Abstract
DNA methylation plays important roles via regulation of numerous cellular mechanisms in diverse organisms, including humans. The paradigm bacterial methyltransferase (MTase) HhaI (M.HhaI) catalyzes the transfer of a methyl group from the cofactor S-adenosyl-l-methionine (AdoMet) onto the target cytosine in DNA, yielding 5-methylcytosine and S-adenosyl-l-homocysteine (AdoHcy). The turnover rate (kcat) of M.HhaI, and the other two cytosine-5 MTases examined, is limited by a step subsequent to methyl transfer; however, no such step has so far been identified. To elucidate the role of cofactor interactions during catalysis, eight mutants of Trp41, which is located in the cofactor binding pocket, were constructed and characterized. The mutants show full proficiency in DNA binding and base-flipping, and little variation is observed in the apparent methyl transfer rate kchem as determined by rapid-quench experiments using immobilized fluorescent-labeled DNA. However, the Trp41 replacements with short side chains substantially perturb cofactor binding (100-fold higher and ) leading to a faster turnover of the enzyme (10-fold higher kcat). Our analysis indicates that the rate-limiting breakdown of a long-lived ternary product complex is initiated by the dissociation of AdoHcy or the opening of the catalytic loop in the enzyme.
INTRODUCTION
DNA methylation plays important roles in the regulation of numerous cellular mechanisms in diverse organisms ranging from bacteriophages to humans. DNA cytosine-5 MTases (C5-MTases) transfer a methyl group from the cofactor S-adenosyl-l-methionine (AdoMet) onto the carbon-5 of cytosine in DNA. In higher organisms, DNA methylation is important for transcriptional regulation, genomic imprinting and silencing of retroviruses (1). Bacterial MTases are usually smaller than those from eukaryotes, and are often found as components of restriction–modification systems (2). Both classes of enzymes share a set of 10 conserved sequence motifs, some of which can be identified in most other AdoMet-dependent enzymes (3). Given the high-structural homology of C5-MTases (2), it is likely that these enzymes, including the eukaryotic paralogs, also share many mechanistic features among themselves.
The paradigm model for structural and mechanistic studies is the HhaI MTase (M.HhaI), a component of a type II restriction–modification system from Haemophilus haemolyticus. M.HhaI recognizes the tetranucleotide sequence GCGC and methylates the inner cytosine residue (underlined). Crystallographic studies of M.HhaI revealed that DNA binds in a cleft of the bilobal protein, leading to dramatic conformational changes in both the DNA and the enzyme itself (4,5). An MTase-mediated rotation of the target nucleotide out of the DNA helix (base-flipping) serves to deliver the base into a concave catalytic site in the enzyme. Base-flipping is accompanied by a massive movement of the catalytic loop (residues 81–99) towards the DNA, which locks the flipped-out base for methylation (see Figure 1A). The enzymatic catalysis involves the transient formation of a Michael adduct between the catalytic cysteine (Cys81 in M.HhaI) and C6 of the cytosine, permitting a direct SN2 transfer of the methyl group onto C5. The reaction cycle is completed by β-elimination at the C5–C6 bond of the resulting dihydro-cytosine intermediate, the reversal of the conformational changes in both protein and DNA, and the dissociation of products, AdoHcy and methylated DNA. Pre-steady and steady-state kinetic analyses of M.HhaI demonstrated that the rate of covalent methyl transfer, kchem = 0.26 s−1, is considerably faster than the enzymatic turnover, kcat = 0.04 s−1 (6,7). Interestingly, a pre-steady burst was also observed for two other C5-MTases, the bacterial MTase MspI (8) and the mouse Dnmt1 MTase (9), indicating that, in all cases examined, a step subsequent to covalent methylation determines the rate of the reaction cycle. However, the exact molecular event responsible for the slow turnover of the C5-MTases has not been identified.
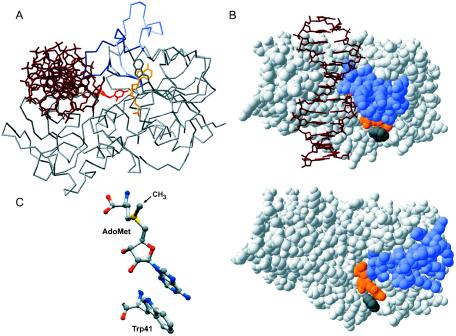
Figure 1.
Cofactor interactions in HhaI MTase. (A) The Cα trace of the HhaI–DNA–AdoHcy complex (PDB code 3mht); the closed catalytic loop is marked in blue and the open catalytic loop (from HhaI–AdoMet; 2hmy) is in light blue; DNA shown as dark red and flipped-out base is red; AdoHcy is orange. (B) Space filling models of the HhaI–DNA–AdoHcy complex (top) and the HhaI–AdoMet complex (bottom) revealing the position of the catalytic loop (blue) in the closed and open conformation, respectively, with respect to cofactor (orange) and Trp41 (gray). (C) A stick model of the AdoMet–Trp41 stacking interaction in the binary complex (2hmy); atom coloring: C is in gray, O in red, N in blue and S in yellow.
The cofactor AdoMet binds to the larger catalytic domain of M.HhaI in a pocket facing the active site cleft (7,10,11). Residues from conserved motifs I–V contribute to the binding pocket. The outer wall of the pocket is formed by the protruding Trp41 (motif II), whose indole ring intimately stacks to the adenine ring of the bound cofactor (Figure 1B and C). Studies with synthetic AdoHcy analogs suggested that the molecular anchor for cofactor binding is its adenosyl moiety (12). Recently, conserved residues Phe18 (conserved motif I), Trp41 (motif II), Asp60 (motif III) and Leu100 (motif V) involved in cofactor binding have been subjected to site-directed mutagenesis and biochemical studies (13,14). However, only conservative replacements were analyzed in some detail, and the functional significance of these residues could not be fully evaluated. In addition to its primary function as the methyl group donor, the cofactor AdoMet (and its product AdoHcy) was shown to play important roles in stabilizing the locked conformation of the reaction complexes (6,7,15–17).
