Abstract
Using a microarray that tiles all known yeast non-coding RNAs, we compared RNA from wild-type cells with RNA from mutants encoding known and putative RNA modifying enzymes. We show that at least five types of RNA modification (dihydrouridine, m1G, , m1A and ) catalyzed by 10 different enzymes (Trm1p, Trm5, Trm10p, Dus1p-Dus4p, Dim1p, Gcd10p and Gcd14p) can be detected by virtue of differential hybridization to oligonucleotides on the array that are complementary to the modified sites. Using this approach, we identified a previously undetected m1A modification in GlnCTG tRNA, the formation of which is catalyzed by the Gcd10/Gcd14 complex.
INTRODUCTION
Many cellular RNAs are subject to covalent modification, providing a means to expand the chemical repertoire of the four bases. The modifications are diverse and include methylation of base and sugar functional groups (1,2), deamination of adenosine and cytosine residues (3), conversion of double to single bonds (4) and changing the nature of glycosyl and hydrogen bond functional groups (5). Modified RNAs include rRNA, tRNA, mRNA, snRNA and snoRNAs (6,7). Among these, tRNAs are the most heavily modified; in the yeast Saccharomyces cerevisiae, every one of the 34 tRNAs that have been chemically sequenced contains at least nine modified nucleotides within its ∼76 nucleotide sequence, and at least 25 different modifications to either base or sugar moieties of tRNAs have been identified (8).
Genome sequencing has revealed potential new RNA modifying enzymes in many species. Even in yeast the full complement of modifying enzymes and cognate modified sites remains an open issue; novel modification enzymes are still being described (9). Discovery of RNA modification sites is difficult in part because traditional methods used to locate and study modifications [typically primer extension assays and/or chromatographic analysis (high-performance liquid chromatography or thin-layer chromatography) of purified RNAs] have limited throughput. To facilitate large-scale exploration of RNA modification events, it would be beneficial to have a method to analyze modifications across all non-coding RNAs in a single assay.
In a previous study using microarrays to analyze processing of non-coding RNA, we observed that loss of dihydrouridine modification at tRNA positions 16 and 17 in a dus1-Δ mutant resulted in increased binding specifically to the two tRNA probes on the array that were complementary to those nucleotides. This suggested that the presence of the modification interfered with binding to the array, such that the difference in affinity between mutant and wild-type could be monitored by microarray (10). To study such events on a much broader scale, we subsequently designed a higher density array of 21 939 fifteen to twenty-five base long oligonucleotides that begin every 5 bases along all known and predicted yeast non-coding RNAs in the S.cerevisiae genome, as well as introns and the 3′ ends of mRNAs for which processing sites are known (for details see Table 1 and Materials and Methods). This new microarray contains oligos complementary to 70 genomic tRNA transcripts comprising all 42 unique tRNA species (including 14 that have not previously been analyzed). We used this array to analyze mutants in each of the four yeast dihydrouridine synthase enzymes, and used the differential binding of mutant and wild-type tRNAs to make specificity assignments for the four proteins (9).
Table 1.
Known and predicted yeast non-coding RNAs included on the microarray
| ncRNA | Number of transcripts | Tiling frequency |
|---|---|---|
| 35S pre rRNA | 1 | 5 |
| 5S rRNA | 1 | 5 |
| Genomic tRNAs | 70 | 5 |
| snoRNAs | 84 | 5 |
| snRNAs | 6 | 5 |
| RNase P | 1 | 5 |
| RNase MRP | 1 | 5 |
| SRP RNA | 1 | 5 |
| Telomerase RNA | 1 | 5 |
| RUFs | 8 | 5 |
| Introns | 236 | 20 |
| Spliced junctions | 236 | 5 |
| Mitochondrial genome features | 44 | 15 |
| mRNA 3′ ends | 8 | 20 |
Here, we have examined in detail whether any of the 17 different types of RNA modifications can be detected using oligonucleotide microarrays. We used the 21 939-probe array to perform 25 different mutant versus wild-type comparisons, following a protocol in which the RNA from the wild-type cell (which carries the modification) and RNA from the mutant cell (which lacks the modification) are labeled and hybridized to the array in the two separate channels (Cy3 and Cy5). In this way, differential hybridization caused by the RNA modifications would be detected as a change in ratio of Cy3/Cy5 signal from probes complementary to modified nucleotides. We successfully detected the following modifications: dihydrouridine (catalyzed by Dus1p-Dus4p), m1G (catalyzed by Trm1p), (catalyzed by Trm1p), m1A (catalyzed by the Gcd10p-Gcd14p complex) and (catalyzed by Dim1p). Our results establish a simple rule: with the exception of dihydrouridine (which severely perturbs nucleotide architecture), modifications to the Watson–Crick surface impact microarray hybridization. In addition to observing hundreds of known modification events, our data suggested several new modification sites, one of which we subsequently verified by primer extension analysis. This demonstrates the general utility of microarrays as a genome-wide tool for RNA modification detection.
