Version Changes
Revised. Amendments from Version 1
This version 2 highlights an additional question of how the parasite metabolism changes during the day to Section 5, as this was missing from the previous version.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Cynthia He Yingxin, Department of Biological Sciences, Centre for BioImaging Sciences, National University of Singapore, Singapore, 117543, Singapore
Christian Tschudi, Department of Epidemiology of Microbial Diseases, School of Public Health, Yale University, New Haven, CT, USA
Kenneth Stuart, Center for Infectious Disease Research, Seattle, WA, USA
Julius Lukes, Biology Centre, Institute of Parasitology, Czech Academy of Sciences, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, Ceske Budejovice, Czech Republic; Canadian Institute for Advanced Research, Toronto, Canada
Abstract
Cellular metabolic activity is a highly complex, dynamic, regulated process that is influenced by numerous factors, including extracellular environmental signals, nutrient availability and the physiological and developmental status of the cell. The causative agent of sleeping sickness, Trypanosoma brucei, is an exclusively extracellular protozoan parasite that encounters very different extracellular environments during its life cycle within the mammalian host and tsetse fly insect vector. In order to meet these challenges, there are significant alterations in the major energetic and metabolic pathways of these highly adaptable parasites. This review highlights some of these metabolic changes in this early divergent eukaryotic model organism.
Keywords: Trypanosoma brucei, metabolism, adaptations
1. Introduction
Trypanosoma brucei is a unicellular protozoan parasite, transmitted by the bite of tsetse flies ( Glossina genus). Different species/subspecies of trypanosomes infect a variety of different vertebrates, including animals and humans. Human African trypanosomiasis (HAT), also known as sleeping sickness, is caused by two subspecies: Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense. In recent years, the number of reported cases of HAT has decreased steadily, falling to just about 6,000 in 2013 1. Other trypanosome species infect both domestic and wild animals, causing animal African trypanosomiasis. The infection of livestock has a major impact on the African economy, limiting the production of milk and meat and the development of agriculture in areas otherwise amenable to animal husbandry 2.
Trypanosomatids are also of intrinsic scientific interest as they separated early (>600 million years ago) and have evolved differently from other well-studied eukaryotes 3. T. brucei brucei (here called T. brucei), a subspecies non-infectious to human, is by far the best characterised. In the mammalian host, T. brucei parasites colonise the blood and interstitial spaces of several tissues, including the brain, adipose tissue and skin 4– 6. The presence of parasites in the brain is associated with the appearance of the sleep disorder and neurological symptoms characteristic of later stages of the disease 1.
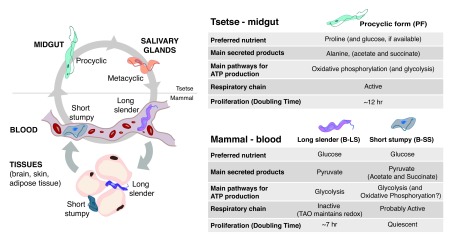
In the mammalian host, parasites exist in two stages: bloodstream long slender form (B-LS), which doubles every 7 hours by binary fission, and short stumpy form (B-SS), which is terminally cell cycle–arrested ( Figure 1). The differentiation from B-LS to B-SS is irreversible and is triggered by a quorum-sensing mechanism 7. The B-SS form is pre-adapted to life in the tsetse fly midgut 7. These pre-adaptions probably help in the efficient differentiation into the replicative procyclic forms (PFs). Eventually, PFs migrate from the midgut to the proventriculus, where they further differentiate into epimastigotes and later into metacyclics in the salivary glands ( Figure 1). The latter are cell cycle–arrested and are able to re-colonise/re-infect a mammalian host when a tsetse fly takes a blood meal.
Figure 1. Changes in metabolism during the life cycle of Trypanosoma brucei.
T. brucei life cycle spans two hosts: a mammal (human, cattle, wild animals) and the tsetse fly. As this protozoan parasite is extracellular, it adapts its metabolism to the available extracellular nutrients. The two stages that have been better characterised in terms of metabolism are the bloodstream long slender and procyclic forms, which mainly catabolise glucose and proline, respectively. Fewer studies have studied bloodstream short stumpy forms. In the mammalian host, parasites accumulate in the interstitial spaces of several tissues, mainly the brain, skin and visceral adipose tissue (adipocytes are shown as an example). The metabolism of parasites in these tissues remains mostly unknown, except for the activation of fatty acid β-oxidation in parasites resident of the adipose tissue. Metabolism of metacyclic stage has not been characterised to date. TAO, trypanosome alternative oxidase.