To assess the functional importance of cofactor interactions during catalysis, Trp41 in M.HhaI was replaced by residues of a different size. The effects of individual replacements on methylation activity, catalytic parameters, DNA binding and base-flipping were studied using a protection assay, fluorescence spectroscopy and steady-state kinetic analysis. Rapid-quench single-turnover kinetic experiments were performed using a newly developed procedure that utilizes immobilized fluorescent-labeled DNA substrates. We found that the Trp41 mutations, which selectively perturb binding of AdoMet and AdoHcy, facilitate enzymatic turnover. Our analyses suggest that the breakdown of the ternary product complex could be initiated by the dissociation of AdoHcy or the opening of the catalytic loop in the enzyme, which for the first time identifies the rate-limiting events in the catalytic cycle of a DNA cytosine-5 MTase.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were obtained from MWG-Biotech AG (HPSF grade) or Fermentas (gel-purified). Oligonucleotides for electrophoretic binding experiments were 5′-32P- labeled using a DNA labeling kit (Fermentas). DNA duplexes for kinetic and biochemical studies were produced by annealing appropriate oligonucleotides (Table 1) as described previously (18).
Table 1.
Structure and abbreviation of DNA duplexesa
| DNA | Abbreviation |
|---|---|
| 37mer hemimethylated duplexes | |
| 5′-GACTGGTACAGTATCAGGCGCTGACCCACAACATCCGTGACCATGTCATAGTCCGMGACTGGGTGTTGTAGGCT-5′ | GCGC/GMGC |
| 5′-GACTGGTACAGTATCAGGPGCTGACCCACAACATCCGTGACCATGTCATAGTCCGMGACTGGGTGTTGTAGGCT-5′ | GPGC/GMGC |
| 5′-GACTGGTACAGTATCAGGAGCTGACCCACAACATCCGTGACCATGTCATAGTCCGMGACTGGGTGTTGTAGGCT-5′ | GAGC/GMGC |
| Labeled 35/34mer duplex | |
| 5′-biotin-ACTGGTACAGTATCAGGCGCTGACCCACAACATCT-JOEGACCATGTCATAGTCCGCGACTGGGTGTTGTAGA-5′ | biotin-GCGC-JOE |
| 24mer duplex for the H5-exchange assay | |
| 5′-TAATAATGCGCTAATAATAATAATATTATTACGCGATTATTATTATTA-5′ | GCGC |
aHhaI recognition site is in boldface, nucleotide at the target position is underlined; M, 5-methylcytosine; P, 2-aminopurine; and JOE, 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein.
Site-directed mutagenesis
Trp41 mutations were introduced by the Kunkel method as described previously (19) using the following oligonucleotides (Fermentas, gel-purified) (5′…3′, the modified codon-41 is in boldface, silent mutations creating diagnostic restriction endonuclease sites are italicized): ATATTTATCGAATTCATTAGAATAAACGC (Phe), GTGCATATTTATCC(C,G)CTTCATTCGAATAAACGC (Gly, Ala) and GTGCATATTTATCTA(C,G,T)TTCATTCGAATAAACGC (Val, Leu, Ile). All mutations were confirmed by complete sequencing of the genes.
Protein expression and purification
M.HhaI and its variants were expressed in Escherichia coli ER1727 cells containing the pHH553 plasmid (19). MTases were selectively enriched exploiting a high salt (0.4 M NaCl) back-extraction from the cell debris. Following extensive dialysis to remove bound AdoMet, MTases were purified by passing through a pre-column of Q-Sepharose followed by column chromatography on S-Sepharose (18). All proteins were purified to >95% purity as judged on Coomassie-stained polyacrylamide gels (data not shown). Protein concentrations were estimated using a Coomassie G-250 assay with BSA as standard and further refined by active site titration with a 37mer fluorescent duplex GPGC/GMGC (see below). The molecular mass of each mutant was verified by ESI–mass spectrometry (ESI–MS).
Electrophoretic gel-mobility shift assays
Dissociation constants were estimated by titrating 10 or 50 pM 5′-32P-labeled hemimethylated 37mer DNA duplex GCGC/GMGC (see Table 1) with increasing protein concentrations (0.01–100 nM). Ternary complex formation was monitored with 1 or 5 pM DNA and 0.2 pM–10 nM MTase in the presence of 500 μM AdoHcy. Samples of 20 μl were preincubated in the reaction buffer (10 mM Tris–HCl, pH 7.4, 0.5 mM EDTA, 50 mM NaCl and 2 mM 2-mercaptoethanol) containing 0.2 mg/ml of BSA and 10% glycerol for 30 min at room temperature. Aliquots were loaded onto an 8% polyacrylamide gel and fractionated by electrophoresis in 45 mM Tris–borate, pH 8.3 and 1 mM EDTA at 10 V/cm for 1 h. Dried gels were analyzed using a Cyclone PhosphorImager (Packard Instruments). Bound and free bands were quantified with Optiquant software and data were fit to the full quadratic equation for single-site binding using Grafit software (20).
Fluorescence spectroscopy
Measurements of 2-aminopurine fluorescence were performed at 25°C on a Perkin-Elmer LS-50B luminescence spectrometer equipped with a Xe lamp and a 4-mm rectangular quartz cell as described previously (18). The 200 nM 37mer GPGC/GMGC duplex (Table 1) was titrated by an incremental addition of 2 μM M.HhaI in the reaction buffer. Fluorescence intensity was recorded at λEx = 320 nm and λEm = 370 nm with excitation and emission bandwidths of 2.5 and 5 nm, respectively. Emission spectra (340–420 nm) were recorded at an excitation wavelength (λEx) of 320 nm, and excitation spectra (260–350 nm) were recorded at an emission wavelength (λEm) of 370 nm. At least three scans were averaged for each spectrum. Control scans were collected under identical conditions except that the GAGC/GMGC duplex was used instead of fluorescent DNA. The corrected spectra were obtained by subtracting the controls to eliminate contributions from the protein and other components (21).