MATERIALS AND METHODS
Array construction
Oligonucleotide sequences are contained in the Supplemental Material. Oligos were designed to be complementary to known non-coding RNA sequences and flanking regions and were tiled at 5 nt intervals for most RNAs (intron-containing mRNAs were tiled every 20 nt and mitochondrial RNAs every 15 nt; see Table 1 for details). Probe lengths were adjusted to have a melting temperature of ∼41°C (11). Ink-jet microarrays were manufactured by Agilent Technologies (Palo Alto, CA).
Strains
Homozygous deletion mutants (12) were obtained from Research Genetics. TetO7-promoter alleles were constructed as described previously (13). gcd14-1ts (14) and gcd14-Δ were kindly provided by Mercedes Tamame; the dim1-Y131G strain was provided by Denis Lafontaine. The gcd14-Δ strain used for primer extension analysis overexpresses IMT4, which suppresses the lethal phenotype of the deletion (M. Tamame, personal communication).
RNA isolation and array analysis
Isogenic wild-type and mutant strains were grown in parallel at 30°C in SC medium (with the exception of gcd14-1 which was grown in YPD+Ade) with shaking in baffled flasks (Bellco) to final cell concentrations matched as closely as possible to 107cells/ml. TetO7-promoter strains were exposed to 10 μg/mldoxycycline for a total of 20–24 h. Cells were harvested and RNA extracted as described previously (10). An aliquot of 10 μg of DNase I-treated RNA was labeled with Alexa Fluor 546 or 647 according to the manufacturer's instructions (Molecular Probes ‘Ulysis’ kit), ethanol-precipitated and hybridized to the array as described previously (15). Formamide was added to the final concentrations of 25 or 33%, as described previously (15). Hybridizations were carried out in a rotating incubator at 42°C for 16–20 h and washed as described previously (15). Arrays were scanned on an Axon 4000B instrument.
Image processing, array normalization and data visualization
Scanned images were quantitated with GenePix (Axon Instruments). Individual channels were spatially detrended (i.e. overall correlations between spot intensity and position on the slide removed) by high-pass filtering [(16); http://www.psi.utoronto.ca/~ofer/detrendingReport.pdf] using 10% outliers. Dye bias was corrected in each slide using the Lowess smoother from the MAANOVA package (written by Hao Wu) with 0.3 smoother span. After these steps, the normalized intensities were converted in log2 ratios of mutant expression versus wild-type.
Primer extension analysis
Bulk RNA isolated from either wild-type or gcd14-Δ cells was used as a template for primer extension assays using a 5′ 32P-labeled primer (5′-GGAGGTCCCACCCGG-3′) that is specific for tRNAGlnCTG RNA. Approximately 5–10 μg bulk RNA and primer (0.1 μM) were annealed by heating to 95°C for 3 min and then cooling to room temperature in a 5 μl reaction containing 50 mM Tris–HCl, 15 mM NaCl and 10 mM DTT, pH 7.5. Then 2 μl of the annealed mixture of primer and RNA was used in a primer extension reaction of 10 μl containing 0.4 mM of each dNTP and 4 U AMV-reverse transcriptase (20 U/μl; Promega). Sequencing reactions also contained 0.2 mM of each individual ddNTP (ddG, ddA, ddT or ddC). The reactions were terminated after 1 h of incubation at 37°C by the addition of an equal volume of formamide/50 mM EDTA loading dye; subsequently the samples were resolved on a 15% acrylamide/4 M urea gel and visualized by phosphorImager.
Data availability
Oligonucleotide sequences on the arrays, and all microarray data are available at (hugheslab.med.utoronto.ca/Hiley). Spreadsheets containing the data displayed in Figures 1B and 2A are also available on the website.
Figure 1.
