Abstract
Tissue damage or pathogen invasion triggers the auto-proteolysis of an initiating serine protease (SP), rapidly leading to sequential cleavage activation of other cascade members to set off innate immune responses in insects. Recently, we presented evidence that Manduca sexta hemolymph protease-1 zymogen (proHP1) is a member of the SP system in this species, and may activate proHP6. HP6 stimulates melanization and induces antimicrobial peptide synthesis. Here we report that proHP1 adopts an active conformation (*) to carry out its function, without a requirement for proteolytic activation. Affinity chromatography using HP1 antibodies isolated from induced hemolymph the 48 kDa proHP1 and also a 90 kDa band (detected by SDS-PAGE under reducing conditions) containing proHP1 and several serpins, as revealed by mass spectrometric analysis. Identification of tryptic peptides from these 90 kDa complexes included peptides from the amino-terminal regulatory part of proHP1, indicating that proHP1* was not cleaved, and that it had formed a complex with the serpins. As suicide inhibitors, serpins form SDS-stable, acyl-complexes when they are attacked by active proteases, indicating that proHP1* was catalytically active. Detection of M. sexta serpin-1, 4, 9, 13 and smaller amounts of serpin-3, 5, 6 in the complexes suggests that it is regulated by multiple serpins in hemolymph. We produced site-directed mutants of proHP1b for cleavage by bovine blood coagulation factor Xa at the designed proteolytic activation site, to generate a form of proHP1b that could be activated by Factor Xa. However, proHP1b cut by Factor Xa failed to activate proHP6 and, via HP6, proHP8 or proPAP1. This negative result is consistent with the suggestion that proHP1* is a physiological mediator of immune responses. Further research is needed to investigate the conformational change that results in conversion of proHP1 to active proHP1*.
Keywords: clip domain, insect immunity, phenoloxidase, serine protease cascade, zymogen activation
Graphical abstract
1. Introduction
Serine protease (SP) pathways are conserved in evolution as a strategy to mediate rapid defense responses to tissue damage or microbial infection (Jiang and Kanost, 2000; Krem and Di Cera, 2002). In such cascade pathways, a series of preexisting, inactive zymogens become active SPs through specific recognition and sequential proteolysis. The potency and duration of these responses are often controlled by SP inhibitors including serpins (Gubb et al., 2010; Meekins et al., 2017). Serpins are suicide inhibitors that form stable acyl-enzyme complexes with cognate proteases, only when the active SPs attack the serpins at their reactive center loop (Gettins, 2002). In the last two decades, genetic and biochemical studies have generated much knowledge on the constitution and regulation of immune SP pathways in Drosophila, mosquitoes, beetles, moths, and other insects (Barillas-Mury, 2007; Jiang et al., 2010; Park et al., 2010; Veillard et al., 2016). Many pathway members contain an amino-terminal regulatory clip domain, a linker, and then an SP catalytic domain (Kanost and Jiang, 2015). After cleavage, the catalytic domain remains attached to the clip domain and linker via an interchain disulfide bridge. Clip-domain serine protease homologs (SPHs) also participate in insect immune responses (e.g. phenoloxidase (PO) mediated melanization), but the cleavage occurs between Cys-3 and Cys-4 of the clip domain in these SPHs (Kwon et al., 2000; Wang and Jiang, 2004; Yu et al., 2003).
Melanization pathway has been well studied in Manduca sexta (Kanost and Jiang, 2015). A modular serine protease, HP14, is auto-activated in the presence of β-1,3-glucan and its recognition protein βGRP1/2 (Wang and Jiang, 2006 and 2010) or in the presence of diaminopimelic acid-peptidoglycan (DAP-PG), microbe-binding protein (MBP), and PG recognition protein-1 (PGRP1) (Wang and Jiang, 2017). Active HP14 then cuts proHP21 to form HP21 (Wang and Jiang, 2007), which in turn activates proPAP2 and proPAP3 (PAP for proPO activating protease) (Gorman et al., 2007). ProPAP1 is activated by HP6 (An et al., 2009). PAP1, PAP2 or PAP3 cleaves proPO to PO in the presence of a complex of SPH1 and SPH2. The proHP6 cleavage activation may involve proHP1 (Yang et al., 2016). Besides proPAP1, HP6 cuts proHP8 to form HP8, HP8 activates pro-Spätzle, and Spätzle binds a Toll receptor to trigger an intracellular pathway that induces the synthesis of antimicrobial peptides (AMPs) (An et al., 2009 and 2010).
Proteolytic processing is a common, posttranslational modification and, in the case of zymogen activation, it rapidly converts inactive precursors to active enzymes. Zymogen activation can also occur in the absence of an activating protease. Prophenoloxidases (proPOs), isolated from the tobacco hornworm Manduca sexta, are activated by cetylpyridinium chloride (CPC, a cationic detergent) (Hall et al., 1995). Concentration of proPAP1 in a centrifugal device produced some active PAP1 (i.e. proPO activating protease-1), even though PAP1 does not usually activate proPAP1 by itself (Wang et al., 2001).
We recently reported the CPC-induced auto-proteolysis of proHP1, which can then cleave proHP6 (Yang et al., 2016). HP1 is the first clip-domain SP cloned from M. sexta and a member of the CLIPD subfamily (Cao et al., 2015; Jiang et al., 1999; Yang et al., 2016). ProHP1 is present in plasma at an estimated concentration of 27 μg/ml as a mixture of two closely related proteins, proHP1a and proHP1b (89.7% identical; 95.1% similar), at a ratio of approximately 7:1. For simplicity, we use proHP1* to denote the active proHP1 in plasma after microbial elicitation. The proenzymes are produced in granular hemocytes but not in fat body, and injection of bacteria into larvae led to differential expression of the two genes (Yang et al., 2016). Incubation of CPC with proHP1a/b or catHP1a/b (the catalytic domain) yielded an amidase activity that hydrolyzed N-acetyl-Leu-Asp-Leu-His-p-nitroanilide (LDLHpNa), a substrate synthesized to match the sequence preceding the proteolytic activation site of M. sexta proHP6 (An et al., 2009). The recombinant proHP1a/b or catHP1a/b elicited proPO activation in cell-free hemolymph, and catHP1a-treated plasma contained the proteolytically activated forms of HP6, HP8, and PAP1 (Yang et al., 2016). Based on these results, we hypothesize that proHP1* may be a physiological activator of proHP6.