Steady-state kinetics
Methylation reactions were carried out in the methylation buffer (50 mM Tris–HCl, pH 7.4, 0.5 mM EDTA, 50 mM NaCl, 2 mM 2-mercaptoethanol and 0.2 mg/ml of BSA) at 37°C for 10 min. measurements were performed with constant (50 or 500 pM) MTase and poly(dG–dC) DNA (500 nM) and varying [methyl-3H]AdoMet concentration from 2 nM to 200 μM. Reactions were stopped by adding HCl to 0.5 N concentration. Duplicate samples were spotted onto DE-81 filters and processed as described previously (7). The quenching of 3H counts on DE-81 filters was estimated with poly(dG-5-[3H]methyldC) and the obtained quenching factor of 1.6 was used to correct the counter readings. Data were analyzed by non-linear regression fitting to a Michaelis–Menten equation using the Grafit (20) or DynaFit (22) software packages.
Hydrogen exchange assay
Reactions containing 12 μM of both MTase and unlabeled 24mer GCGC duplex (Table 1) were incubated for 30 min at 22°C in the reaction buffer containing 80% D2O. Samples were then treated with Nuclease P1 and alkaline phosphatase and analyzed on an integrated HPLC/ESI–MS Hewlett-Packard 1100 series system. A sample of 200–300 pmol of total nucleosides was injected into a Discovery C18 column and was eluted with 20 mM ammonium formate, pH 3.5, for 5 min followed by a linear gradient to 55% methanol over 15 min at a flow rate of 0.3 ml/min at 45°C. The dCyd peak (elution time 3.4 ± 0.1 min) was analyzed in a mass detector operating at a capillary voltage of 5000 V. Spectra were collected in the positive ion mode and mass range of 108–600 m/z to monitor the presence of Cyt–Na+ (134 m.u.) and dCyd–Na+ (250 m.u.) ions.
Equilibrium dialysis
Fifty microliter of enzyme solution (30–200 μM) in a Slide-A-Lyzer® Mini Dialysis Unit (10 000 MWCO) was equilibrated against 1 ml of 2–50 μM [methyl-3H]-AdoMet (4–5 concentration points for each mutant) in the reaction buffer containing 200 mM NaCl, by gently mixing at 8°C for 48 h. Duplicate aliquots from both chambers were withdrawn and radioactivity was determined in 2 ml of Rotiszint Eco Plus scintillation cocktail (Roth) using a Beckman LS 1801 liquid scintillation spectrometer. Protein concentration in each equilibrated sample was verified using a Coomassie G-250 assay with BSA as standard. The amount of bound AdoMet was calculated from the difference of radioactivity measured in protein–AdoMet and AdoMet chambers, and the data were fitted to a single-site ligand binding model using DynaFit (22).
Pre-steady-state kinetics
Single-turnover reactions were performed at 37°C in the reaction buffer containing 0.2 mg/ml of BSA. The 34mer biotin-GCGC-JOE duplex (1 μM) preincubated with WT or mutant M.HhaI (2 μM) was mixed rapidly with 500 μM AdoMet (final concentrations), and after a specified period, were quenched with HCl (final 0.5 N) in a Rapid-Quench-Flow instrument RQF-3 (KinTek). The reaction mixture was neutralized with a 580 mM Tris base and 1% SDS buffer (final concentrations), loaded into streptavidin-coated microplate wells (high capacity, 96-well, 350 μl/well; Sigma) and incubated for 1 h at room temperature. The wells were washed three times with TPST buffer (25 mM Tris–HCl, pH 7.4, 2.5 mM KCl, 140 mM NaCl and 0.05% Tween). After DNA cleavage with the Hin6I restriction endonuclease (Fermentas), the samples were transferred into white microplates (96-well; Porvair Sciences) for JOE fluorescence measurements. Fluorescence was quantified with a Perkin-Elmer LS-50B luminescence spectrometer equipped with a well plate reader accessory. Three readings were taken at an excitation wavelength of 526 nm and emission wavelength of 550 nm and averaged for each well. Reaction progress was analyzed by fitting data to a single exponential equation using Grafit (20) or DynaFit (22).
Analysis of a rate-limiting step
The dependence was analyzed by fitting experimental values with the Grafit program (20) to the mechanisms described in Figure 5A and B according to the following equations:
| 1 |
| 2 |
Experimental sets of kcat and KM from each mutant where used to refine global values for kchem, kunlock and f.
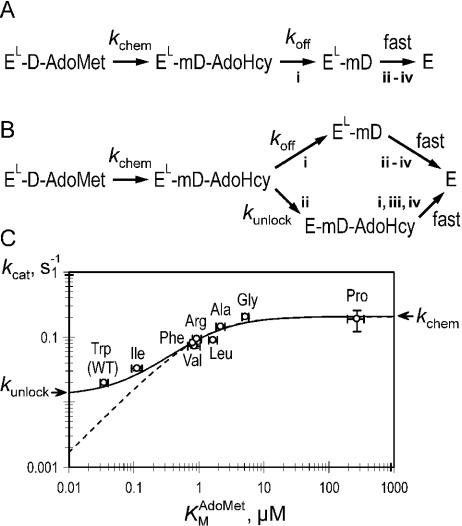
Figure 5.
Breakdown of the ternary product complex. The minimal (A) and extended (B) kinetic mechanisms of the breakdown of the ternary product complex is shown. Acronyms describing molecular species are: E and EL, open and closed (locked) forms of M.HhaI, respectively; D and mD, unmethylated and methylated DNA, respectively. Designation of events: dissociation of AdoHcy (i), opening of the catalytic loop (ii), departure (flip-back) of the target base from the active site (iii) and dissociation of the DNA (iv). (C) A log–log plot of versus kcat with non-linear fits to mechanism A (broken line) and B (solid line).
