fig. 5a). Similar results were obtained with a separate human cell line, U2OS osteosarcoma (Supplementary Fig. 5b). As expected, UV-induced apoptosis was reversed in ADAR1 knocked down cells by transfecting a FLAG-tagged ADAR1p110-WT expression construct. Interestingly, ADAR1p110-E912A, a deamination defective mutant, was also able to rescue the ADAR1 knocked down cells from apoptosis (Fig. 5b, Supplementary Fig. 5c). These results indicate that the function of ADAR1p110 and not its A-to-I editing function but most likely the dsRNA binding function are responsible for suppression of stress-induced apoptosis.
Figure 5. ADAR1p110 protects stressed cells from apoptosis.
(a-d) The extent of apoptosis was evaluated by ApoTox-Glo Triplex apoptosis assay (Promega), which simultaneously measures cell viability, cytotoxicity, and apoptosis (caspase3/7 activities). The signal intensity of relative luminescent units corresponding to caspase 3/7 activities was normalized with relative fluorescent units representing the cell viability measurements (400 nm/505 nm). More details in ONLINE METHODS. (a) ADAR1 knockdown induced apoptosis in UV-irradiated A172 cells at a much higher rate than in control cells. ADAR1p150 specific knockdown did not cause UV-induced apoptosis. For ADAR1 p150 selective knockdown, a specific siRNA targeting the human ADAR1 mRNA (NM_001111.4) exon1 region was used. Positive apoptosis controls: un-irradiated A172 cells treated with Camptothecin (20 μM) for 48 hrs or Doxorubicin (1 μM) for 24 hrs. (b) Apoptosis induced in UV irradiated cells by ADAR1 knockdown was rescued by transfection of both wildtype (WT) and editing deficient mutant (E912A) of ADAR1p110 expression vectors. For selective knockdown of endogenous ADAR1, siRNAs corresponding to the human ADAR1 mRNA 3′UTR (siADAR1-2) were designed and used here. (c) Apoptosis was induced in UV irradiated cells by Xpo5 knockdown but not by TRN1 knockdown. (d) Apoptosis induced in UV irradiated cells by ADAR1 knockdown was not rescued by transfection of synthetic I:U dsRNAs, MDA5 knockdown, or MAVS knockdown. (a-d) Knockdown efficiency by each siRNA was confirmed by immunoblotting (left panels). Data are mean ± S.D. (n = 4, cell cultures); significant differences were identified by two-tailed Student's t-tests: *P <0.05, **P < 0.01, ***P < 0.001, n.s., not significant. Source data are available online in Source Data Set 1. (e) The extent of apoptosis induced in UV-irradiated A172 cells was evaluated by western blotting analysis of cleaved PARP (89 kDa) and Caspase-3 (17 and 19 kDa) fragments. (a-e) Source data for western blotting images are available online in Source Data Set 2.
Finally, we found that knockdown of Xpo5 but not TRN1 also induced apoptosis in UV irradiated cells (Fig. 5c), indicating that nuclear export of certain proteins such as ADAR1p110 is critical for the mechanism that suppresses stress-induced apoptosis.
The MDA5-MAVS-IFN signaling independent function of cytoplasmic ADAR1p110
Aberrant activation of the dsRNA sensing mechanism mediated by melanoma differentiation associated protein-5 (MDA5)-mitochondrial antiviral signaling protein (MAVS)-interferon (IFN) signaling underlies the embryonic lethality of Adar-/- mutant mice14-16. ADAR1p150 suppresses this MDA5-MAVS-IFN pathway by editing its triggers, the 3′UTR long dsRNAs made from inverted repeats such as SINE and Alu, and consequently making them undetectable by the dsRNA sensing mechanism15. The 3′UTR long dsRNAs edited extensively by ADAR1p150 may even inhibit binding of the unedited 3′UTR dsRNAs to MDA514-16. In support of this hypothesis, synthetic dsRNAs that contain multiple I:U wobble base pairs and mimic extensively A-to-I edited dsRNAs (I:U dsRNAs) were shown to be capable of preventing activation of the MDA5-MAVS-IFN pathway14,41. Accordingly, we investigated the relationship between the stress induced anti-apoptotic function of cytoplasmic ADAR1p110 and the MDA5-MAVS-IFN pathway. We found that transfection of I:U dsRNA could not prevent UV-induced apoptosis in ADAR1 knockdown cells (Fig. 5d). Furthermore, knockdown of neither MDA5 nor MAVS could suppress UV-induced apoptosis of ADAR1 knocked down cells (Fig. 5d). On the other hand, a known trigger of the MDA5-MAVS-IFN pathway, synthetic long dsRNA poly(I:C), induced strong IFN responses, indicating that the MDA5-MAVS mediated dsRNA sensing mechanism remains intact in A172 cells (Supplementary Fig. 5d). These results together indicate that the anti-apoptotic function of ADAR1p110 translocated to the cytoplasm in response to stress is independent of its A-to-I editing activity and also independent of the MDA5-MAVS signaling pathway.
ADAR1p110 suppresses Staufen1-mediated decay of mRNAs containing 3′UTR dsRNAs
In order to obtain insights into the function of ADAR1p110 exported to the cytoplasm, we then investigated global mRNA expression profile in UV irradiated and ADAR1 knocked down cells by high throughput sequencing analysis (RNA-seq). We found that the expression of many gene transcripts was significantly reduced in UV-irradiated and ADAR1 knocked down cells (Supplementary Table 1), but noted no significant upregulation of interferon-stimulated genes (ISGs) (Supplementary Table 2), in contrast to the dramatic increase of ISG expression levels noted in Adar-/- mouse embryos14-16. This confirms that the stress-induced cytoplasmic ADAR1p110 function is independent of MDA5-MAVS-IFN signaling. Interestingly, we noted that most of these ADAR1 regulated genes contain 3′UTR dsRNAs made primarily from inverted Alu repeats, of which some were A-to-I edited (Supplementary Fig. 6 and Supplementary Table 3).