Throughout the life cycle, parasites encounter and adapt to very different environments. In the mammalian host, such adaptations include avoidance of the host immune system (by employing antigenic variation) as well as metabolic adaptations to use available nutrients. For example, the brain glucose levels is normally 10–20% of blood levels 8, whereas adipose tissue may be a better source of lipids. In the tsetse fly vector, the parasites face a proteolytic rather than immune challenge and also have to adapt to an environment that is free of glucose but rich in amino acids, particularly proline 9. T. brucei re-programmes its metabolism in order to benefit from the nutrients available in the environment. In this review, we will compare the metabolic differences that take place during the T. brucei life cycle, highlighting the questions that remain unanswered. To facilitate the understanding of this review by a non-metabolism expert, we will first summarise the main metabolic pathways present in most eukaryotic cells.
2. Basics of eukaryote metabolism
2.1. Multiple carbon sources for energy production
All living organisms use adenosine triphosphate (ATP) as an intracellular energy source. ATP is generated by the catabolism (breakdown) of nutrients. The most common nutrients or carbon sources are carbohydrates (such as glucose), fatty acids and amino acids.
Most organisms derive energy from the breakdown of glucose, by a process known as glycolysis, a universal and evolutionarily ancient metabolic pathway, which converts glucose (6-carbon) into pyruvate (3-carbon). Under aerobic conditions, pyruvate can undergo further breakdown to acetyl coenzyme A (acetyl-CoA) (2-carbon) and subsequently to carbon dioxide (CO 2) via the tricarboxylic acid (TCA) cycle with the concomitant production of reducing equivalence (NADH and FADH 2) and GTP. Transfer of electrons from these reduced cofactors to oxygen via an electron transport chain generates a proton electrochemical gradient across the mitochondrial inner membrane that is used to generate ATP by a membrane-bound ATP synthase, collectively a process termed oxidative phosphorylation (OXPHOS). The complete oxidation of each glucose molecule leads to the production of about 36 ATP molecules, showing how OXPHOS is a very efficient mechanism of producing energy.
In the absence of oxygen, the glycolysis product (pyruvate or phosphoenolpyruvate) can be further metabolised by fermentation into excreted end-products, such as lactate (for instance, in humans during running) and ethanol (for instance, in yeast) in the cytoplasm, leading to the net production of two ATP molecules per molecule of glucose consumed. Although the flux through fermentation can be very high, the pathway is energetically inefficient in terms of ATP production. In the absence of oxygen, some microorganisms use nitrate ions, sulfate ions, and carbon dioxide as final electron acceptors, in a process named anaerobic respiration. For example, the final product of glycolysis could be converted into acetyl-CoA, which enters the TCA cycle or is converted into acetate. The electrons are then donated to the final acceptor through the mitochondrial electron transport chain.
In many organisms, fatty acids can be catabolised via β-oxidation in the mitochondria to again generate the 2-carbon unit acetyl-CoA, which feeds into the TCA cycle and OXPHOS. Fatty acid β-oxidation of a palmitate molecule (a fatty acid with 16 carbons that is very abundant in mammalian adipocytes) can generate 106 ATP molecules. The balance between making and breaking down fatty acids is tightly regulated.
Amino acids can also contribute to total energy production by oxidation to urea and CO 2. The first reaction is the removal of the amino group by transaminases. While the amino group enters the urea cycle, the ketoacid carbon skeletons typically enter the TCA cycle and fuel OXPHOS.
The relative abundances of sugars, amino acids, and fatty acids along with the availability of sufficient oxygen to use OXPHOS influence which metabolic pathways are preferentially used to produce ATP. Thus, the metabolic profile of a cell is a consequence of the regulated expression of pathway-specific proteins and associated transporters in response to extracellular nutritional and environmental conditions 10.
2.2. Metabolic adaptations in eukaryotes
Textbooks on metabolism explain that in nutrient-rich conditions, model unicellular organisms undergoing exponential growth often use fermentation 11. Proliferating cells in a multicellular organism also metabolise glucose primarily through glycolysis, secreting ethanol, lactate, or another organic acid such as acetate. When unicellular organisms are starved of nutrients, they switch and rely primarily on oxidative metabolism, as do terminally differentiated cells in a multicellular organism. It is no surprise that there are many exceptions to these generalised concepts, and as we will describe below (Section 4), T. brucei is a quintessential example of these exceptions.
The metabolism of cells is a highly regulated process that is influenced by numerous extracellular factors. For example, yeast uses glucose from the environment as their preferred carbon source. Even in the presence of oxygen, glucose is converted into excreted ethanol, with a low yield of ATP production. Although this process may seem wasteful, it is a highly efficient way to support exponential growth. When glucose levels are low, yeast undergoes a diauxic switch to another carbon source, ethanol, which requires an alteration of its mitochondrial metabolism. By using this alternative carbon source, cells are able to continue to grow and divide, but at a significantly reduced rate 12.
In mammals, most non-proliferating differentiated cells use glycolysis and OXPHOS to generate ATP and convert glucose to CO 2 and H 2O. However, most proliferating cancer cells convert glucose into pyruvate and lactate (3-carbon) even under aerobic conditions, a phenomenon known as the Warburg effect, named after its discoverer 11, 13. Although this fermentation-like process is intrinsically energetically less efficient, these cells use far higher rates of glycolysis to meet their higher metabolite demand as they divide faster. This metabolic reprogramming allows cancer cells to rapidly produce the building blocks and increase total biomass for a faster propagation time 14.