RESULTS
Construction and in vivo characterization of Trp41 mutants
As noted above, Trp41 forms the outer wall of the cofactor pocket and stacks against the adenine ring of the bound AdoMet (Figure 1C). Substitutions at residue 41 were chosen to vary the size of the side chain up to its complete elimination. Site-directed mutagenesis with degenerate primers was used to replace Trp41 with the following amino acid residues: Phe, Leu, Ile, Val, Ala and Gly. Sequencing of the clones produced additionally revealed the Pro and Arg variants, which were also included in further experiments. The functional ability of the mutant MTases to modify the target GCGC sites in vivo was assessed by the resistance of plasmid DNA isolated from the corresponding clones against cleavage with a methylation-sensitive restriction endonuclease (21). All eight substitutions at position-41 lead to protection levels comparable to that of the WT MTase (data not shown). The mutants and WT proteins were overexpressed and purified to near homogeneity as described previously (18).
DNA binding and base-flipping activity of Trp41 mutants
We have determined the binding constants of the Trp41 mutant MTases to a 37mer duplex substrate containing a unique hemimethylated GCGC site using electrophoretic gel-mobility shift titration experiments. All but one of the mutants bound the DNA substrate with <2-fold difference in affinity from that of the WT M.HhaI (Table 2). The W41R mutant binds several fold better to DNA than the WT enzyme most probably due to the positive charge of its side chain.
Table 2.
DNA-binding affinity of the Trp41 mutants
| (nM)a | (pM)b | |
|---|---|---|
| Trp (WT) | 0.25 ± 0.04 | 0.57 ± 0.14 |
| Ile | 0.46 ± 0.10 | 0.79 ± 0.31 |
| Val | 0.13 ± 0.03 | 0.47 ± 0.14 |
| Phe | 0.27 ± 0.08 | 1.91 ± 0.45 |
| Arg | 0.04 ± 0.01 | 0.72 ± 0.25 |
| Leu | 0.53 ± 0.13 | 0.94 ± 0.83 |
| Ala | 0.11 ± 0.05 | 0.58 ± 0.35 |
| Gly | 0.12 ± 0.02 | 0.62 ± 0.17 |
| Pro | 0.16 ± 0.04 | 1.16 ± 0.77c |
abin, binary complex, MTase-DNA.
bter, ternary complex, MTase-DNA–AdoHcy.
cOwing to smearing of the ‘bound DNA’ band, the ‘free DNA’ band alone was used for quantification.
Similar experiments were performed in the presence of the cofactor AdoHcy to measure MTase–DNA interaction in the ternary complex. Previous studies of the WT enzyme showed that the addition of AdoHcy leads to the formation of a stable dead-end ternary M.HhaI–DNA–AdoHcy complex. All mutant MTases bound DNA in the presence of 500 μM AdoHcy to the same extent as the WT enzyme (Table 2). The unaltered ability of these mutants to bind DNA in the binary and ternary complexes indicates that the Trp residue is of little importance for MTase–DNA interactions.
The capacity to flip out the target base upon interaction with DNA was examined in two ways. First, titrations with the hemimethylated duplex containing 2-aminopurine at the target site were performed. As expected, the addition of saturating amounts of the MTases results in a strong increase of 2-aminopurine fluorescence intensity (18). Fluorescence titration amplitudes and corrected excitation and emission spectra were identical to those of the WT enzyme (data not shown), suggesting that the mutations affect neither the efficiency of flipping nor the environment of the flipped 2-aminopurine base. Second, we also examined the mutants using a mechanism-based property of M.HhaI to catalyze the exchange of the hydrogen-5 atom on the target cytosine for protons of water in the absence of cofactor (23). The exchange reaction requires the formation of a transient covalent bond to the cytosine ring, which is only possible when the target cytosine is flipped out and bound in the catalytic site of the enzyme. The incorporation of D from deuterated solvent into the target cytosines in a 24mer unmethylated GCGC duplex (see Table 1) was monitored using mass-spectrometric analysis. The exchange reaction was ∼90% complete at the 30 min time point with the WT enzyme and the mutants (data not shown), indicating that their cytosine-flipping rates differ not more than 8-fold. Our observations thus clearly indicate that the mutant MTases are proficient in base-flipping.
AdoMet binding
Previously, M.HhaI–cofactor interactions were studied using intrinsic Trp41 fluorescence (7), but for obvious reasons this approach was not possible for the Trp41 mutants. We therefore examined the stability of binary complexes for the mutant MTases using an equilibrium dialysis technique. Binding equilibria at several concentration points of labeled [methyl-3H]AdoMet for each mutant were determined and KD values derived by fitting data to a single-site binding mechanism. The derived KD values for the WT (4.4 μM) and W41I mutant (14 μM) are in good agreement with those determined previously by fluorescent spectroscopy (6,7,13) or isothermal calorimetry (13,14). Overall, for the mutant MTases were higher than those of the WT protein by 1–2 orders of magnitude (four orders of magnitude for W41P) and increased in the order: WT < W41I < W41F, W41R, W41V < W41L, W41A < W41G ≪ W41P (Figure 3A). Consistent with our structural prediction, replacing Trp41 with smaller residues reduces the affinity of M.HhaI for the cofactor AdoMet.
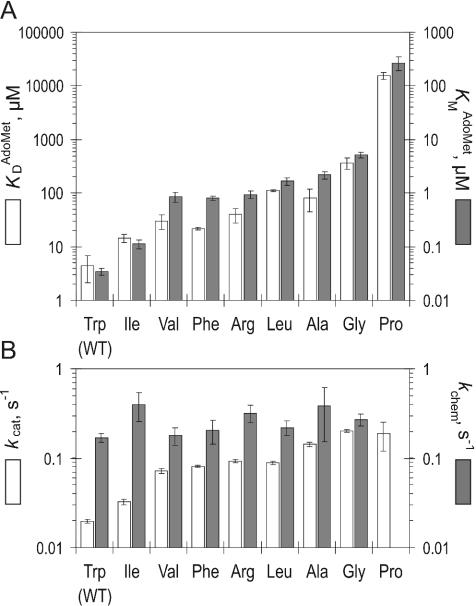
Figure 3.
Cofactor binding and kinetic parameters of Trp41 mutants. (A) Comparison of binary () and ternary () MTase–cofactor interactions in the Trp41 mutants. (B) Comparison of a steady-state turnover rate kcat and the rate of covalent methyl transfer kchem in the Trp41 mutants.
