Abstract
Gene transcription in a set of 49 human primary lung adenocarcinomas and 9 normal lung tissue samples was examined using Affymetrix GeneChip technology. A total of 3442 genes, called the set MAD, were found to be either up- or down-regulated by at least 2-fold between the two phenotypes. Genes assigned to a particular gene ontology term were found, in many cases, to be significantly unevenly distributed between the genes in and outside MAD. Terms that were overrepresented in MAD included functions directly implicated in the cancer cell metabolism. Based on their functional roles and expression profiles, genes in MAD were grouped into likely co-regulated gene sets. Highly conserved sequences in the 5 kb region upstream of the genes in these sets were identified with the motif discovery tool, MoDEL. Potential oncogenic transcription factors and their corresponding binding sites were identified in these conserved regions using the TRANSFAC 8.3 database. Several of the transcription factors identified in this study have been shown elsewhere to be involved in oncogenic processes. This study searched beyond phenotypic gene expression profiles in cancer cells, in order to identify the more important regulatory transcription factors that caused these aberrations in gene expression.
INTRODUCTION
The transformation of normal lung tissue into lung adenocarcinomas involves, among other characteristic features, a hallmark process by which the cell loses control of its replication process (an accelerated cell cycle) (1). Adenocarcinomas have a high incidence of fatality in patients in US, and a similar trend is developing in other countries (2). At present, lung cancer studies generally incorporate two main objectives: providing an early and sensitive diagnosis, and trying to understand the molecular basis underlying the disease formation. Recently, the availability of the human genome sequence (3) and gene expression profiling techniques (4) have provided new insights, narrowing the gap to achieve these objectives. The challenges that lie ahead include systematically identifying the functions of all cancer associated genes, and continuing the efforts to decipher their regulatory networks. This information will provide a much deeper understanding of the mechanism of cancer cell formation and development, and assist in the identification of potent therapeutic targets for disease control and eradication.
Computational methods that are employed to identify cancer associated genes from megabytes of noisy microarray data still require further development. Data normalization procedures may have an important effect on the succeeding downstream data analysis (5–8). Using human housekeeping genes as the least variable set of gene expression profiles is one accepted method (9). Many computational methods have been introduced to determine marker genes for cancer from gene expression datasets (10,11). These methodologies aim to stratify samples into tissue classes or phenotypes based on the ability of sets of differentially regulated genes to discriminate among the samples. Methods such as recursive partitioning (12), expression ratio analysis (13), principal component analysis (14), partial least squares (15), and independent component analysis (16) have been used to identify the minimum set of genes that can achieve this classification. However, the usually small number (tens) of (tissue) samples per class and the large number (tens of thousands) of features (genes) in these datasets cast doubt on the statistical significance of genes identified as discriminating between normal or cancer tissues or cancer subtypes. The effects on the detection of cancer marker genes due to these constraints, which can lead to genes being classified as markers by chance, have been investigated (17).
Recently, the use of computational methods to identify regulatory elements has become increasingly important (18). This is partly because the alternative of experimental determination of cis-regulatory elements can be inaccurate, and is often slow and laborious (19). A common way to analyze regulatory relationships among genes using microarray data is to cluster the genes, based on their expression profiles, into sets of putatively co-regulated genes. This assumes that co-regulated genes are likely to have cis-regulatory elements in common (20). However, searching for common sequence signals in genomic regions near these genes can lead to the detection of spurious cis-regulatory elements, as many genes may show similar expression profiles for reasons other than co-regulation (20). Many studies have shown that biologically relevant cis-regulatory elements often occur in groups (21,22). Following this rationale, conserved regulatory motifs correlated to gene expression were discovered by fitting a linear regression model to the expression arrays from Saccharomyces cerevisiae (23) and an extension of this technique was used to identify binding motifs of the transcription factors ROX1p and YAP1p (24). In this work, we performed a microarray based study of a set of normal lung tissues and a set of primary lung adenocarcinomas. Our aims were, first, to distinguish the broadest set of genes (MAD) that showed differential expression levels across the two tissue types and investigate the correlation of their gene expression profiles with the tissue type. Second, we wished to examine the division of genes with the same functional annotation between the MAD set and the remaining genes on the microarray to find functional groups disproportionately represented in MAD. Finally, we attempted to identify the transcription factors, as well as their corresponding binding sites, which regulate the observed expression differences of the genes in the MAD set.
The rationale for the first two aims was that, we could make use of the knowledge accumulated by scientists on genes in the MAD set, by using functional annotations assigned through Gene Ontology terms, to investigate the nature of the biological processes that were actually perturbed in cancer cells. It was expected that some functional classes would preferentially be found in the MAD gene set. Instead of clustering genes based solely on their expression profiles, genes were first selected by sharing a gene ontology term and then clustered by an expression profile. The reasoning behind this was that genes with the same function and similar expression profiles were more likely to be under the same regulatory control than genes with differing functions but similar expression profiles. ‘In biblio’ analysis of genes' neighborhoods has been long advocated as an efficient means to permit inductive reasoning by using the knowledge accumulated by the worldwide community of researchers (25). A motif finding algorithm developed by us, MoDEL (26), was used to discover highly conserved DNA regions associated with the genes in a cluster, before these sequences were scanned against the TRANSFAC 8.3 database to detect plausible oncogenic transcription factor binding sites.
MATERIALS AND METHODS
Primary lung adenocarcinoma dataset
Tissue samples for the complete cohort of this study were collected, with informed consent, by the Department of Pathology, The University of Hong Kong, Queen Mary Hospital, Pokfulam, Hong Kong. A total of 58 patients gave samples with normal lung tissue (n = 9) and primary lung adenocarcinomas (n = 49). Identifier code numbers were assigned to each tissue sample and its correlated clinical data. The link between the code numbers and all patient identifiers was destroyed, rendering the samples and clinical data completely anonymous. Clinical data from hospital records included the age and sex of the patient, smoking history, type of resection, post-operative pathological staging, post-operative histopathological diagnosis, patient survival information, time of last follow-up interval or time of death (when known), and site of disease recurrence (when known). Information for the entire dataset is provided as Supplementary Material at http://bioinfo.hku.hk/~daniely/lung_microarray/. It is noted that the numbers do not always add to 58, as complete information could not be found for all samples.
The gender composition of the cohort was 25 males and 33 females. The reported smoking history of the patients was 24 non-smokers, 10 smoking at least 40 packs per year, seven ex-smokers and nine passive smokers. Post-operative pathological staging of these samples revealed 26 stage I, 8 stage II, 14 stage III and 1 stage IV tumors.
Tissue samples were snap-frozen in liquid nitrogen within 30 min after dissection and kept at −70°C until use. Tumor samples were examined before use to ensure at least 70% of tumor by area. RNA was extracted following standard protocols and hybridized to Affymetrix HG-U133A GeneChips. Expression values from a total of 22 283 transcript probe sets were collected using Affymetrix scanners and analysis software (Microarray Suite 5.0.1). The raw dataset is publicly available at ArrayExpress (public repository for microarray data www.ebi.ac.uk/arrayexpress; accession number: E-MEXP-231) (27,28); or can be downloaded at http://bioinfo.hku.hk/~daniely/lung_microarray/.
Data re-scaling and feature selection
The raw expression data from each sample was rescaled (normalized) to account for systematic differences in signal intensities among the microarrays, using standard procedures in Affymetrix Microarray Suite 5.0.1. Expression values from each microarray were multiplied by a scaling factor to make the average intensity of a set of house keeping genes on each microarray equal to an arbitrarily defined target intensity of 500.
To identify genes that are tissue phenotype related, the mean expression level of all genes in normal tissues and in adenocarcinoma tissues were calculated. If the ratio of the average expression levels of a gene between the two tissue classes exceeded 2-fold, the genes were included in the set MAD.
Gene to tissue correlation
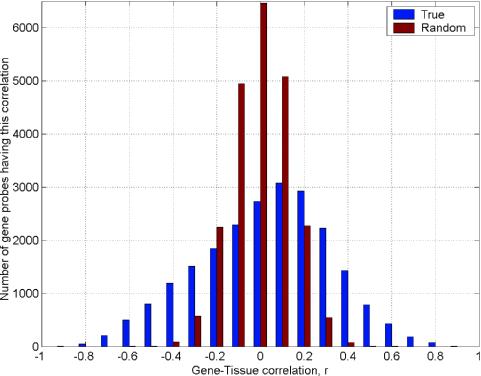
The tissue type distinction is represented by an idealized expression pattern (a vector with size 1 × 58), in which the expression is labeled uniformly high (value = 1) in adenocarcinoma tissue type and labeled uniformly low (value = 0) in normal tissue class. Correlation coefficients were calculated for the comparison of this vector with the expression profiles of each gene in MAD. The distribution of correlation coefficients was counted in bins of 0.2. The result was compared to the corresponding distribution obtained for ten random permutations of the idealized tissue labels to give the average random correlation coefficients for each gene (Figure 1).
Figure 1.