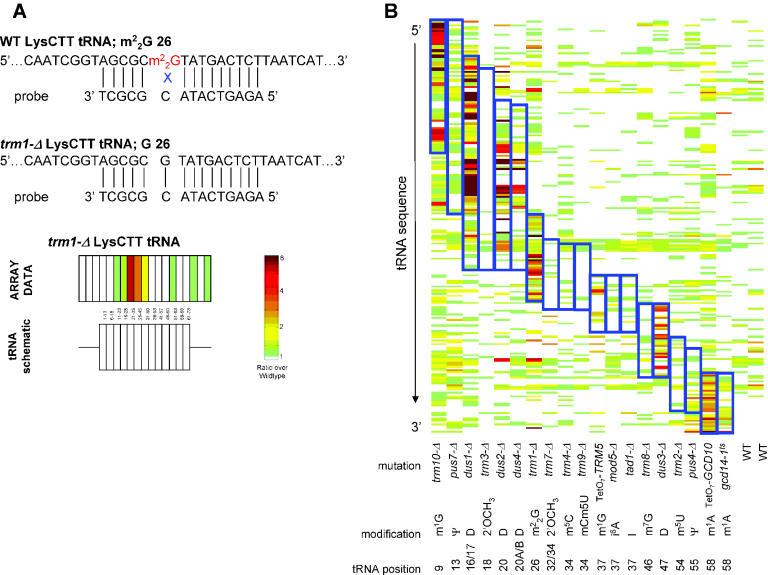
Detection of covalent modification by microarray. (A) Modification disrupts base pairing between RNA and probe. Wild-type tRNA LysCTT (top) contains a dimethylguanosine residue at position 26, which disrupts pairing with the probe. trm1-Δ tRNA LysCTT (bottom) lacks this modification and can pair completely with the probe (see also Figure 2B). A schematic diagram of the tRNA is shown below. Rectangles represent probes complementary to tRNA sequence, and thin lines represent probes complementary to 5′ and 3′ genomic flanking regions. The relative fluorescence of each probe is indicated by color-coded rectangles above the schematic diagram (according to the scale on the right); the tRNA nucleotides covered by each oligo are shown. (B) Analysis of strains defective for tRNA modification. tRNA oligos (ordered from 5′ to 3′) versus individual experiments (described below the figure) are plotted. Oligos to which there was significantly better binding in the mutant tRNA samples are indicated by red color, as shown by the color-bar in (A). Groups of probes covering tRNA nucleotides modified by each enzyme are outlined in blue rectangles. The type of modification and positions known to be modified by each enzyme are shown. Only tRNA probes with ratios at least 2-fold above wild type are shown.
Figure 2.
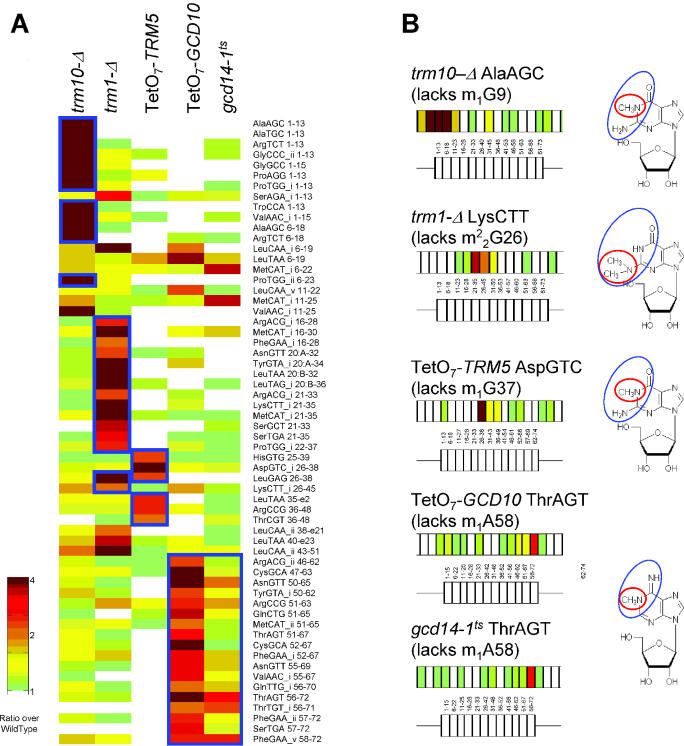
tRNA methylation analyzed by microarray. (A) Three different types of detectable methylation. Unique tRNA probes with ratios of at least 2 are color-coded according to the scale shown and displayed from 5′ to 3′ of the tRNA sequence. The tRNA isoforms and specific nucleotides covered are shown to the right of the figure. Oligos predicted to be affected in the each experiment are outlined with blue rectangles. (B) Schematic representation of selected tRNAs. One tRNA from each of the experiments in which the methylation defect was detected is shown in schematic form as described in Figure 1A. Functional groups involved in Watson–Crick base pairing are circled in blue; modifications are circled in red.
RESULTS
RNA modifications detected by microarray
In order to test the general applicability of detecting and mapping RNA modifications by microarray, we analyzed total RNA from a set of 25 yeast modifying enzymes. At least one enzyme that catalyzes each of the 17 known modifications was examined (Table 2). In each experiment, RNA isolated from wild-type cells (labeled with Cy3) and RNA isolated from mutant yeast cells (labeled with Cy5) were hybridized to a microarray. The fluorescence in each channel was measured and compared as a ratio [(mutant RNA fluorescence)/(wild-type RNA fluorescence)] (for details see Materials and Methods). Figure 1A shows an example of a detectable modification: the presence of at position 26 of LysCTT tRNA appears to interfere with binding to the probe sequence, because in probes overlapping position 26 there is a relative increase in binding of the tRNA in the trm1-Δ mutant strain (which lacks the modification) compared with the wild-type. Probes complementary to tRNAs outside of the modified region, however, show no difference in binding affinity. Below, the tRNA is shown in schematic form with rectangles representing tRNA sequence and thin lines representing flanking sequence. The relative fluorescence of each probe is shown above, color-coded according to the scale shown.
Table 2.