Steady-state kinetic analysis
To better understand the role of Trp41 in cofactor interactions, steady-state kinetic analysis was performed to determine KM for AdoMet and the multiple turnover rates (kcat) for the mutant enzymes. Measurements were carried out at constant saturating poly(dG–dC) and varied [methyl-3H]AdoMet concentrations as described previously, and maintaining substrate consumption <10 and 3% for DNA and the cofactor, respectively (Figure 2A). Both kinetic parameters measured for the WT enzyme (; kcat = 0.02 s−1) are in excellent agreement with those reported previously (19). values for the mutant MTases were significantly higher and showed a very wide variation (Figure 3A). The was 3- to 50-fold higher for the mutant MTases with fairly large residues (W41I, W41F, W41R, W41V, W41L), but the variants with small side chains (W41A, W41G) showed even higher increases (60- and 150-fold, respectively), indicating a dramatic decrease in the affinity for cofactor as the side chain is shortened. Replacement of Trp41 with a structurally constrained proline leads to the largest change in KM value for AdoMet (>1000-fold). Limitations in accessible concentrations of radioactive AdoMet precluded the measurement of high end points, and thus KM and kcat were derived by fitting the lower half of the curve (Figure 2A).
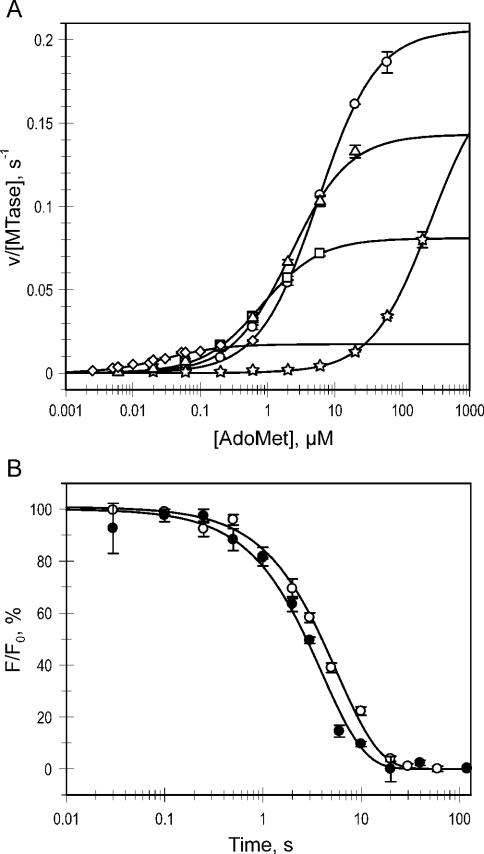
Figure 2.
Kinetic analysis of Trp41 mutants. (A) Steady-state velocities for the mutants (circles, W41G; triangles, W41A; squares, W41F; diamonds, WT; stars, W41P) were determined at constant saturating poly(dG–dC) and varied [methyl-3H]AdoMet concentrations and the data were fitted to a Michaelis–Menten equation to reveal and kcat (shown in Figure 3). (B) Single-turnover methylation reactions were performed by incubation of a biotin- and JOE-labeled 34mer DNA duplex with excess MTase and AdoMet in a rapid-quench-flow device. The DNA was immobilized and digested with R.Hin6I to release fluorescent fragments from unmethylated DNA. Fluorescence time courses (open circles, WT; filled circles, W41G) were fitted to a single exponential equation (solid lines) to reveal kchem.
Unexpectedly, the multiple turnover kcat values for the mutant MTases proved to be higher when compared with the WT enzyme (Figure 3B). Replacements of Trp41 with large hydrophobic homologs (Ile, Phe, Arg, Val, Leu) gave MTases that were 1.5- to 5-fold faster than WT protein; however, the W41A and W41G mutants exhibited 7- and 10-fold higher multiple turnover rate. The observed trend points to the importance of cofactor interactions during catalysis.
Pre-steady-state kinetic analysis
In light of the higher turnover rate of some of the mutants, we examined if the rate of the methyl transfer step, kchem, is affected by the Trp41 replacements. To avoid high experimental concentrations of radioactively labeled AdoMet (because of the high KM values of the mutants), single-turnover kinetic measurements were performed under saturating concentrations of protein and unlabeled AdoMet, using a newly developed procedure. The procedure employs an unmethylated 35/34mer duplex (biotin-GCGC-JOE, see Table 1) that contains a biotin anchor on the 5′ end and a fluorescence label on the 3′ end. The methylation was analyzed by monitoring the release of fluorescent label from streptavidin-immobilized DNA after its digestion with the R.Hin6I endonuclease, which only cleaves unmethylated GCGC sites. The fluorescence signal followed single exponential kinetics to reveal the methylation rate constant kchem (Figure 2B). The measured kchem = 0.17 s−1 for WT M.HhaI with unmethylated DNA was similar to previously reported values obtained with hemimethylated DNA [0.26 s−1, (6,7)]. The Trp41 mutants showed uniform methyl transfer rates within error (0.2–0.4 s−1) (Figure 3B). This observation is consistent with the high proficiency of the mutants in DNA binding and base-flipping noted above. Although the variations of the side chain at residue-41 alter the stability of the cofactor–MTase complexes, they do not perturb the conformation of the bound cofactor in the active site pocket.