Histogram of the cancer associated genes (MAD) correlation to the tissue labels (normal or lung adenocarcinomas). The average histograms generated from 10 separate random permutations of the cancer labels in the original lung adenocarcinoma dataset is also displayed.
Determination of overrepresentation of gene ontology terms in the set MAD
GeneOntology (http://www.geneontology.org/) terms, which classify a gene according to its molecular function, biological process, cellular component and chromosomal localization, were collected for each gene on the Affymetrix HG-U133A microarray from the Affymetrix library files. By using the hypergeometric distribution (Equation 1), genes with each of these functional annotations could be assessed to see if they are overrepresented in the set MAD. Given G annotated genes on a microarray, of which A have a certain function (gene ontology term), and a set of k genes selected independently of the functional annotations (MAD), the probability that n or more of the set of k genes have this function can be calculated by Equation 1 (23). If the P-value of observing the number of genes with a particular gene ontology term in the set MAD was <0.001, the term was considered to be significantly overrepresented in the set MAD. DNA-Chip Analyzer (dChip) (29) was used to perform this task.
| 1 |
Constructing gene relationship trees for overrepresented gene ontology terms
For all possible combinations of gene pairs that belong to each gene ontology term overrepresented in MAD the correlation coefficient, r, of their expression profiles was calculated. A pairwise gene distance matrix Mdistance, using the distance 1-r was formed for the genes. The neighbor-joining algorithm (NJ) (30) was used to construct a gene relationship tree from pairwise gene distance matrix. This was performed to identify gene neighbors whose expression values followed a common trend. The NJ algorithm is a special case of the star decomposition method. Starting from a star tree, the final relationship tree is constructed systematically by linking the least distant pair of nodes (genes in this case). The main advantage of the algorithm is that it permits lineages with largely different branch lengths. The programming script for computing r was implemented in the MatLab technical programming language and the tree was calculated using MEGA2 (31).
Extraction of the upstream regions for putatively co-regulated gene sets
Putatively co-regulated genes from each gene ontology term that was overrepresented in MAD were selected in accordance with two criteria: (i) a distance metric cutoff value (di,j < 0.20) for all pairwise gene distances within the selected N members of the gene set; and (ii) the minimum mean aggregated pairwise distances [] for the selected N members of the gene set. The rationale for choosing these criteria was to find a single most correlated gene cluster that minimizes the total branch length di,j. For instance, if there are two gene clusters (each constituted of four and five gene members, respectively) in the tree topology found to be satisfying criterion one, i.e. get sets in which all pairwise gene distances (4C2 = 6 and 5C2 = 10 distances, respectively) satisfy the distance metric cutoff value <0.2, the final gene set selected should be the one with the minimum mean aggregated pairwise distances (criterion two). As a result, a different numbers of genes will be selected from each gene ontology term based on these criteria. For each of the selected genes, the corresponding 5 kb region located directly upstream of the transcription start site was extracted as described previously (32). Several sequence features including sequence gaps, continuity, consistency between the two distinct drafts of human genomes (3,33,34) were taken into consideration. Detailed information can be found in (32).
Identification of conserved regions and detection of associated transcription factors
All 5 kb unaligned DNA sequences associated with each gene ontology term group overrepresented in MAD, were searched using MoDEL (26), to reveal possible highly conserved DNA regions. MoDEL employs an evolutionary algorithm and hill-climbing optimization for global and local exploration of two targeted search spaces, respectively (all possible words and all possible ungapped local multiple alignments). This heuristic algorithm has been shown to have more efficient optimization capabilities than other motif discovery tools (26). The word size was set to be 50 bp in the present study because we found that the conserved regions identified by MoDEL remained rather consistent with different sizes of word or segment length. A 50 bp segment length (the longest implemented in MoDEL) also allows a larger window, whereby the most conserved motifs can be captured together with their less similar surrounding residues. The information content for all conserved regions identified was calculated based on the Kullback–Leibler divergence (relative entropy).
All conserved regions identified by MoDEL were scanned against all vertebrate transcription factor position weight matrix profiles contained in the TRANSFAC database version 8.3 (35) to identify all previously known transcription binding sites. To incorporate stronger matches of transcription factor binding sites, stringent settings for the Match program (36) were employed. Both the core matrix and overall matrix similarity were required to be least 0.9 to be considered a match.
RESULTS
Selection of the cancer associated gene set MAD
A total of 3442 genes were found to be either up- or down-regulated by more than 2-fold between the normal and adenocarcinoma tissue sets (Table 1). These genes formed the cancer associated gene set MAD. Of these genes, 1294 showed down-regulation and 2148 showed up-regulation of gene expression levels in adenocarcinomas. At the extreme ends of the fold change range, the receptor for advanced glycation end product (RAGE) was found to be repressed by >32-fold in adenocarcinomas while the D G antigen (GAGED2) was found to be up-regulated by >128-fold. Real-time quantitative RT–PCR analysis (Supplementary Materials) to verify the mRNA transcript levels for carbonic anhydrase IV (CA4) and RAGE were performed in 14 independent tissue samples (seven samples from each tissue phenotype). The abundance of mRNA transcripts for both genes was extremely low in the adenocarcinoma samples. If a gene is not expressed or expressed at very low levels in a sample, then fold change values may become large due to the low denominator. Fold change values must be considered in conjunction with expression levels.
Table 1.
Genes that were identified to be down- or up-regulated in adenocarcinomas
| Gene description (Gene down-regulated in lung AD) | Probe set | Fold log(AD/N) | Mean expression for normal lung | Mean expression for AD lung |
|---|---|---|---|---|
| Consensus sequence for Homo sapiens mRNA for receptor for Advanced Glycation End Product (RAGE) | 217046_s_at | −5.523 | 942.82 | 20.51 |
| Homo sapiens fatty acid binding protein 4, adipocyte (FABP4) | 203980_at | −4.768 | 3365.42 | 123.48 |
| Human alpha-globin gene with flanks | 217414_x_at | −4.419 | 9787.41 | 457.42 |
| Homo sapiens mRNA; cDNA DKFZp564N0582 (from clone DKFZp564N0582) | 209074_s_at | −4.294 | 678.28 | 34.58 |
| Homo sapiens carbonic anhydrase IV (CA4) | 206208_at | −4.276 | 275.78 | 14.24 |
| Homo sapiens RAGE mRNA for advanced glycation endproducts receptor, whole CDS | 210081_at | −4.261 | 1593.08 | 83.06 |
| Homo sapiens ficolin (collagen fibrinogen domain-containing) 3 (Hakata antigen) (FCN3) | 205866_at | −4.166 | 1790.33 | 99.70 |
| Human sickle cell beta-globin mRNA | 209116_x_at | −4.155 | 14733.26 | 827.29 |
| Consensus includes gb:BF939489 | 209469_at | −4.028 | 330.68 | 20.27 |
| Homo sapiens hemoglobin, gamma A (HBG1) | 204848_x_at | −3.922 | 264.79 | 17.47 |
| Homo sapiens adipose specific 2 (APM2) | 203571_s_at | −3.898 | 3042.43 | 204.08 |
| Homo sapiens hypothetical protein FLJ10970 (FLJ10970) | 219230_at | −3.884 | 1521.30 | 103.06 |
| Consensus includes gb:T50399/UG=Hs.251577 hemoglobin, alpha 1 | 214414_x_at | −3.874 | 13447.88 | 917.38 |
| Homo sapiens colony stimulating factor 3 (granulocyte) (CSF3) | 207442_at | −3.873 | 145.31 | 9.92 |
| Homo sapiens mutant beta-globin (HBB) gene | 217232_x_at | −3.864 | 15087.91 | 1036.22 |
| Gene description (Gene up-regulated in lung AD) | Probe Set | Fold log(AD/N) | ||
| Homo sapiens XAGE-1 protein (XAGE-1) | 220057_at | 7.311 | 4.79 | 760.58 |
| Human alpha-1 type XI collagen (COL11A1) | 37892_at | 6.208 | 6.10 | 451.07 |
| Consensus includes gb:AI697108;/UG=Hs.102482 mucin 5, subtype B, tracheobronchial | 213432_at | 6.192 | 4.84 | 354.11 |
| Homo sapiens dipeptidyl peptidase IV (DPP4) | 203716_s_at | 5.932 | 6.81 | 415.78 |
| Consensus includes gb:AU159942;/UG=Hs.156346 topoisomerase (DNA) II alpha (170 kDa) | 201291_s_at | 5.620 | 3.24 | 159.61 |
| Homo sapiens serine protease inhibitor, Kazal type 1 (SPINK1);/UG=Hs.181286 serine protease inhibitor, Kazal type 1 | 206239_s_at | 5.274 | 51.74 | 2002.30 |
| Consensus includes gb:X98568;/UG=Hs.179729 collagen, type X, alpha 1 (Schmid metaphyseal chondrodysplasia) | 217428_s_at | 4.991 | 21.98 | 698.84 |
| Consensus includes gb:AW192795;/UG=Hs.103707 apomucin | 214303_x_at | 4.969 | 5.26 | 164.65 |
| Human nephropontin mRNA;/UG=Hs.313 secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 209875_s_at | 4.851 | 121.29 | 3499.34 |
| Homo sapiens matrix metalloproteinase 1 (interstitial collagenase) (MMP1) | 204475_at | 4.806 | 25.07 | 701.09 |
| Homo sapiens neuromedin U (NMU) | 206023_at | 4.776 | 5.80 | 158.88 |
| Homo sapiens cytokine receptor-like factor 1 (CRLF1) | 206315_at | 4.737 | 14.22 | 379.16 |
| Homo sapiens, serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 3 | 209720_s_at | 4.597 | 2.13 | 51.63 |
| Homo sapiens multidrug resistance-associated protein homolog MRP3 (MRP3);/UG=Hs.90786 ATP-binding cassette, sub-family C (CFTRMRP), member 3 | 209641_s_at | 4.570 | 14.73 | 350.01 |
| Consensus includes gb:BE791251;/UG=Hs.25640 claudin 3 | 203953_s_at | 4.462 | 6.82 | 150.39 |
The description of each gene, its probe set in HG-U133A GeneChip and log fold change are given in the table. The complete table can be downloaded at http://bioinfo.hku.hk/~daniely/lung_microarray.
