Abstract
ArrayOme is a new program that calculates the size of genomes represented by microarray-based probes and facilitates recognition of key bacterial strains carrying large numbers of novel genes. Protein-coding sequences (CDS) that are contiguous on annotated reference templates and classified as ‘Present’ in the test strain by hybridization to microarrays are merged into ICs (ICs). These ICs are then extended to account for flanking intergenic sequences. Finally, the lengths of all extended ICs are summated to yield the ‘microarray-visualized genome (MVG)’ size. We tested and validated ArrayOme using both experimental and in silico-generated genomic hybridization data. MVG sizing of five sequenced Escherichia coli and Shigella strains resulted in an accuracy of 97–99%, as compared to true genome sizes, when the comprehensive ShE.coli meta-array gene sequences (6239 CDS) were used for in silico hybridization analysis. However, the E.coli CFT073 genome size was underestimated by 14% as this meta-array lacked probes for many CFT073 CDS. ArrayOme permits rapid recognition of discordances between PFGE-measured genome and MVG sizes, thereby enabling high-throughput identification of strains rich in novel genes. Gene discovery studies focused on these strains will greatly facilitate characterization of the global gene pool accessible to individual bacterial species.
INTRODUCTION
To date, the entire genomic sequence of more than 180 bacterial strains has been determined. Based on comparative analysis of multiple genomes of the same species, it is increasingly apparent that some bacteria possess an extremely plastic genome (1–4). Foreign DNA segments, acquired via horizontal gene transfer, result in a genomic mosaic that reflects the lifestyle of the bacterium, pathogenic traits, adaptation to particular ecological niches and evolutionary history (5). This ‘optional’ genomic repertoire, which we refer to as the ‘mobilome’ (mobile genome), includes episomal plasmids, transposons, integrons, prophages and a growing list of genomic islands (GIs) (6,7). Pathogenicity islands, the virulence-associated subset of GIs, have now been identified in many bacterial species and are undoubtedly recognized as major players in the moulding of pathogenic traits. The high cost of genome sequencing has been a barrier to high-throughput prospecting of the mobilome (8). Even costly mega-scale metagenomics projects do not facilitate this process as the derived data are largely skewed towards abundant DNA sequences and low-prevalence mobilome sequences would rarely fit within a wider genomic context (9). A rapid and more cost-effective approach to discovering strain-specific DNA sporadically dispersed among hundred of members of the same species remains a major challenge (10).
Since DNA microarrays were first used to compare the genomes of Mycobacterium bovis BCG strains with that of Mycobacterium tuberculosis strain H37Rv to reveal several strain-specific deletions (11), comparative genomic hybridization technology has been extensively applied to investigate genome diversity among distinct isolates of many bacterial species including Bacillus anthracis (12), Brucella spp. (13), Campylobacter jejuni (14,15), Escherichia coli (8,16–18), Helicobacter pylori (19), Mycobacterium spp. (20,21), Porphyromonas gingivalis (22), Pseudomonas aeruginosa (23), Rickettsia prowazekii (24), Salmonella spp. (25), Shewanella spp. (26), Xylella fastidiosa (27) and Yersinia spp. (28,29). Indeed, microarray-based comparative genomic indexing (CGI) (18) has become a powerful generic tool in the study of bacterial pathogens.
In particular, E.coli appears to be a front-runner in the genome diversity stakes comprising isolates with a total genome size of 4.5–5.5 Mb (30). The common E.coli chromosomal ‘backbone’ of ∼3.6 Mb is punctuated by numerous integrated GIs (1,3,16). The ability of E.coli to tap into a large mobile gene pool has probably led to its remarkable success as both an innocent commensal and a virulent pathogen, with its near ubiquitous distribution in the environment and in animal hosts. To date, human pathogenic E.coli are classified into at least 10 distinct pathotypes including, for reasons of genetic relatedness, the four species of Shigella that are now considered to be subspecies of E.coli (31).
CGI data identifies the subset of genes common to both the microarray used and the genome of the strain under investigation. This complement of genes represents an entity, which we define as the microarray-visualized genome (MVG). In this paper, we describe, test and validate a new program, ArrayOme v1.0, which provides an accurate estimate of the size of the MVG of a bacterium based on CGI data alone. Using an approach we have termed Microarray-Assisted mobilome Prospecting (MAmP), we plan to exploit this tool to screen large numbers of isolates and identify E.coli and Shigella strains that are rich in novel genetic material for further detailed analyses. The MAmP approach combines CGI, ArrayOme and pulsed-field gel electrophoresis (PFGE) to predict the size of the novel, non-microarray-borne mobilome in a test strain (Figure 1). ArrayOme calculates the sizes of the MVGs of strains based on their possession and presumed reference template-like organization of CGI-defined subsets of microarray-borne genes. The sizes of novel mobilomes borne by individual strains can be estimated by comparing MVG sizes with PFGE-measured chromosome sizes. The degree of size-discordance between PFGE-measured physical genomes and MVGs can be used to identify isolates rich in novel genetic material that are likely to carry unique GIs or prophages. Ochman and Jones (16) had estimated that the amount of unique DNA present in four E.coli strains investigated ranged from 65 to 1183 kb based on differences in chromosome length and gene content relative to E.coli K-12 MG1655.
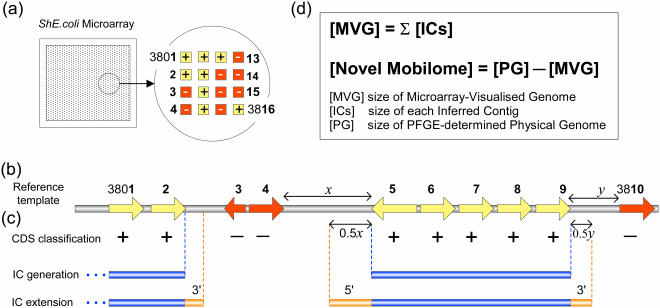
Figure 1.