RNA modification enzymes and their targets
| Modification | ORF name | Gene name | Target | Detected by microarray? | |
|---|---|---|---|---|---|
| Methylation | YDR120C | TRM1 | tRNA 26 | Yes | |
| m5U | YKR056W | TRM2 | tRNA 54 | No | |
| 2′O CH3 | YDL112W | TRM3 | tRNA 18 | No | |
| m5C | YBL024W | TRM4 | tRNA 34, 40, 38, 49 | No | |
| m1G | YHR070W | TRM5 | tRNA 37 | Yes | |
| 2′O CH3 | YBR061C | TRM7 | tRNA 32, 34 | No | |
| m7G | YDL201W | TRM8 | tRNA 46 | No | |
| mcm5U/mcm5s2U | YML014W | TRM9 | tRNA 34 | No | |
| m1G | YOL093W | TRM10 | tRNA 9 | Yes | |
| m1A | YNL062C | GCD10 | tRNA 58 | Yes | |
| m1A | YJL125C | GCD14 | tRNA 58 | Yes | |
| YPL266W | DIM1 | 18S rRNA 3′ terminal loop | Yes | ||
| 2′O CH3 | YCL054W | SPB1 | 25S rRNA 2918 | No | |
| m5C | YNL061W | NOP2 | Unknown | No | |
| 2′O CH3 | YDL014W | NOP1 | Unknown | No | |
| Dihydrouridylation | D | YML080W | DUS1 | tRNA 16/17 | Yes |
| D | YNR01W | DUS2 | tRNA 20 | Yes | |
| D | YLR401C | DUS3 | tRNA 47 | Yes | |
| D | YLR405W | DUS4 | tRNA 20:A/20:B | Yes | |
| Pseudouridylation | Ψ | YPL212C | PUS1 | tRNA 27 | No |
| Ψ | YGL063W | PUS2 | Unknown | No | |
| Ψ | YFL001W | PUS3 | tRNA 38, 39 | No | |
| Ψ | YNL292W | PUS4 | tRNA 55 | No | |
| Ψ | YLR165C | PUS5 | Mitochondrial 21S rRNA 2819 | No | |
| Ψ | YGR169C | PUS6 | tRNA 31 | No | |
| Ψ | YOR243C | PUS7 | Unknown | No | |
| Ψ | YLR175W | CBF5 | rRNA | No | |
| Adenosine deamination | YGL243W | TAD1 | tRNA 37 | No | |
| i6A formation | YOR274W | MOD5 | tRNA 37 | No |
Enzymes in bold were examined in this study.
tRNA modifications
tRNAs are an ideal target for the analysis of covalent modification by microarray: they are abundant, and they are subject to a wide variety of covalent modifications at different positions. The results from microarray analysis of 19 tRNA modifying enzymes are summarized in Figure 1B. The relative fluorescence of tRNA-specific oligonucleotide probes (tiled from the 5′ to the 3′ end of the transcripts) versus individual experiments is plotted. The ratio of fluorescence (mutant versus wild-type) to each probe is color-coded according to the scale shown, with red indicating more efficient binding of the mutant tRNA to the microarray. The nature of the modifications and the nucleotides known to be modified are shown for each experiment, and probes complementary to nucleotides known to be modified in at least one tRNA are outlined with blue rectangles. Whereas Figure 1A shows all oligos corresponding to a single tRNA (and emphasizes the specificity of the technique), this Figure summarizes the differential hybridization to all 70 tRNA sequences across 21 experiments (and shows the ability of technique to detect trends that emerge across all experiments). For clarity, we have included only probes that display at least a 2-fold difference in binding between mutant and wild-type in one or more experiments. There is a concentration of red probes within the blue rectangles, indicating that probes complementary to modified nucleotides are more efficiently bound by mutant tRNAs than by their wild-type counterparts in nine of the experiments: trm10-Δ, trm1-Δ, TetO7-TRM5, TetO7-GCD10, gcd14-1ts and the four dihydrouridine synthases.
Figure 2A shows a detailed view of the successfully detected methylation modifications. Probes complementary to known modified nucleotides are outlined in blue and the identities of the probes are listed. The trm10-Δ array specifically detects an increase in mutant binding (i.e. positive ratios) for oligos covering tRNA nucleotides 1–13 and 6–22, both of which span position 9 where the m1G is absent in the mutant. Oligos complementary to these positions in Ser and Leu tRNAs, which do not contain the m1G modification (8), do not show differential binding in mutant and wild-type, demonstrating that this technique is specific for individual tRNA isoforms. The same specificity is demonstrated by the exclusion of His and Asp tRNAs from the trm1-Δ array data and Lys tRNA from the TetO7-TRM5 data.
Gcd10p and Gcd14p form a complex to catalyze the formation of m1A at tRNA position 58. Microarrays with conditional alleles of both of these essential genes show high-ratio probes spanning position 58. The gcd14-1ts experiment produces smaller ratios and affects fewer oligos. Coupled with the fact that the TetO7-GCD10 strain has a more severe growth defect than the gcd14-1ts strain (data not shown) indicates that the gcd14-1ts allele is weaker than the tetracycline-regulated allele of GCD10.