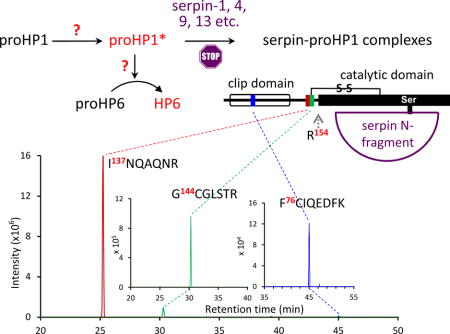
Another line of evidence for the current hypothesis arose from the study of M. sexta serpin-4 and -5, which suppress proPO activation in plasma by forming covalent complexes with HP1, HP6, and other HPs (Tong et al., 2005; Tong and Kanost, 2005). The complex of HP1 and serpin-4 migrated as a 90 kDa band in SDS-PAGE under reducing condition, substantially higher than 75 kDa or the typical size of a serpin-protease complex. This observation led us to test whether the 90 kDa band contains a region before the predicted cleavage activation site of proHP1 [(A/P)QGR↓VF(G/D)S]. In addition, we substituted AQGR of proHP1a (or PQGR of proHP1b) with IEGR – the recognition sequence of bovine blood coagulation factor Xa, to test whether proHP1a/b mutant cleaved by Factor Xa can function as a proHP6 activating enzyme. We also tested a hypothesis that α-helical antimicrobial peptides (detergent-like AMPs) or other plasma proteins may mimic the role of CPC by inducing the proHP1* conformation that can cleave proHP6 at the LDLH↓ILGG site. To address these questions, we have performed a series of experiments, demonstrating the existence of proHP1*, and discuss implications of the positive and negative results.
2. Materials and methods
2.1. Insect rearing, immune challenge, and hemolymph collection
M. sexta eggs were purchased from Carolina Biological Supply and larvae were reared on an artificial diet (Dunn and Drake, 1983). Each day 2, 5th instar larva was injected with a mixture of killed bacteria (Yang et al., 2016). Induced hemolymph (IH) was collected from cut prolegs of three larvae 24 h later and centrifuged to remove hemocytes. Similarly, control hemolymph (CH) samples were prepared from naïve day 2, 5th instar larvae.
2.2. Immuno-affinity purification of proHP1 and its associated proteins
Cell-free hemolymph samples (10 ml) from immune-challenged larvae were incubated with Micrococcus luteus (100 μg) for 30 min at room temperature to activate the SP-SPH system. To avoid melanization and protein crosslinking, 10 mM diethylthiocarbonate (a Cu2+ chelator that inhibits PO) and 1 mM 1-phenyl-2-thiourea (another PO inhibitor) were added to the reaction mixture. Rabbit antiserum against HP1 (4.8 ml) was coupled to 2.4 ml Protein A-Sepharose beads (Sigma) according to the manufacturer’s instructions. For isolating serpin-HP1 complexes, the plasma activated by M. luteus was mixed with the antibody-coupled beads for 8 h at 4°C with gentle agitation. The suspension was loaded into a Poly-Prep column (Bio-Rad) and then washed with 20 ml of 1 M NaCl and 20 ml of 10 mM sodium phosphate, pH 6.8 to remove unbound proteins. Bound proteins were eluted from the column with 50 mM glycine-HCl, pH 2.5. Fractions (0.5 ml each) were instantly neutralized with 50 μl, 1 M Tris-HCl, pH 8.0 in each collection tube. The elution fractions were separated by SDS-PAGE, followed by light staining with Coomassie Blue or immunoblot analysis.
2.3. In-gel trypsinolysis and LC-MS/MS analysis
The 80–95 kDa region excised from the SDS-PAGE gel described above was cut into small pieces before extensive destaining with 50% acetonitrile in 50 mM NH4HCO3, pH 8.0. The proteins in the gel pieces were reduced with tris(2-carboxyethyl) phosphine (TCEP), alkylated by iodoacetamide at Cys residues, and digested with sequencing grade trypsin for 16 h at 37°C, as described before (He et al., 2016). The tryptic peptides were extracted from the gel pieces using 1% trifluoroacetic acid for LC-MS/MS analysis on an LTQ-OrbitrapXL mass spectrometer (Thermo Scientific) in the DNA/Protein Resource Facility at Oklahoma State University. The MS/MS spectra were searched against Msexta_060614.fasta (He et al., 2016) for protein identification. Centroided ion masses were extracted by extract_msn.exe utility from Bioworks (v3.3.1) for database searching. Scaffold (v4.2.0, Proteome Software Inc.) was used to validate MS/MS-based peptide or protein identifications. Scan numbers were extracted from Scaffold Viewer to locate HPLC peaks of specific tryptic peptides. Xcalibur Qual Browser (v3.0 Thermo Scientific) was used to find the retention time along with base peak from the raw file. To generate a peak map, parameters were set to: mass tolerance (10.0 ppm); scan filter (FTMS+p NSI full ms [360.00–1400.00]; plot type (base peak); detector (MS); peak algorithm (ICIS); delay (0.00 min); ranges (m/z numbers of the peptide). For selected peptides, 2nd mass spectra were directly extracted from Scaffold.
2.4. Tests for the generation of proHP1* and HP6 in vitro
To obtain heat-stable plasma proteins including AMPs, CH and IH (Section 2.1) stimulated with 5 mg/ml M. luteus at 25°C for 10 min were separately heated for 5 min at 95°C and centrifuged at 10,000×g for 10 min to remove precipitated proteins. Each supernatant (1 μl) or synthetic moricin (1 μl, 0.2 μg/μl) was incubated with proHP6 (1 μl, 100 ng/μl) in the presence or absence of recombinant proHP1a (1 μl, 100 ng/μl) (Yang et al., 2016) in 1 mM CaCl2, 20 mM Tris-HCl, pH 7.4 (total volume: 20 μl) for 3 h at 37°C and separated by SDS-PAGE. Possible cleavage of proHP6 was analyzed using 1:1000 diluted HP6 antiserum as the primary antibody and 1:1000 diluted goat-anti-rabbit IgG conjugated with alkaline phosphatase (GAR-AP) (Bio-Rad) as the secondary antibody.
2.5. Site-directed mutagenesis of proHP1a/b cDNA
The proHP1a cDNA and mutagenic primer J1470 (5′-AAGCACGCGAATAGAAGGGCGGGTATTCGACTCCCGC) were used to construct proHP1aXa/pGEM-T as described before for other clip domain proteases (An et al., 2009). The underlined part encodes IEGR (the recognition sequence of bovine clotting factor Xa) to substitute for AQGR, the putative activation site of proHP1a. Similarly, proHP1bXa/pGEM-T was generated using proHP1b cDNA and J1468 (5′-AAGCACGCGAATAGAAGGGCGGGTATTCGACTCCCGC) to change PQGR to IEGR. After sequence verification, the EcoRI-XhoI fragments were directionally cloned into the same sites in pMFH6 (Lu and Jiang, 2008) to yield plasmids proHP1aXa/pMFH6 and proHP1bXa/pMFH6.
2.6. Production of M. sexta proHP1a and proHP1b mutants in the baculovirus system
The recombinant plasmids were used to generate bacmids, baculoviruses, and virus-infected Sf9 cell cultures (Yang et al., 2016). The proHP1a or proHP1b mutants in the conditioned media (500 ml) were purified by ion exchange chromatography on dextran sulfate-Sepharose, Ni2+-NTA affinity chromatography, and Superdex-100 gel filtration chromatography. After concentration and buffer exchange on Amicon ultracentrifugal filter devices (30K MWCO, Millipore), the proteins were stored at −80°C in 20 mM Tris-HCl, pH 7.4, 50 mM NaCl.