Functional annotation groups significantly overrepresented in MAD
Down- and up-regulated genes in MAD were treated separately to detect functional annotation groups that may be overrepresented in adenocarcinoma associated genes. Tables 2 and 3, respectively, give the gene ontology terms significantly overrepresented (P < 0.001) in down- and up-regulated genes of MAD. The tables give the number of genes with that gene ontology term on the HG-U133A microarray, the number found, and the P-value of finding at least that number of genes (by random chance) in MAD.
Table 2.
The gene ontology terms overrepresented in the set of genes down-regulated by at least 2-fold in adenocarcinomas
| Annotation term | Total | Found | Expected | P-value |
|---|---|---|---|---|
| GeneOntology terms | ||||
| Globin | 17 | 12 | 0.0E+00 | 0.0E+00 |
| Rhodopsin-like receptor activity | 384 | 49 | 3.8E−04 | 1.0E−06 |
| G-protein chemoattractant receptor activity | 34 | 8 | 2.8E−02 | 8.4E−04 |
| Peptide receptor activity | 139 | 23 | 1.7E−03 | 1.2E−05 |
| G-protein-coupled receptor binding | 52 | 21 | 0.0E+00 | 0.0E+00 |
| Defense/immunity protein activity | 230 | 39 | 0.0E+00 | 0.0E+00 |
| Antimicrobial peptide activity | 32 | 8 | 1.7E−02 | 5.4E−04 |
| Complement activity | 32 | 8 | 1.7E−02 | 5.4E−04 |
| Signal transducer activity | 2558 | 253 | 0.0E+00 | 0.0E+00 |
| Receptor activity | 1542 | 162 | 0.0E+00 | 0.0E+00 |
| Transmembrane receptor activity | 1083 | 121 | 0.0E+00 | 0.0E+00 |
| G-protein coupled receptor activity | 467 | 61 | 0.0E+00 | 0.0E+00 |
| Chemokine receptor activity | 34 | 8 | 2.8E−02 | 8.4E−04 |
| Receptor binding | 592 | 72 | 0.0E+00 | 0.0E+00 |
| Cytokine activity | 253 | 39 | 0.0E+00 | 0.0E+00 |
| Heavy metal binding | 23 | 8 | 9.4E−04 | 4.1E−05 |
| Sugar binding | 132 | 28 | 0.0E+00 | 0.0E+00 |
| Extracellular | 1085 | 138 | 0.0E+00 | 0.0E+00 |
| Extracellular space | 457 | 72 | 0.0E+00 | 0.0E+00 |
| Hemoglobin complex | 18 | 12 | 0.0E+00 | 0.0E+00 |
| Plasma membrane | 2297 | 219 | 0.0E+00 | 0.0E+00 |
| Integral to plasma membrane | 1702 | 176 | 0.0E+00 | 0.0E+00 |
| Oxygen and reactive oxygen species metabolism | 65 | 15 | 4.6E−04 | 7.0E−06 |
| Calcium ion homeostasis | 26 | 8 | 2.9E−03 | 1.1E−04 |
| Cell motility | 414 | 50 | 1.2E−03 | 3.0E−06 |
| Chemotaxis | 133 | 39 | 0.0E+00 | 0.0E+00 |
| Muscle contraction | 202 | 25 | 1.3E−01 | 6.2E−04 |
| Response to stress | 1025 | 143 | 0.0E+00 | 0.0E+00 |
| Defense response | 1031 | 169 | 0.0E+00 | 0.0E+00 |
| Inflammatory response | 218 | 50 | 0.0E+00 | 0.0E+00 |
| Immune response | 950 | 153 | 0.0E+00 | 0.0E+00 |
| Humoral immune response | 235 | 38 | 0.0E+00 | 0.0E+00 |
| Antimicrobial humoral response (sensu Invertebrata) | 145 | 24 | 1.2E−03 | 8.0E−06 |
| Cellular defense response | 139 | 45 | 0.0E+00 | 0.0E+00 |
| Cell communication | 3667 | 326 | 0.0E+00 | 0.0E+00 |
| Cell adhesion | 658 | 84 | 0.0E+00 | 0.0E+00 |
| Heterophilic cell adhesion | 97 | 20 | 9.7E−05 | 1.0E−06 |
| Signal transduction | 2947 | 254 | 0.0E+00 | 0.0E+00 |
| Cell surface receptor linked signal transduction | 1124 | 117 | 0.0E+00 | 0.0E+00 |
| G-protein coupled receptor protein signaling pathway | 657 | 77 | 0.0E+00 | 0.0E+00 |
| Cytosolic calcium ion concentration elevation | 49 | 10 | 3.2E−02 | 6.5E−04 |
| Cell-cell signaling | 689 | 64 | 3.7E−01 | 5.4E−04 |
| Development | 1920 | 150 | 1.5E+00 | 8.1E−04 |
| Histogenesis and organogenesis | 125 | 18 | 7.5E−02 | 6.0E−04 |
| Muscle development | 167 | 27 | 5.0E−04 | 3.0E−06 |
| Respiratory gaseous exchange | 36 | 11 | 2.2E−04 | 6.0E−06 |
| Chemokine activity | 52 | 21 | 0.0E+00 | 0.0E+00 |
| Circulation | 142 | 22 | 7.2E−03 | 5.1E−05 |
| Peptide receptor activity/G-protein coupled | 139 | 23 | 1.7E−03 | 1.2E−05 |
| Response to external stimulus | 1591 | 210 | 0.0E+00 | 0.0E+00 |
| Response to biotic stimulus | 1126 | 179 | 0.0E+00 | 0.0E+00 |
| Response to wounding | 356 | 91 | 0.0E+00 | 0.0E+00 |
| Response to pest/pathogen/parasite | 596 | 123 | 0.0E+00 | 0.0E+00 |
| Response to bacteria | 19 | 6 | 1.3E−02 | 7.1E−04 |
| Response to abiotic stimulus | 577 | 71 | 0.0E+00 | 0.0E+00 |
| Morphogenesis | 1119 | 101 | 4.9E−02 | 4.4E−05 |
| Organogenesis | 1029 | 91 | 2.2E−01 | 2.2E−04 |
| Cellular process | 7140 | 534 | 0.0E+00 | 0.0E+00 |
| Membrane | 4225 | 356 | 0.0E+00 | 0.0E+00 |
| Integral to membrane | 3220 | 281 | 0.0E+00 | 0.0E+00 |
| Cell growth | 97 | 17 | 7.3E−03 | 7.5E−05 |
| Humoral defense mechanism (sensu Invertebrata) | 145 | 24 | 1.2E−03 | 8.0E−06 |
| Cell–cell adhesion | 220 | 30 | 6.8E−03 | 3.1E−05 |
| Antimicrobial humoral response | 145 | 24 | 1.2E−03 | 8.0E−06 |
| Cytolysis | 20 | 8 | 2.4E−04 | 1.2E−05 |
| Cytokine binding | 80 | 14 | 2.6E−02 | 3.2E−04 |
| Chemokine binding | 34 | 8 | 2.8E−02 | 8.4E−04 |
| Carbohydrate binding | 133 | 28 | 0.0E+00 | 0.0E+00 |
| Chemoattractant activity | 52 | 21 | 0.0E+00 | 0.0E+00 |
| Response to chemical substance | 206 | 48 | 0.0E+00 | 0.0E+00 |
| Peptide binding | 213 | 26 | 1.3E−01 | 6.1E−04 |
| Taxis | 133 | 39 | 0.0E+00 | 0.0E+00 |
| Chemokine receptor binding | 52 | 21 | 0.0E+00 | 0.0E+00 |
| Innate immune response | 220 | 50 | 0.0E+00 | 0.0E+00 |
| Eicosanoid biosynthesis | 25 | 7 | 1.4E−02 | 5.7E−04 |
| Protein domain | ||||
| Vertebrate metallothionein | 12 | 7 | 2.4E−05 | 2.0E−06 |
| Aspartic acid and asparagine hydroxylation site | 143 | 21 | 3.5E−02 | 2.5E−04 |
| Rhodopsin-like GPCR superfamily | 289 | 42 | 0.0E+00 | 0.0E+00 |
| Endothelin receptor | 6 | 4 | 1.3E−03 | 2.1E−04 |
| Small chemokine, C-C subfamily | 26 | 11 | 0.0E+00 | 0.0E+00 |
| Fos transforming protein | 13 | 6 | 9.5E−04 | 7.3E−05 |
| Thrombospondin, type I | 52 | 13 | 7.8E−04 | 1.5E−05 |
| Globin | 16 | 12 | 0.0E+00 | 0.0E+00 |
| Small chemokine, C-X-C subfamily | 18 | 6 | 1.1E−02 | 6.0E−04 |
| C-type lectin | 95 | 19 | 5.7E−04 | 6.0E−06 |
| Alpha crystallin | 8 | 4 | 7.2E−03 | 9.0E−04 |
| Myelin proteolipid protein (PLP) | 7 | 6 | 0.0E+00 | 0.0E+00 |
| Zn-binding protein, LIM | 95 | 18 | 2.2E−03 | 2.3E−05 |
| Small chemokine, interleukin-8 like | 48 | 20 | 0.0E+00 | 0.0E+00 |
| EGF-like calcium-binding | 147 | 21 | 5.3E−02 | 3.6E−04 |
| Heat shock protein Hsp20 | 8 | 4 | 7.2E−03 | 9.0E−04 |
| Fibrinogen, beta/gamma chain, C-terminal globular | 38 | 10 | 3.4E−03 | 8.9E−05 |
| P2 purinoceptor | 21 | 7 | 4.3E−03 | 2.1E−04 |
| Myoglobin | 9 | 6 | 3.6E−05 | 4.0E−06 |
| Beta haemoglobin | 8 | 7 | 0.0E+00 | 0.0E+00 |
| Alpha haemoglobin | 6 | 5 | 3.6E−05 | 6.0E−06 |