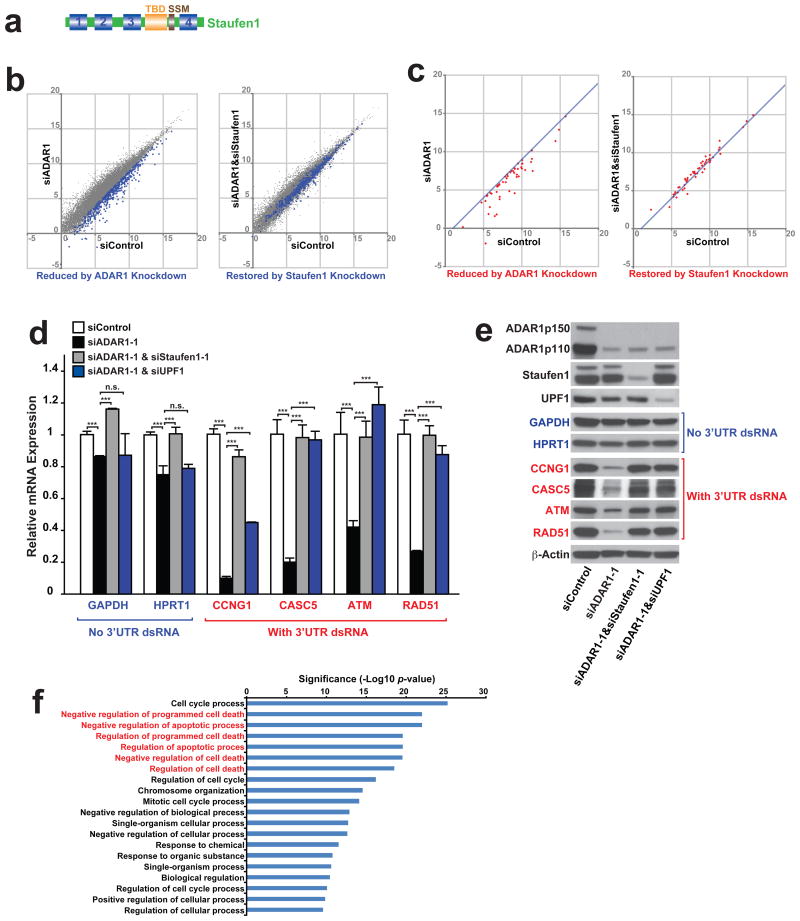
Association of 3′UTR Alu dsRNA with Staufen1 has been reported42,43. Staufen1 is also a dsRNA binding protein, like ADAR1, containing four dsRNA binding domains (Fig. 6a). Staufen1 binds to Staufen1 binding sites (SBSs), recruits UPF1 RNA helicase, and promotes degradation of its target mRNAs (Staufen1-mediated mRNA decay or SMD)22,44. Staufen1 almost exclusively localizes in the cytoplasm regardless of UV irradiation, and co-localized with the ADAR1p110 newly translocated to the cytoplasm in UV-irradiated A172 cells (Supplementary Fig. 7). Accordingly, we reasoned that the cytoplasmic ADAR1p110 may protect certain mRNAs containing 3′UTR Alu dsRNAs, which are otherwise degraded by SMD. We first confirmed that knockdown of Staufen1 and/or UPF1 did not activate apoptosis in UV-irradiated cells (Fig. 5e, Supplementary Fig. 5e). Most importantly, however, knockdown of Staufen1 and/or UPF1 did indeed prevent UV-irradiation induced apoptosis in ADAR1 knocked down cells (Fig. 5e, Supplementary Fig. 5b, 5e). RNA-seq data analysis revealed that simultaneous Staufen1 knockdown restored the mRNA levels of 488 genes significantly downregulated by ADAR1 knockdown (Fig. 6b and Supplementary Table 1). On the other hand, a majority of genes downregulated by ADAR1 knockdown but restored by Staufen1 knockdown was not significantly affected by Staufen1 knockdown alone (Supplementary Table 1). We selected a set of genes (total 52 genes), which contain dsRNA structures within their already annotated 3′UTRs or, in some case, extended 3′UTRs revealed in this study through analysis of RNA-seq reads. Reduced or restored expression of these transcripts by ADAR1 knockdown alone or by simultaneous ADAR1 and Staufen1 knockdown, respectively, was confirmed (Fig. 6c). We further tested those selected gene transcripts containing a 3′UTR dsRNA structure, such as Cyclin G1 (CCNG1), Cancer Susceptibility Candidate 5 (CASC5)/Kinetochore Scaffold 1, ATM, and RAD51, by qRT-PCR analysis to evaluate the effect of Staufen1 knockdown more quantitatively (Fig. 6d and Supplementary Table 3). We confirmed that the expression of most of the 52 genes was significantly reduced by ADAR1 knockdown but restored by Satufen1 knockdown. Furthermore, we confirmed with select genes that UPF1 knockdown also has rescue effects very similar to that of Staufen1 knockdown (Fig. 6d). In addition, we analyzed pre-mRNA levels of four SMD target genes containing 3′UTR Alu dsRNAs (Supplementary Fig. 8). We found that the expression of CCNG1 pre-mRNAs containing 3′UTR Alu dsRNAs was slightly increased, instead of decreased, by ADAR1 knockdown. No additional effects of Staufen1 or UPF1 were detected. We do not know the reason for this upregulation of CCNG1 gene transcription by ADAR1 knockdown. The pre-mRNA levels of the remaining SMD target genes were unchanged. The results indicate that the reduced expression of mRNAs containing 3′UTR Alu dsRNAs are very likely caused post-transcriptionally by SMD. Western blotting analysis confirmed reduced and restored protein levels of those genes containing 3′UTR Alu dsRNAs by ADAR1 knockdown and Staufen1 or UPF1 knockdown, respectively (Fig. 6e). We were puzzled by a slight reduction by ADAR1 knockdown and restoration by Staufen1 knockdown also noted with GAPDH and HPRT1 mRNA levels, since these two housekeeping genes lacking 3′UTR dsRNA structures were used as our negative controls (Fig. 6d). However, analysis of their pre-mRNA levels revealed that the changes in their mRNA levels perhaps reflected their transcriptional rate changes (Supplementary Fig. 8). In any case, the very slight changes of their mRNA levels may be physiologically irrelevant, unlike those of the SMD target mRNAs, as both GAPDH and HPRT1 protein levels remained unchanged (Fig. 6e).
Figure 6. ADAR1 protects mRNAs containing 3′UTR dsRNA from Staufen1-mediated decay.