A few high-ratio oligos exist outside of the expected tRNA regions in the trm1-Δ experiment and both experiments in which the Gcd10-Gcd14 complex was disrupted. In the case of the Gcd10–Gcd14 complex, these probes correspond to MetCAT oligos, examined in detail in Figure 4D. Probes with unusual behavior on the trm1-Δ microarray have previously been observed as false-positives [see (9) Figure 3, dus2-Δ]. It is possible that the loss of modification causes changes in the secondary structure of the tRNAs, which impacts hybridization to the array.
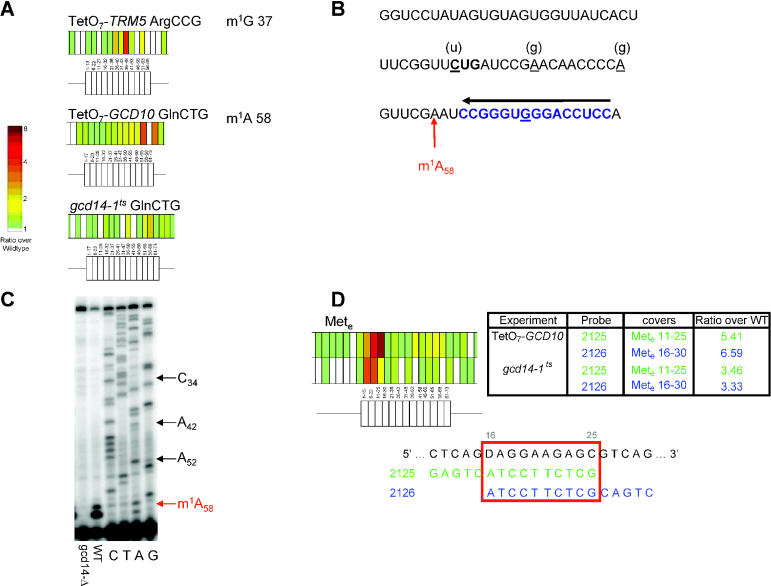
Figure 4.
Novel modification events. (A) Potential new targets for Trm5 and the Gcd10/Gcd14 complex. Modifications and target sites are proposed for three tRNAs whose RNA sequences have not been published and RNA modification profiles are unknown. (B) Demonstration of m1A58 modification in tRNAGlnCUG. Inferred RNA sequence of tRNAGlnCUG showing the position of the primer used to detect m1A modification at position 58 (highlighted in blue). The four positions that are underlined are the residues of this minor tRNA species that differ from the sequence of the other two previously characterized tRNAGln isoforms (both tRNAGlnUUG). The residues found at those positions in tRNAGlnUUG are shown in parentheses above. (C) Primer extension analysis of RNA derived from either gcd14-Δ (lane 1) or wild-type cells (lane 2). Lanes C, T, A and G are sequencing lanes of the primer extended RNA. (D) Two elongation-specific tRNA Met oligos show formamide-dependent differential hybridization in GCD10/GCD14 mutants. A schematic diagram and the corresponding values in the chart show that probes covering Mete nucleotides 11–25 and 16–30 exhibited high ratios in both experiments targeting the GCD10–GCD14 complex. tRNA and probe sequences are shown below; the overlapping region, Mete 16-25, is outlined in red. The table to the right shows the difference in ratio of representative probes for two formamide concentrations.
Figure 3.
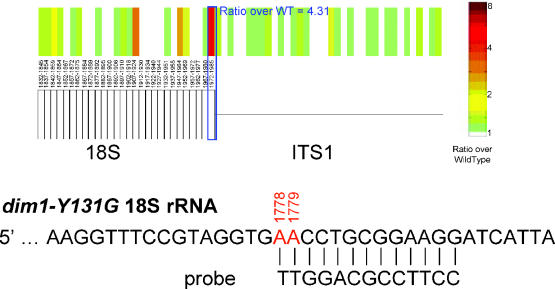
18S rRNA modification by Dim1p. A schematic diagram of the 3′ portion of 18S RNA from the dim1-Y131G microarray is shown. The 18S oligo with the highest ratio was 11157, complementary to the modified adenosines in the 3′ terminal loop of the RNA, shown below.
Selected tRNAs from each of these mutant strains are shown in detail in Figure 2B. We observe between one and three high-ratio oligos for each tRNA, suggesting that additional factors beyond the simple presence or absence of the modified nucleotide can affect binding to the array (see Discussion). However, only oligos complementary to modified nucleotides show increased binding to the mutant tRNAs, confirming the ability of the microarray to detect the oligos complementary to the modified nucleotides with good specificity.
The site and nature of each modification is shown on the nucleotide base diagrams to the right of the tRNAs; the modified functional group is outlined with a red circle, and functional groups involved in Watson–Crick base pairing are circled in blue. All of the successfully-detected modifications involve methylation of a functional group required for formation of canonical Watson–Crick base-pairs.