| Pi haemoglobin | 6 | 5 | 3.6E−05 | 6.0E−06 |
| Small chemokine, C-X-C/Interleukin 8 | 18 | 8 | 1.1E−04 | 6.0E−06 |
| Metallothionein superfamily | 12 | 7 | 2.4E−05 | 2.0E−06 |
| Orphan nuclear receptor | 9 | 5 | 9.1E−04 | 1.0E−04 |
| Immunoglobulin C-2 type | 223 | 31 | 6.2E−03 | 2.8E−05 |
| Immunoglobulin subtype | 368 | 48 | 3.7E−04 | 1.0E−06 |
| PMP-22/EMP/MP20 family | 8 | 4 | 7.2E−03 | 9.0E−04 |
| L1 transposable element | 8 | 4 | 7.2E−03 | 9.0E−04 |
| EGF-like domain | 431 | 46 | 1.4E−01 | 3.3E−04 |
| Type I EGF | 169 | 23 | 6.7E−02 | 4.0E−04 |
| AIG1 family | 6 | 4 | 1.3E−03 | 2.1E−04 |
| BRICHOS domain | 13 | 8 | 0.0E+00 | 0.0E+00 |
| Immunoglobulin-like | 678 | 75 | 6.8E−04 | 1.0E−06 |
| LST-1 | 6 | 6 | 0.0E+00 | 0.0E+00 |
| Thrombospondin, subtype 1 | 27 | 8 | 5.0E−03 | 1.8E−04 |
| Saposin-like type B, 2 | 7 | 5 | 1.3E−04 | 1.9E−05 |
| Saposin B | 12 | 5 | 6.5E−03 | 5.4E−04 |
| Pathway | ||||
| GPCRs_Class_A_Rhodopsin-like | 212 | 34 | 0.0E+00 | 0.0E+00 |
| Peptide_GPCRs | 88 | 20 | 0.0E+00 | 0.0E+00 |
| MAP00590//Prostaglandin and leukotriene metabolism | 41 | 9 | 3.2E−02 | 7.7E−04 |
| GPCRs_Class_B_Secretin-like | 34 | 10 | 9.2E−04 | 2.7E−05 |
| Chromosomal location | ||||
| 12p | 301 | 32 | 2.4E−01 | 8.1E−04 |
| 8p21 | 117 | 18 | 1.8E−02 | 1.5E−04 |
| 17q23 | 68 | 14 | 2.2E−03 | 3.2E−05 |
| 16q13 | 37 | 12 | 3.7E−05 | 1.0E−06 |
For each gene ontology term, the total number of genes with this term in the HG-U133A GeneChip, the total number of genes carrying that term in MAD, the P-value of this and the expected number of genes are tabulated. The member genes for each gene ontology term can be downloaded at http://bioinfo.hku.hk/~daniely/lung_microarray.
Table 3.
The gene ontology terms overrepresented in the set of genes up-regulated by at least 2-fold in adenocarcinomas
| Annotation term | Total | Found | Expected | P-value |
|---|---|---|---|---|
| Gene Ontology term | ||||
| DNA replication and chromosome cycle | 233 | 54 | 0.0E+00 | 0.0E+00 |
| Cell cycle checkpoint | 50 | 17 | 5.5E−04 | 1.1E−05 |
| S phase of mitotic cell cycle | 183 | 38 | 1.1E−02 | 6.1E−05 |
| M phase of mitotic cell cycle | 149 | 46 | 0.0E+00 | 0.0E+00 |
| Nucleotide binding | 1737 | 235 | 2.1E−01 | 1.2E−04 |
| Mitotic cell cycle | 421 | 97 | 0.0E+00 | 0.0E+00 |
| M phase | 201 | 52 | 0.0E+00 | 0.0E+00 |
| Nuclear division | 195 | 50 | 0.0E+00 | 0.0E+00 |
| Chromatin | 117 | 29 | 1.8E−03 | 1.5E−05 |
| Nucleosome | 60 | 16 | 3.0E−02 | 5.0E−04 |
| Cytokinesis | 85 | 24 | 6.8E−04 | 8.0E−06 |
| Catalytic activity | 4887 | 638 | 0.0E+00 | 0.0E+00 |
| Carboxypeptidase A activity | 18 | 8 | 5.5E−03 | 3.1E−04 |
| Extracellular matrix structural constituent | 89 | 21 | 4.0E−02 | 4.5E−04 |
| Collagen | 54 | 23 | 0.0E+00 | 0.0E+00 |
| ATP binding | 1280 | 177 | 4.0E−01 | 3.1E−04 |
| Extracellular matrix | 345 | 65 | 2.1E−03 | 6.0E−06 |
| Collagen | 59 | 24 | 0.0E+00 | 0.0E+00 |
| Fibrillar collagen | 23 | 14 | 0.0E+00 | 0.0E+00 |
| Chromosome | 147 | 32 | 1.3E−02 | 8.8E−05 |
| Spindle | 64 | 21 | 1.3E−04 | 2.0E−06 |
| Intermediate filament | 76 | 19 | 2.9E−02 | 3.8E−04 |
| DNA metabolism | 606 | 97 | 3.1E−02 | 5.1E−05 |
| DNA replication | 178 | 36 | 2.9E−02 | 1.6E−04 |
| DNA dependent DNA replication | 94 | 23 | 1.3E−02 | 1.4E−04 |
| DNA replication initiation | 25 | 11 | 6.3E−04 | 2.5E−05 |
| Amino acid and derivative metabolism | 240 | 54 | 0.0E+00 | 0.0E+00 |
| Amino acid metabolism | 197 | 43 | 1.2E−03 | 6.0E−06 |
| Oncogenesis | 521 | 84 | 6.6E−02 | 1.3E−04 |
| Cell cycle | 871 | 145 | 0.0E+00 | 0.0E+00 |
| Chromosome segregation | 35 | 11 | 2.9E−02 | 8.4E−04 |
| Mitosis | 145 | 45 | 0.0E+00 | 0.0E+00 |
| Regulation of mitosis | 35 | 12 | 6.9E−03 | 2.0E−04 |
| Mitotic checkpoint | 16 | 7 | 1.3E−02 | 8.3E−04 |
| Ectoderm development | 98 | 26 | 1.1E−03 | 1.1E−05 |
| Cell proliferation | 1356 | 190 | 1.2E−01 | 8.9E−05 |
| Epidermal differentiation | 80 | 22 | 2.2E−03 | 2.8E−05 |
| Glutamine family amino acid metabolism | 46 | 15 | 2.9E−03 | 6.3E−05 |
| Amine metabolism | 283 | 60 | 0.0E+00 | 0.0E+00 |
| Histogenesis | 131 | 28 | 4.3E−02 | 3.3E−04 |
| Glucuronosyltransferase activity | 18 | 8 | 5.5E−03 | 3.1E−04 |
| Transferase activity | 1634 | 224 | 1.3E−01 | 8.1E−05 |
| Transferase activitytransferring glycosyl groups | 225 | 42 | 7.0E−02 | 3.1E−04 |
| Transferase activitytransferring hexosyl groups | 148 | 34 | 2.5E−03 | 1.7E−05 |
| Other carbon–nitrogen ligase activity | 25 | 9 | 2.1E−02 | 8.3E−04 |
| Purine nucleotide binding | 1723 | 233 | 2.3E−01 | 1.4E−04 |
| Adenyl nucleotide binding | 1292 | 179 | 3.4E−01 | 2.6E−04 |
| Intermediate filament cytoskeleton | 76 | 19 | 2.9E−02 | 3.8E−04 |
| Protein domain | ||||
| Fibrillar collagen, C-terminal | 23 | 14 | 0.0E+00 | 0.0E+00 |
| Endoplasmic reticulum targeting sequence | 76 | 19 | 2.0E−02 | 2.6E−04 |
| Epsin N-terminal homology | 16 | 7 | 1.1E−02 | 6.9E−04 |
| MCM family | 11 | 8 | 2.2E−05 | 2.0E−06 |
| Prolyl oligopeptidase | 12 | 6 | 8.6E−03 | 7.1E−04 |
| Intermediate filament protein | 67 | 17 | 3.0E−02 | 4.4E−04 |
| von Willebrand factor, type D | 9 | 6 | 7.7E−04 | 8.6E−05 |
| UDP-glucoronosyl/UDP-glucosyl transferase | 15 | 7 | 6.4E−03 | 4.3E−04 |
| Prolyl endopeptidase, serine active site | 8 | 5 | 4.4E−03 | 5.5E−04 |
| Immunoglobulin V-type | 146 | 32 | 6.3E−03 | 4.3E−05 |
| Cyclin, C-terminal | 19 | 9 | 1.0E−03 | 5.4E−05 |
| Disulphide isomerase | 12 | 7 | 8.4E−04 | 7.0E−05 |
| Cyclin | 44 | 14 | 4.7E−03 | 1.1E−04 |
| Cyclin, N-terminal domain | 34 | 12 | 3.7E−03 | 1.1E−04 |
| Histone core | 25 | 9 | 1.7E−02 | 6.7E−04 |
| Collagen triple helix repeat | 106 | 28 | 3.2E−04 | 3.0E−06 |
| Collagen helix repeat | 69 | 22 | 6.9E−05 | 1.0E−06 |
| Pathway | ||||
| Cell_cycle | 133 | 43 | 5.7E+00 | 4.3E−02 |
| Glutamate_metabolism | 27 | 12 | 3.2E−01 | 1.2E−02 |
| MAP00251//glutamate metabolism | 43 | 15 | 6.5E−01 | 1.5E−02 |
| Androgen_and_estrogen_metabolism | 15 | 7 | 1.1E−01 | 7.0E−03 |
| MAP00150//androgen and estrogen metabolism | 32 | 11 | 3.5E−01 | 1.1E−02 |
| Chromosomal location | ||||
| 7 | 961 | 129 | 8.7E−01 | 9.1E−04 |
| 1q | 920 | 126 | 4.4E−01 | 4.8E−04 |
| 8q | 388 | 67 | 5.8E−03 | 1.5E−05 |
Details are as described in Table 2.