2.7. Treatment of proHP1bXa, proHP6 or both with bovine clotting factor Xa
The purified proHP1bXa, proHP6, or both were incubated with different amounts of Factor Xa (Haematologic Technologies) in 1 mM CaCl2, 20 mM Tris-HCl, pH 7.4, as indicated in Fig. 6 legend. The reaction mixtures and controls were heated at 95°C for 5 min in 1×SDS sample buffer with or without dithiothreitol (DTT) and separated by SDS-PAGE. Following electrotransfer onto a nitrocellulose membrane, the blot was blocked with 3% dry milk in Tris-buffered saline (TBS) and incubated overnight with 1:1000 diluted rabbit polyclonal antiserum in TBST (TBS supplemented with 0.05% Tween-20) as the primary antibody. After washing with TBST three times, 1:1000 diluted GAR-AP was incubated with the membrane for 4 h before washing and development using AP Conjugate Substrate Kit (Bio-Rad).
Fig. 6. Examination of proHP6 proteolytic processing by Factor Xa-treated proHP1bXa·.
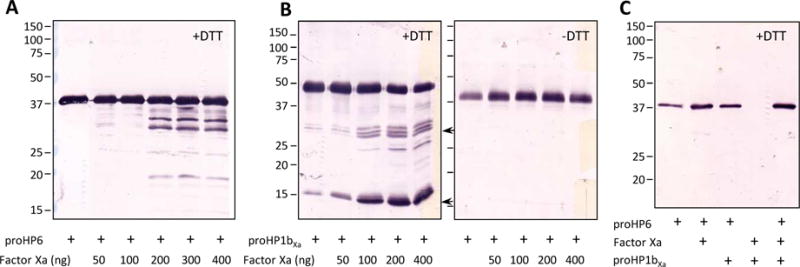
(A) Proteolysis of proHP6 by Factor Xa at different levels. As controls, aliquots of proHP6 (1 μl, 100 ng/μl) were incubated with 0 to 400 ng Factor Xa at 37°C for 90 min. After 12% SDS-PAGE, immunoblot analysis was performed using 1:1000 diluted HP6 antisera as the primary antibody. (B) Cleavage of proHP1bXa by Factor Xa. One μl of 150 ng/μl proHP1bXa was incubated with 1 μl Factor Xa at 0 to 400 ng at 37°C for 90 min, treated with SDS-sample buffer with (left panel) or without (right panel) DTT, and separated by 12% SDS-PAGE gel followed by immunoblotting using HP1 antibodies. The predicted 30 kDa catalytic and 16 kDa regulatory domains are indicated with arrows. (C) Processing of proHP6 by proHP1bXa mutant and Factor Xa. The three proteins (50 ng each) were incubated at 37°C for 90 min and separated by reducing SDS-PAGE. Diluted HP6 antiserum was used for immunoblot analysis. Positions and sizes of the Mr markers are indicated on the left.
3. Results and discussion
Six M. sexta serpins are known to inhibit serine proteases in larval hemolymph by forming covalent complexes with HPs during immune responses (An and Kanost, 2010; An et al., 2011; Christen et al., 2012; Jiang et al., 2003; Ragan et al., 2010; Suwanchaichinda et al., 2013; Tong et al., 2005; Zhu et al., 2003; Zou and Jiang, 2005). These SDS-stable complexes contain the N-terminal most part (40–45 kDa typically) of the serpin and the C-terminal catalytic domain (about 30 kDa) of the protease. For instance, serpin-4 has a typical size of 45 kDa and forms 75 kDa acyl-enzyme complexes with the catalytic domains of HP6, HP21, and two unknown HPs (Tong et al., 2005). Surprisingly, serpin-4 was also detected in a complex with a form of HP1 at 90 kDa (Tong et al., 2005). This unusually high Mr suggests that the entrapped protease contains at least a part of the pro-region and lack of proteolysis at putative cleavage activation site. Since this interpretation contradicts the usual concept of SP zymogen activation (i.e. specific proteolysis at the activation site), we decided to further analyze the protein composition of the 90 kDa HP1-serpin complexes to test for the presence of peptides from the pro-region.
3.1. Isolation and characterization of the 90 kDa proHP1-serpin complexes formed in larval plasma
We prepared an HP1 antibody column to isolate proHP1 and associated proteins from M. luteus-stimulated plasma by immunoaffinity chromatography (Fig. 1). ProHP1 was relatively abundant in plasma (25–30 μg/ml), enriched on the column, and eluted as a major protein that migrated as a band at 48 kDa on the Coomassie blue-stained gel. In comparison, a 90 kDa band was hardly visible after light staining but was strongly recognized by the HP1 polyclonal antibodies (Fig. 1A, 1B). Enrichment of this putative proHP1-serpin complex by the affinity chromatography was obvious, since it was only detected as a faint band in plasma. A fainter immunoreactive band at 75 kDa was uncharacterized and could be a proteolytic fragment of the proHP1-serpin complexes (Tong et al., 2005, Christen et al., 2012).
Fig. 1. Electrophoretic separation of the affinity purified M. sexta proHP1 and its associated hemolymph proteins followed by Coomassie staining (A) or immunoblot analysis (B).
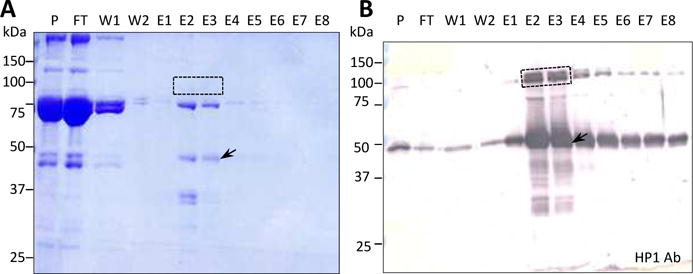
Protein samples from the HP1 antibody column, along with the plasma control, were resolved by 10% SDS-PAGE and stained by Coomassie blue R-250. Proteins in the duplicate gel were electro-transferred onto a nitrocellulose membrane for detection using polyclonal HP1 antibodies and alkaline phosphatase-conjugated secondary antibodies. P, 1 μl plasma sample before loading onto the affinity column; FT, 2 μl flow-through fraction; W1, 10 μl washing fraction in 1 M NaCl; W2, 10 μl washing fraction in 20 mM sodium phosphate, pH 6.8; E1–E8, 10 μl elution fractions. As described in Section 2.3, a gel slice containing the 90 kDa band (in lanes E2 and E3), marked with a dashed box, was treated for LC-MS/MS analysis. The proHP1 band is indicated by an arrow. Positions of the Mr markers are shown as short bars with their sizes marked on the left.