Microarray-Assisted mobilome Prospecting (MAmP): a method for determining the discrepancy between the physical genome size and that accounted for by known genes represented on an expanded species-specific microarray. (a) A schematic representation of a microarray-based CGI output for an hypothetical region of the genome in a strain under investigation by the MAmP technique. Scanned raw data are normalized permitting classification of microarray-represented genes as ‘Present (+)’ or ‘Absent (−)’ in individual test strains. (b) The arrows represent the genetic organization of these CDS within E.coli K-12 MG1655, E.coli K-12 W3110, E.coli O157 EDL933, S.flexneri 2a Sf301 or other virulence-associated gene clusters included on the MG1655, W3110 or ShE.coli microarrays. (c) Contiguous CDS classified as ‘Present’ are merged into an IC with intergenic non-coding segments between the contiguous CDS included in the corresponding IC. Each IC was then extended in both directions by lengths equal to half the flanking 5′ and 3′ intergenic segments, as indicated by the double-headed arrows of lengths 0.5x and 0.5y in the examples shown. When the ShE.coli meta-array sequences were used, each probe was mapped onto a single source reference template to allow for the generation of specific ICs. (d) The size of the MVG was calculated as the sum of all IC lengths. Consequently, the size of the non-microarray-borne novel mobilome was estimated as equal to the discrepancy between the pulsed-field gel electrophoresis-determined physical genome size and the MVG size. Figure not drawn to scale.
Previously reported experimental CGI data and in silico hybridization data derived from analysis of various E.coli and Shigella strains ‘hybridized’ against E.coli K-12 or the newly developed ShE.coli metagenome microarray are used to test ArrayOme v1.0. This generic bioinformatics program can be easily used to analyse any CGI data set for which a reference annotated template exists.
MATERIALS AND METHODS
Experimental CGI data
Experimental CGI data versus the E.coli K-12 MG1655 or the E.coli K-12 W3110 microarrays that had previously been interpreted were taken from the papers of Anjum et al. (18) and Fukiya et al. (8), respectively. The MG1655 microarray featured 4264 of the 4289 annotated protein-coding sequences (CDS) of MG1655, while the W3110 microarray bore probes specific for 4071 of 4390 W3110-annotated CDS. The microarray-borne DNA probes were amplified by PCR using CDS-specific primers and spotted onto slides as described previously (8,18). Following the reported microarray hybridization and data normalization protocols, the original authors had classed the status of individual CDS in test strains as ‘Present’ or ‘Absent’. Data associated with probes yielding low-reference signals relative to background were excluded from further interpretation. In this study, we have grouped CDS mapped onto these latter probes and CDS that were not represented on the microarray as being of ‘Indeterminate’ status.
Method to estimate the size of the MVG
A heuristic method was employed to estimate the size of the MG1655 MVG based upon the genetic organization of all 4289 annotated CDS present on the MG1655 reference template. The complete nucleotide sequence and annotation of MG1655 downloaded from GenBank (accession number U00096) served as the reference template. The method used is schematically shown in Figure 1. First, each of the 4289 annotated MG1655 CDS was flagged as ‘Present’ or ‘Absent’ in a test strain based on the results of a microarray hybridization experiment. Next, contiguous CDS classified as ‘Present’ were merged into an inferred contig (IC) with intergenic non-coding segments between these CDS included in the corresponding IC. Each IC was then extended in both directions. The 5′ end of each IC was extended by a length equal to half of that of the intergenic segment between the 5′ end of the IC and the adjacent ‘Absent’ CDS. Occasionally, prior to extension, the 5′end of the IC overlapped the flanking ‘Absent’ CDS. In such cases, the 5′ end of the IC was extended by 64 bp, a length approximately equal to half the average intergenic distance in the reference template MG1655. These same steps were employed for the extension of the 3′ ends of ICs as well. Finally, the lengths of all extended ICs were summated to yield the MVG size. Details of individual ICs could also be generated as optional output files. A computer program, named ArrayOme v1.0, was encoded using the programming language C++ to implement this algorithm. ArrayOme is freely available on request for academic purposes.
In silico CGI
Simulated CGI data corresponding to E.coli and Shigella strains with fully sequenced genomes were obtained using an in silico DNA–DNA ‘hybridization’ approach. The DNA sequences of the following genomes were downloaded from GenBank: E.coli K-12 MG1655 (accession number U00096) (32), E.coli O157:H7 EDL933 (AE005174) (1), E.coli O157:H7 Sakai (BA000007) (2), uropathogenic E.coli CFT073 (AE014075) (3), Shigella flexneri 2a Sf301 (AE005674) (4) and S.flexneri 2a 2457T (AE014073) (33).
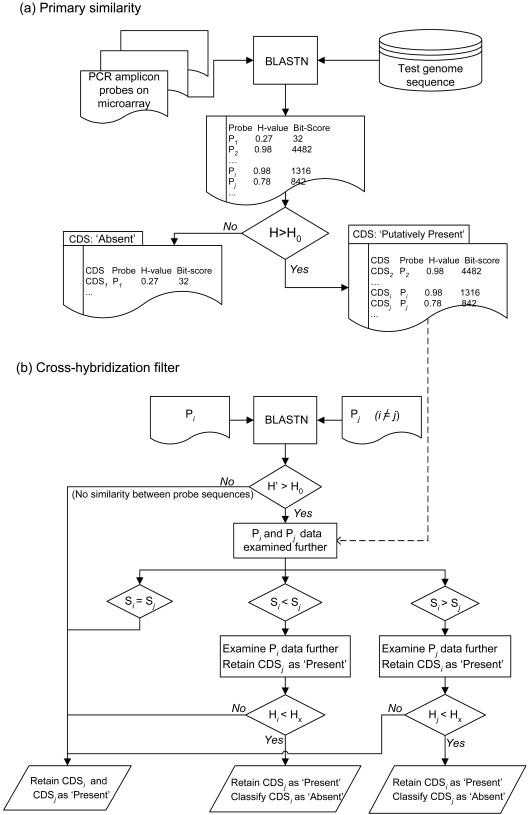
In a microarray experiment, ‘hybridization is a complex process that is not fully understood’ (34), in which bound DNA probes on the microarray and labelled targets in solution form heteroduplexes via Watson–Crick base pairing (34). Adjusting the temperature, salt concentrations and the concentration of labelled target molecules can alter the stringency of hybridization. In the in silico approach presented here, we utilize the degree and extent of DNA sequence similarity between the probe and target molecules as a surrogate measure for the likelihood of formation of stable heteroduplexes between these entities. The in silico CGI procedure applied included the following steps (Figure 2):
Each of the DNA sequences of the probes was used as a query in a similarity search against a test genome sequence using a locally installed version of BLASTN (35) and default NCBI BLASTN parameters.