Modifications to other ncRNAs
Covalent modification is an important feature of ribosomal RNA as well as tRNA. Mature ribosomal RNA in S.cerevisiae contains over one hundred modified nucleotides; 60% of these occur in functionally important regions including the peptidyl transferase centre and the A, P and E sites (17). One of these modifications is the formation at consecutive nucleotides in the 3′ terminal of 18S rRNA (17) by Dim1p, as was successfully detected by microarray analysis of a catalytic knockout (Y131G) of DIM1 (Figure 3). However, the majority of the modifications to rRNA, snoRNAs and snRNA are pseudouridine and 2′O-methylation [44 and 54 respective occurrences, (7,18)], neither of which were detected in the experiments presented here, presumably because they do not strongly affect Watson–Crick base paring.
Previously undocumented modification events
In addition to the probes complementary to nucleotides known to be modified, additional high-ratio probes were observed on the TetO7-TRM5, TetO7-GCD10 and gcd14-1ts arrays (see below). The specificity demonstrated for known sites of modification led us to speculate that the differential binding to these probes may indicate previously undocumented sites of modification. In order to separate potential modification sites from noise in the data, we looked at the intensity of each of these probes, as well as the behavior (ratio and intensity) of overlapping probes in the same region. We reasoned that at least two high-ratio, overlapping probes with intensities >10-fold above background may indicate the presence of a novel site of modification.
Our microarray contains probes complementary to 14 tRNAs whose RNA sequences are not contained in the publicly available Bayreuth database (8). At least two of these tRNAs show hybridization patterns consistent with modification at positions known to be modified in other tRNAs: m1G 37 in ArgCCG and m1A 58 in GlnCTG isolated from GCD10/14 complex mutants. Schematic diagrams of each of these tRNAs with ratios from the relevant array are shown in Figure 4A. In order to confirm the microarray observations, these tRNAs were examined using primer extension analysis.
We confirmed the modification at position 58 of GlnCTG by primer extension analysis. To rule out the possibility of contamination by hybridization to the major tRNA GlnUUG species (which were already known to contain m1A 58), we used a probe for primer extension that spanned a region at the 3′ end which overlaps one of the positions that differs between tRNAUUG and tRNACUG species (Figure 4B). Using this primer, a block is observed at position A59 in the RNA from wild-type cells (Figure 4C, lane 2), consistent with the presence of m1A 58; this block is absent in RNA from mutant cells (Figure 4C, lane 1), which extends to the 5′ end of the tRNA. The sequencing reactions demonstrate the specificity of the primer for this tRNA species (Figure 4C, lanes C, T, A and G) since the sequence at the positions indicated by arrows are all those of the tRNAGlnCUG isoform (C34, A42 and A52). The ArgCCG tRNA failed to yield visible primer extension products, possibly because it is a single-copy tRNA and of low abundance.
The Gcd10/Gcd14 complex is responsible for methylating the N1 group at A58 in many yeast tRNAs. We noted that in addition to position 58-specific probes, oligos specific for the elongator Met tRNA nucleotides 11–25 and 16–30 had high ratios in both the TetO7-GCD10 and gcd14-1ts arrays (Figure 4C). Unlike other high-ratio oligos, differential binding to these probes was sensitive to the formamide concentration in the hybridization buffer; the ratios of probes 2125 and 2126 were significantly higher on arrays hybridized in 33% formamide than 25% (Figure 4C; see text below). We were unable to use primer extension analysis to determine whether one or more of the nucleotides in the overlapping region (tRNA nucleotides 16–25) is modified in this tRNA because the proximity of the known modification to the proposed site of modification interfered with binding of the probe (data not shown). When analyzed by denaturing gel electrophoresis and northern blotting, elongator Met tRNA from wild-type and conditional GCD10/14 complex mutants did not show any differential hybridization to probe sequences complementary to nucleotides 16–25 (data not shown). Taken together with the microarray results, this suggests that the differential binding observed at the low formamide concentration is caused by a secondary structure effect rather than a covalent modification. Although this phenomenon has not been fully characterized, it is intriguing that the effect is specifically observed in the Met tRNA because of the well-established, essential role of the Gcd10/14 complex in catalyzing the maturation of the initiator Met tRNA (14).
DISCUSSION
We have designed a tiling microarray to assay all known and several predicted non-coding RNAs in S.cerevisiae. We used this array to demonstrate that a subset of covalent RNA modifications could be detected by microarray: loss of modification in mutant strains enabled specific portions of tRNAs to bind more efficiently to the array than the modified, wild-type tRNA. Modifications that were successfully detected include both dihydrouridine and methylation of base functional groups directly involved in Watson–Crick base pairing. We further confirmed the utility of the array in detecting previously undocumented modifications, one of which we confirmed with primer extension experiments.