(a) Schematic domain structure of Staufen1. TBD, tubulin-binding domain. SSM, Staufen-swapping motif. (b) Gene expression comparison by RNA-seq analysis between UV irradiated A172 cells treated with siControl versus siADAR1 or siControl versus siADAR1&siStaufen1. Numbers indicates logarithm of FPKM to base 2. All genes detected by RNA-seq (gray dots) and 488 selected genes (blue dots) whose expression levels decreased more than 2 fold by ADAR1 knockdown and rescued more than 1.5 fold by ADAR1&Staufen1 knockdown (Supplementary Table 1) are shown. (c) Expression levels of genes selected manually from Supplementary Table 1 because of the presence of 3′UTR dsRNA structures are compared. The 3′UTR dsRNA structure was identified using USCS genome browser and mfold algorism. Numbers indicates logarithm of FPKM to base 2. The line shows 2-fold decrease compared to siControl. Expression levels of the selected genes were also examined by quantitative RT-PCR (Supplementary Table 3). (d) Quantitative RT-PCR of non-structured 3′UTR mRNAs (Blue) and dsRNA-containing 3′UTR mRNAs (Red) was used to determine the effects of ADAR1 only, ADAR1&Staufen1 double-knockdown, or ADAR1&UPF1 double-knockdown in UV irradiated A172 cells. ADAR1 knockdown decreased significantly levels of mRNAs containing 3′UTR dsRNA, and Staufen1 or UPF1 knockdown restored their expression levels. Data are mean ± S.D. (n = 3 for GAPDH, 4 for other genes, technical replicates); significant differences were identified by two-tailed Student's t-tests: ***P < 0.001, n.s., not significant. Source data are available online in Source Data Set 1. (e) Western blotting analysis confirmed that changes in the mRNA levels are reflected in the protein levels. Source data are available online in Source Data Set 2. (f) The selected 488 genes (described in b) was subjected to GOrilla gene ontology enrichment analysis.
Ontology analysis of these 488 SMD target genes specifically protected by ADAR1p110 revealed that they are particularly enriched in anti-apoptotic functions (Fig. 6f and Supplementary Table 4), perhaps explaining why stressed cells undergo apoptosis in the absence of ADAR1p110.
ADAR1p110 competes with Staufen1 for binding to 3′UTR dsRNAs and antagonizes SMD
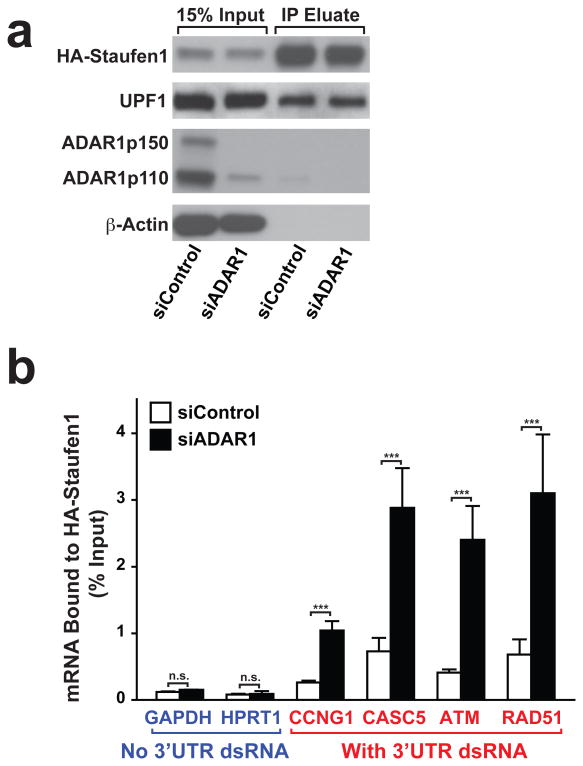
Apoptosis induced in UV irradiated and ADAR1 knocked down cells was rescued by ADAR1p110-E912A catalytically defective mutant (Fig. 5b), indicating that the SMD regulatory function of ADAR1p110 is independent of its A-to-I editing activity, but most likely dependent on its dsRNA binding activity. We, therefore, next conducted RNA immunoprecipitation (RIP)-qRT-PCR analysis (Fig. 7a) for selected genes, whose expression was suppressed by ADAR1 knockdown but rescued by Staufen1 knockdown. We first confirmed that Staufen1 did not bind to 3′UTRs containing no dsRNA structure such as GAPDH and HPRT1 mRNAs (Fig. 7b) in UV irradiated and ADAR1 knocked down cells (Supplementary Table 5). In contrast, Staufen1 bound to the 3′UTR dsRNAs of CCNG1, CASC5, ATM, and RAD51 mRNAs, and its binding was increased significantly by ADAR1 knockdown (Fig. 7b). Taken together, our results clearly indicate that ADAR1p110 protects only those mRNAs containing a dsRNA structure in their 3′UTRs by competitively inhibiting the binding of Staufen1 and suppressing their Staufen1 mediated degradation (Fig. 8).
Figure 7. ADAR1 inhibits binding of Staufen1 to the 3′UTR dsRNA structure.
(a) Formaldehyde crosslinking of RNA-protein complex and immunoprecipitation by HA-tagged Staufen1 was conducted in siControl or siADAR1-treated and UV-irradiated A172 cells. Input and IP eluate fractions were monitored by immunoblotting analysis. Source data are available online in Source Data Set 2. (b) Quantitative RT-PCR of mRNAs without structured 3′UTR (Blue) and mRNAs containing 3′UTR dsRNA (Red) were performed using RNAs purified from immunoprecipitated RNA-HA-Staufen1 complex. Data are mean ± S.D. (n = 4, technical replicates); significant differences were identified by two-tailed Student's t-tests: ***P < 0.001, n.s., not significant. Source data are available online in Source Data Set 1.
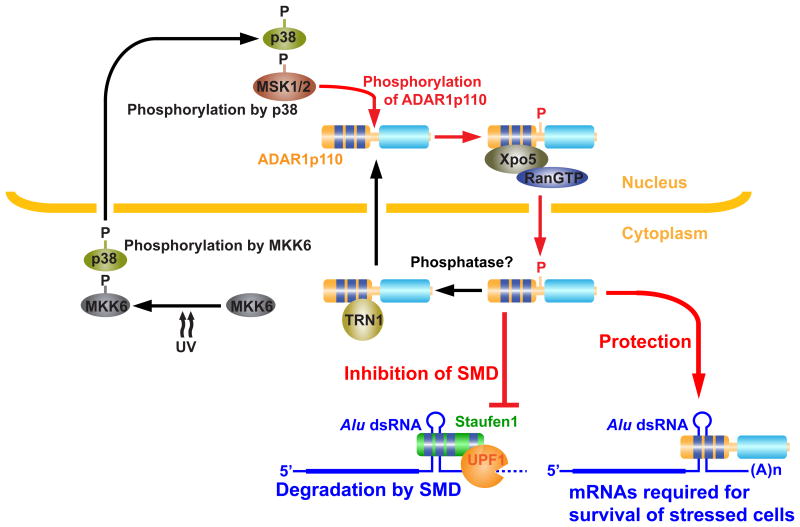
Figure 8. Stress response function of ADAR1p110 regulated by MKK6-p38-MSK1&2 MAP kinases.