For genes down-regulated in adenocarcinomas, several gene ontology terms related to immune responses were overrepresented, indicating that there appeared to be a depression in defense mechanisms in general, for the adenocarcinoma tissue samples (Table 2). In addition, genes associated with ‘signal transducer activity’ (e.g. TEK tyrosine kinase, G protein-coupled receptor kinase) were also identified to be significantly overrepresented in down-regulated genes in MAD, suggesting the blockage of signal transduction genes in adenocarcinoma cells. Many gene ontology terms that were overrepresented in the up-regulated genes of MAD were associated with the cell cycle and cell replication machinery (Table 3) as might be expected from accelerated cancer cell proliferation.
Construction of relationship trees and determination of putatively co-regulated genes
After obtaining the constituent member genes for each gene ontology term overrepresented in MAD, we investigated their pairwise gene expression relationships. Supplementary Material figure 2 shows an example of such a study for the gene ontology term ‘DNA replication and chromosomal cycle’ with the GenBank accession numbers for each tree branch corresponding to the genes in MAD that are assigned this ontology term. The branch distances displayed were used to derive the putatively co-regulated gene set (marked by an asterisk) according to the two criteria stated in the Materials and Methods section. In this example, the putatively co-regulated genes were: (i) MCM2—mini-chromosome maintenance deficient 2; (ii) replication factor C (activator 1) 4; and (iii) CDC45—cell division cycle 45-like.
Identification of conserved DNA motifs and transcription factors associated with a GO term
Conserved regions, within 5 kb of the transcription start site, of the putatively co-regulated genes associated with each gene ontology term overrepresented in MAD were identified using MoDEL (30). Example results from four gene ontology terms: (i) DNA replication and chromosomal cycle; (ii) nuclear division; (iii) cellular defense response and (iv) signal transduction, are shown in Table 4. The first two terms are associated with genes that were up-regulated in adenocarcinoma tissues, whereas the latter two terms are associated with down-regulated genes. Conserved regions are presented using IUPAC uncertainty codes, with highly conserved residues shown in bold, along with their start position relative to the transcription start site. The occurrence of each of these 50mers in regions 5 kb upstream of all human genes (32) is shown along with the proportion of those genes that have the same GO term and regulation pattern of the gene in the table. The final column reports the transcription factors (from TRANSFAC 8.3) that may bind to the conserved region based on matches to their binding site motifs. The complete data for Table 4 can be found at http://bioinfo.hku.hk/~daniely/lung_microarray/.
Table 4.
Highly conserved DNA regions, detected with MoDEL, in regions 5 kb directly upstream of the transcription start site in putatively co-regulated gene sets
| Gene Ontology | Locationb | Frequencyc | Similar regulationd | Putative transcription factorse | |||
|---|---|---|---|---|---|---|---|
| Gene up-regulated in lung adenocarcinomas cells | |||||||
| DNA replication and chromosome cyclea | |||||||
| NM 004526 | gggcgt GGTGGCTCACGCCTGTAATCCTAGCACTATGGGAGGCCAAGGCAGGCGGAt ccact | −2467 | 1 | 100% | Whn,AhR,GATA-1,PITX2,HIF-1 | ||
| NM 002916 | aggcac GGTGGCTCACACCTGTAATCCCAGCACTTTGGGAGGCCAAGGTGGGTGGAt gccga | −2920 | 7 | 75% | MyoD,E47,AREB6,E12,USF,GATA-1,PITX2 | ||
| NM 003504 | gggtgc GGTGGCTCACGCCTATAATCCCAGCACTTTGGGAGGCAGAGGCAGGTGGA ttacct | −3179 | 1 | 100% | Whn,AhR,GATA-1,PITX2,Major,MyoD,E47,TFII-I,AREB6,TAL1,E12,USF,HIF-1 | ||
| Profile | RGGYRY GGTGGCTCACRCCTRTAATCCYAGCACTWTGGGAGGCMRAGGYRGGYGGA TBMMSW | ||||||
| Nuclear divisiona | |||||||
| NM 004701 | agtagt CCCAGCTACTCGGGTGGCTGAGGTAGGAGGATCACTTGAGCCCGGGGGAT tgaggc | −3735 | 1 | 100% | HES1,SREBP-1,TFII-I,DEC,USF,Nkx2-5,GATA-1 | ||
| NM 001211 | tatagt CCCAGCTACATGGGAGGATGAGGCAGGAAGATCGCTTGAACCTGGGAGGT ggaggt | −3705 | 1 | 100% | TFII-I,Elk-1,NERF1a,c-Ets-1(p54),68,AREB6 | ||
| NM 020242 | tgtagt CCCAGCTGCTTGAGAGGCTGAGGCAGGAGGATCACTTGAGCCCAGGAGGT caaggc | −2016 | 1 | 100% | TAL1,HEB,TFII-I,Zta,c-Ets-1(p54),DEC,SREBP-1,USF,Nkx2-5,RORalpha1 | ||
| NM 022346 | ctgtaa CCCAGCTACTTGGGAGACTGAGGCGGGAGAATCGCTTCAACCCGGGAGGC agaggt | −2071 | 1 | 100% | |||
| NM 003981 | tgtaat CTGAGCTACTTGGGAGGCTGAAGCAGGAGAATCACTTGAACCTGGGAGGC ggaggt | −866 | 1 | 100% | TFII-I,DEC,Nkx2-2,SREBP-1,USF,Nkx2-5,AREB6,HIF-1 | ||
| NM 001237 | ttgaac TGCAAGAACAGCCGCCGCTCCGGCGGGCTGCTCGCTGCATCTCTGGGCGT ctttgg | −167 | 1 | 100% | |||
| NM 001813 | tgtaat CCCAGCTACTGGGGAGGCTGAGGCAGGAGAATCACTTGAATGCAGGAGGT ggaggc | −715 | 1 | 100% | TFII-I,Zta,DEC,Nkx2-2,SREBP-1,USF,TTF1,Nkx2-5,c-Ets-1,c-Ets-1(p54),AREB6 | ||
| NM 002358 | tgtagt CTCAGCTACTTGGGAGTCCGAGGCAGGAGAATTGCTTGAACCTGGGAGGC agaggt | −4881 | 1 | 100% | TFII-I,C/EBPdelta,AREB6 | ||
| NM 022346 | ctgtaa CCCAGCTACTTGGGAGACTGAGGCGGGAGAATCGCTTCAACCCGGGAGGC agaggt | −2071 | 1 | 100% | |||
| Profile | HDKWRH YBSARSWRCWBSVGHSDMYSMRGYRGGMDRMTYRCTKSADYBYDGGRSRY NDWKGB | ||||||
| Gene down-regulated in lung adenocarcinomas cells | |||||||
| Cellular defense responsea | |||||||
| NM 005874 | cttgat GGTCCCGGGACCCTGTGGCATCTCACCTCTGGCCTCTGTTCTTTCTTGTG agtccg | −715 | 1 | 100% | USF,AREB6,GR | ||
| NM 000265 | tggcag GATCTCGGCTCACTGCAACCTCCACCTCCCTGGTTCAAGTGATTCTCCTG tcttac | −3997 | 2 | 100% | RFX1,TFII-I,AREB6,DEC,SREBP-1,Nkx2-2,Nkx2-5,USF, HIF-1 | ||
| NM 016382 | tggcgt GATCTCGGCTCACTGCAACCTCCACCTCCTGGATTCAAGTGATTCTCCTG cctcag | −3907 | 1 | 100% | RFX1,TFII-I,AREB6,c-Ets-1(p54),GATA-1,DEC,TTF1,SREBP-1,Nkx2-2,Nkx2-5,USF,Zta | ||
| Profile | YKKSRK GRTCYCGGSWCMCTGYRRCMTCYMMCYYCYKGVYTCWRKTSWTTCTYSTG HSTYMS | ||||||
| Signal transductiona | |||||||
| NM 000459 | tcagga GGCTGAGGCAGAAGAATCGCTTGAACCCAGGAGGCGGACGTTGCAGTGAG ccgaga | −2597 | 1 | 100% | Zta,c-Ets-1(p54),RFX1 | ||
| NM 005308 | ttggga GGCTGAAGTACAAGAATCATTTGAACCTGGGAGGCCGAGGTTGCAGTGAG ccgaga | −2401 | 1 | 100% | GATA-1,AREB6,RFX1 | ||
| NM 000115 | ttggga GGCTGAGGCAGGAGAATCACTTGAACCTGGGAGCCGGAGGTTGCAGTGAG ctgaga | −1967 | 2 | 50% | TFII-I,Zta,DEC,SREBP-1,Nkx2-2,USF,Nkx2-5,AREB6,c-Ets-1(p54) | ||
| NM 005424 | gccagt GGTGGCAAGAGGTGGAGCAACGGGTGCCAGGGCAGGGAGAGGTGAGTCTG ggaggg | −1039 | 1 | 100% | AREB6,v-Myb,MAZ,Hand1:E47 | ||
| NM 003991 | tgggcg CGCTGCGGGAGCTGTAGCTCAGCCAGCCAGGGAGTAGCGGCTTTCATCCG ccggga | −340 | 1 | 100% | SMAD-3 | ||
| NM 005795 | ttggga GGTTGAGGCAGGAGAATTGCTTGAACCCGGGAGGTGGAGGTTGCAGTGAG ctgaga | −1930 | 1 | 100% | Zta,TFII-I,C/EBPdelta,c-Ets-1(p54),AREB6 | ||