Major metabolic changes also take place when immune cells are activated and initiate proliferation. When T cells are activated upon infection or inflammation, gene expression is reprogrammed, resulting in rapid growth, proliferation and the acquisition of new effector functions. Effector T cells, like cancer cells, rely upon aerobic glycolysis when proliferating 11. In contrast, T cells destined to become memory cells maintain an oxidative metabolism, which allows them to keep their quiescence and longevity. Regulatory T (Treg) cells predominantly use OXPHOS for development and survival 15, 16, while activated B cells show increased glucose uptake and induction of glycolysis 17. These examples demonstrate how malleable metabolism is, as cells react to environmental factors and signals to acquire new functions.
3. Trypanosomes have unusual metabolic features
3.1. Trypanosomes have a single mitochondrion and glycosomes
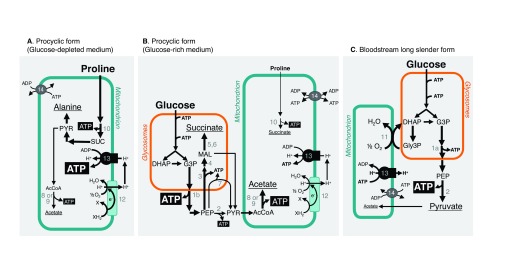
Trypanosomes are characterised by the presence of a dense network of circularised interlocking rings of mitochondrial DNA termed the kinetoplast, located within the large, single mitochondrion of the cell. The single mitochondrion in PFs has a highly defined branched structure with discoid cristae, whereas in the B-LS forms the organelle is a less well-developed narrow tubular structure with an acristate morphology similar to that of the promitochondrion of anaerobic yeast 18, 19. T. brucei also contains peroxisome-like organelles, named glycosomes, which contain the first six (PF) or seven (B-LS) glycolytic enzymes 3. Since the glycosomal membrane is impermeable to ATP, no net ATP production occurs inside these organelles. Thus, net ATP production from glycolysis occurs during the cytoplasmic steps ( Figure 2).
Figure 2. Multiple pathways to produce ATP in trypanosomes.
Catabolism of the most abundant carbon sources in procyclic form grown in glucose-depleted ( A) or in glucose-containing ( B) conditions and in bloodstream long slender form ( C). Excreted end-products from glucose and proline degradation (pyruvate, acetate, succinate and alanine) are underlined. Arrows with different thicknesses tentatively represent the metabolic flux at each enzymatic step. In ( B), the direction of ADP/ATP exchange between the cytosol and the mitochondrion (step 14) is unknown and is represented by double arrows. Key enzymatic steps: 1a, glycosomal phosphoglycerate kinase; 1b, cytosolic phosphoglycerate kinase; 2, pyruvate kinase; 3, phosphoenolpyruvate carboxykinase; 4, glycosomal malate dehydrogenase; 5, cytosolic fumarase (for simplification this reaction is placed in the glycosome); 6, glycosomal NADH-dependent fumarate reductase; 7, pyruvate phosphate dikinase; 8, acetate:succinate coenzyme A-transferase, or ASCT; 9, acetyl-coenzyme A thioesterase; 10, succinyl-coenzyme A synthetase; 11, trypanosome alternative oxidase; 12, respiratory chain; 13, F 0F 1-ATP synthase; 14, mitochondrial ADP/ATP exchanger. AcCoA, acetyl-coenzyme A; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde 3-phosphate; Gly3P, glycerol 3-phosphate; MAL, malate; PEP, phosphoenolpyruvate; PYR, pyruvate; SUC, succinate.
In contrast, ADP and ATP molecules can be exchanged between the cytosolic and mitochondrial compartments through the TbMCP5 mitochondrial ADP/ATP exchanger 20. This exchanger is required because oxidative phosphorylation does not occur in B-LS and thus no ATP is generated inside this organelle. To maintain the mitochondrial proton electrochemical gradient across the mitochondrial membrane, the F1F0-ATPase operates in the reverse direction, hydrolysing ATP into ADP ( Figure 2). This unusual way of generating a mitochondrial potential also implies that a functional phosphate/H + exchanger must be present.
Significant differential expression of mitochondrial and glycosomal proteins occurs during the life cycle 21. Indeed, during the differentiation of B-LS into PFs, degradation of glycosomes probably via autophagy is enhanced and new glycosomes with different enzymatic contents are produced, so that parasites become rapidly metabolically adapted to the new host environment 22. In fact, the differential expression of glycosomal and mitochondrial proteins is a clear indicator of the difference in metabolic life styles between the two major life-cycle stages of T. brucei.