DISCUSSION
Role of Trp41 in cofactor binding
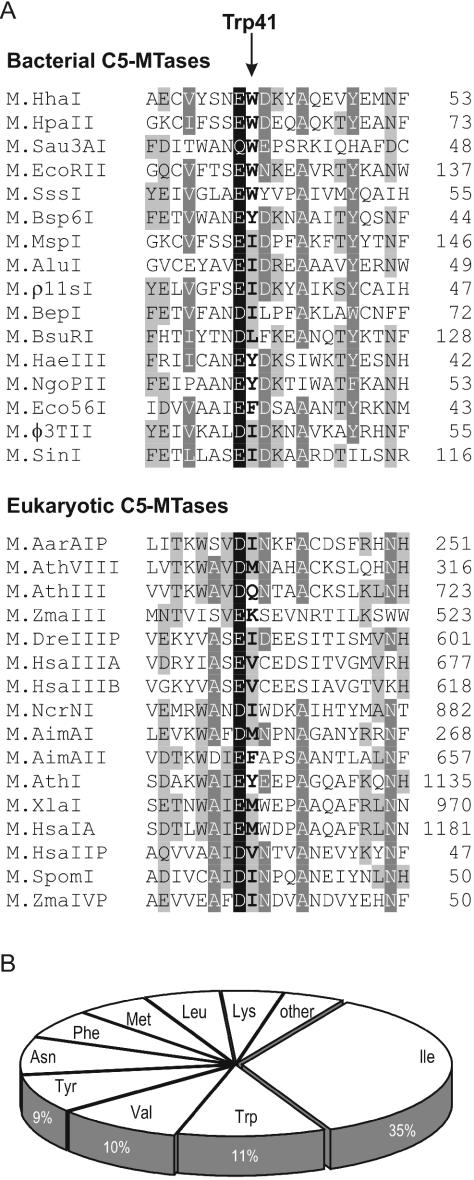
Trp41 is located on the surface of the protein and its side chain makes the bulk of the outer wall of the cofactor pocket. The adenine ring of bound AdoMet (or AdoHcy) stacks against the indole ring of Trp41 (see Figure 1C). Therefore, it is not surprising that the mutations of Trp41 affect the cofactor binding affinity both in the binary (KD) and ternary (KM) complexes (Figure 3A). In fact, both affinities decrease in concert in the order: Trp > Ile > Phe, Val > Arg, Leu, Ala > Gly ≫ Pro. This finding suggests that the same elementary step (cofactor binding) is probed by the mutations. Trp is the largest side chain and renders the best binding for the cofactor (Figure 3A), including the highest efficiency (kcat/KM) of catalysis in M.HhaI. However, other bulky residues can substitute for Trp41 and still permit efficient catalysis. From a structural viewpoint, the ability of the side chain to support the adenine ring of the bound cofactor may be the most important aspect of the residue occupying position-41. Therefore, branching early in the side chain (at Cβ as in Ile, Phe, Val) appears more important than the overall bulk (Leu, Arg) of the side chain (Figure 3A). Indeed, phylogenetic analysis of DNA C5-MTases (Figure 4) indicates that a bulky hydrophobic residue is usually found at this position, but Ile, Trp, Val and Tyr appear most often, whereas Met and Leu appear at a lower frequency. Clearly, the occurrence of Trp with its face-to-face stacking to the adenine ring of the cofactor appears to be a peculiarity of the M.HhaI system that adds efficiency, but is not essential. The most severe kinetic perturbations of the enzyme are observed in the proline mutant (Figure 3A). Reference to available structures (5,11) suggests a significant steric repulsion arising between Cγ, Cδ of the structurally constrained side chain of the proline and the N3, C2 atoms of the adenine ring of the bound cofactor. This unfavorable interaction apparently leads to a more dramatic effect than a mere shortening or removal of the outer wall of the pocket. Consistent with this, we found no occurrences of Pro at this position of C5-MTases.
Figure 4.
Amino acid conservation of the Trp41 position (HhaI) in DNA C5-MTases is depicted, based on an alignment of 330 DNA C5-MTase sequences from the Pfam database (http:www.sanger.ac.uk/Software/Pfam/). (A) Representative sequences of the conserved motif II. Trp41 position (HhaI) is shown in boldface. Four degrees of conservation in descending order: black background with white text, dark gray background with white text, gray background with black text and white background with black text. (B) Distribution of amino acids found in the Trp41 position (HhaI) in DNA C5-MTases.
Calorimetric analyses showed that the binding of cofactor to M.HhaI is exothermic (ΔΔG = −6.7 and −7.8 kcal/mol for AdoMet and AdoHcy, respectively) and is driven by a favorable enthalpic change (14). Solution binding studies with truncated AdoHcy analogs showed that the thermodynamic contribution of the amino acid side chain is in the order of 2.3–2.6 kcal/mol (12). Interestingly, the removal of the outer wall of the pocket (in W41G) leads to a decline in binding of a similar magnitude (∼100-fold; Figure 3A), indicating that the interactions of the adenine ring are worth 2.5–3 kcal/mol . Although these fractional contributions perhaps are not fully additive, it appears that each of the three parts (the amino acid part, the ribose ring and the adenine base; Figure 1C) may have comparable thermodynamic weights for cofactor recognition. It comes as no surprise that the adenosyl moiety alone (adenine + ribose) serves as a molecular anchor for cofactor binding (12), and, therefore, AdoMet analogs, in which the methylsulfonium center is replaced by another reactive (e.g. azaridine) group, can be efficiently utilized by MTases (24).
On the other hand, we find that the Trp41 replacements in M.HhaI lead to minor effects on DNA binding [ and , Table 2] (except for the positively charged Arg), base-flipping or the catalytic methyl transfer (Figure 3B, kchem). These observations are consistent with the remote position of Trp41 with respect to the bound DNA and the flipped-out cytosine (Figure 1). The Trp mutations thus selectively perturb (weaken) the interactions of AdoMet and AdoHcy with the protein, and thus can be used as valuable probes for elucidating the role of the cofactor in the catalytic mechanism.