| NM 003357 | ttggga GGCTGAGGCAGGAGAATCGTCTGAACTCGGGAGGTGGAGGTTGCAGTGAG ccgctg | −4624 | 1 | 100% | Zta,TFII-I,c-Ets-1(p54),AREB6,RFX1 | ||
| NM 005856 | ttgtga GGCTGAGGCAGGAGAATCGCTTGAATCCAGGAGGTGGAGGTTGCAGTGAG ccggga | −3601 | 3 | 100% | Zta,TFII-I,GATA-1,c-Ets-1(p54),AREB6,RFX1 | ||
| NM 004844 | acagga GGCTGAGGCAGGAGAATAGCTTGAACCCAGGAGGCGGAGGTTGCAGTGAG ctaaca | −4824 | 3 | 100% | Zta,TFII-I,c-Ets-1(p54) | ||
| Profile | DBVDSD SGYKGMRRBASVWGDAKHDHHKSVWBYYRGGRVVBVGMSRBKKBMRTSHG SBRVBR | ||||||
The complete table can be downloaded at http://bioinfo.hku.hk/~daniely/lung_microarray.
aOverrepresented gene ontology terms.
bLocation of conserved sequence upstream of the transcription start site.
cFrequency of occurrence of conserved sequence in 5 kb upstream regions of genes given in (32).
dPercentage of the genes with a matched upstream sequence and their expression trends.
DISCUSSION
This study first identified a large set of genes (MAD) showing a 2-fold differential behavior in adenocarcinoma cells when compared with normal lung tissue. Of these genes, 2528 genes (73.45%) were also identified passing the t-test criteria (P < 0.005, complete t-test gene list available at http://bioinfo.hku.hk/~daniely/lung_microarray/). Transcription factors with binding site motifs that matched conserved DNA regions upstream of genes in MAD were then identified, as these may be the factors that regulate the oncogenic process. This was achieved by incorporating both experimentally determined gene expression data and bioinformatic tools. Below, we will discuss the functional annotation groups (gene ontology terms) that were overrepresented in the cancer associated genes and their putative regulatory transcription factors. Only some salient findings can be presented due to the size of the dataset and full details are provided as Supplementary Material.
In a separate study, we identified 88 lung cancer associated genes (data not shown) from our microarrays, using a feature partitioning method we developed earlier (37). However, here, we aimed to identify the broadest set of cancer associated genes (MAD) by using fold-ratio analysis, and to examine their functional annotations in order to understand the biological processes that are altered in cancer when compared with normal tissue. A broad gene set was important to ensure statistical validity when determining the functional groups (gene ontology terms) that were overrepresented in the gene population in MAD. More than three thousand genes were found to be up- or down-regulated by >2-fold and all 88 cancer associated genes identified using the earlier method (37) were found in this set.
In previous works (38–40), differential gene expression in cancer was reported but relatively little elaboration of the genes' functions, or the regulatory cascades and biological processes underlying the observations was made. Here, we found that many gene ontology terms disproportionately occurred (P < 0.001) among the sets of genes that were either substantially up- or down-regulated in adenocarcinomas. This gave evidence of the systematic up- or down-regulation of several biological processes directly linked to oncogenesis. Such processes included increased cell multiplication, angiogenesis, vascularization, and glucose and amino acid metabolism.
Glucose metabolism is crucial because cancer cell growth depends on glucose availability, rather than respiration, for biomass construction (41). Increased expression of glycolytic enzymes, including pyruvate carboxylase, citrate synthase, aconitate hydratase, oxalosuccinate decarboxylase, glucose-6-phosphate isomerase, fructose-bisphosphate aldolase, glucose transporter (GLUT) and l-lactate dehydrogenase were observed in the microarray data. This is consistent with the fermentation metabolism (needed for ATP synthesis in the absence of efficient respiration), and with entry into a tricarboxylic acid pathway for glutamate and aspartate synthesis (i.e. biomass construction) rather than respiration.
Unlike mostly resting normal cells, where oxygen is used in oxidative phosphorylation for ATP synthesis and cell maintenance, cancer cells metabolize glucose at a much higher rate, in order to generate ATP and use pyruvate as the substrate to generate lactate to replete the NAD pool (Warburg's effect), while stopping the cycling of the tricarboxylic acid pathway (42,43). The major outcome of this metabolic shift is, by preventing the tricarboxylic acid pathway cycling, to produce biomass rather than energy. This effect, overlooked for some time, was discovered >70 years ago (41). Much effort has been initiated to identify the transcription factor(s) that facilitate this change of course in cancer cells (from aerobic slow growth or resting state into anaerobic use of glucose while growing) by up-regulating the expression and activity of all enzymes directly related to this essential metabolic pathway. In recent publications, several transcription factors [hypoxia inducible factor 1 (HIF-1) (44); Myc (45); Ras (46); v-SRC(47); p53(48) and pVHL(49)] were reported to play a role in the regulation of the expression of these glycolytic enzymes.
From the genes in MAD associated with each overrepresented gene ontology term, a subset of genes with more consistent expression profiles was identified and the upstream regions of these genes were searched for conserved elements. Such conserved DNA regions, if they exist, are likely to be evolutionarily significant (50–54). Wasserman et al. (55) showed that a large proportion (>98%) of experimentally defined transcription factor binding sites are restricted to the most conserved residues within their own promoter regions. Earlier studies have used databases such as TRANSFAC to search for transcription factor binding sites in the upstream regions of genes; however, this can lead to many false positives (56,57). Clustering of genes based on expression profiles has been used to select sets of genes more likely to be co-regulated (20); however, with increasing numbers of genes in the clusters, the number of false positive identifications increases. One reason for this is the inclusion of genes in the cluster that are not actually co-regulated, hampering the correct detection of conserved DNA regions by most motif discovery tools (21,22). Methods to evaluate putative regulatory sites and newly detected motifs have also been proposed (58).