The stretch of sequence from the test genome with the highest Bit score for each query sequence was retrieved and a homology score (H-value) calculated for each probe in turn. This homology score had been proposed by Fukiya et al. (8) and reflected the degree of similarity between the matching test genome sequence and the probe itself in terms of the length of match and the percentage sequence identity at the DNA level. For each query, the H-value was calculated as follows (8): H = (i × l)/lq, where i is the level of identity of the region with the highest Bit score expressed as a frequency of between 0 and 1, l, the length of the highest scoring matching sequence (including gaps) and lq, the query length. If there were no matching sequences with a BLASTN E-value < 0.01, the H-value assigned to that query sequence was defined as zero (8). Therefore, H belonged to the set, H ∈ [0,1].
For each probe, a threshold value of H (H0) was used to determine whether the corresponding CDS was to be classed as ‘Putatively Present’ or ‘Absent’. As mentioned above, each probe had been assigned an H-value. If H > H0 for a particular probe, the CDS mapped onto that probe was considered to be ‘Putatively Present’ and the probe itself further scrutinized to minimize misclassification of CDS as ‘Present’ due to signals arising from cross-hybridization events. The threshold value, H0, was selected to optimize sensitivity and specificity of the in silico approach versus true experimental CGI analysis (see Results and Discussion).
To increase the specificity of the in silico CGI output, cross-hybridization analysis was performed by a BLASTN search of each probe sequence against the remaining probe sequences in turn. For instance, a BLASTN similarity search was performed using the sequence of probe Pi as a query against the sequence of probe Pj (i ≠ j) and an H-value designated as H′ was obtained. If H′ > H0, the probes Pi and Pj were considered as a potential cross-hybridization pair and the original BLASTN results linked to these probes further examined. The CDS corresponding to either probe Pi or Pj would be re-classified as ‘Absent’ if the results of the BLASTN search against the test genome satisfied both of the following conditions: (i) it had a lower Bit score than that associated with the second probe and (ii) it had a H-value less than Hx, a second H threshold value that was also determined following review of the real hybridization results of E.coli EDL933 versus the MG1655 microarray (see Results and Discussion). The remaining ‘Putatively Present’ CDS were now designated as ‘Present’ resulting in a final in silico CGI data set of CDS classed as ‘Present’ or ‘Absent’ only. A Perl program was written to perform the BLASTN-based in silico DNA–DNA hybridization steps with a C++ program used to implement the remaining steps and produce the final in silico CGI output file.
Figure 2.
A flow diagram of the logic steps used for in silico CGI. CDS, P, H and S denote CDS mapped onto specific probes, individual microarray-borne amplicon probes, H-values and Bit scores, respectively. Pi and Pj denote distinct individual probes, with the subscripts i and j identify matching CDS, H-values and Bit scores. The steps involved in the primary similarity search are shown in (a), while the cross-hybridization algorithm that re-categorizes CDS from ‘Putatively Present’ to ‘Present’ or ‘Absent’ are shown in (b). A direct comparison of in silico generated data and true experimental CGI data for E.coli EDL933 obtained using the MG1655 microarray was used to validate the algorithm. The key threshold values, H0 = 0.40 and Hx = 0.96, were set to optimize the sensitivity and specificity of identifying genes as ‘Present’ when using the in silico as compared to experimental approach (see Figure 3).
RESULTS AND DISCUSSION
Analysis of E.coli K-12 microarray data using ArrayOme v1.0
In a ‘compact’ bacterial genome, CDS occupy the majority of the chromosome. For example, 88% of the E.coli K-12 MG1655 chromosome contains CDS. The average CDS length in MG1655 is 953 bp, while the mean distance between CDS is 128 bp. We have utilized previously reported CGI data derived from the E.coli K-12 MG1655 microarray (18) and the complete chromosomal sequence of MG1655 as a reference template to predict the MVG sizes of a range of pathogenic E.coli strains associated with diarrhoeal diseases. The fully sequenced enterohaemorrhagic E.coli (EHEC) strain EDL933 was included in this set. Of the 4264 MG1655 CDS represented as probes on the microarray, 3775 were identified as ‘Present’ in EDL933, while the status of a further 114 was ‘Indeterminate’ (see Materials and Methods). Inputs of these data into ArrayOme produced 113 or 193 ICs (see Figure 1) scattered along the MG1655 template chromosome depending on whether the ‘Indeterminate’ CDS were classified as ‘Present’ or ‘Absent’, respectively. After 5′ and 3′ extension of each of these ICs, the MG1655-MVG size of EDL933 was calculated as the sum of all IC lengths resulting in an estimate of 4141–4254 kb. The size of the genome occupied by non-MG1655 genes was then deduced by subtracting this MVG size from its known physical size based on the available EDL933 genome sequence to yield an estimate of 1275–1387 kb for the non-MG1655 mobilome present in EDL933. This number compared well with the previously reported total length (1.34 Mb) of species-specific ‘O-islands’ identified in the EDL933 deduced following pairwise similarity alignment between the EDL933 and MG1655 genomes (1). Ochman and Jones (16) had proposed that the size of the novel mobilome of a strain could be estimated based on the differences in chromosome lengths and gene contents between a test strain and a reference strain, but had not stipulated in detail the procedure used to calculate the amount of ‘missing DNA’ relative to the reference strain. Importantly, the ArrayOme approach takes account of likely lost intergenic sequences (Figure 1). As mentioned above, the average length of intergenic sequences in MG1655 was 128 bp. Hence, as there were 400–514 missing CDS in EDL933 the size of its non-MG1655 mobilome would have been underestimated by ∼51–66 kb using a method based on the total size of missing CDS alone. To improve prediction accuracy, we used the strategy of creating ICs and then subjecting these virtual entities to rule-defined 5′ and 3′ extensions to allow the inclusion of likely conserved intergenic stretches within the MVG. Based on this algorithm, reference template-borne non-coding sequences flanking CDS identified as ‘Absent’ were also attributed as missing from the final virtual genome.