Stress such as UV irradiation and heat shock activates MKK6-p38-MSK1&MSK2 signaling cascade. Stress induced phosphorylation of ADAR1p110 by MSK1 and MSK2 in the nucleus promotes its binding to Xpo5 and export to the cytoplasm. ADAR1p110, perhaps dephosphorylated by a currently unknown phosphatase, binds to TRN1, and is imported to the nucleus. Once translocated to the cytoplasm, ADAR1p110 inhibits binding of Staufen1 to Alu dsRNA present in 3′UTRs of many anti-apoptotic mRNAs and promotes survival of stressed cells.
Discussion
Messenger RNA degradation is an important mechanism to regulate swiftly the expression of many genes required for cells to respond to enviromental changes or stress24,45. In the present study, we identified ADAR1p110 as a stress response protein and established its anti-apoptotic function which is required for survival of stressed cells. We believe that this is the first identification of cytoplasmic ADAR1p110 as a stress response protein. The newly identified stress response function of ADAR1p110 is regulated through phosphorylation by MKK6-p38-MSK1&2 MAP kinases. Phosphorylation of T808, T811, S814, S823, and S825 facilitates binding of ADAR1p110 to the nuclear exporter Xpo5 and consequently its export to the cytoplasm (Fig. 8). These phosphorylation sites highly conserved are all located in the unstructured hinge region connecting dsRBD3 and the deaminase domain (Fig. 3d). Negative charges resulting from phosphorylation likely cause significant conformational changes, and may also affect the interaction between dsRNA and dsRBD3, and consequently ADAR1p110 binding to Xpo5.
Most importantly, the newly exported ADAR1p110 then protects a set of mRNAs, those containing a 3′UTR dsRNA structure mostly made from inverted Alu repeats, from SMD by competitively inhibiting binding of Staufen1 to the 3′UTR dsRNA (Fig. 8). Interestingly, mRNAs of many anti-apoptotic genes contain 3′UTR dsRNAs and are protected by the cytoplasmic ADAR1p110, which is most likely the explanation for the apoptotic death of stressed cells in the absence of ADAR1p110. For instance, both ATM and RAD51, which are protected by ADAR1p110 (Fig. 6d, 7b) are involved in the DNA repair and cell cycle check point mechanisms, but their function in protection of stressed cells against apoptosis has also been repeatedly reported. For instance, ATM is required for suppression of apoptosis and survival of cells exposed to oxidative stress such as ionizing radiation46. Similarly, the protective function of RAD51 against DNA-damage-induced apoptosis has also been reported47. However, we currently do not know exactly which gene transcripts protected by ADAR1p110 are essential for survival of UV-irradiated cells. A few gene transcripts may each play a critical role in promoting survival of UV-irradiated cells. Alternatively, the cumulative effects of many or all gene transcripts (488 genes) protected by ADAR1p110 may be of importance.
It is not clear whether the ADAR1p110 function in inhibiting SMD is specific for this isoform. However, it seems that the endogenous ADAR1p150 expression levels are very low (Fig. 5a) and likely insufficient to potect all Staufen1 target mRNAs, which may be the reason why ADAR1p110 needs to be exported to the cytoplasm under stressed conditions. Although a fraction of ADAR1p150 moves to stress granules, Staufen1 stays in the cytoplasm prior and upon stress, excluding the possibility of their inetraction in stress granules.
It appears that ADAR1p110 outcompetes the binding of Staufen1 to its target 3′UTR Alu dsRNA almost completely. The reason why ADAR1p110 outcompetes Staufen1 is currently not clear, since a very similar dsRNA binding affinity of these two dsRNA binding proteins has been reported48. Interestingly, however, the interaction of Staufen1 with dsRNA is more dynamic, whereas that of ADAR1 is very static48, which possibly explains the dominant negative effects of ADAR1p110 on binding of Staufen1 to 3′UTR Alu dsRNAs.
The MAP kinase p38 is involved in the mechanism that regulates death or survival of stressed cells39,49. p38 kinase activated by MKK3 and/or MKK6 mediated phosphorylation translocates to the nucleus and phosphorylates many transcription activators and suppressors as well as downstream kinases such as MSK1 and MSK2, which in turn rapidly changes expression of many genes required in response to stress. The p38 signaling pathway also affects cellular distribution of many target gene products. The fate of stressed cells seems to be determined by a balance between survival and death mechanisms, both of which p38 regulates19,28,40. Here, we demonstrated that p38 and its targets MSK1 and MSK2 regulate the nuclear export of ADAR1p110, which then promotes survival of stressed cells in the cytoplasm (Fig. 8).
Recent studies on anaylysis of Adar-/- and crosses of Adar-/- and Ifih1-/- (MDA5 null) or Mavs-/- led to identification of a function of ADAR1p150 in suppression of the dsRNA sensing mechanism mediated by MDA5-MAVS-IFN signaling14-16. ADAR1p150 suppresses the dsRNA sensing initiation step by extensively editing cytoplasmic and endogenous trigger dsRNAs such as 3′UTR inverted Alu repeats. The activation of this pathway also results in apoptotic cell death due to overwhelming interferon responses in Adar-/- mouse embryos14-16. In this study, however, we found that the stress response function of the cytoplasmic ADAR1p110 is A-to-I editing independent and also MDA5-MAVS pathway independent, revealing a function of ADAR1p110 distinct from that of ADAR1p150.
Although crosses of Adar-/- and Ifih1-/- or Mavs-/- mice survive to birth, they die within 2 to 3 days despite the suppression of the dsRNA sensing mechanism and interferon responses in these new born mice14,15, indicating that ADAR1 functions other than suppression of the MDA5-MAVS-IFN pathway are required for adult life. Interestingly, crosses of Adar p150-/- and Mavs-/- mice not only survive to birth but also beyond weaning15. Furthermore, although AdarE861A mice carrying an editing-deficient knock-in mutation also die at E13.5 with activated interferon and dsRNA sensing pathways, crosses of AdarE861A and Ifih1-/- mice live completely normal adult lives16. These findings together suggest that the A-to-I editing independent and also MDA5-MAVS-IFN signaling independent functions of ADAR1p110, in particular, are required for normal postnatal development. Histological inspection of Adar p150-/-:Mavs-/- mice and rare surviving Adar-/-:Mavs-/- mice revealed the importance of the MDA5-MAVS-IFN pathway independent functions of each ADAR1 isoform for development and homeostasis of particular organs and tissues, e.g. ADAR1p150 for intestinal homeostasis and B cell development and ADAR1p110 for kidney paterning15.