3.2. Trypanosomes have specific pathways and unique enzymes
Kinetoplastida are of great intrinsic scientific interest as they have diverged very early compared with most studied eukaryotic models (for example, yeast, plants and animals), on which the foundations of molecular, biochemical and cellular biology have been built. Cytochrome oxidase (COX) is the terminal oxidase of the mammalian electron transport chain and is responsible for the reduction of oxygen to water. However, T. brucei possesses an additional plant-like, non-energy-conserving terminal oxidase called alternative oxidase (TAO). Indeed, B-LS forms are unique in the sense that they do not use COX but rely on TAO (step 11 in Figure 2C). TAO is 100-fold more expressed in B-LS than PFs and thus is considered a potential drug target 23.
Sphingolipids are a class of lipids important in cell recognition and signal transmission. To date, T. brucei is the only organism known to make all three types of sphingolipids (sphingomyelin, inositolphosphoceramide and ethanolamine-phosphoceramide). These lipids are synthesised via four sphingolipid synthases (SLSs) that are encoded by genes organised in a tandem array. Sphingolipid synthesis is highly controlled during development: a dedicated inositolphosphoceramide synthase (SLS1) is highly upregulated in B-SS parasites and maintained in PFs 24. As a consequence of more ceramide being used for inositolphosphoceramide synthesis, the synthesis of sphingomyelin is reduced, causing an alteration in the levels of phosphatidylinositol species.
4. Metabolic adaptations during the Trypanosoma brucei life cycle
The bloodstream of a mammalian host is a very rich environment, containing 5 mM of glucose, 95% to 99% oxygen saturation levels and 0.6 to 0.8 g/mL proteins, including lipoproteins (low-density lipoprotein and high-density lipoprotein). In contrast, when parasites are ingested by the tsetse during a blood meal, they end up in a glucose-poor but amino acid-rich environment that is very different from the mammalian bloodstream. Given that we can mimic these growing conditions in vitro, most of our knowledge about metabolic changes during the T. brucei life cycle originates from the comparison between B-LS and PFs.
4.1. Amino acids: an abundant carbon source in the midgut of the fly
The midgut of the tsetse fly has a temperature of about 28°C and a variable pH and contains hardly any glucose, but is rich in amino acids, such as proline (about 100 μM) 9. It is well accepted that, in a glucose-depleted environment, PFs primarily use proline for their energy production 25, 26 ( Figure 1), but catabolism of other amino acids, such as threonine and leucine, is also used 27, 28. These latter amino acids feed fatty acid biosynthesis and/or enter into the mevalonate pathway to produce the building blocks to generate essential lipids, including isoprenoids and sterols. Proline is catabolised within the mitochondrion and excreted from the cell as the end-product alanine, with the production of several reduced cofactors, which are reoxidised in the respiratory chain for ATP production by OXPHOS ( Figure 2A). However, if glucose is provided, PFs adjust their metabolism and produce most of their ATP via glucose degradation (glycolysis), even in the presence of proline ( Figure 2B) 25, 29. These findings highlight that these parasites, like most other eukaryotes, are extremely flexible at adapting their central metabolism to their environment.
4.2. Glucose: differences in consumption rate and efficiency of ATP production
So far, the only carbon source for ATP production described for replicative bloodstream parasites is glucose, which is converted via glycolysis ( Figure 1 and Figure 2C). Unlike proliferative yeast and tumour cells, B-LS does not undergo fermentation per se. In fact, instead of being metabolised and generating ethanol or lactate, most pyruvate in B-LS is immediately excreted, and only about 1% is fermented into succinate 30, 31. To oxidise the NADH produced during glycolysis back to NAD +, B-LS consume large amounts of oxygen that act as an electron acceptor in a reaction catalysed by the unusual TAO. This type of glucose metabolism is uncommon and does not fit textbook knowledge. Interestingly, B-LS cells also tolerate anaerobic conditions where they convert glucose to equimolar amounts of glycerol and pyruvate, with a two-fold reduction of the ATP production rate.
As mentioned above, although PFs rely upon proline in vivo, they prefer glucose to produce ATP 25. Interestingly, the rate of glucose degradation is about 10-fold higher in B-LS than in PFs 30, 32. This considerable difference is probably due to metabolic adaptations developed by B-LS in response to a much higher ATP demand compared with PFs. Firstly, B-LS replicate faster than PFs (doubling times of about 7 and about 12 hours, respectively), which means that theoretically B-LS should show a 1.5-fold higher rate of ATP production. Secondly, the estimated number of ATP molecules produced per glucose consumed is about two-fold lower in B-LS. This difference is explained by the different strategies used by B-LS and PFs to degrade glucose into excreted end-products, which are mainly pyruvate in B-LS ( Figure 2C) and acetate plus succinate in PFs ( Figure 2B) 33. Indeed, at the end of glycolysis, PFs convert pyruvate into acetate and ATP by the acetate:succinate CoA-transferase (ASCT)/succinyl-CoA synthetase cycle 34, 35. This pathway accounting for about 70% of the glycolytic flux in PFs is reduced to 5% in B-LS 30. In addition, the glycosomal succinate fermentation pathway (steps 3–6 in Figure 2B), pyruvate phosphate dikinase (step 7), and cytosolic localisation of phosphoglycerate kinase (step 1b in Figure 2B) improve the rate of ATP production within the cytosol of PFs 29, 36.