Similar calculations were performed with the remaining CGI data reported by Anjum et al. (18) for the 25 other pathogenic E.coli strains. However, because of the large number of CDS classified as ‘Indeterminate’ for many of these strains, we applied a set of ArrayOme-implemented logic rules in an attempt to better assign these genes based on biological realities. First according to Rule I, all ‘Indeterminate’ CDS that were conserved at a nucleotide level across all six fully sequenced E.coli and Shigella genomes were re-classified as ‘Present’. In this study, we chose a threshold of H > 0.64 as it reflected a degree of nucleotide conservation equivalent to 80% identity over 80% of the CDS length. Selection of H-values ranging from 0.4 to 0.8 resulted only in minor differences in the number of CDS regarded as conserved. Next according to Rule II, single and paired ‘Indeterminate’ CDS that were directly flanked by genes identified as ‘Present’ were also deemed to be ‘Present’, while the reverse was applied to CDS flanked by ‘Absent’ CDS. When ‘Indeterminate’ CDS were present in clusters of three or more, the implicated CDS were classified as ‘Absent’ regardless of the status of the flanking CDS. Indeed, if an intermediate category was used in the interpretation of CGI data, many such CDS would be likely to represent truly divergent genomic sequences. An additional sub-rule based on the G+C-content of the ‘Indeterminate’ CDS cluster as compared to the genome average could also be applied to clusters of ≥3 CDS to vary the final interpretation (see instructions for ArrayOme program). The choice of rules and the order in which these were to be implemented were entirely user-defined. Depending on the interpretation criteria applied, many CDS classed as ‘Indeterminate’ following experimental CGI analysis might represent examples of gene sequence polymorphism resulting in weaker, poorly interpretable microarray hybridization signals. Indeed, when wide ranges of inferred MVG sizes are obtained following analysis of un-reconciled microarray data these could reflect the extent of genomes occupied by microarray-related but divergent genes. The MVG sizes for all 26 E.coli strains predicted using the reconciled CGI data and ArrayOme are available in the Supplementary Material. The estimated number of ‘Absent’ MG1655 CDS ranged from 222 to 888, while the MVG size estimates spanned 3.7–4.4 Mb. The chromosome sizes of natural E.coli strains vary by as much as 1 Mb and are currently recognized to range from 4.5 to 5.5 Mb (30). Based on these limits, we speculate that as much as 1.8 Mb (5.5−3.7 = 1.8 Mb) of the genome of one of these E.coli isolates may differ from that of E.coli K-12 and that this mobilome alone could harbour up to 1800 non-MG1655 CDS.
Recently, Fukiya et al. (8) reported the CGI results of 22 pathogenic E.coli and Shigella strains using an E.coli K-12 strain W3110 microarray that bore probes for 4071 of 4390 annotated CDS. We have estimated the MVG sizes of these test strains using their reported CGI data. The 4 641 433 bp E.coli K-12 W3110 chromosome was used as the reference template for this analysis (http://gib.genes.nig.ac.jp/). The enteropathogenic E.coli strain E2348/69 was found to possess 3568 W3110 CDS (8). CDS categorized as ‘Indeterminate’ were re-classified based on the logic rules described above. Entry of the reconciled E2348/69 CGI data into ArrayOme resulted in the generation of 164 or 165 ICs and an estimated MVG size of 4096–4125 kb. Consequently, as the physical chromosome size of E.coli E2348/69 had previously been estimated by PFGE as ∼4.7 Mb (17), it was predicted to possess a non-W3110-related genome complement of ∼0.6 Mb, suggesting an accessory gene content of about 600 relative to W3110. The soon to be available E2348/69 genome sequence (http://www.sanger.ac.uk/Projects/Escherichia_Shigella/) would allow validation of these predictions. The estimated W3110-MVG sizes for all 22 strains are available as Supplementary Material.
Determination of threshold values of H to be used in in silico CGI
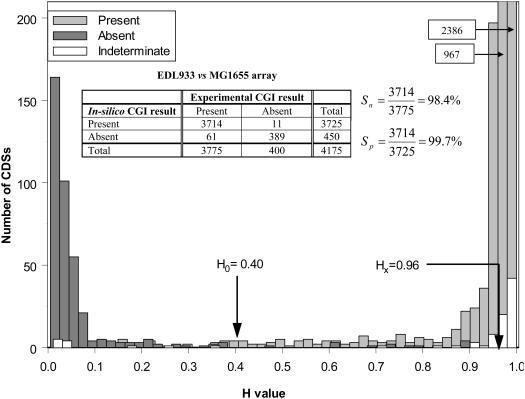
To classify CDS as ‘Present’ or ‘Absent’ using a BLASTN-facilitated in silico DNA–DNA hybridization approach, suitable threshold values of H (see Materials and Methods) were required. Based on the experimental microarray data of Anjum et al. (18) for EDL933, 400 CDS were classified as ‘Absent’, 3775 CDS as ‘Present’ and a further 89 as ‘Indeterminate’ out of a total of 4264 MG1655-CDS probes spotted onto the microarray. BLASTN-based in silico hybridization of the 4264 microarray probes versus the E.coli EDL933 genome sequence resulted in similarity Bit scores and the H-values for each of the amplicon probe sequences. The distribution of H-values corresponding to probes was bipolar with the average H-value for the 400 ‘Absent’ CDS (0.087; SD = 0.009) being significantly less than that for the 3775 CDS identified as ‘Present’ (0.962; SD = 0.110) and the 89 microarray-represented CDS of ‘Indeterminate’ status (0.852; SD = 0.309) (Figure 3). The H-values associated with the 89 ‘Indeterminate’ calls were predominantly clustered with those of probes mapped onto CDS categorized as ‘Present’, with only 12 having an H-value of ≤0.40. Based on the observed spread of H-values corresponding to the true experimental data, the threshold value, H0 = 0.40, was selected as the primary discriminant between CDS designated as ‘Putatively Present’ and those classed as ‘Absent’ (Figures 2 and 3). BLASTN data corresponding to CDS classed as ‘Putatively Present’ were then further scrutinized using the cross-hybridization filter described in Materials and Methods. The Hx threshold value was set at a high value of 0.96 to minimize the likelihood of miscalling a CDS as ‘Present’ when sequence similarity existed between its probe and a second microarray-borne probe(s) mapped onto an alternate CDS (Figure 2). With the EDL933 data set, this algorithm correctly predicted 389/400 ‘Absent’ and 3714/3775 ‘Present’ CDS for EDL933, yielding satisfactory sensitivity (98.4%) and specificity rates (99.7%) in the in silico CGI classification of CDS as ‘Present’ (Figure 3). By using these same H-value parameters, ∼86% (77/89) of the ‘Indeterminate’ CDS were classed as ‘Present’ by in silico simulation. This number was similar to the percentages of experimentally defined ‘Indeterminate’ CDS that were re-categorized as ‘Present’ following application of our logic rules concerning species-wide CDS conservation and the status of flanking CDS and reaffirms the value of these rules for the purpose of MVG sizing. In subsequent in silico hybridization simulations, CDS were judged to be ‘Present’ or ‘Absent’ using the algorithm shown in Figure 2 with the threshold values set at H0 = 0.40 and Hx = 0.96.