To address this issue, we combined the gene expression correlation coefficients and gene functional classes of all the cancer-associated genes (MAD) to select a more consistent set of likely co-regulated genes. These genes not only had a consistent expression pattern with the highest possible pairwise gene correlation, but also shared the same functional role. No limit was placed on the number of genes that would be selected from each functional group, and all genes with expression profiles within a cutoff value (d < 0.20) were selected. These criteria were motivated by there being many examples, which show that transcription factors have multiple target genes, of which a significant portion is involved in a common metabolic pathway. For instance, the CAP transcription factor in Escherichia coli has been shown to mediate the regulation of dozens of genes involved in glucose metabolism (59,60). In humans, the GATA binding protein 1 (globin transcription factor 1, GATA-1) plays an important role in erythroid development by regulating hemoglobin production (61). The majority of genes that are regulated by this transcription factor contain the gene ontology term ‘hemoglobin’. Moreover, growth factor independent 1 (Gfi-1) acts on a subset of genes involved in the differentiation of the hematopoietic lineage (62).
MoDEL, the motif discovery program used here, has been demonstrated extensively and compared with other existing motif finding algorithms by analyzing sets of complex natural amino acid sequences (e.g. HTH protein motifs) and artificial datasets (planted motifs) (26). It was shown to have a more efficient optimization method than other local multiple alignment methods. Unlike algorithms that search for motifs by exhaustive enumeration of overrepresented words (63), MoDEL looks for a set of conserved occurrences based on information content (26). The objective of MoDEL is to identify exactly one occurrence per sequence in such a way that all chosen occurrences are maximally similar across the sequence set. A validation of MoDEL on the CAP-mediated gene set (59) in bacteria successfully extracted the conserved regions that incorporate the CAP binding sites (Supplementary Material).
Having identified conserved DNA regions associated with genes with the same functional annotation and similar expression profiles, in silico pattern-based scanning against the TRANSFAC 8.3 database for transcription factors with binding site motifs in these conserved DNA regions was performed. Among the transcription factors identified as putative regulatory factors for these genes (Table 4), some had been reported in previous publications to promote or suppress cancer formation, whereas the remaining transcription factors have generally not been sufficiently characterized in vivo. Four of these appear to be particularly significant, namely: HIF-1, Gfi-1, nuclear factor TG-interacting factor (TGIF) and erythroid transcription factor (GATA-1).
HIF-1 is a regulatory heterodimer consisting of two subunits; HIF-1β is constitutively expressed in all conditions, whereas HIF-1α is rapidly degraded under normal conditions but is stabilized under hypoxia (64). Despite an average up-regulation of this protein (HIF-1α) by ∼30% in our dataset, our initial screening for cancer gene markers did not reveal this protein because the expression change was too small to be selected. From our microarray findings, the up-regulation of this protein did not result in a systematic activation of gene clusters with a specific function. However, the fact that HIF-1 binding sites were found to be enriched in some down-regulated genes that belonged to the cellular defense response gene ontology term (Table 4), suggested that this protein might be one of the cellular components responsible for the suppression of the defense response of hypoxic cancer cells. Other genes related to growth factor, protease and apoptosis pathways, e.g. epidermal growth factor receptor, carbonic anhydrase IX, p53-, matrix metalloproteinase 9, that were known to be dependent on HIF-1α for their activation (65) had fold changes of 2.41, 2.8, 6.5 and 2.51, respectively, in our dataset.
Gfi-1 is a zinc finger protein that binds DNA and functions as a transcriptional repressor through its unique repressor domain, SNAG (66). In our arrays, this gene was down-regulated in adenocarcinoma cells by an average of 69%, and it was observed that genes that contain activation sites for Gfi-1 were mostly up-regulated in adenocarcinoma cells. One example is the pro-apoptotic regulator gene Bax which was up-regulated by 2.3-fold in adenocarcinoma cells but was shown to be down-regulated by Gfi-1 in immortalized T-cell lines and primary transgenic thymocytes (67).
TGIF is a transcriptional core-repressor that directly associates with Smad (Sma- and Mad-related protein) proteins and inhibits Smad-mediated transcriptional activation (68). The gene responses activated by Smad underlie both proliferative and anti-proliferative events that contribute to cancer (69,70). Originally, TGIF was isolated as a ubiquitously expressed homeodomain protein that can bind to the retinoid X receptor (RXR) response element (71). Based on our analysis, this gene was up-regulated in lung cancer cells by an average of 2.6-fold while the RXR gene was repressed by an average of 25%.
GATA-1 is a factor that had been shown to be important in the regulation of globin and non-globin genes in erythroid, megakaryocytic and mast cell lineages (72). From our arrays, this gene was down-regulated by an average of ∼40% in cancer cells. This is consistent with our findings that members in globin gene family (α, β and γ) were all repressed in adenocarcinomas, despite their weak association with primary lung cancers (Table 2).
In conclusion, by investigating the statistical distribution of the functional annotations attached to cancer associated genes (MAD) derived from lung tissue microarrays, we have identified functions, corresponding to several key biological systems, which are overrepresented in cancer associated genes (Tables 2 and 3). The congruence of these functions with known cancer cell oncogenic processes suggests the up- or down-regulation of genes in MAD is linked to cancer-related metabolism processes. Subsequently, we clustered the genes in MAD into putatively co-regulated gene sets by assuming that co-regulated genes will share common functional roles and exhibit very similar expression profiles. Conserved DNA segments in the upstream regions of these putatively co-regulated gene sets were found and transcription factors that recognize these DNA regions were identified (Table 4). A literature search on these transcription factors, which are putative regulatory factors in adenocarcinoma development, substantiated that the majority had been previously documented experimentally to be oncogenic transcription factors. These transcription factors, together with their conserved binding sites, suggest new candidates for therapeutic intervention in the treatment of lung adenocarcinomas.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
Indispensable support was provided by a doctoral fellowship from The University of Hong Kong and the Hong Kong Innovation and Technology Fund (ITF) BIOSUPPORT Programme. The microarray experiments are supported by the HKSAR RGC grants 7486/03M, 7468/04M. Work at the Genetics of Bacterial Genomes Unit is supported by the Centre National de la Recherche Scientifique (CNRS, URA 2171).
REFERENCES
- 1.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Murray T., Samuels A., Ghafoor A., Ward E., Thun M.J. Cancer statistics, 2003. CA Cancer J. Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Ramaswamy S., Golub T.R. DNA microarrays in clinical oncology. J. Clin. Oncol. 2002;20:1932–1941. doi: 10.1200/JCO.2002.20.7.1932. [DOI] [PubMed] [Google Scholar]
- 5.Geller S.C., Gregg J.P., Hagerman P., Rocke D.M. Transformation and normalization of oligonucleotide microarray data. Bioinformatics. 2003;19:1817–1823. doi: 10.1093/bioinformatics/btg245. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann R., Seidl T., Dugas M. Profound effect of normalization on detection of differentially expressed genes in oligonucleotide microarray data analysis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0033. RESEARCH0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quackenbush J. Microarray data normalization and transformation. Nature Genet. 2002;32(Suppl.):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 8.Zien A., Aigner T., Zimmer R., Lengauer T. Centralization: a new method for the normalization of gene expression data. Bioinformatics. 2001;17(Suppl. 1):S323–331. doi: 10.1093/bioinformatics/17.suppl_1.s323. [DOI] [PubMed] [Google Scholar]
- 9.Cope L.M., Irizarry R.A., Jaffee H.A., Wu Z., Speed T.P. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–331. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- 10.Brazma A., Vilo J. Gene expression data analysis. Microbes Infect. 2001;3:823–829. doi: 10.1016/s1286-4579(01)01440-x. [DOI] [PubMed] [Google Scholar]
- 11.Krajewski P., Bocianowski J. Statistical methods for microarray assays. J. Appl. Genet. 2002;43:269–278. [PubMed] [Google Scholar]
- 12.Zhang H., Yu C.Y., Singer B., Xiong M. Recursive partitioning for tumor classification with gene expression microarray data. Proc. Natl Acad. Sci. USA. 2001;98:6730–6735. doi: 10.1073/pnas.111153698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theilhaber J., Bushnell S., Jackson A., Fuchs R. Bayesian estimation of fold-changes in the analysis of gene expression: the PFOLD algorithm. J. Comput. Biol. 2001;8:585–614. doi: 10.1089/106652701753307502. [DOI] [PubMed] [Google Scholar]
- 14.Horn D., Axel I. Novel clustering algorithm for microarray expression data in a truncated SVD space. Bioinformatics. 2003;19:1110–1115. doi: 10.1093/bioinformatics/btg053. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen D.V., Rocke D.M. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics. 2002;18:39–50. doi: 10.1093/bioinformatics/18.1.39. [DOI] [PubMed] [Google Scholar]