Thirdly and probably the most important reason for a higher rate of glucose degradation in B-LS is that some biological processes require more ATP in B-LS compared with PFs. This is the case of endocytosis, which is at least about 10-fold upregulated in B-LS compared with PFs and other trypanosomatids 37, 38. The high endocytic activity observed in B-LS is required for rapid recycling of cell-surface glycosylphosphatidylinositol (GPI)-anchored variant surface glycoprotein (VSG) for internalisation and removal of bound antibodies, facilitating escape from the host immune defences, but also for nutrient scavenging from the mammalian host. Knockdown of actin resulted in a significant decrease (>70%) in endocytic activity and clearance of anti-VSG antibodies by B-LS forms, but did not significantly affect cellular ATP levels 38, 39. Surprisingly, measurement of the rates of pyruvate production and oxygen consumption, under conditions identical to those employed for the ATP and transferrin uptake assays, revealed a decrease of about four-fold in both rates after a knockdown of 15 hours (D. P. Nolan, unpublished data). Although the consumption of glucose was not measured, these data suggest that membrane trafficking in the B-LS may represent a significant additional ATP demand compared with the PFs and even more surprisingly that the rate of ATP utilisation may also influence its rate of production via glycolysis. However, knockdown of actin also led to a rapid cessation in cell division and eventual cell death, so the implications of these preliminary metabolic investigations may not be so straightforward.
Interestingly, although B-SS live in a glucose-rich environment, they undergo morphological and gene expression alterations that are consistent with a preparation to survive within the tsetse midgut environment 7, 24 ( Figure 1). These adaptions also include increased sensitivity to specific environmental cues that signal entry to the tsetse fly vector, as well as resistance to extracellular acidic and proteolytic stress 40, 41. Given that B-SS are non-proliferative and existent only in pleomorphic strains, it is more difficult to obtain large and pure quantities of this life-cycle stage in vitro. As a result, its metabolism has been less characterised. Nevertheless, we know that B-SS consume glucose and produce pyruvate and intermediate levels of acetate 42, suggesting that metabolism is being pre-adapted to the conditions in which procyclic forms will live within the tsetse midgut. Transcriptomic studies have confirmed the downregulation of several genes that encode for components of the glycosomes and are involved in glucose uptake and breakdown 24. Genes upregulated in B-SS include TAO, fructose-2,6- biphosphatase, specific membrane proteins, and specific lipid biosynthesic genes, including TbSLS1 involved in inositolphosphoceramide synthesis. Further biochemical studies will be necessary to characterise and allow a better understanding of B-SS metabolism.
4.3. Lipids: responding to a great demand for myristate
In the bloodstream, trypanosomes are able to survive extracellularly in the mammalian host as they are coated by a dense homogenous layer of GPI-anchored VSGs. VSG coats are periodically exchanged by a mechanism of antigenic variation, protecting parasites against the host’s innate and adaptive immune responses 43. In B-LS forms, GPI anchors exclusively contain two myristate molecules (14 carbon fatty acid); however, myristate is present at very low levels within the mammalian bloodstream, which could not sustain the B-SL demand 44. Thus, it was initially thought that de novo synthesis of myristate occurred via a type II prokaryotic-like synthase 45, 46, but this synthesis is not sufficient for the GPI requirement 47. Hence, it has been discovered that T. brucei uses four microsomal elongases, with different specificities, to synthesise fatty acids in a stepwise manner 47. These elongases are responsible for the majority of de novo fatty acid synthesis in T. brucei. In bloodstream forms, downregulation of the elongase 3 in the pathway (which converts C14 to C18) explains the high production of myristate required for GPI anchors.
Trindade et al. have shown that at the RNA level, parasites in adipose tissue and blood are quite different and that many genes associated with metabolism are differentially expressed 6. Using pulse-chase experiments with stable isotope-labelled myristate, the authors showed that the first three enzymatic steps of fatty acid β-oxidation were active in parasites occupying the adipose tissue. In this pathway, a fatty acid molecule is converted into acetyl-CoA and a shorter fatty acid. The fate of each of these molecules remains unknown. The most ‘classic’ scenario would be that fatty acids undergo several cycles of β-oxidation, releasing acetyl-CoA, which could enter the TCA cycle, leading to the production of NADH and FADH 2, which could result in the production of ATP by OXPHOS. This hypothesis, however, implies that complexes III and IV of the respiratory chain are active, and this has not been observed so far in T. brucei mammalian forms. An alternative scenario is that the fatty acid chains (normally, 16 carbon palmitate) released by adipocytes are shortened through β-oxidation to enter an anabolic process in order to produce complex lipids. The resulting acetyl-CoA could be converted into acetate and lead to the production of one ATP molecule by the action of ASCT enzyme.