Figure 3.
The distribution of H-values corresponding to 4264 amplicon probes spotted onto the MG1655 microarray obtained following a BLASTN similarity search against the EDL933 chromosomal sequence are shown. Interval-grouped H-values are plotted with the data stratified into the experimental CGI categories of ‘Present’ (3775), ‘Absent’ (400) and ‘Indeterminate’ (89). The selected threshold values for in silico CGI, H0 = 0.40 and Hx = 0.96, are as indicated. The numbers in the boxes at the top right corner correspond to the ‘Number of CDS’ associated with the two bars that extend beyond the limits of the graph. The inset table shows a direct comparison of experimental CGI data derived by Anjum et al. (18) and in silico CGI data for the CDS classified as ‘Present’ or ‘Absent’ only. The sensitivity (Sn) and specificity (Sp) of identifying genes as ‘Present’ when using the in silico as compared to experimental approach, are shown on the right-hand side.
Estimated MVG sizes based on in silico CGI data generated using the ShE.coli meta-array
The recently developed PCR amplicon-based ShE.coli microarray contains many more E.coli CDS than the first generation E.coli K-12 microarrays. The ShE.coli meta-array probe sequences used in this study represented 6239 CDS comprising 4264 E.coli K-12 MG1655 CDS, 1101 E.coli EDL933 CDS, 516 S.flexneri 2a Sf301 CDS and a further 358 virulence-associated E.coli CDS derived from strains representative of different pathotypes of E.coli, particularly enteropathogenic E.coli (EPEC) and enterotoxigenic E.coli (ETEC). Of the 358 virulence-associated CDS, 132 were located on E.coli chromosomes while the remaining 226 mapped onto a range of plasmids based on published literature and GenBank submissions. The 25 MG1655 CDS not represented on the microarray were assigned based on logic rules described in the text with any remaining ‘Indeterminate’ CDS classed as ‘Present’ for simplicity. The ShE.coli metagenome microarray sequences presently constitute the most comprehensive representation of the global gene pool available to E.coli and Shigella strains.
In this study, we have used the five completed E.coli and Shigella genomes, MG1655, Sakai, CFT073, Sf301 and 2457T, to assess the ArrayOme-based estimation of MVG sizes with data derived by in silico CGI using sequences of probes present on the MG1655 and ShE.coli microarrays. Results of the analysis with EDL933 were also shown. However, it should be noted that as the EDL933 experimental data were used to train the in silico CGI analysis, the simulated output for this strain was of minimal significance. The total sets of MG1655 and ShE.coli microarray DNA probes were subjected to BLASTN analysis against each of the six completed genomes and the results interpreted using the logic steps shown in Figure 2. Given the in silico CGI algorithm applied, all microarray-represented CDS were classified as either ‘Present’ or ‘Absent’ with no ‘Indeterminate’ category. When the EHEC O157:H7 Sakai genome was analysed using the ShE.coli microarray, 5082 CDS were deduced to be ‘Present’, which comprises 3780 MG1655 CDS, 1083 EDL933 CDS, 164 Sf301 CDS, 50 other E.coli chromosomal virulence-associated CDS and 5 plasmid-borne virulence genes (Table 1).
Table 1.
The ArrayOme-predicted MVG sizes of E.coli and Shigella strains based on data derived by in silico hybridization of the complete genomes against the ShE.coli microarray amplicon probe sequences
| Strain | Size of the complete chromosome (kb)a | No. of CDS classified as ‘Present’b | Size of the ShE.coli-MVG [MG1655-MVG] (kb)c | Discrepancy between the chromosome length and size of the ShE.coli-MVG (kb) (% error)d | ||||
|---|---|---|---|---|---|---|---|---|
| MG1655 CDS (n = 4264 + 25) | EDL933-specific CDS (n = 1101) | Sf301-specific CDS (n = 516) | Other chromosomal CDS (n = 132) | Plasmid-borne CDS (n = 226) | ||||
| E.coli O157:H7 EDL933 | 5528 | 3783 | 1097 | 162 | 50 | 5 | 5566 [4192] | +38 (+0.7) |
| E.coli K-12 MG1655 | 4639 | 4288 | 58 | 59 | 17 | 8 | 4771 [4639] | +132 (+2.8) |
| E.coli O157:H7 Sakai | 5498 | 3780 | 1083 | 164 | 50 | 5 | 5556 [4191] | +58 (+1.1) |
| S.flexneri 2a Sf301 | 4607 | 3541 | 122 | 515 | 16 | 14 | 4519 [3882] | −88 (−1.9) |
| S.flexneri 2457T | 4599 | 3534 | 123 | 499 | 17 | 14 | 4507 [3882] | −92 (−2.0) |
| E.coli UPEC CFT073 | 5231 | 3638 | 278 | 174 | 26 | 9 | 4498 [4013] | −733 (−14.0) |
aThe lengths of the complete chromosomes, shown to the nearest kilobase (kb), are based on genome sequences lodged with GenBank.
bA total of 4264 E.coli K-12 MG1655 CDS, 1101 E.coli EDL933 CDS, 516 S.flexneri 2a Sf301 CDS, 132 other E.coli chromosomal virulence-associated CDS and 226 plasmid-borne E.coli virulence genes are classified as ‘Present’ or ‘Absent’ based on in silico CGI analysis. The 25 MG1655 CDS not represented on the microarray were assigned based on logic rules described in the text with any remaining ‘Indeterminate’ CDS classed as ‘Present’ for simplicity, leading to the classification of all 4289 MG1655 CDS.
cThe data corresponding to plasmid-borne genes was omitted when calculating MVG sizes as these CDS would normally be considered to be borne on episomal entities other than the main chromosome given their original identified location. The ArrayOme-predicted sizes of ShE.coli-MVGs (left) and MG1655-MVGs (right, square brackets) are shown to the nearest kilobase (kb).
dThe percentage errors between the reported lengths of the chromosomes and the sizes of the ShE.coli-MVGs are shown within parentheses.