Defective development of certain organs and tissues detected in adult Adar mutant mice likely results from, at least in part, the deficiency in the stress response function of ADAR1p110 (A-to-I editing independent and MDA5-MAVS mediated dsRNA sensing-independent) described in this study as well as the Dicer promoting and thus miRNA processing promoting function of ADAR1p110, which is also A-to-I editing independent34. Our future studies will be focused on better understanding the A-to-I editing independent functions of ADAR1p110 in postnatal development and adult life.
Online Methods
Cell culture, RNA interference knockdown, and plasmid transfection
In vivo (tissue culture) experiments were done with A172 human glioblastoma (ATCC CRL-1620) or U2OS human osteosarcoma cells (ATCC HTB-96). A172 and U2OS cells were free mycoplasma contamination. RNA silencing experiments were done using lipofectamine RNAiMax (Life technologies) at 2.5 nM as the final siRNA concentration. All siRNAs used in this study are listed in Supplementary Table 6. Plasmid transfection was carried out using the Amaxa cell line nucleofector kit V (Lonza) with program U-29 of the Nucleofector device. UV irradiation (40 J/m2) was done for cells washed twice with PBS. For p38 MAPK inhibition, the treatment of the cell culture with 10 μM of SB203580 (Selleckchem) started before UV irradiation and continued till recovery of cells. For heat shock, cells were incubated at 44°C for 30 min, and then placed at 37°C for 1 hour.
Apoptosis assay
Assessment of cell apoptosis was carried out in 96-well plates using ApoTox-Glo Triplex assay kit (Promega), which simultaneously measures cell viability, cytotoxicity, and apoptosis (caspase3/7 activities). One day before the assay, 1 × 104 cells were subjected to siRNA transfection. One set of plate was irradiated with 40 J/m2 UVC 60 hrs post transfection, and Apotox-Glo Triplex assay was performed 12 hrs post UV irradiation. The signal intensity of relative luminescent units (RLUs) for caspase 3/7 activity was normalized with relative fluorescent units (RFUs) from the viability measurements (400 nm/505 nm). I:U dsRNA41 synthesized at IDT was used at 10 nM or 50 nM for transfection with Lipofectamin RNAiMax. Synthetic siRNAs corresponding to the human ADAR1 mRNA 3′UTR sequence or ADAR1 mRNA (NM_001111.4) exon1 sequence (Supplementary Table 6) were used for knockdown of the endogenous ADAR1 (both p150 and p110) or only ADAR1p150, respectively. Apoptotic cells were analyzed also by fluorescence microscopy using an AnnexinV conjugated with Alexa594 (ThermoFischer) and CellEvent™ Caspase-3/7 Green apoptosis detection system (ThemoFischer), which utilizes a fluorogenic substrate for activated Caspase-3 and -7. Apoptosis was also determined by immunoblotting analysis using mAbs specific to cleaved PARP (89 kDa) and Caspase-3 (17 and 19 kDa) fragments (Supplementary Table 7).
Plasmid construction
Two fusion constructs used for fluorescence microscopy, mCherry-ADAR1p110 and mCherry-ADAR1p150, were constructed using ADAR1p110 and ADAR1p150 as template plasmids34. Constructs carrying mutations at multiple phosphorylation sites (T808, T811, S814, S823, and S825) were prepared by PCR-based mutagenesis using wildtype plasmids and mutation primers (Supplementary Table 6). EGFP-α-Tubulin (pIRESneo-EGFP-alpha Tubulin) was purchased from Addgene. Baculovirus constructs, FLAG-ADAR1p110 and HAT-ADAR1p110, were described previously34. Their phosphorylation site mutants were prepared by PCR. The deamination defective mutant FLAG-ADAR1p110-E912A was prepared by PCR using the template plasmid FLAG-ADAR1p150-E912A50. Constitutively active MAPKK plasmids, MKK1*, MKK6*, and JNKK* were obtained from Zain Paroo and Melanie Cobb. The Staufen1 expression constructs hSTAU1(R)-FLAG and HA-Staufen151 were kind gifts of Lynn Maquat. Staufen1-EGFP was prepared by PCR using hSTAU1(R)-FLAG as the template plasmid.
Preparation of cytoplasmic and nuclear proteins
Un-irradiated or UV-irradiated A172 cells cultured in 6-well plates were harvested after washing with ice-cold PBS. The cell pellets were resuspended in 300 μl of Cytoplasmic Extract buffer containing 10 mM Na2HPO4 pH7.4, 137 mM NaCl, 2.7 mM KCl, 1 mg/ml Digitonin (SIGMA), complete EDTA-free protease inhibitor cocktail (Roche), and PhosStop phosphatase inhibitor cocktail (Roche) and incubated at 4°C for 10 min. After centrifugation at 6,000 g for 10 min at 4°C, the supernatants were collected to give the cytoplasmic fractions. Nuclear pellets were washed with Cytoplasmic Extract buffer and resuspended in RIPA buffer containing 50 mM Tris pH8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, complete EDTA-free protease inhibitor cocktail, PhosStop phosphatase inhibitor cocktail. After incubation for 10 min at 4°C and centrifugation at 20,000 g for 30 min at 4°C, the supernatants were collected to give the nuclear fractions.
Preparation of ADAR1 mutant proteins and in vitro editing assay
Preparation of the baculovirus construct, HAT-ADAR1p110, was described previously34. Constructs with phosphorylation site mutations of HAT-ADAR1p110 were prepared by PCR using mutagenesis primers (Supplementary Table 6). HAT-ADAR1p110 recombinant proteins were purified from Sf9 cell extracts as described previously34. The A-to-I RNA editing activities of ADAR1p110 mutants were determined as described previously52.
Preparation of Dicer-ADAR1 complexes and pre-miRNA cleavage assay
The FLAG-Dicer-HAT-ADAR1p110 mutant complexes were purified by two consecutive TALON and Flag Ab affinity column chromatography, and the pre-let-7a cleavage assay was done as described previously34.