We excised the 80–95 kDa region in lanes E2 and E3, thoroughly treated it with TCEP, iodoacetamide, and trypsin, and analyzed the resulting peptides by nanoLC-MS/MS. We identified from this band HP1 and several serpins (serpin-1, 3, 4, 5, 6, 9, 13, and 15B), which are presumably present as serpin-protease complexes. Serpin-9, 13, and 15B had not previously been identified in hemolymph but were annotated recently in the M. sexta genome (Kanost et al., 2016). In addition to HP1 and serpins, several other proteins were identified in this band (Table 1), including 1) apolipoproteins and hexamerins, which are inevitable because of their high levels and Mr’s close to 80 kDa; 2) defense proteins (>70 kDa) (e.g. hemicentin, hemocytin, Hsp90, nimrod B, proPOs); 3) defense proteins (30–70 kDa) (e.g. Hsp25.4d, immulectins, PAP3), as parts of high Mr immune complexes (He et al., 2016); 4) Remarkably, about 2/3 of the 36 proteins are involved in immune responses, suggesting that they may interact with proHP1 and were thus enriched by the HP1 immunoaffinity chromatography.
Table 1.
A list of 36 identified M. sexta hemolymph proteins in the 80–95 kDa region*
| Protein name | Mr (kDa) | TSC | Protein name | Mr (kDa) | TSC | Protein name | Mr (kDa) | TSC |
|---|---|---|---|---|---|---|---|---|
| hemocytin-1 | 467 | 65 | HP1b | 43 | 16 | apolipophorin I and II | 367 | 140 |
| HP1a | 43 | 47 | serpin-15 | 41 | 13 | Met-rich storage protein | 90 | 77 |
| serpin-4 | 46 | 35 | C04954.5.0.COO2W_XNI | 186 | 12 | carboxylesterase | 75 | 68 |
| serpin-1 | 44 | 34 | nimrod B | 72 | 12 | α-arylphorin | 84 | 50 |
| proPO1 | 79 | 30 | serpin-6 | 47 | 8 | fibulin 1 | 150 | 18 |
| proPO2 | 80 | 29 | IML3 | 34 | 5 | apolipophorin-like | 475 | 13 |
| serpin-13 | 51 | 28 | PAP3 | 46 | 5 | collagen α-2IV protein | 183 | 11 |
| hemicentin-1 | 209 | 26 | Hsp25.4d | 29 | 5 | atlastin-like-1 | 86 | 10 |
| serpin-9 | 46 | 21 | Hsp90NI | 82 | 4 | Hp29 | 48 | 7 |
| serpin-3 | 51 | 18 | IML5 | 37 | 4 | β-arylphorin | 84 | 6 |
| inter-α-trypsin inh. H4NI | 101 | 18 | transferrin-1 | 75 | 4 | Hp KGM_13066 | 76 | 4 |
| twelve Cys protein-1NI | 78 | 17 | serpin-5 | 45 | 3 | α-NAGase | 97 | 2 |
Most proteins in the first two columns are related to immune responses or induced upon immune challenge (proteins marked NI are not induced; C04954.5.0.COO2W_X: MCOT ID). The calculated molecular masses (Mr’s) and total spectral counts (TSCs) are also listed.
Besides protein identification, total spectral counts (TSCs) of the mass spectrometric analysis, which reflect relative levels of the proteins, are also informative (Table 1). For instance, proHP1a was more abundant than PAP3 or the individual serpins, and proHP1b level was much lower than proHP1a. To confirm by another method the presence of the detected HPs and serpins, we used immunoblotting and the available antibodies to validate that distinct immunoreactive bands existed in the range of 80–95 kDa. Serpin-1, 3, 4, 5, 6, 9, and 13 were indeed detected at about 90 kDa (Fig. 2A). (No antibody was available yet for serpin-15B.) Additionally, serpin-1, 4, 5, 6, 9, and 13 were also found in 70–75 kDa, likely as complexes with other HPs, which were somehow associated with proHP1. Sequence alignment of their reactive center loop regions shows these serpins all have a positively charged P1 residue (Lys in serpin-3 and Arg in others) (Fig. 2C). This finding agrees well with the predicted trypsin-like specificity of proHP1* (Yang et al., 2016), and the predicted cleavage of these serpins at the P1 residue by proHP1*, leading to detection of proHP1*-serpin complexes identified by mass spectrometry and immunoblot analysis. Observing multiple serpin-proHP1* complexes was also consistent with high intensity of the 90 kDa band and the presence of proHP1 at a relatively high abundance.
Fig. 2. Ininiunoblot analyses of the affinity purified proteins separated by 10% SDS-PAGE under reducing (A) and non-reducing (B) conditions and sequence alignment of the identified serpins (C).
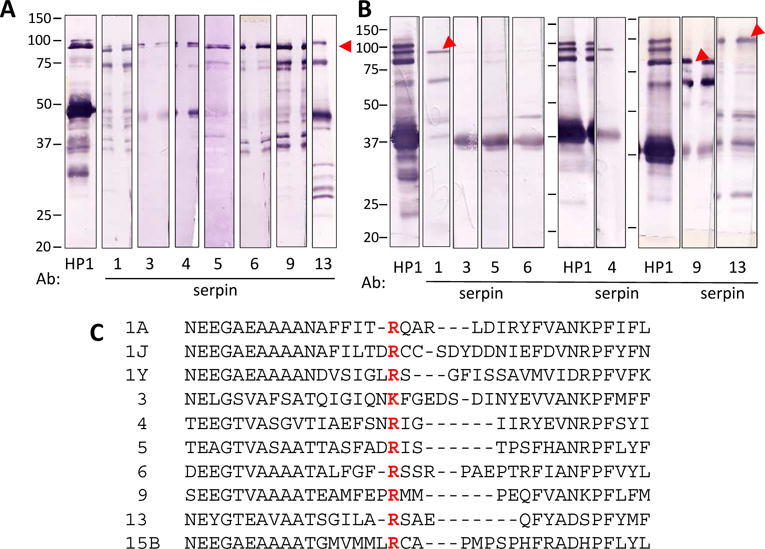
The E2 fraction (10 μl/lane) was treated with SDS sample buffer with (A) or without (B) dithiothreitol (DTT) and separated by electrophoresis. After electro-transfer, the nitrocellulose membrane blotted with the proteins was cut into strips which, after blocking, were individually incubated with 1:1000 diluted antisera against M. sexta HP1, serpin-1, 3–6, 9 or 13, as indicated under each strip. Positions and sizes (in kDa) of the Mr standards are indicated. The 90 kDa SECs are indicated by red arrowheads. (C) Amino acid sequences of the reactive site loop in serpin-1, 3–6, 9, 13, and 15B were aligned and the region that can be cut by protease was shown. P1 amino acid was in red and bold font.
Because resolution of the 90 kDa band was insufficient for distinguishing different proHP1-serpin complexes, we repeated the immunoblot analysis after non-reducing SDS-PAGE (Fig. 2B). The HP1 antibodies detected three intense bands between 80 and 95 kDa. The top one was also recognized by serpin-13 antibodies; the middle by serpin-4 and serpin-1 antibodies; the bottom band by serpin-9 antibodies. Partly due to the low levels, serpin-3, 5 and 6 antibodies failed to detect the unreduced complexes with HP1. Redox state of the proteins may have impacted conformation, gel mobility, and accessibility of antigenic determinants of the serpins in their complexes with proHP1*.