MVG sizes were deduced using in silico ShE.coli microarray data in a manner similar to that described for the E.coli K-12 microarrays. In addition to the MG1655 chromosome, we used the annotated chromosomal sequences of E.coli EDL933 and S.flexneri Sf301 as reference templates for creating ICs. The genetic organizations of all other E.coli genes represented on the microarray were inferred from sequence and annotation details available from GenBank and related publications. When CGI data derived using meta-arrays are analysed, ArrayOme generates a set of linear ICs using as the reference template a single genome or GenBank sequence for each IC. The choice of template for each IC is directed by the contents of the user-generated Microarray Index file that specifies mapping details among probes, CDS and reference templates. Each probe is mapped onto a single CDS, which in turn is mapped onto a single template only. Further details of the algorithm used are available in the Supplementary Material. Comparisons of actual chromosome sizes and ArrayOme-estimated MVG sizes derived following the analysis of in silico CGI data are shown in Table 1. For five of the six test genomes, the discrepancy between the physical chromosome size and the MVG size as calculated using ShE.coli-derived in silico data was <2.8%. However, with CFT073 the margin of error was significantly larger; ArrayOme underestimated the true genome size of CFT073 by 733 kb or 14.0% of the complete genome. This was entirely consistent with the fact that the ShE.coli microarray lacked probes specific for at least 1501 CFT073-borne genes [(3) and H.-Y. Ou and K. Rajakumar, unpublished data]. As expected, analysis of MG1655 microarray data yielded significantly lower estimates of MVG sizes for all strains except MG1655, consistent with the known incomplete coverage of these genomes on this single-strain microarray (Table 1).
In order to further validate the ArrayOme tool, we generated a set of virtual derivative genomes. These derivatives were constructed from the six intact E.coli and Shigella genomes analysed above following defined ‘deletions’ of tRNA gene-borne GIs previously identified by the Islander algorithm (36). These virtual genomes were analysed by in silico comparison with the ShE.coli microarray sequences and the resulting data processed using ArrayOme (Table 2). Eight archetypal GIs encompassing 235 511 bp were deleted from the EHEC Sakai genome. Taking precise account of likely island boundaries, we constructed a virtual genome of 5 262 939 bp in length. The predicted MVG size of this derivative (Sakai-V8) was 5333 kb; a discrepancy of ∼1.3% compared with the known length of its virtual genome. A similar level of precision was obtained with four of the five other virtual derivatives but with CFT073-V6 the margin of error was significantly larger. However, the degree of discordance between the ‘physical’ length and the MVG size had fallen from 14.0% for the parent strain to 9.9% for the CFT073-V6 hypothetical construct, consistent with the fact that the deletion derivative had lost 311 CFT073-borne genes not represented on the ShE.coli microarray [(3) and H.-Y. Ou and K. Rajakumar, unpublished data].
Table 2.
The MVG sizes of ‘virtual genomes’ constructed by precise deletion of Islander-defined GIs
| Virtual strain designation | Virtual genome | No. of CDS classified as ‘Present’b | Size of the MVG (kb)c | Discrepancy between the sizes of genome and MVG (kb) (% error)d | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of GIs deleteda | Total length of deletion (bp) | Size of the derivative virtual genome (kb) | MG1655 CDS (n = 4264 + 25) | EDL933-specific CDS (n = 1101) | Sf301-specific CDS (n = 516) | Other chromosomal CDS (n = 132) | Plasmid-borne CDS (n = 226) | |||
| EDL933-V9 | 9 (398) | 321 129 | 5207 | 3776 | 905 | 144 | 48 | 3 | 5351 | +144 (+2.8) |
| MG1655-V3 | 3 (81) | 65 825 | 4573 | 4219 | 53 | 55 | 17 | 5 | 4705 | +132 (+2.9) |
| Sakai-V8 | 8 (327) | 235 511 | 5263 | 3774 | 888 | 142 | 48 | 3 | 5333 | +70 (+1.3) |
| Sf301-V3 | 3 (70) | 75 968 | 4531 | 3538 | 112 | 469 | 16 | 11 | 4445 | −86 (−1.9) |
| 2457T-V4 | 4 (68) | 82 476 | 4517 | 3527 | 113 | 448 | 17 | 11 | 4424 | −93 (−2.1) |
| CFT073-V6 | 6 (435) | 395 611 | 4836 | 3606 | 214 | 138 | 20 | 6 | 4357 | −479 (−9.9) |
The ArrayOme-facilitated MVG size prediction was based on data derived by in silico CGI hybridization of the complete genomes against the ShE.coli microarray amplicon probe sequences.
aThe GIs that were deleted from the corresponding complete genomes were based on the Islander-derived data of Mantri and Williams (36). The details of these GIs along with the precise boundaries were downloaded from the database of Islander (www.indiana.edu/~islander) (36). The total numbers of CDS contained within the deleted GIs are shown within parentheses.
bA total of 4264 E.coli K-12 MG1655 CDS, 1101 E.coli EDL933 CDS, 516 S.flexneri 2a Sf301 CDS, 132 other E.coli chromosomal virulence-associated CDS and 226 plasmid-borne E.coli virulence genes are classified as ‘Present’ or ‘Absent’ based on in silico CGI analysis. The 25 MG1655 CDS not represented on the microarray were assigned based on logic rules described in the text with any remaining ‘Indeterminate’ CDS classed as ‘Present’ for simplicity.
cThe data corresponding to plasmid-borne genes was omitted when calculating MVG sizes as these CDS would normally be considered to be borne on episomal entities other than the main chromosome given their original identified location. ArrayOme-predicted MVGs are shown to the nearest kilobase (kb).
dThe percentage errors between the lengths of virtual chromosomes and the sizes of the ShE.coli-MVGs are shown within parentheses.