Tryponosoma cruzi also infects adipocytes (specialised cells of adipose tissue) and is capable of consuming stored lipids 48. It is unclear how T. brucei, an extracellular parasite, accesses lipids that are stored inside adipocytes, which constitute the largest storage of lipids in a mammalian host. Inside adipocytes, the stored triglycerides can be converted via lipolysis into fatty acids and glycerol, which are eventually secreted. During a T. brucei infection, animals typically lose weight and serum shows hyperlipidaemia 6, 49. We speculate that during a T. brucei infection, lipolysis is increased, leading to the secretion of fatty acids, which could be readily taken up and used by parasites.
5. Concluding remarks and future perspectives
Fields such as immunometabolism emerged because of the development of highly sensitive metabolomic approaches, including untargeted metabolomic analysis, stable isotope labelling, mass spectrometry, and nuclear magnetic resonance 50– 52. Researchers discovered that during immune cell activation, the levels of many metabolites undergo alterations and these changes are directly linked to immune cell effector functions. In a way, the life cycle of a pathogen is a series of irreversible differentiation steps, in which the cells adapt to a new environment to perform new functions. In addition, the use of modern metabolomics approaches has revealed that T. brucei uses an incomplete TCA cycle in PFs 53 and that proline has a different fate in PFs depending upon high- or low-glucose availability in the medium and has allowed the identification of the carbon and nitrogen sources of essential metabolites 53– 56.
However, much remains to be discovered. Some questions for the future are the following:
-
•
Besides the known glucose and proline transporters, what are the transporters of other essential nutrients?
-
•
What is the signalling cascade that coordinates metabolism remodelling? In most eukaryotes, two key kinases are involved in nutrient sensing: target of rapamycin protein (TOR) and AMP-activated kinase (AMPK). T. brucei, the eukaryote with the most complex network of TOR proteins described so far, is composed of four TOR proteins 57, which are necessary for cell proliferation. Interestingly, knockdown of one of these proteins, TbTOR4, which appears to be kinetoplastid-specific, caused irreversible differentiation of the B-LS form into a quiescent form with properties very similar to those of the B-SS form, which suggested that TbTOR4 negatively regulates the slender-to-stumpy transition 58. Activation of AMPK also triggers differentiation to the quiescent B-SS forms 59. It is likely that some of these kinases are directly involved in remodelling metabolism during the T. brucei life cycle. It is interesting that changes in inositol metabolism lead to perturbations in VSG gene expression, suggesting that inositol metabolites are important for the control of this B-LS-specific process 60.
-
•
What is the metabolism of other stages of the life cycle? With the possibility of generating in vitro multiple stages of tsetse life cycle by overexpressing RBP6 61, it may be possible in the near future to understand metabolism of epimastigotes and metacyclics.
-
•
Do other trypanosome species undergo similar or different metabolic adaptations during their life cycle as they encounter different environments?
-
•
What is the metabolism of slender and stumpy forms when these occupy other tissues within the mammalian host? Trindade et al. have shown that at the RNA level, parasites in adipose tissue and blood are quite different and that many genes associated with metabolism are differentially expressed 6. It will be important in the future, perhaps for drug development, to confirm these observations at protein and metabolite levels not only in visceral adipose tissue but also in the skin and importantly in the brain.
-
•
What are the consequences of a T. brucei infection in the host? Trypanosomiasis is characterised by decreased levels of aromatic amino acids, especially tryptophan, in the host and these levels are accompanied by the excretion of abnormal amounts of aromatic ketoacids, such as indole-3-pyruvate 62. It was recently shown that B-LS generates indole-3-pyruvate by transamination of tryptophan and secretes significant amounts of this aromatic ketoacid. Indole-3-pyruvate appears to be able to modulate the host inflammatory responses, which may prolong host survival and thereby potentiate transmission of the parasite to the tsetse fly vector and ensure completion of the life cycle 62. The roles of other secreted parasite metabolites that can possibly modify the host’s metabolism remain to be established.
-
•
How does the parasite metabolism change during the day? We have recently shown that, in T. brucei, many genes that encode for metabolic enzymes are circadianly regulated 63. This cycling expression pattern leads to two peaks of intracellular ATP concentration, indicating that metabolism is indeed under circadian control. How is this control achieved? Which metabolic pathways are affected? Does this adaptation reflect the circadian variation of nutrients in the host?
The interplay between the host and pathogen and the influences upon their respective metabolisms is likely to be complex but is probably very significant since adaptation to nutrient availability is a major driving force during evolution. With the help of new and more sensitive biochemical and metabolic methodologies, it should be possible to use systems biology approaches to simultaneously characterise the metabolic changes undergone by parasites and host during an infection and within different tissues.