Purification of Xpo5, RanGTP, and TRN1 recombinant proteins
The expression constructs, Exportin-5 (pQE60-Exp5)53 and RanQ69L (pET-RanQ69L)54, were purchased from Addgene. The Transportin-1 expression plasmid (pET28-His-TRN1)55 was a gift from Gideon Dreyfuss. E. coli cells transformed with constructs for expression of His-fusion proteins were grown at 37°C in LB media supplemented with 2% (vol/vol) ethanol to an A600 of 0.6 and induced with 1 mM IPTG for His-Transportin-1 and His-Ran(Q69L) and 100 mM IPTG for His-Exportin-5 at 20°C overnight. Harvested E. coli cells were sonicated, and the soluble fraction was incubated with 1 mM CaCl2 and 400 units/ml micrococcal nuclease for 12 min at room temperature. After adjusting the NaCl concentration to 500 mM, the extracts were applied to nickel-nitrilotriacetic acid beads (QIAGEN) for 1 h at 4 °C. Beads were washed twice with Buffer A (20 mM Hepes, pH 8.0, 500 mM NaCl, 5 mM imidazole, 20% glycerol, 7 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 1× Proteinase inhibitor cocktail), twice with Buffer A containing 2 M NaCl, and then in Buffer A containing 100 mM NaCl. His-tagged proteins were eluted in Buffer A containing 150 mM imidazole, and dialyzed against Buffer B (20 mM Hepes, pH 8.0, 100 mM NaCl, 20% glycerol, and 7 mM β-mercaptoethanol).
Xpo5 in vitro binding assay
VA1 RNA derivative (VARdm), which lacks the short bulge in the middle of the dsRNA structure, was used for preassembly of the Xpo5-RanGTP-RNA ternary complex as described previously38. VARdm was synthesized by T7 in vitro transcription using a partial dsDNA composed of an antisense DNA oligonucleotide containing VARdm and T7 promoter sequences and a sense DNA oligonucleotide containing T7 promoter sequence (Supplementary Table 6) with MEGAscript T7 transcription kit (ThermoFisher Scientific). The binding assay mixture containing 80 nM of His-Xpo5, 80 nM of His-RanQ69L, 2 mM GTP, and 80 nM of VA1 RNA derivative VARdm in a total volume of 25 μl binding buffer, 20 mM HEPES-KOH, pH7.0, 50 mM NaCl, 1 mM MgCl2, 0.01% NP-40, 5% Glycerol, 1 mM EGTA, 1 mg/ml BSA, and complete EDTA-free proteinase inhibitor cocktail (Roche), was preincubated for 10 min at 37°C. In parallel, 80 nM of FLAG-ADAR1p110 was prepared in the binding buffer in a total volume of 25 μl. After pre-incubation, the Xpo5-RanGTP-VA1 RNA complex was added to FLAG-ADAR1p110 and incubated further for 10 min at 37°C. The mixture was then added to 10 μl of packed anti-FLAG M2 magnetic beads (Sigma), which had been washed and blocked with binding buffer containing 10 mg/mL BSA. After incubating for 15 min at 4°C with gentle agitation, beads were captured using a magnetic beads separator (ThermoFisher Scientific), and then washed with the binding buffer five times. The beads were dissolved in Laemmli buffer and boiled at 95°C for 5 min. Samples were loaded on SDS-PAGE, and immunoblotting was performed with anti-Xpo5, anti-Ran, and anti-ADAR1 antibodies (Supplementary Table 7).
TRN1 in vitro binding assay
The reaction mixture of 80 nM of FLAG-ADAR1p110 in a total volume of 25 μl of the binding buffer containing 20 mM HEPES-KOH, pH7.0, 50 mM NaCl, 1 mM MgCl2, 0.05% NP-40, 5% Glycerol, 1 mM EGTA, 1 mg/ml BSA, and complete EDTA-free proteinase inhibitor cocktail (Roche) was pre-incubated for 15 min at 37°C, and then 80 nM of His-TRN1 in 25 μl of the binding buffer was added and incubated for 10 min at 37°C followed by anti-FLAG M2 magnetic beads capture, washing and elution procedures as described above for the Xpo5 binding assay. For immunoblotting, anti-TRN1 antibody (Supplementary Table 7) was used.
Fluorescence Microscopy
Transfection of fluorescent-protein-tagged plasmids or siRNAs into A172 cells was carried out as described above. After electroporation, cells were seeded on μ-Dish 35 mm high (IBIDI) and incubated at 37°C for 24-36h. Cells were then washed with PBS, irradiated with UV (40 J/m2). Microscopic analysis was done in normal medium containing 1 μg/ml Hoechst 33342 (Life technologies) for nuclear staining. Microscopic images were obtained by using a Leica TCS SP5 DMI6000 CS Confocal Microscope (Leica), equipped with ultraviolet 405 Diode, Argon, DPS3561, HeNe594 Lasers. Fluorescent images were captured with a 63.0 × lens zoomed in 1-3× with a 1,024×1,024 frame and 400 Hz scanning speed. For multicolor experiments, the following wavelength settings were used; mCherry (Ex 594 nm/Em 604-766 nm), EGFP (Ex 488 nm/Em 498-567 nm), YFP (Ex 514 nm/Em 520-590 nm), dsRed (Ex 561 nm/Em 571 -723 nm), and Hoechst (Ex 405 m/Em 414-475 nm). Images were further analyzed using Leica LAS AF software (Leica) and Image J (NIH).
Quantitative RT-PCR
Total RNA samples were extracted by miRNeasy kit (QIAGEN), and quantification of select mRNAs as well as pre-mRNAs was done by qRT-PCR. Reverse transcription was done using 1 μg of total RNA with Transcriptor First strand cDNA synthesis Kit (Roche), and the resultant cDNA equivalent to 20 ng of total RNA was used for qPCR with Power SYBR Green Master Mix (ThermoFisher Scientific) and 7900HT Fast Real-Time PCR system (Applied Biosystems), and each experiment was conducted in quadruplicate. Primers used are listed in Supplementary Table 6.
Combined RIP and qRT-PCR analysis
The RIP procedure was done as described previously56 with some modifications. A172 cells were transfected first with siControl or siADAR1-1 and then with HA-Staufen1. The cells were fixed with 0.1% formaldehyde in PBS at room temperature for 5 min. The cytoplasmic fraction was sonicated, and the lysate was incubated for 6 h at 4 °C with 100 μl (1mg) of anti-HA magnetic beads (ThermoFisher) prewashed and blocked with 20% BSA in lysis buffer. The RNA-Protein complexes captured on beads were washed five times with high salt buffer (50 mM Tris-HCl pH7.6, 1 M NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS, 1 mM EDTA), and then with the suspension buffer (50 mM Tris-HCl pH7.6, 150 mM NaCl, 1% NP-40). At the final washing step, beads were aliquoted into two fractions, one for RNA extraction and the other for protein preparation. RNA was reverse-crosslinked as described previously57, and purified with TRIZOL LS (Themofisher). Quantitative RT-PCR was performed with primers designed at the non-double stranded region adjacent to the predicted dsRNA structure in each mRNA 3′UTR (Supplementary Table 6).