To further characterize the serpin-protease complexes, we closely examined the primary and secondary mass spectra of tryptic peptides from the identified proteins (Table S1), in particular HP1. We detected three peptides in the amino-terminal regulatory region of proHP1a (Fig. 3) and six peptides in the catalytic domain. F76CIQEDFK83 resides between Cys-2 and Cys-4 of the clip domain (lowest intensity); I137NQAQNR143 (highest intensity) and G144CGLSTR150 are adjacent peptides in the linker between the clip and catalytic domains. We deduced that the putative proteolytic activation site was uncut because, otherwise, 75 kDa serpin-proHP1* complexes would have been detected on the immunoblots after SDS-PAGE under reducing condition. For proHP1b, two peptides (I137SQAQNR143 and G144CGLSTR150P151QGR154) in the linker were detected (Table S1, Fig. 5). The first peptide had S138 replacing N138 in proHP1a; the second was longer than G144CGLSTR150 in proHP1a, as trypsin cannot efficiently cleave between R150 and P151 to release P151QGR154 from proHP1b. Trypsin likely cut out A151QGR154 from proHP1a but the 371.3 Da peptide was filtered out by the mass spectrometer with the low limit set at 400.0 Da. In summary, we have obtained solid evidence that proHP1*, lacking cleavage at the zymogen activation site, formed covalent complexes with multiple serpins and thus must be catalytically active. In previous work, identifying proteases inhibited by serpin-4 and serpin-5, HP1 was identified in a 90 kDa band, but no peptide from the pro-region was identified by peptide fingerprint analysis (Tong et al., 2005). A difference in the current study is that we reduced the sample with TCEP (a reducing agent stronger than DTT), which may have led to more complete digestion of the alkylated proteins with trypsin, particularly in the clip domains, and we analyzed the resulting peptides on a significantly more sensitive mass spectrometer.
Fig. 3. Mass spectrometric (MS) analysis of three trypsinolytic peptides in the clip domain and linker region of M. sexta proHP1a.
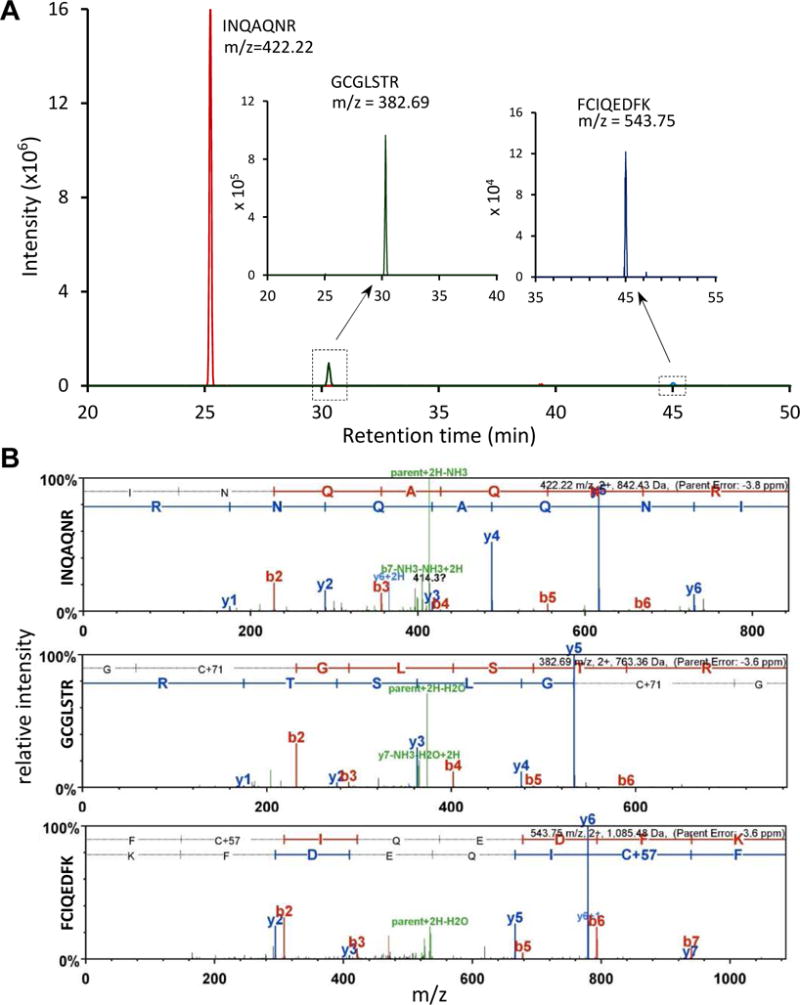
(A) Mass peaks corresponding to the peptides are displayed with their elution times on nano-LC. The second and third peaks are lower in intensity and, therefore, shown in insets with smaller scales on the y-axis. The m/z value and sequence of each peak are indicated. (B) 2nd MS spectra of the peptides. Peaks for forward (red, b) and reverse (blue, y) fragments were labeled with their order and number (e.g. y4 in the top panel means fragment of the first 4 residues in reverse order, A140QNR143).
Fig. 5. Site-directed mutagenesis of proHP1 for in vitro activation by Factor Xa.
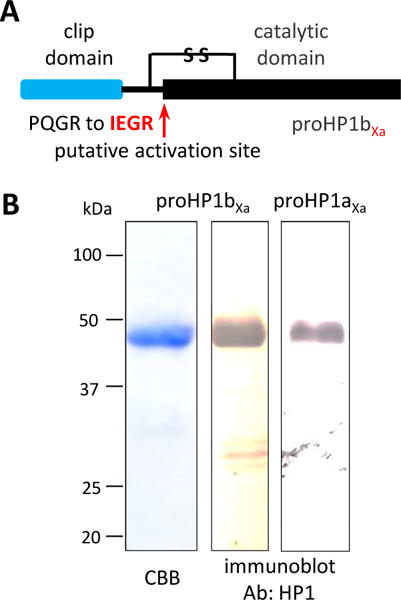
(A) Domain structure and mutant design. Based on the conventional mechanism, an unknown protease in M. sexta activates proHP1 via cleavage next to AQGR in proHP1a (not shown) or PQGR in proHP1b. By changing the sequence to IEAR, active HP1 would be generated by the commercially available enzyme that cuts after IEAR to activate proHP6. (B) 12% SDS-PAGE followed by Coomassie staining or immunoblot analysis. Left lane: purified proHP1b (1.0 μg) on the stained gel. Middle and right lanes: 0.2 μg total proteins on the immunoblots detected using HP1 polyclonal antibodies. Positions and sizes of the Mr markers are shown on the left.