Rate-limiting step
The enzymatic cycle of HhaI MTase involves a series of molecular events, such as binding of substrates, dramatic conformational rearrangements in both the DNA (base-flipping) and protein (closure of the catalytic loop), covalent transformations in the catalytic site of enzyme (transient covalent activation of the cytosine and methyl transfer from bound AdoMet), followed by a breakdown of the ternary complex to the release of products and the enzyme. Steady-state (15) and pre-steady-state (6,7) kinetic analyses demonstrated that a step following the covalent methyl transfer determines the rate of catalytic turnover kcat. Structural evidence indicates that the methyl transfer occurs in the tightly closed covalent ternary complex, resulting in the ternary product EL–mD–AdoHcy (Figure 5A). In all likelihood, the ternary product complex is the most stable intermediate on the downhill pathway dominating the scene at a steady state with a half-life of ∼30 s. The X-ray structures of the ternary product complexes involving AdoHcy and covalently bound (5) or non-covalently bound (25) methylated DNA show intimate contacts between the flipped-out methylated cytosine, the closed catalytic loop in the enzyme and the AdoHcy molecule. It should be noted that the ternary product complex is considerably less stable than the ternary dead-end complex involving AdoHcy and unmethylated or hemimethylated DNA (15), which has a half-life extending into hours (7). The key structural difference between the two complexes is the presence of a methyl group on the C5-position of the flipped-out target cytosine (25). The methyl group is bent out of the plane of the cytosine ring and is in tight contact with the sulfur atom of AdoHcy (separation of 2.9 Å) (25). This tight steric interaction may account for a slight dislocation (>0.5 Å) of the sulfur atom towards the exterior, as evident (data not shown) from comparing the corresponding structures (1MHT or 4MHT against 3MHT or 5MHT). The latter two structures contain cytosine in the catalytic site, and therefore, no steric repulsion is exerted towards the sulfur atom, which in turn may adopt a more favorable conformation.
The breakdown of the ternary product will determine the rate of dissociation of the enzyme and thus the catalytic turnover. Structural considerations above suggest that the breakdown of the closed complex can be initiated by the dissociation of AdoHcy via a separate channel (i) yielding a short-lived closed binary EL–mD complex (Figure 5A). Owing to the dynamic nature of binary M.HhaI–DNA complexes (15,17), it is obvious that the opening of the catalytic loop (ii), departure (flip-back) of the target base from the active site (iii) and dissociation of the DNA (iv) would be fast. Alternatively, the opening of the catalytic loop (ii) would greatly facilitate the release of AdoHcy (i) from the cofactor pocket (see Figure 1B) and subsequent fast decay of the complex (iii and iv) (Figure 5B). Evidence in favor of step (i) being rate-limiting comes from our findings that the enhanced mobility of cofactor in the W41 mutants leads to a faster turnover (higher kcat) of the enzyme. Since the rate of the covalent methyl transfer kchem is not affected by the mutations, the increase in kcat must derive from a faster breakdown of the ternary product complex. A log plot of versus kcat is hyperbolic where kcat approaches kchem as the mobility of cofactor increases (Figure 5C). This means that kchem becomes the rate-limiting step in the latter mutants, and that all subsequent steps are faster. This, in turn, implies that the rate of reversal of the transient covalent bond at C6 in the 5,6-dihydrocytosine adduct via elimination of H5, which occurs after methyl transfer and must precede the opening of the loop, is faster than kchem and thus can be disregarded in kinetic analyses. This idea is consistent with recent analyses by others (26). Assuming that the rate of cofactor release from the ternary complex is proportional to KM (koff ∼ f·KM), one can fit the observed values to a simple sequential kinetic mechanism shown in Figure 5A. However, the fit was not satisfactory at the lowest KM values which, interestingly, were associated with the amino acids present in nearly half of C5-MTases: Trp and Ile (see Figure 4B). The Trp and Ile variants had larger kcat values than expected (compare with broken line in Figure 5C), meaning that their turnover rates are higher than expected if the AdoHcy dissociation from the ternary product complex were rate-limiting. This discrepancy suggests that a second decay path of the ternary product complex may be operational. Structural considerations above hint that such a path is likely the opening (unlocking) of the catalytic loop that occurs independently of the cofactor release (step ii), and which is followed by fast events (i), (iii) and (iv) (Figure 5B). This mechanism invokes an additional rate constant, kunlock. Similar analysis gives a very good fit to the experimental data (Figure 5C, solid line), yielding kchem = 0.21 ± 0.02 s−1 and kunlock = 0.013 ± 0.02 s−1 for all the variants, and the rate of AdoHcy release for the WT enzyme koff = 0.008 ± 0.02 s−1. The calculated value for kchem is in excellent agreement with that obtained experimentally. Although derived with substantial uncertainties, the rates for the two competing decay paths (kunlock and koff) are of the same magnitude, suggesting that both routes may be operational and contribute to the turnover rate.
If the opening of the catalytic loop indeed occurs independently of cofactor release, it can perhaps be probed in a similar manner by targeted mutations in or around the catalytic loop. Alterations that destabilize the closed conformation of the loop in the ternary complex would be expected to speed up the decay of the ternary product. Two such mutations in which stabilizing contacts to the loop are removed have recently been described. The Q237L mutation in the target recognition domain of M.HhaI eliminates a H-bond to Ser87 in the closed loop (23). Indeed, the mutant has a several-fold higher kcat and substantially higher . A similar effect was observed by us (data not shown) and others (26) when another stabilizing contact to the loop is removed. The 2-amino group of the outer guanine residue on the target strand, which makes a H-bond to the backbone oxygen of Ile86 (5), is not present if poly(dI–dC) instead of poly(dG–dC) is used as a substrate. As expected, this replacement leads to a faster turnover of the enzyme and the disappearance of a pre-steady-state burst. Interestingly, both the murine and human Dnmt1 MTases show an even greater acceleration of the steady-state turnover with poly(dI–dC) versus poly(dG–dC) (27,28). Since a pre-steady-state burst has also been observed for the mouse enzyme (9), it cannot be excluded that the faster opening of the catalytic loop is also responsible for this effect.
In aggregate, our analysis of M.HhaI indicates that the rate-limiting breakdown of the ternary product complex is initiated either by dissociation of AdoHcy or by conformational unlocking of the complex via opening of the catalytic loop. Obviously, variations in the DNA substrate, buffer and other conditions may affect the relative rates of these competing steps. The reversal of the covalent bond to C6 via β-elimination and all other subsequent steps are fast, with negligible contributions to the turnover rate. In light of the noted structural and mechanistic similarity, many of these mechanistic observations are likely to be valid for the whole family of C5-MTases, including the physiologically important eukaryotic enzymes.