Predicted size of mobilomes in E.coli and Shigella strains based on in silico CGI data generated using the E.coli K-12 MG1655 microarray
The sequenced genomes of E.coli O157:H7 Sakai, uropathogenic E.coli CFT073 and S.flexneri Sf301 were used to test the proposed method of predicting the size of the non-MG1655 mobilome using results obtained via simulated CGI analysis versus the MG1655 microarray (Table 3). For the Sakai, CFT073 and Sf301 genomes, there were 3822, 3662 and 3553 CDS classified as ‘Present’, respectively. Based on this in silico CGI data and the rule-based assignment of the 25 unrepresented MG1655 CDS as described earlier, the size of the MVG was estimated for each strain in turn. The discrepancies between the true chromosome and the MVG lengths were then used to compute the size of individual strain-borne mobilomes. These sizes were then compared with the sizes of strain-specific sequences reported by the authors of the original genome sequences published. For example, in CFT073 there were 191 ICs and the MVG size was estimated to be 4013 kb. Thus, the size of novel mobilome was calculated as ∼1218 kb, which was comparable with the total length of non-MG1655 sequences (1306 kb) reported by Welch et al. (3). The results of this analysis for all three strains are shown in Table 3.
Table 3.
ArrayOme-predicted MVG sizes of three sequenced E.coli and Shigella genomes based on in silico CGI data derived using the E.coli K-12 MG1655 microarray
| Strain | Sequenced genome | MVG | |||
|---|---|---|---|---|---|
| Size of chromosome (bp) | Total size of non-MG1655 sequences (bp)a | No. of CDS classified as ‘Present’b | Size of the MVG (kb) | Size of the non-MG1655 mobilome (kb) (% error)c | |
| E.coli O157:H7 Sakai | 5 498 450 | 1 393 070 | 3822 | 4191 | 1307 (−6.2) |
| E.coli UPEC CFT073 | 5 231 428 | 1 306 391 | 3662 | 4013 | 1218 (−6.7) |
| S.flexneri 2a Sf301 | 4 607 203 | ∼700 000 | 3553 | 3882 | ∼725 (∼+3.6) |
aThe total lengths of strain-specific sequences with respect to E.coli K-12 MG1655 genome were reported by the authors of the published sequences. The extent of non-MG1655 sequences present in S.flexneri Sf301 was reported by Jin et al. (4) as ∼0.7 Mb.
bThe 4264 annotated E.coli K-12 MG1655 CDS represented on the microarray were classified as ‘Present’ or ‘Absent’ based on in silico CGI analysis described in the text. The 25 MG1655 genes not represented on the microarray were assigned based on logic rules described in the text with any remaining ‘Indeterminate’ CDS classed as ‘Present’ for simplicity.
cThe percentage errors between the reported lengths of the non-MG1655 sequences and the sizes of the non-MG1655 mobilomes as determined using ArrayOme are shown within parentheses.
Contribution of IS elements to MVG size estimates
In all calculations performed to this point, we have included the contribution of IS-associated CDS present in MG1655, EDL933 and Sf301, where these have been identified as ‘Present’ based on experimental or in silico CGI data. The MG1655 genome contains 52 annotated IS-associated CDS, particularly comprising IS1, IS2, IS3 and IS5, contributing to up to 44 kb of additional DNA. In addition, ShE.coli probes mapped onto IS-associated CDS on EDL933 and SF301 templates accounted for up to 73 kb of the MVG size. However, ArrayOme could be run using an option that assigned all IS-associated CDS as ‘Absent’ resulting in reduced MVG size estimates (Supplementary Material). Empirical or experimentally guided adjustments could then be made to account for the estimated IS content of individual strains. ArrayOme also offers the option of accounting for other amplified CDS that may be borne on duplicated regions of the genome in its calculation of MVG sizes.
CONCLUSION
In summary, the ArrayOme program that we have developed offers a powerful and simple means of transforming microarray data into an accurate estimate of bacterial genome size. The ArrayOme program is versatile and can process any CGI data set for which a suitable reference genomic template(s) is available, thus offering to add significant value to both pre-existing and newly generated data sets. We have applied ArrayOme to analyse freely available CGI data derived using a two-strain H.pylori array (19) and a Salmonella enterica array (25), and have included our findings in the Supplementary Material. Coupled with physical genome sizing, researchers will be able to exploit information on MVG sizes for high-throughput identification of strains rich in novel genetic material. Indeed, the ability of ArrayOme to predict the genome size of a strain to within ∼2% when CGI data are derived using a comprehensive species-related meta-array, suggests that the accuracy of PFGE-based chromosome sizing would be the major limitation to Microarray-Assisted mobilome Prospecting (MAmP). PFGE-based chromosome sizing typically gives rise to errors of ∼5–10% depending on the detailed strategy used and the presence or absence of confounding episomal replicons (37). However, we are currently developing a PFGE strategy based on the sizing of linearized bacterial chromosomes. Together with the use of standard DNA size markers and accurate calibration standards generated from the chromosomes of various sequenced strains, an accuracy of ±2% should be achievable. In addition to the identification of strains for gene prospecting studies, MAmP would be invaluable for selecting strains for future genome sequencing projects, ensuring maximum return of novel, non-redundant data per sequenced genome.
It is clear that even with current high-throughput genomic sequencing facilities, it will not be feasible to sequence hundreds of isolates to identify and decode the global gene pool accessible to a single bacterial species. However, the application of a strategy such as MAmP, underpinned by ArrayOme and CGI technology, followed by either random or targeted gene-discovery studies focused on selected strains bearing mobilome-rich regions would make a major contribution to this effort.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank The´rèse Hall for assistance in preparing the manuscript. We are also grateful to the two anonymous referees for their valuable and constructive comments and for the suggested refinements to ArrayOme. This study was supported by a mediSearch grant from The Leicestershire Medical Research Foundation, UK to K.R. and M.R.B., and by the BBSRC Core Strategic Grant to J.H. Funding to pay the Open Access publication charges for this article was provided by mediSearch.