Immunoblotting analysis
Cell lysates were prepared in Laemmli buffer (Boston BioProducts) containing Benzonase Nuclease (Sigma, E8263), Complete EDTA-free proteinase inhibitor cocktail (Roche), and PhosStop phosphatase inhibitor cocktail (Roche) and fractionated by 4%–15% SDS-PAGE. For detection of phosphorylated ADAR1 proteins, 6 % SDS-polyacrylamide gel containing 25 μM of Phos-tag acrylamide (Wako) and 125 μM of MnCl2 was used. For confirmation of phosphorylation, cell lysates were treated with 4 units/μl of Lambda phosphatase (NEB) at 30°C for 30 min prior to Phos-tag immunoblotting analysis. Proteins were transferred to Immobilon-P nylon membrane (Millipore). Blots were blocked with 1% Blocker BSA buffer (ThermoFisher Scientific) and incubated with primary antibodies (Supplementary Table 7) overnight at 4°C. After the incubation with the secondary antibodies, membranes were developed with ECL (GE Healthcare).
LC-MS/MS analysis of phosphorylated ADAR1p110
Cell lysates were prepared from HEK293T (ATCC CRL-11268) expressing FLAG-ADAR1p110 constitutively in a buffer containing 50 mM Tris-HCl pH7.6, 150 mM NaCl, 0.5% NP-40, complete EDTA-free proteinase inhibitor cocktail (Roche), and PhosStop phosphatase inhibitor cocktail (Roche). FLAG-ADAR1p110 proteins were purified on anti-FLAG M2-monoclonal antibody-agarose beads (Sigma). The eluted fraction was concentrated with ethanol precipitation and stored at -80°C overnight. The concentrated F-ADAR1p110 (∼30 μg from 4 × 108 cells) was loaded on 7% NuPAGE Tris-Aetate gel (ThermoFisher Scientific) or 6 % SDS-polyacrylamide gel containing 25 μM of Phos-tag acrylamide and 125 μM of MnCl2. Gels were stained with Colloidal Blue Staining kit (ThermoFisher Scientific), and the band corresponding to ∼110 kDa was excised, and proteins in the gel were digested with trypsin (Promega). Reverse phase liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis was performed using a Q Exactive HF mass spectrometer (ThermoFisher Scientific) coupled with a Nano-ACQUITY UPLC system (Waters). MS/MS spectra were screened with MaxQuant 1.5.1.258 against the human UniProt protein database (July 2014) using full tryptic specificity with up to two missed cleavages, static carboxamidomethylation of Cys, and variable oxidation of Met, protein N-terminal acetylation, deamidation of Asn, and phosphorylation on Ser, Thr and Tyr. Modified peptides were required to have a minimum score of 40. Consensus identification lists were generated with false discovery rates of 1% at protein, peptide and site levels.
Generating ADAR1 dsRBD3 model
To generate a model of the human ADAR1 dsRBD3 region (710 – 830), we used the software Protein Homology/analogY Recognition Engine V 2.0 (Phyre 2), which produces a model of the protein of interest based on sequence alignment to known structures59. Phyre 2 generated several models of this region of human ADAR1 corresponding to dsRNA binding protein motifs (dsRBM) with 100% confidence. From the list of models generated by Phyre 2 we selected the solution structure of the Rattus norvegicus ADAR2 dsRBM in complex with dsRNA60. The two proteins have 25% sequence identity and are both involved in dsRNA binding. The model was further refined by applying geometry minimization in Phenix61. The final model was used to generate figures in Pymol62.
RNA-seq analysis
A172 cells were transfected with siControl, siADAR1-1, siStaufen1-1, or siADAR1-1 & siStaufen1-1, and then incubated for 60 hrs. Cells were washed with PBS and irradiated with UV (40 J/m2) and harvested at 6 hrs post irradiation. Total RNA was extracted by miRNeasy kit. Cultures were prepared in duplicate for each condition. Preparation of RNA-seq samples was carried out using strand-specific SENSE mRNA library preparation kit V2 (Lexogen). Paired end 151 bp sequencing was performed with NextSeq 500 (Illumina).
Raw sequencing RNA-seq reads were trimmed 10 bp at the 5′ end and 30 bp at the 3′ end and aligned using bowtie2 algorithm allowing for up to 2 mismatches. RSEM v1.2.12 software63 was used to estimate fragment counts and FPKM values on gene level using the transcriptome table for hg19 genome version downloaded from UCSC browser (http://genome.ucsc.edu, tables hg19.knownGene). EdgeR64 was used to estimate significance of differential expression between two experimental groups. Edited sites were determined by using samtools v0.1.1965. Only A->G editing events occurring in any sample were considered, and coordinates for all found events were used to obtain direct A/C/G/T counts across all samples. The sum of counts between replicates were calculated and significance of differences in editing between any two conditions was tested using Fisher Exact Test. Genes with at least one edited site that showed significant reduction (P <0.05) in siADAR1 samples compared to siControl samples were considered as edited by ADAR1.
Statistical analysis
Experiments were run in triplicate or quadruplicate and repeated in a minimum of three independent trials. Image quantitation was done using ImageQuant image analysis software (GE Healthcare). Data are presented as means ± standard deviation (S.D.) or standard error of the mean (S.E.M.). Two-tailed t-tests were conducted where the minimum level of significance was P < 0.05.