Although proHP1* differs from a cleavage-activated SP, we expect that the serpins inhibit the active zymogen via the same mechanism. As long as the carboxyl-terminal domain is in the active conformation, it doesn’t seem to matter whether or not the amino-terminal clip domains is attached by a peptide bond at the “cleavage activation site”. Despite that, an unknown protein modulator in the hemolymph may still interact with the clip domain of proHP1 and that provides specificity for the shift of proHP1 to proHP1*. Besides, such conformation modulator has to somehow associate with pathogen infection or tissue damage to ensure the defense response occurs only when needed.
The tight regulation of proHP1* formation suggests the importance of HP6. Factor Xa-cleaved M. sexta HP6Xa converted proPAP1 to PAP1 for proPO activation in larval hemolymph (An et al., 2009). The HP6Xa also processed proHP8 to form HP8 for pro-Spätzle and Toll pathway activation that induces AMP synthesis (An et al., 2010). The fact that CPC-treated proHP1 (▾) hydrolyzed LDLHpNa led us to hypothesize that proHP1* cleaves proHP6 in vivo at LDLH↓ILGG (Yang et al., 2016). After establishing the existence of active zymogen, we attempted to induce proHP1* in vitro and test if it can directly activate proHP6.
3.2. Exploration of mechanisms for generating proHP1* that may activate proHP6 by cleaving at the correct site
Because cationic detergents (e.g. CPC, CTAB) induced an amidase activity of proHP1 ▾ that cut itself and proHP6 at a wrong site (Yang et al., 2016), we tested whether or not endogenous AMPs induce proHP1 to correctly process proHP6. Like detergents, some AMPs (e.g. cecropins, moricin) (He et al., 2015) are detergent-like, amphipathic α-helical molecules that compromise the integrity of bacterial cell membrane. Incubation of M. sexta moricin-1 with the two proHPs did not induce any proteolytic cleavage of proHP6 (Fig. 4A), although this synthetic peptide has a positively charged tail. We then heated four hemolymph samples at 95°C and harvested heat-stable molecules in the supernatants, hoping that a mixture of endogenous AMPs would work better than moricin-1 alone. After proHP6 had been incubated with proHP1 and heat-stable components of the plasma, there was no proHP6 cleavage either (Fig. 4B).
Fig. 4. Exploration of various conditions to generate proHP1* in vitro.
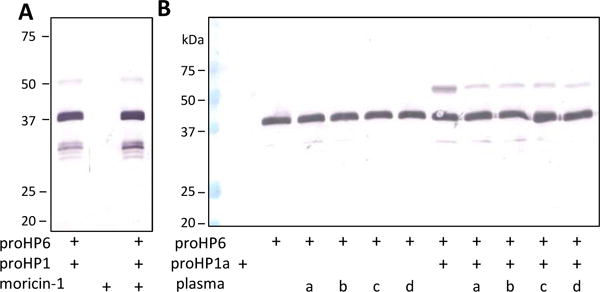
As described in Section 2.4, aliquots of proHP6 (1 μl, 100 ng/μl) were incubated with the recombinant proHP1a (1 μl, 100 ng/μl) in the absence or presence of moricin (1 μl, 200 ng/μl) (A) or supernatants (1 μl) of heated plasma (B) from naïve (a, b) and immune challenged (c, d) larvae at 37°C for 3 h. Prior to heat treatment and centrifugation, samples b and d were stimulated by 5 μg/μl M. luteus for 10 min at 25°C. All the reaction mixtures and controls (moricin, proHP1a, proHP6, proHP1a and proHP6, or proHP6 and supernatant a/b/c/d) were separated by 10% SDS-PAGE followed by immunoblot analysis using HP6 polyclonal antibodies.
We have tested other ways to convert proHP1 to proHP1*, such as crosslinking proHP1a with 5,6-dihydroxyindole (Zhao et al., 2011), incubating proHP1a with β1,3-glucan and βGRP2 (Wang and Jiang, 2006) or with DAP-PG, MBP and PGRP1 (Wang and Jiang, 2017), and treating proHP1a with HP14 and a complex of SPH1 and SPH2. To our disappointment, after further reacting these reaction mixtures with proHP6, the proenzyme was not processed (data not shown). Apparently, the in vitro assays failed to mimic the conditions in plasma for proHP1a* generation.
Protease zymogens are often activated by other proteases in a cascade mode that results in amplification or localization to specific sites. One exception to the rule includes initiation proteases that activate themselves via auto-proteolysis (Davie et al., 1979; Stennicke et al., 1999; Wang and Jiang, 2006; Kobayashi et al., 2014). Another exception involves staphylocoagulase, streptokinase, and, perhaps, other non-protease proteins secreted by bacterial pathogens (Friedrich et al., 2003; Renatus et al., 1997), which bind and activate precursors of thrombin, tissue-type plasminogen activator and other proteases, respectively. While the proHP1-proHP1* transition in response to a physiological or pathological cue is poorly understood, the above cases of zymogen activation by non-catalytic protein/subunit provide useful insights for future exploration of proHP1* structure, function, and activation mechanism.
3.3. Possible activation of proHP6 by Factor Xa-cleaved proHP1 mutant
After trying various ways to generate proHP1* without any success, we tested whether or not artificially cleaved HP1 could activate proHP6 because, if yes, an unknown activating enzyme may exist to cleave proHP1 under certain conditions and active HP1 can then be used to confirm its putative role as a proHP6 activating enzyme. If not, the concept of active zymogen would be further supported so that we can focus on finding other in vitro conditions for converting proHP1 to proHP1*. In either way, the experiment would inform us how proHP1 is activated.
We designed and expressed proHP1 mutants by site-directed mutagenesis (Fig. 5A). The four residues preceding the putative cleavage site A151QGR154 in proHP1a and P151QGR154 in proHP1b were substituted with I151EGR154, the recognition sequence of bovine clotting factor Xa. Their apparent Mr’s were close to 48 kDa (Fig. 5B), partly due to N-linked glycosylation (Yang et al., 2016). The proHP1aXa expression level and product purity were a lot lower than proHP1bXa under the same conditions of cell culturing and protein purification (data not shown). Therefore, we had to use the purified proHP1bXa to test conditions for Factor Xa cleavage and proHP6 activation by HP1bXa.
As a control, we first tested if Factor Xa itself could cleave wild-type proHP6 and found that Factor Xa effectively cut proHP6 at a molar ratio of ≥ 1.6 : 1 (Xa : proHP6) and yielded two major bands around 30 kDa (Fig. 6A). We also reacted the recombinant proHP1bXa with different amounts of Factor Xa and found that Factor Xa more efficiently cut proHP1bXa at a molar ratio of ≥ 1 : 1.5 (Xa : proHP1bXa) (Fig. 6B). In the presence of DTT, cleavage of proHP1bXa led to a decrease in the proenzyme band and increase of 30 kDa triplets (likely due to differential glycosylation) and a 16 kDa band. Since all these species came together as a part of the broad band with an apparent Mr of 40 kDa under non-reducing condition, we suggested the 30 and 16 kDa bands corresponded to the catalytic and regulatory domains of HP1 (Fig. 5A), respectively. We then tested whether or not Factor Xa-cleaved proHP1bXa was able to cleave proHP6. At the molar ratio of 1 : 1 : 0.8 (Xa : proHP1bXa : proHP6), where Factor Xa is sufficient to activate proHP1bXa but not proHP6, HP1bXa did not activate proHP6 at all (Fig. 6C, last lane). This result is consistent with our conclusion that it is proHP1*, instead of HP1, which may cut proHP6 to form HP6 that generates HP8 and PAP1 to trigger the proPO and Toll pathways.