- 16.Lee S.I., Batzoglou S. Application of independent component analysis to microarrays. Genome Biol. 2003;4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somorjai R.L., Dolenko B., Baumgartner R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics. 2003;19:1484–1491. doi: 10.1093/bioinformatics/btg182. [DOI] [PubMed] [Google Scholar]
- 18.Cora D., Di Cunto F., Provero P., Silengo L., Caselle M. Computational identification of transcription factor binding sites by functional analysis of sets of genes sharing overrepresented upstream motifs. BMC Bioinformatics. 2004;5:57. doi: 10.1186/1471-2105-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tullai J.W., Schaffer M.E., Mullenbrock S., Kasif S., Cooper G.M. Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J. Biol. Chem. 2004;279:20167–20177. doi: 10.1074/jbc.M309260200. [DOI] [PubMed] [Google Scholar]
- 20.Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M. Systematic determination of genetic network architecture. Nature Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 21.Liu X.S., Brutlag D.L., Liu J.S. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 22.Zheng J., Wu J., Sun Z. An approach to identify over-represented cis-elements in related sequences. Nucleic Acids Res. 2003;31:1995–2005. doi: 10.1093/nar/gkg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bussemaker H.J., Li H., Siggia E.D. Regulatory element detection using correlation with expression. Nature Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 24.Conlon E.M., Liu X.S., Lieb J.D., Liu J.S. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl Acad. Sci. USA. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitschke P., Guerdoux-Jamet P., Chiapello H., Faroux G., Henaut C., Henaut A., Danchin A. Indigo: a World-Wide-Web review of genomes and gene functions. FEMS Microbiol. Rev. 1998;22:207–227. doi: 10.1111/j.1574-6976.1998.tb00368.x. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez D., Gras R., Appel R. MoDEL: An efficient strategy for ungapped local multiple alignment. Comput. Biol. Chem. 2004;28:119–128. doi: 10.1016/j.compbiolchem.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Brazma A., Parkinson H., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocca-Serra P., Brazma A., Parkinson H., Sarkans U., Shojatalab M., Contrino S., Vilo J., Abeygunawardena N., Mukherjee G., Holloway E., et al. ArrayExpress: a public database of gene expression data at EBI. C R Biol. 2003;326:1075–1078. doi: 10.1016/j.crvi.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 29.Li C., Wong W. DNA-Chip Analyzer (dChip) In: Parmigiani G., Garrett E.S., Irizarry R., Zeger S.L., editors. The Analysis of Gene Expression Data: Methods and Software. Springer; 2003. pp. 120–141. [Google Scholar]
- 30.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 32.Aach J., Bulyk M.L., Church G.M., Comander J., Derti A., Shendure J. Computational comparison of two draft sequences of the human genome. Nature. 2001;409:856–859. doi: 10.1038/35057055. [DOI] [PubMed] [Google Scholar]
- 33.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 34.Cravchik A., Subramanian G., Broder S., Venter J.C. Sequence analysis of the human genome: implications for the understanding of nervous system function and disease. Arch. Neurol. 2001;58:1772–1778. doi: 10.1001/archneur.58.11.1772. [DOI] [PubMed] [Google Scholar]
- 35.Wingender E. TRANSFAC, TRANSPATH and CYTOMER as starting points for an ontology of regulatory networks. In Silico Biol. 2004;4:55–61. [PubMed] [Google Scholar]
- 36.Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yap Y., Zhang X., Ling M., Wang X., Wong Y., Danchin A. Classification between normal and tumor tissues based on the pair-wise gene expression ratio. BMC Cancer. 2004;4:72. doi: 10.1186/1471-2407-4-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beer D.G., Kardia S.L., Huang C.C., Giordano T.J., Levin A.M., Misek D.E., Lin L., Chen G., Gharib T.G., Thomas D.G., et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharjee A., Richards W.G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fardin P., Bahler J., Capanni P., Inglese E., Ricciardi A., Ferrara G.B. Gene expression analysis in non small cell lung cancer (NSCLC) using microarray technology. Hum. Immunol. 2003;64:S116. [Google Scholar]
- 41.Warburg O. The Metabolism of Tumours. London: Arnold Constable; 1930. [Google Scholar]
- 42.Goel A., Mathupala S.P., Pedersen P.L. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J. Biol. Chem. 2003;278:15333–15340. doi: 10.1074/jbc.M300608200. [DOI] [PubMed] [Google Scholar]
- 43.Lee M.G., Pedersen P.L. Glucose metabolism in cancer: importance of transcription factor-DNA interactions within a short segment of the proximal region of the type II hexokinase promoter. J. Biol. Chem. 2003;278:41047–41058. doi: 10.1074/jbc.M307031200. [DOI] [PubMed] [Google Scholar]
- 44.Carmeliet P., Dor Y., Herbert J.M., Fukumura D., Brusselmans K., Dewerchin M., Neeman M., Bono F., Abramovitch R., Maxwell P., et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 45.An W.G., Kanekal M., Simon M.C., Maltepe E., Blagosklonny M.V., Neckers L.M. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 46.Mathupala S.P., Heese C., Pedersen P.L. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J. Biol. Chem. 1997;272:22776–22780. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 47.Rempel A., Mathupala S.P., Griffin C.A., Hawkins A.L., Pedersen P.L. Glucose catabolism in cancer cells: amplification of the gene encoding type II hexokinase. Cancer Res. 1996;56:2468–2471. [PubMed] [Google Scholar]
- 48.Gnarra J.R., Zhou S., Merrill M.J., Wagner J.R., Krumm A., Papavassiliou E., Oldfield E.H., Klausner R.D., Linehan W.M. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl Acad. Sci. USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis B.C., Shim H., Li Q., Wu C.S., Lee L.A., Maity A., Dang C.V. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol. Cell. Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akiyama Y., Hosoya T., Poole A.M., Hotta Y. The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc. Natl Acad. Sci. USA. 1996;93:14912–14916. doi: 10.1073/pnas.93.25.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu E.S., Lee A.S. Common sets of nuclear factors binding to the conserved promoter sequence motif of two coordinately regulated ER protein genes, GRP78 and GRP94. Nucleic Acids Res. 1991;19:5425–5431. doi: 10.1093/nar/19.19.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh H., Sen R., Baltimore D., Sharp P.A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 53.Siomi H., Matunis M.J., Michael W.M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasula S.M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R.A., Robbins P.D., Fernandes-Alnemri T., Shi Y., et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 55.Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. Human-mouse genome comparisons to locate regulatory sites. Nature Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 56.Fickett J.W., Hatzigeorgiou A.G. Eukaryotic promoter recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- 57.Claverie J.M., Audic S. The statistical significance of nucleotide position-weight matrix matches. Comput. Appl. Biosci. 1996;12:431–439. doi: 10.1093/bioinformatics/12.5.431. [DOI] [PubMed] [Google Scholar]
- 58.Jakt L.M., Cao L., Cheah K.S., Smith D.K. Assessing clusters and motifs from gene expression data. Genome Res. 2001;11:112–123. doi: 10.1101/gr.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gosset G., Zhang Z., Nayyar S., Cuevas W.A., Saier M.H., Jr Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 2004;186:3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magasanik B., Neidhardt F.C. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Vol. 2. Washington, DC: American Society for Microbiology; 1987. Regulation of carbon and nitrogen utilization; pp. 1318–1325. [Google Scholar]
- 61.Trainor C.D., Evans T., Felsenfeld G., Boguski M.S. Structure and evolution of a human erythroid transcription factor. Nature. 1990;343:92–96. doi: 10.1038/343092a0. [DOI] [PubMed] [Google Scholar]
- 62.Duan Z., Horwitz M. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc. Natl Acad. Sci. USA. 2003;100:5932–5937. doi: 10.1073/pnas.1031694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Helden J., Andre B., Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 64.Huang L.E., Gu J., Schau M., Bunn H.F. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl Acad. Sci. USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swinson D.E., Jones J.L., Cox G., Richardson D., Harris A.L., O'Byrne K.J. Hypoxia-inducible factor-1alpha in non small cell lung cancer: relation to growth factor, protease and apoptosis pathways. Int. J. Cancer. 2004;111:43–50. doi: 10.1002/ijc.20052. [DOI] [PubMed] [Google Scholar]
- 66.Zweidler-Mckay P.A., Grimes H.L., Flubacher M.M., Tsichlis P.N. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grimes H.L., Gilks C.B., Chan T.O., Porter S., Tsichlis P.N. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc. Natl Acad. Sci. USA. 1996;93:14569–14573. doi: 10.1073/pnas.93.25.14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wotton D., Lo R.S., Lee S., Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 69.Eppert K., Scherer S.W., Ozcelik H., Pirone R., Hoodless P., Kim H., Tsui L.C., Bapat B., Gallinger S., Andrulis I.L., et al. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 70.Lagna G., Hata A., Hemmati-Brivanlou A., Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 71.Bertolino E., Reimund B., Wildt-Perinic D., Clerc R.G. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- 72.Orkin S.H. Globin gene regulation and switching: circa 1990. Cell. 1990;63:665–672. doi: 10.1016/0092-8674(90)90133-y. [DOI] [PubMed] [Google Scholar]
- 73.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.