Funding Statement
TKS is supported by the Wellcome Trust, the Biotechnology and Biological Sciences Research Council, the Engineering and Physical Sciences Research Council, the Medical Research Council (MR/M020118/1) and the European Community Seventh Framework Programme under grant agreement 602773 (Project KINDRED). FB is supported by the Centre National de la Recherche Scientifique (CNRS), the Université de Bordeaux, the Agence Nationale de la Recherche (ANR) through the GLYCONOV grant (ANR-15-CE15-0025-01) of the ‘Générique’ 2015 call and the Laboratoire d’Excellence (LabEx) ParaFrap (grant ANR-11-LABX-0024). DPN is supported by the Wellcome Trust and Science Foundation Ireland. LMF is supported by the Howard Hughes Medical Institute (55007419) and the Fundação para a Ciência e a Tecnologia (FCT) (PTDC/BIM-MET/4471/2014).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 4 approved]
References
- 1. Franco JR, Simarro PP, Diarra A, et al. : Epidemiology of human African trypanosomiasis. Clin Epidemiol. 2014;6:257–75. 10.2147/CLEP.S39728 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Morrison LJ, Vezza L, Rowan T, et al. : Animal African Trypanosomiasis: Time to Increase Focus on Clinically Relevant Parasite and Host Species. Trends Parasitol. 2016;32(8):599–607. 10.1016/j.pt.2016.04.012 [DOI] [PubMed] [Google Scholar]
- 3. Gualdrón-López M, Brennand A, Hannaert V, et al. : When, how and why glycolysis became compartmentalised in the Kinetoplastea. A new look at an ancient organelle. Int J Parasitol. 2012;42(1):1–20. 10.1016/j.ijpara.2011.10.007 [DOI] [PubMed] [Google Scholar]
- 4. Caljon G, van Reet N, De Trez C, et al. : The Dermis as a Delivery Site of Trypanosoma brucei for Tsetse Flies. PLoS Pathog. 2016;12(7):e1005744. 10.1371/journal.ppat.1005744 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Capewell P, Cren-Travaillé C, Marchesi F, et al. : The skin is a significant but overlooked anatomical reservoir for vector-borne African trypanosomes. eLife. 2016;5: pii: e17716. 10.7554/eLife.17716 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Trindade S, Rijo-Ferreira F, Carvalho T, et al. : Trypanosoma brucei Parasites Occupy and Functionally Adapt to the Adipose Tissue in Mice. Cell Host Microbe. 2016;19(6):837–48. 10.1016/j.chom.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. MacGregor P, Szöőr B, Savill NJ, et al. : Trypanosomal immune evasion, chronicity and transmission: an elegant balancing act. Nat Rev Microbiol. 2012;10(6):431–8. 10.1038/nrmicro2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Vries MG, Arseneau LM, Lawson ME, et al. : Extracellular glucose in rat ventromedial hypothalamus during acute and recurrent hypoglycemia. Diabetes. 2003;52(11):2767–73. 10.2337/diabetes.52.11.2767 [DOI] [PubMed] [Google Scholar]
- 9. Balogun RA: Studies on the amino acids of the tsetse fly, Glossina morsitans, maintained on in vitro and in vivo feeding systems. Comp Biochem Physiol A Comp Physiol. 1974;49(2A):215–22. 10.1016/0300-9629(74)90110-8 [DOI] [PubMed] [Google Scholar]
- 10. Thompson CB: Rethinking the regulation of cellular metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:23–9. 10.1101/sqb.2012.76.010496 [DOI] [PubMed] [Google Scholar]
- 11. Vander Heiden MG, Cantley LC, Thompson CB: Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Galdieri L, Mehrotra S, Yu S, et al. : Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS. 2010;14(6):629–38. 10.1089/omi.2010.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Warburg O: On the origin of cancer cells. Science. 1956;123(3191):309–14. 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 14. Ward PS, Thompson CB: Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michalek RD, Gerriets VA, Jacobs SR, et al. : Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4 + T cell subsets. J Immunol. 2011;186(6):3299–303. 10.4049/jimmunol.1003613 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 16. Shi LZ, Wang R, Huang G, et al. : HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of T H17 and T reg cells. J Exp Med. 2011;208(7):1367–76. 10.1084/jem.20110278 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Doughty CA, Bleiman BF, Wagner DJ, et al. : Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood. 2006;107(11):4458–65. 10.1182/blood-2005-12-4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vickerman K: Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull. 1985;41(2):105–14. 10.1093/oxfordjournals.bmb.a072036 [DOI] [PubMed] [Google Scholar]