3.4. Summary
As supported by results from this and the previous (Yang et al., 2016) studies, activation of M. sexta proHP1, a member of the CLIPD subfamily, adopts an unusual approach: the conformational change from inactive to active is not induced by proteolytic cleavage at the specific activation site. This finding at least partly explained the difficulties experienced in the last two decades during the functional analysis of HP1, as we did not even know what we were looking for. While the nature of such change is still unclear at present, this phenomenon is validated by thorough characterization of the 90 kDa SDS-stable complexes of proHP1 with the endogenous serpins and the suicide inactivation mechanism of serpins. The failure of proHP6 activation by artificially generated HP1b further supports the mechanism. Otherwise, we need to demonstrate HP1* may activate proHP6 under certain conditions and proHP1* may do so under other conditions. Future research is necessary to first demonstrate the association of proHP6 activation in the proHP1-containing hemolymph fractions before mechanistic research is performed to examine the conversion of proHP1 to proHP1* upon recognition of invading microbes. More efforts will be made to explore other conditions and their combinations to establish proHP1* as the activator of proHP6 in vitro. Identification of the seven serpins in the 90 kDa serpin-protease complexes clearly demonstrated the importance of proHP1* in terms of system activation.
Supplementary Material
Highlights.
Detection of proHP1 in the SDS-stable complexes with unusually high Mr of 90 kDa
Identification of seven serpins as physiological regulators of active proHP1
No proHP6 activation by proHP1b mutant cleaved at the designed activation site
ProHP1 can be activated without being cleaved.
Acknowledgments
This work was supported by National Institutes of Health Grants GM58634 and AI112662 (to H. Jiang). This article was approved for publication by the Director of the Oklahoma Agricultural Experiment Station and supported in part under project OKLO2450. We thank Dr. Michael R. Kanost at Kansas State University for his critical comments on the manuscript.
Abbreviations
- AMP
antimicrobial peptide
- βGRP1/2
β1,3-glucan recognition protein-1/2
- DAP
diaminopimelic acid
- DHI
5,6-dihydroxyindole
- MBP
microbe binding protein
- NP and IP
cell-free hemolymph from naïve or induced larvae
- CPC
cetylpyridinium chloride
- DTT
dithiothreitol
- HP
hemolymph (serine) protease
- HP1
formerly hemocyte protease-1
- PO and proPO
phenoloxidase and its precursor
- PAP
proPO activating protease
- PGRP1
peptidoglycan recognition protein-1
- SP and SPH
serine protease and its non-catalytic homolog
- TCEP
tris(2-carboxyethyl)phosphine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J Biol Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Jiang H, Kanost MR. Proteolytic activation and function of the cytokine Spätzle in innate immune response of a lepidopteran insect, Manduca sexta. FEBS J. 2010;277:148–162. doi: 10.1111/j.1742-4658.2009.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Kanost MR. Manduca sexta serpin-5 regulates prophenoloxidase activation and the Toll signaling pathway by inhibiting hemolymph proteinase HP6. Insect Biochem Mol Biol. 2010;40:683–689. doi: 10.1016/j.ibmb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Ragan EJ, Kanost MR. Serpin-1 splicing isoform J inhibits the proSpatzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta. Dev Comp Immunol. 2011;35:135–141. doi: 10.1016/j.dci.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillas-Mury C. CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends Parasitol. 2007;23:297–299. doi: 10.1016/j.pt.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Cao X, He Y, Hu Y, Zhang X, Wang Y, Zou Z, Chen Y, Blissard GW, Kanost MR, Jiang H. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem Mol Biol. 2015;62:51–63. doi: 10.1016/j.ibmb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen JM, Hiromasa Y, An C, Kanost MR. Identification of plasma proteinase complexes with serpin-3 in Manduca sexta. Insect Biochem Mol Biol. 2012;42:946–955. doi: 10.1016/j.ibmb.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie EW, Fujikawa K, Kurachi K, Kisiel W. The role of serine proteases in the blood coagulation cascade. Adv Enzymol Relat Areas Mol Biol. 1979;48:277–318. doi: 10.1002/9780470122938.ch6. [DOI] [PubMed] [Google Scholar]
- Dunn PE, Drake DR. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm Manduca sexta. J Invertebr Pathol. 1983;41:77–85. [Google Scholar]
- Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Gorman MJ, Wang Y, Jiang H, Kanost MR. Manduca sexta hemolymph proteinase 21 activates prophenoloxidase activating proteinase 3 in an insect innate immune response proteinase cascade. J Biol Chem. 2007;282:11742–11749. doi: 10.1074/jbc.M611243200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubb D, Sanz-Parra A, Barcena L, Troxler L, Fullaondo A. Protease inhibitors and proteolytic signalling cascades in insects. Biochimie. 2010;92:1749–1759. doi: 10.1016/j.biochi.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Hall M, Scott T, Sugumaran M, Soderhall K, Law JH. Proenzyme of Manduca sexta phenol oxidase: purification, activation, substrate specificity of the active enzyme, and molecular cloning. Proc Natl Acad Sci USA. 1995;92:7764–7768. doi: 10.1073/pnas.92.17.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Cao X, Zhang S, Rogers J, Hartson S, Jiang H. Changes in the plasma proteome of Manduca sexta larvae in relation to the transcriptome variations after an immune challenge: evidence for high molecular weight immune complex formation. Mol Cell Proteomics. 2016;15:1176–1187. doi: 10.1074/mcp.M115.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Cao X, Li K, Hu Y, Chen Y, Blissard G, Kanost MR, Jiang H. A genome-wide analysis of antimicrobial effector genes and their transcription patterns in Manduca sexta. Insect Biochem Mol Biol. 2015;62:23–37. doi: 10.1016/j.ibmb.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Four serine proteinases expressed in Manduca sexta haemocytes. Insect Mol Biol. 1999;8:39–53. doi: 10.1046/j.1365-2583.1999.810039.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Jiang H. Clip-domain serine proteases as immune factors in insect hemolymph. Curr Opin Insect Sci. 2015;11:47–55. doi: 10.1016/j.cois.