We do observe a significant number of false-negative probes, i.e. not all probes covering the modified nucleotides were more efficiently bound by the mutant tRNAs. This may be caused by the potential secondary structure in either the probes or the tRNAs themselves. In some cases, these may be overcome by changing hybridization stringency and/or probe length; however, we note that regardless of the RNA sample, hybridization stringency, or oligo probe length, we observed more efficient binding of the 5′ and 3′ ends of tRNAs compared with binding to probes covering the middle sections of tRNAs (Supplementary Material and data not shown).
A clear pattern emerged regarding the ability of the microarray to detect any given modification: modifications that involve the placement of one or more methyl groups on a functional group involved in Watson–Crick base pairing were efficiently detected, and those that involve methylation of the opposite face of the base or the sugar were not. The simplest explanation for this is that the presence of the methyl group directly interferes with base pairing to the probe sequences; absence of the methyl group in the mutant improves binding. This explanation is consistent with the observation that single-base mismatches are efficiently detected by microarray (15,19). The exception to this is the exceptionally good detection of dihydrouridine modifications described previously (9). Dihydrouridine contains two additional hydrogen atoms, at positions five and six of the base. The presence of these hydrogens not only changes the sugar-pucker of the ribose, but removes the double bond between C5 and C6, seriously perturbing the architecture of the six-membered ring and moving the N3 and O4 functional groups out of alignment for Watson–Crick base pairing (4).
If the disruption of Watson–Crick base pairing is sufficient for the detection of modification, we would expect to have successfully detected the modifications catalyzed by Tad1p and Mod5p. Tad1p catalyzes the deamination of adenosine to inosine, converting the hydrogen bond donor N6 to oxygen. Because this modification involves the substitution of NH2 for O, rather than the addition of a bulky methyl group, it is likely that it is less disruptive to Watson–Crick base pairing. The presumably small difference in binding efficiency may be detected if the RNAs were hybridized in a different combination of salt and formamide concentrations or at a different temperature. In addition, this modification only occurs at nucleotide 37 of tRNA AlaAGC, a position covered by only three probes on the array. Mod5p catalyzes the addition of an isopentyl group to the N6 position of tRNA A37. It is expected that the attachment of this bulky group to the Watson–Crick face of adenosine would interfere with base pairing and therefore be detectable by microarray; however, an i6A-modified nucleotide retains one amino proton available for hydrogen-bonding, and can form a base pair. Consistent with this, the i6A modification is not a primer extension block (J. Jackman and E. Phizicky, unpublished data). Furthermore, this modification occurs in only a few tRNA isoforms (Ser, Cys and Tyr) and none of the (relatively few) oligos on the array that are complementary to these sequences is detected above background. Taken together, these data suggest that it is the disruption of base pairing potential, rather than the simple presence of a methyl group on the Watson–Crick face of a nucleotide, that renders a modification detectable by microarray.
While it is logical that the addition of one or more bulky methyl groups to the Watson–Crick face of a nucleotide interferes with the ability of that nucleotide to form a base pair, the predicted effect of pseudouridine modification on base pairing is not as straightforward. Pseudouridine contains a C–C glycosyl bond linking base and sugar, and an additional hydrogen bond donor in the free N1H group. Although these changes do not directly affect the Watson–Crick face of the base, they do act to increase local base stacking in both single- and double-stranded regions (5) and might be expected to increase the affinity of the modified RNA to the microarray. We examined the differences in hybridization between wild-type RNA and RNA from two pseudouridine synthase mutants (pus4-Δ and pus7-Δ) and were not able to detect any differences in base pairing efficiency, positive or negative.
Historically, one of the most common ways to detect modified nucleotides at specific positions was via primer extension analysis. For several modifications (e.g. m1G and ), the presence of the modified nucleotide is sufficient to disrupt elongation of the template and cause a primer extension stop at the site of modification. Individual modifications are assigned to specific enzymes when the stop is not present in RNA isolated from strains with mutant alleles of the enzyme responsible for the modification (20,21). Other modifications are not sufficiently disruptive to the polymerase and can only be detected by primer extension after chemical modification of the modified nucleotide. Methods have been developed to detect both pseudouridine and dihydrouridine in this way (9,22). It may be possible to apply a similar strategy for the detection of additional modifications by microarray; the attachment of a bulky group, such as CMCT specifically to pseudouridine residues, may disrupt base pairing to the probe and allow detection of the modification by microarray. Similar chemical strategies may be possible for the detection of 2′OMe and other modifications, including DNA methylation, which has been shown to be an important epigenetic silencing method (23,24). DNA replication in Escherichia coli is regulated by methylation of N6 of A residues (which are involved in Watson–Crick base pairs) and could in principle be detectable by microarray (25).
Recent years have seen the roles played by RNA molecules in the cell increase from the simple translator between of DNA and protein, to include regulation of gene expression (26), both structural and catalytic roles in protein synthesis (27) as well as roles in processing of other RNA molecules (2). It remains to be seen how many newly discovered RNA classes will also feature modifications. The method presented here for directly detecting the modification events provides the first example of a genome-wide screen for RNA modifications, and further optimization should extend the scope of the technique, contributing to our understanding of basic RNA functions in the cell.