Acknowledgments
The authors are grateful to Rūta Gerasimaitė for her help with developing the fluorescence-based MTase assay. We also thank Prof. Elmar Weinhold for numerous discussions. This work was in part supported by grants from the Howard Hughes Medical Institute and the Volkswagen-Stiftung and the Lithuanian State Science and Study foundation. The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.Robertson K.D., Jones P.A. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 2.Cheng X., Roberts R.J. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujnicki J.M., Feder M., Ayres C.L., Redman K.L. Sequence–structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng X., Kumar S., Posfai J., Pflugrath J.W., Roberts R.J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 5.Klimasauskas S., Kumar S., Roberts R.J., Cheng X. Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 6.Lindstrom W.M., Flynn J., Jr, Reich N.O. Reconciling structure and function in HhaI DNA cytosine-C-5 methyltransferase. J. Biol. Chem. 2000;275:4912–4919. doi: 10.1074/jbc.275.7.4912. [DOI] [PubMed] [Google Scholar]
- 7.Vilkaitis G., Merkiene E., Serva S., Weinhold E., Klimasauskas S. The mechanism of DNA cytosine-5 methylation. Kinetic and mutational dissection of HhaI methyltransferase. J. Biol. Chem. 2001;276:20924–20934. doi: 10.1074/jbc.M101429200. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya S.K., Dubey A.K. Kinetic mechanism of cytosine DNA methyltransferase MspI. J. Biol. Chem. 1999;274:14743–14749. doi: 10.1074/jbc.274.21.14743. [DOI] [PubMed] [Google Scholar]
- 9.Flynn J., Glickman J.F., Reich N.O. Murine DNA cytosine-C5 methyltransferase: pre-steady- and steady-state kinetic analysis with regulatory DNA sequences. Biochemistry. 1996;35:7308–7315. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S., Horton J.R., Jones G.D., Walker R.T., Roberts R.J., Cheng X. DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res. 1997;25:2773–2783. doi: 10.1093/nar/25.14.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Gara M., Zhang X., Roberts R.J., Cheng X. Structures of a binary complex of HhaI methyltransferase with S-adenosyl-l-methionine formed in the presence of a short non-specific DNA oligonucleotide. J. Mol. Biol. 1999;287:201–209. doi: 10.1006/jmbi.1999.2608. [DOI] [PubMed] [Google Scholar]
- 12.Pignot M., Pljevaljcic G., Weinhold E. Efficient synthesis of S-Adenosyl-l-homocysteine natural product analogues and their use to elucidate the structural determinant for cofactor binding of the DNA methyltransferase M.HhaI. Eur. J. Org. Chem. 2000:549–555. [Google Scholar]
- 13.Sankpal U.T., Rao D.N. Mutational analysis of conserved residues in HhaI DNA methyltransferase. Nucleic Acids Res. 2002;30:2628–2638. doi: 10.1093/nar/gkf380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swaminathan C.P., Sankpal U.T., Rao D.N., Surolia A. Water-assisted dual mode cofactor recognition by HhaI DNA methyltransferase. J. Biol. Chem. 2002;277:4042–4049. doi: 10.1074/jbc.M109237200. [DOI] [PubMed] [Google Scholar]
- 15.Wu J.C., Santi D.V. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 16.Klimasauskas S., Roberts R.J. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res. 1995;23:1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klimasauskas S., Szyperski T., Serva S., Wüthrich K. Dynamic modes of the flipped-out cytosine during HhaI methyltransferase–DNA interactions in solution. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holz B., Klimasauskas S., Serva S., Weinhold E. 2-Amino-purine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daujotyte D., Vilkaitis G., Manelyte L., Skalicky J., Szyperski T., Klimasauskas S. Solubility engineering of the HhaI methyltransferase. Protein Eng. 2003;16:295–301. doi: 10.1093/proeng/gzg034. [DOI] [PubMed] [Google Scholar]
- 20.Leatherbarrow R.J. Graphit version 3.0. Staines, UK: Erithacus Sortware Ltd; 1992. [Google Scholar]
- 21.Vilkaitis G., Dong A., Weinhold E., Cheng X., Klimasauskas S. Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem. 2000;275:38722–38730. doi: 10.1074/jbc.m005278200. [DOI] [PubMed] [Google Scholar]
- 22.Kuzmic P. Analysis and simulation of chemical kinetic, biochemical kinetic, and pharmacokinetic data. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 23.Daujotyte D., Serva S., Vilkaitis G., Merkiene E., Venclovas C., Klimasauskas S. HhaI DNA methyltransferase uses the protruding Gln237 for active flipping of its target cytosine. Structure. 2004;12:1047–1055. doi: 10.1016/j.str.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Pljevaljcic G., Pignot M., Weinhold E. Design of a new fluorescent cofactor for DNA methyltransferases and sequence-specific labeling of DNA. J. Am. Chem. Soc. 2003;125:3486–3492. doi: 10.1021/ja021106s. [DOI] [PubMed] [Google Scholar]
- 25.O'Gara M., Klimasauskas S., Roberts R.J., Cheng X. Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaI methyltransferase–DNA–AdoHcy complexes. J. Mol. Biol. 1996;261:634–645. doi: 10.1006/jmbi.1996.0489. [DOI] [PubMed] [Google Scholar]
- 26.Svedruzic Z.M., Reich N.O. The mechanism of target base attack in DNA cytosine carbon 5 methylation. Biochemistry. 2004;43:11460–11473. doi: 10.1021/bi0496743. [DOI] [PubMed] [Google Scholar]
- 27.Pedrali-Noy G., Weissbach A. Mammalian DNA methyltransferases prefer poly(dI–dC) as substrate. J. Biol. Chem. 1986;261:7600–7602. [PubMed] [Google Scholar]
- 28.Vilkaitis G, Suetake I., Klimasauskas S., Tajima S. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J. Biol. Chem. 2005;280:64–72. doi: 10.1074/jbc.M411126200. [DOI] [PubMed] [Google Scholar]