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR, Arrese EL, Cao X, Chen Y-R, Chellapilla S, Goldsmith M, Grosse-Wilde E, Heckel DG, Herndon N, Jiang H, Papanicolaou A, Qu J, Soulages JL, Vogel H, Walters J, Waterhouse RM, Ahn SJ, Almeida FC, An C, Aqrawi P, Bretschneider A, Bryant WB, Bucks S, Chao H, Chevignon G, Christen JM, Clarke DF, Dittmer NT, Ferguson LC, Garavelou S, Gordon KH, Gunaratna RT, Han Y, Hauser F, He Y, Heidel-Fischer H, Hirsh A, Hu Y, Jiang H, Kalra D, Klinner C, König C, Kovar C, Kroll AR, Kuwar SS, Lee SL, Lehman R, Li K, Li Z, Liang H, Lovelace S, Lu Z, Mansfield JH, McCulloch KJ, Mathew T, Morton B, Muzny DM, Neunemann D, Ongeri F, Pauchet Y, Pu LL, Pyrousis I, Rao XJ, Redding A, Roesel C, Sanchez-Gracia A, Schaack S, Shukla A, Tetreau G, Wang Y, Xiong GH, Traut W, Walsh TK, Worley KC, Wu D, Wu W, Wu YQ, Zhang X, Zou Z, Zucker H, Briscoe AD, Burmester T, Clem RJ, Feyereisen R, Grimmelikhuijzen CJ, Hamodrakas SJ, Hansson BS, Huguet E, Jermiin LS, Lan Q, Lehman HK, Lorenzen M, Merzendorfer H, Michalopoulos I, Morton DB, Muthukrishnan S, Oakeshott JG, Palmer W, Park Y, Passarelli AL, Rozas J, Schwartz LM, Smith W, Southgate A, Vilcinskas A, Vogt R, Wang P, Werren J, Yu XQ, Zhou JJ, Brown SJ, Scherer SE, Richards S, Blissard GW. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem Mol Biol. 2016;76:118–147. doi: 10.1016/j.ibmb.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Shiga T, Shibata T, Sako M, Maenaka K, Koshiba T, Mizumura H, Oda T, Kawabata S. The N-terminal Arg residue is essential for autocatalytic activation of a lipopolysaccharide-responsive protease zymogen. J Biol Chem. 2014;289:25987–25995. doi: 10.1074/jbc.M114.586933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- Kwon TH, Kim MS, Choi HW, Joo CH, Cho MY, Lee BL. A masquerade-like serine proteinase homologue is necessary for phenoloxidase activity in the coleopteran insect, Holotrichia diomphalia larvae. Eur J Biochem. 2000;267:6188–6196. doi: 10.1046/j.1432-1327.2000.01695.x. [DOI] [PubMed] [Google Scholar]
- Lu Z, Jiang H. Expression of Manduca sexta serine proteinase homolog precursors in insect cells and their proteolytic activation. Insect Biochem Mol Biol. 2008;38:89–98. doi: 10.1016/j.ibmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins DA, Kanost MR, Michel K. Serpins in arthropod biology. Semin Cell Dev Biol. 2016;62:105–119. doi: 10.1016/j.semcdb.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Kim CH, Rui J, Park KH, Ryu KH, Chai JH, Hwang HO, Kurokawa K, Ha NC, Soderhill I, Soderhill K, Lee BL. Beetle immunity. Adv Exp Med Biol. 2010;708:163–180. doi: 10.1007/978-1-4419-8059-5_9. [DOI] [PubMed] [Google Scholar]
- Ragan EJ, An C, Yang CT, Kanost MR. Analysis of mutually exclusive alternatively spliced serpin-1 isoforms and identification of serpin-1 proteinase complexes in Manduca sexta hemolymph. J Biol Chem. 2010;285:29642–29650. doi: 10.1074/jbc.M110.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M, Engh RA, Stubbs MT, Huber R, Fischer S, Kohnert U, Bode W. Lys 156 promotes the anomalous proenzyme activity of tPA: X-ray crystal structure of single-chain human tPA. EMBO J. 1997;16:4797–4805. doi: 10.1093/emboj/16.16.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- Suwanchaichinda C, Ochieng R, Zhuang S, Kanost MR. Manduca sexta serpin-7, a putative regulator of hemolymph prophenoloxidase activation. Insect Biochem Mol Biol. 2013;43:555–561. doi: 10.1016/j.ibmb.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Jiang H, Kanost MR. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. J Biol Chem. 2005;280:14932–14942. doi: 10.1074/jbc.M500532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Kanost MR. Manduca sexta serpin-4 and serpin-5 inhibit the prophenol oxidase activation pathway: cDNA cloning, protein expression, and characterization. J Biol Chem. 2005;280:14923–14931. doi: 10.1074/jbc.M500531200. [DOI] [PubMed] [Google Scholar]
- Veillard F, Troxler L, Reichhart JM. Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation. Biochimie. 2016;122:255–269. doi: 10.1016/j.biochi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Prophenoloxidase (proPO) activation in Manduca sexta: an analysis of molecular interactions among proPO, proPO-activating proteinase-3, and a cofactor. Insect Biochem Mol Biol. 2004;34:731–742. doi: 10.1016/j.ibmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang H, Kanost MR. Expression and purification of Manduca sexta prophenoloxidase-activating proteinase precursor (proPAP) from baculovirus-infected insect cells. Protein Expr Purif. 2001;23:328–337. doi: 10.1006/prep.2001.1517. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Interaction of β-1,3-glucan with its recognition protein activates hemolymph proteinase 14, an initiation enzyme of the prophenoloxidase activation system in Manduca sexta. J Biol Chem. 2006;281:9271–9278. doi: 10.1074/jbc.M513797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Reconstitution of a branch of Manduca sexta prophenoloxidase activation cascade in vitro: Snake-like hemolymph proteinase 21 cleaved by HP14 activates prophenoloxidase-activating proteinase-2 precursor. Insect Biochem Mol Biol. 2007;37:1015–1025. doi: 10.1016/j.ibmb.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Binding properties of the regulatory domains in Manduca sexta hemolymph proteinase-14, an initiation enzyme of the prophenoloxidase activation system. Dev Com Immunol. 2010;34:316–322. doi: 10.1016/j.dci.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Prophenoloxidase activation and antimicrobial peptide expression induced by the recombinant microbe binding protein of Manduca sexta. Insect Biochem Mol Biol. 2017;2017;83:35–43. doi: 10.1016/j.ibmb.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang Y, He Y, Jiang H. In search of a function of Manduca sexta hemolymph protease-1 in the innate immune system. Insect Biochem Mol Biol. 2016;76:1–10. doi: 10.1016/j.ibmb.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XQ, Jiang H, Wang Y, Kanost MR. Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- Zhao P, Lu Z, Strand MR, Jiang H. Antiviral, anti-parasitic, and cytotoxic effects of 5,6-dihydroxyindole (DHI), a reactive compound generated by phenoloxidase during insect immune response. Insect Biochem Mol Biol. 2011;41:645–652. doi: 10.1016/j.ibmb.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang Y, Gorman M, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J Biol Chem. 2003;278:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]
- Zou Z, Jiang H. Manduca sexta serpin-6 regulates immune serine proteinases PAP-3 and HP8 – cDNA cloning, protein expression, inhibition kinetics, and function elucidation. J Biol Chem. 2005;280:14341–14348. doi: 10.1074/jbc.M500570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