Acknowledgments
We thank Mercedes Tamame and Denis Lafontaine for providing strains and for helpful discussions, Kirsten Krause and Carol Dieckmann for manual annotations to the mitochondrial genes and members of the Hughes laboratory for discussion of results. We are grateful to Dr Rick Collins for critical evaluation of the manuscript. This work was supported by CIHR and CFI grants to T.R.H. and a CIHR post-doctoral fellowship to S.L.H. J.E.J. and E.M.P. were suppored by NIH grant 52347 to E.M.P. Funding to pay the Open Access publication charges for this article was provided by CIHR. Funding to pay the Open Access publication charges for this article was provided by CIHR.
REFERENCES
- 1.Cheng X., Roberts R.J. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 3.Gerber A.P., Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 4.Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 5.Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- 6.Bachellerie J.P., Cavaille J., Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 7.Ma X., Zhao X., Yu Y.T. Pseudouridylation (Psi) of U2 snRNA in S.cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003;22:1889–1897. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xing F., Hiley S.L., Hughes T.R., Phizicky E.M. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- 10.Peng W.T., Robinson M.D., Mnaimneh S., Krogan N.J., Cagney G., Morris Q., Davierwala A.P., Grigull J., Yang X., Zhang W., et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto N., Nakano S., Yoneyama M., Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 13.Mnaimneh S., Davierwala A.P., Haynes J., Moffat J., Peng W.T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Anderson J., Phan L., Cuesta R., Carlson B.A., Pak M., Asano K., Bjork G.R., Tamame M., Hinnebusch A.G. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 16.Shai O., Morris Q., Frey B.J. 2003. Spatial bias removal in microarray images, University of Toronto Technical Report PSI-2003-21.
- 17.Decatur W.A., Fournier M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 18.Benne R., Grosjean H. Modification and Editing of RNA. Washington, DC: ASM Press; 1998. [Google Scholar]
- 19.Salamon H., Kato-Maeda M., Small P.M., Drenkow J., Gingeras T.R. Detection of deleted genomic DNA using a semiautomated computational analysis of GeneChip data. Genome Res. 2000;10:2044–2054. doi: 10.1101/gr.gr-1529r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackman J.E., Montange R.K., Malik H.S., Phizicky E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maden B.E. Mapping 2′-O-methyl groups in ribosomal RNA. Methods. 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- 22.Bakin A., Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 23.Egger G., Liang G., Aparicio A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 24.Jiricny J. DNA repair: bioinformatics helps reverse methylation damage. Curr. Biol. 2002;12:R846–R848. doi: 10.1016/s0960-9822(02)01350-7. [DOI] [PubMed] [Google Scholar]
- 25.Boye E., Lobner-Olesen A., Skarstad K. Limiting DNA replication to once and only once. EMBO Rep. 2000;1:479–483. doi: 10.1093/embo-reports/kvd116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Oligonucleotide sequences on the arrays, and all microarray data are available at (hugheslab.med.utoronto.ca/Hiley). Spreadsheets containing the data displayed in Figures 1B and 2A are also available on the website.
Figure 1.
Detection of covalent modification by microarray. (A) Modification disrupts base pairing between RNA and probe. Wild-type tRNA LysCTT (top) contains a dimethylguanosine residue at position 26, which disrupts pairing with the probe. trm1-Δ tRNA LysCTT (bottom) lacks this modification and can pair completely with the probe (see also Figure 2B). A schematic diagram of the tRNA is shown below. Rectangles represent probes complementary to tRNA sequence, and thin lines represent probes complementary to 5′ and 3′ genomic flanking regions. The relative fluorescence of each probe is indicated by color-coded rectangles above the schematic diagram (according to the scale on the right); the tRNA nucleotides covered by each oligo are shown. (B) Analysis of strains defective for tRNA modification. tRNA oligos (ordered from 5′ to 3′) versus individual experiments (described below the figure) are plotted. Oligos to which there was significantly better binding in the mutant tRNA samples are indicated by red color, as shown by the color-bar in (A). Groups of probes covering tRNA nucleotides modified by each enzyme are outlined in blue rectangles. The type of modification and positions known to be modified by each enzyme are shown. Only tRNA probes with ratios at least 2-fold above wild type are shown.
Figure 2.
tRNA methylation analyzed by microarray. (A) Three different types of detectable methylation. Unique tRNA probes with ratios of at least 2 are color-coded according to the scale shown and displayed from 5′ to 3′ of the tRNA sequence. The tRNA isoforms and specific nucleotides covered are shown to the right of the figure. Oligos predicted to be affected in the each experiment are outlined with blue rectangles. (B) Schematic representation of selected tRNAs. One tRNA from each of the experiments in which the methylation defect was detected is shown in schematic form as described in Figure 1A. Functional groups involved in Watson–Crick base pairing are circled in blue; modifications are circled in red.