REFERENCES
- 1.Perna N.T., Plunkett G., III, Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., Han C.G., Ohtsubo E., Nakayama K., Murata T., et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 3.Welch R.A., Burland V., Plunkett G., III, Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.-R., Boutin A., Hackett J., et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Q., Yuan Z., Xu J., Wang Y., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., et al. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochman H., Lawrence J.G., Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 6.Hacker J., Kaper J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 7.Rajakumar K., Sasakawa C., Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect. Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukiya S., Mizoguchi H., Tobe T., Mori H. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 2004;186:3911–3921. doi: 10.1128/JB.186.12.3911-3921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 10.Malloff C.A., Fernandez R.C., Lam W.L. Bacterial comparative genomic hybridization: a method for directly identifying lateral gene transfer. J. Mol. Biol. 2001;312:1–5. doi: 10.1006/jmbi.2001.4925. [DOI] [PubMed] [Google Scholar]
- 11.Behr M.A., Wilson M.A., Gill W.P., Salamon H., Schoolnik G.K., Rane S., Small P.M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 12.Read T.D., Peterson S.N., Tourasse N., Baillie L.W., Paulsen I.T., Nelson K.E., Tettelin H., Fouts D.E., Eisen J.A., Gill S.R., et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 13.Rajashekara G., Glasner J.D., Glover D.A., Splitter G.A. Comparative whole-genome hybridization reveals genomic islands in Brucella species. J. Bacteriol. 2004;186:5040–5051. doi: 10.1128/JB.186.15.5040-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorrell N., Mangan J.A., Laing K.G., Hinds J., Linton D., Al-Ghusein H., Barrell B.G., Parkhill J., Stoker N.G., Karlyshev A.V., et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–1715. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson B.M., Pin C., Wright J., I'Anson K., Humphrey T., Wells J.M. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 2003;554:224–230. doi: 10.1016/s0014-5793(03)01164-5. [DOI] [PubMed] [Google Scholar]
- 16.Ochman H., Jones I.B. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt U., Agerer F., Michaelis K., Janka A., Buchrieser C., Samuelson M., Svanborg C., Gottschalk G., Karch H., Hacker J. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 2003;185:1831–1840. doi: 10.1128/JB.185.6.1831-1840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anjum M.F., Lucchini S., Thompson A., Hinton J.C., Woodward M.J. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect. Immun. 2003;71:4674–4683. doi: 10.1128/IAI.71.8.4674-4683.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salama N., Guillemin K., McDaniel T.K., Sherlock G., Tompkins L., Falkow S. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl Acad. Sci. USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajakumar K., Shafi J., Smith R.J., Stabler R.A., Andrew P.W., Modha D., Bryant G., Monk P., Hinds J., Butcher P.D., et al. Use of genome level-informed PCR as a new investigational approach for analysis of outbreak-associated Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 2004;42:1890–1896. doi: 10.1128/JCM.42.5.1890-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsolaki A.G., Hirsh A.E., DeRiemer K., Enciso J.A., Wong M.Z., Hannan M., Goguet de la Salmoniere Y.O., Aman K., Kato-Maeda M., Small P.M. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl Acad. Sci. USA. 2004;101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T., Hosogi Y., Nishikawa K., Abbey K., Fleischmann R.D., Walling J., Duncan M.J. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 2004;186:5473–5479. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfgang M.C., Kulasekara B.R., Liang X., Boyd D., Wu K., Yang Q., Miyada C.G., Lory S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge H., Chuang Y.-Y.E., Zhao S., Tong M., Tsai M.-H., Temenak J.J., Richards A.L., Ching W.-M. Comparative genomics of Rickettsia prowazekii Madrid E and Breinl strains. J. Bacteriol. 2004;186:556–565. doi: 10.1128/JB.186.2.556-565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porwollik S., Wong R.M., McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl Acad. Sci. USA. 2002;99:8956–8961. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray A.E., Lies D., Li G., Nealson K., Zhou J., Tiedje J.M. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl Acad. Sci. USA. 2001;98:9853–9858. doi: 10.1073/pnas.171178898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koide T., Zaini P.A., Moreira L.M., Vencio R.Z., Matsukuma A.Y., Durham A.M., Teixeira D.C., El-Dorry H., Monteiro P.B., da Silva A.C., et al. DNA microarray-based genome comparison of a pathogenic and a nonpathogenic strain of Xylella fastidiosa delineates genes important for bacterial virulence. J. Bacteriol. 2004;186:5442–5449. doi: 10.1128/JB.186.16.5442-5449.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinchliffe S.J., Isherwood K.E., Stabler R.A., Prentice M.B., Rakin A., Nichols R.A., Oyston P.C., Hinds J., Titball R.W., Wren B.W. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 2003;13:2018–2029. doi: 10.1101/gr.1507303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou D., Han Y., Song Y., Tong Z., Wang J., Guo Z., Pei D., Pang X., Zhai J., Li M., et al. DNA microarray analysis of genome dynamics in Yersinia pestis: insights into bacterial genome microevolution and niche adaptation. J. Bacteriol. 2004;186:5138–5146. doi: 10.1128/JB.186.15.5138-5146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergthorsson U., Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 31.Lan R., Reeves P.R. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 2002;4:1125–1132. doi: 10.1016/s1286-4579(02)01637-4. [DOI] [PubMed] [Google Scholar]
- 32.Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 33.Wei J., Goldberg M.B., Burland V., Venkatesan M.M., Deng W., Fournier G., Mayhew G.F., Plunkett G., III, Rose D.J., Darling A., et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 2003;71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stekel D. Microarray Bioinformatics. Cambridge, UK: Cambridge University Press; 2003. [Google Scholar]
- 35.Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantri Y., Williams K.P. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 2004;32:D55–D58. doi: 10.1093/nar/gkh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis M.A., Hancock D.D., Besser T.E., Call D.R. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 2003;41:1843–1849. doi: 10.1128/JCM.41.5.1843-1849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.