Data availability
RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE85455. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via MassIVE (accession no. MSV000080146) with the dataset identifier PXD004959. Other data from this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Zain Paroo, Melanie Cobb, Susan Janiki, Gideon Dreyfus, and Lynne Maquat for various recombinant protein expression constructs. We also thank James Hayden for assistance with fluorescence microscopy, Yukiko Sakurai for her technical assistance, and Maureen Murphy and John M. Murray for critical reading of the manuscript. Research reported in this publication was supported by grants from the National Institutes of Health (R01GM040536 and R01CA175058), the Macula Vision Research Foundation, and the Japan Society for the Promotion of Science (S13204) to K.N. C.S. is a member of the Roy and Dianna Vagelos Scholars Program in the Molecular Life Sciences at the University of Pennsylvania. M.S. is supported in part by a fellowship from the Japan Society for the Promotion of Science (JSPS 2010-22). We thank also for services provided by the Animal, Bioinformatics, Genomics, Imaging, Protein Expression, and Proteomics Shared Facilities of The Wistar Cancer Center, which are supported by the National Cancer Institute (P30CA010815). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions: MS, YS, and KN designed the study. MS conducted fluorescence microscopy experiments, apoptosis analysis, and RIP experiments. MS and HO conducted ADAR1-Xpo5 interaction studies. MS and YS performed deep sequencing sample preparation, bioinformatics data analysis, and gene expression analysis. CS prepared mutant ADAR1 constructs. AK, JW, and LCS performed bioinformatics analysis of sequencing dada. ES conducted molecular modeling studies. HYT and DWS performed LC-MS/MS analysis and data interpretation. MS, YS, and KN supervised research, reviewed data, and wrote and edited the paper. All authors agree to the contents of the final manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mannion N, Arieti F, Gallo A, Keegan LP, O'Connell MA. New Insights into the Biological Role of Mammalian ADARs; the RNA Editing Proteins. Biomolecules. 2015;5:2338–62. doi: 10.3390/biom5042338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hood JL, Emeson RB. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr Top Microbiol Immunol. 2012;353:61–90. doi: 10.1007/82_2011_157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol. 2012;13:252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–40. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazak L, Levanon EY, Eisenberg E. Genome-wide analysis of Alu editability. Nucleic Acids Res. 2014;42:6876–84. doi: 10.1093/nar/gku414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramaswami G, et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 2012 doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–88. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, George CX, Patterson JB, Samuel CE. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem. 1997;272:4419–28. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- 11.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 12.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–15. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–61. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 14.Mannion NM, et al. The RNA-Editing Enzyme ADAR1 Controls Innate Immune Responses to RNA. Cell Rep. 2014;9:1482–94. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestal K, et al. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity. 2015;43:933–44. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liddicoat BJ, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–20. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward SV, et al. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc Natl Acad Sci U S A. 2011;108:331–6. doi: 10.1073/pnas.1017241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulthard LR, White DE, Jones DL, McDermott MF, Burchill SA. p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends Mol Med. 2009;15:369–79. doi: 10.1016/j.molmed.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–17. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 20.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong C, Tang Y, Maquat LE. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat Struct Mol Biol. 2013;20:1214–20. doi: 10.1038/nsmb.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 23.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 24.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–59. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng SK, Weissbach R, Ronson GE, Scadden AD. Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules. Nucleic Acids Res. 2013;41:9786–99. doi: 10.1093/nar/gkt750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–83. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 27.Emrick MA, Hoofnagle AN, Miller AS, Ten Eyck LF, Ahn NG. Constitutive activation of extracellular signal-regulated kinase 2 by synergistic point mutations. J Biol Chem. 2001;276:46469–79. doi: 10.1074/jbc.M107708200. [DOI] [PubMed] [Google Scholar]
- 28.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–55. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn YH, et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator-activated receptor gamma2 expression. Mol Cell Biol. 2011;31:4270–85. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barancik M, et al. SB203580, a specific inhibitor of p38-MAPK pathway, is a new reversal agent of P-glycoprotein-mediated multidrug resistance. Eur J Pharm Sci. 2001;14:29–36. doi: 10.1016/s0928-0987(01)00139-7. [DOI] [PubMed] [Google Scholar]
- 31.Wood CD, Thornton TM, Sabio G, Davis RA, Rincon M. Nuclear localization of p38 MAPK in response to DNA damage. Int J Biol Sci. 2009;5:428–37. doi: 10.7150/ijbs.5.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho DS, et al. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278:17093–102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 33.Nishikura K, Sakurai M, Ariyoshi K, Ota H. Antagonistic and stimulative roles of ADAR1 in RNA silencing. RNA Biol. 2013;10:1240–7. doi: 10.4161/rna.25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ota H, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–89. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajdarpasic A, Ruggenthaler P. Analysis of miRNA expression under stress in Arabidopsis thaliana. Bosn J Basic Med Sci. 2012;12:169–76. doi: 10.17305/bjbms.2012.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–99. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 37.Fritz J, et al. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol. 2009;29:1487–97. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gwizdek C, et al. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem. 2004;279:884–91. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- 39.Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–49. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- 40.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–8. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 41.Vitali P, Scadden AD. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat Struct Mol Biol. 2010;17:1043–50. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Lucas S, Oliveros JC, Chagoyen M, Ortin J. Functional signature for the recognition of specific target mRNAs by human Staufen1 protein. Nucleic Acids Res. 2014;42:4516–26. doi: 10.1093/nar/gku073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elbarbary RA, Li W, Tian B, Maquat LE. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27:1495–510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2013;4:423–35. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta. 2012;1819:593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Takao N, Li Y, Yamamoto K. Protective roles for ATM in cellular response to oxidative stress. FEBS Lett. 2000;472:133–6. doi: 10.1016/s0014-5793(00)01422-8. [DOI] [PubMed] [Google Scholar]
- 47.Raderschall E, et al. Formation of higher-order nuclear Rad51 structures is functionally linked to p21 expression and protection from DNA damage-induced apoptosis. J Cell Sci. 2002;115:153–64. doi: 10.1242/jcs.115.1.153. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Vukovic L, Koh HR, Schulten K, Myong S. Dynamic profiling of double-stranded RNA binding proteins. Nucleic Acids Res. 2015;43:7566–76. doi: 10.1093/nar/gkv726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thornton TM, Rincon M. Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int J Biol Sci. 2009;5:44–51. doi: 10.7150/ijbs.5.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai F, Drakas R, Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem. 1995;270:17098–105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 51.Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol. 2013;20:515–24. doi: 10.1038/nsmb.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci. 1989;86:2647–51. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishi K, Nishi A, Nagasawa T, Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013;19:17–35. doi: 10.1261/rna.034769.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pollard VW, et al. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–94. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 56.Ricci EP, et al. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol. 2014;21:26–35. doi: 10.1038/nsmb.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–90. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 58.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 59.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stefl R, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–37. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schrödinger L. The PyMOL Molecular Graphics System. (Version 1.8) [Google Scholar]
- 63.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE85455. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via MassIVE (accession no. MSV000080146) with the dataset identifier PXD004959. Other data from this study are available from the corresponding author upon reasonable request.