- 19. Matthews KR: The developmental cell biology of Trypanosoma brucei. J Cell Sci. 2005;118(Pt 2):283–90. 10.1242/jcs.01649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peña-Diaz P, Pelosi L, Ebikeme C, et al. : Functional characterization of TbMCP5, a conserved and essential ADP/ATP carrier present in the mitochondrion of the human pathogen Trypanosoma brucei. J Biol Chem. 2012;287(50):41861–74. 10.1074/jbc.M112.404699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vertommen D, Van Roy J, Szikora JP, et al. : Differential expression of glycosomal and mitochondrial proteins in the two major life-cycle stages of Trypanosoma brucei. Mol Biochem Parasitol. 2008;158(2):189–201. 10.1016/j.molbiopara.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 22. Herman M, Pérez-Morga D, Schtickzelle N, et al. : Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy. 2008;4(3):294–308. 10.4161/auto.5443 [DOI] [PubMed] [Google Scholar]
- 23. Chaudhuri M, Ott RD, Hill GC: Trypanosome alternative oxidase: from molecule to function. Trends Parasitol. 2006;22(10):484–91. 10.1016/j.pt.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 24. Kabani S, Fenn K, Ross A, et al. : Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics. 2009;10:427. 10.1186/1471-2164-10-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamour N, Rivière L, Coustou V, et al. : Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J Biol Chem. 2005;280(12):11902–10. 10.1074/jbc.M414274200 [DOI] [PubMed] [Google Scholar]
- 26. Mantilla BS, Marchese L, Casas-Sánchez A, et al. : Proline Metabolism is Essential for Trypanosoma brucei brucei Survival in the Tsetse Vector. PLoS Pathog. 2017;13(1):e1006158. 10.1371/journal.ppat.1006158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nes CR, Singha UK, Liu J, et al. : Novel sterol metabolic network of Trypanosoma brucei procyclic and bloodstream forms. Biochem J. 2012;443(1):267–77. 10.1042/BJ20111849 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Millerioux Y, Ebikeme C, Biran M, et al. : The threonine degradation pathway of the Trypanosoma brucei procyclic form: the main carbon source for lipid biosynthesis is under metabolic control. Mol Microbiol. 2013;90(1):114–29. 10.1111/mmi.12351 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Deramchia K, Morand P, Biran M, et al. : Contribution of pyruvate phosphate dikinase in the maintenance of the glycosomal ATP/ADP balance in the Trypanosoma brucei procyclic form. J Biol Chem. 2014;289(25):17365–78. 10.1074/jbc.M114.567230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mazet M, Morand P, Biran M, et al. : Revisiting the central metabolism of the bloodstream forms of Trypanosoma brucei: production of acetate in the mitochondrion is essential for parasite viability. PLoS Negl Trop Dis. 2013;7(12):e2587. 10.1371/journal.pntd.0002587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Creek DJ, Mazet M, Achcar F, et al. : Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 2015;11(3):e1004689. 10.1371/journal.ppat.1004689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ryley JF: Studies on the metabolism of the protozoa. 9. Comparative metabolism of blood-stream and culture forms of Trypanosoma rhodesiense. Biochem J. 1962;85(1):211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bringaud F, Rivière L, Coustou V: Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149(1):1–9. 10.1016/j.molbiopara.2006.03.017 [DOI] [PubMed] [Google Scholar]
- 34. Rivière L, van Weelden SW, Glass P, et al. : Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J Biol Chem. 2004;279(44):45337–46. 10.1074/jbc.M407513200 [DOI] [PubMed] [Google Scholar]
- 35. van Hellemond JJ, Tielens AG: Adaptations in the lipid metabolism of the protozoan parasite Trypanosoma brucei. FEBS Lett. 2006;580(23):5552–8. 10.1016/j.febslet.2006.07.056 [DOI] [PubMed] [Google Scholar]
- 36. Besteiro S, Biran M, Biteau N, et al. : Succinate secreted by Trypanosoma brucei is produced by a novel and unique glycosomal enzyme, NADH-dependent fumarate reductase. J Biol Chem. 2002;277(41):38001–12. 10.1074/jbc.M201759200 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Pal A, Hall BS, Nesbeth DN, et al. : Differential endocytic functions of Trypanosoma brucei Rab5 isoforms reveal a glycosylphosphatidylinositol-specific endosomal pathway. J Biol Chem. 2002;277(11):9529–39. 10.1074/jbc.M110055200 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Engstler M, Pfohl T, Herminghaus S, et al. : Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131(3):505–15. 10.1016/j.cell.2007.08.046 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. García-Salcedo JA, Pérez-Morga D, Gijón P, et al. : A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004;23(4):780–9. 10.1038/sj.emboj.7600094 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Rico E, Rojas F, Mony BM, et al. : Bloodstream form pre-adaptation to the tsetse fly in Trypanosoma brucei. Front Cell Infect Microbiol. 2013;3: 78. 10.3389/fcimb.2013.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nolan DP, Rolin S, Rodriguez JR, et al. : Slender and stumpy bloodstream forms of Trypanosoma brucei display a differential response to extracellular acidic and proteolytic stress. Eur J Biochem. 2000;267(1):18–27. 10.1046/j.1432-1327.2000.00935.x [DOI] [PubMed] [Google Scholar]
- 42. van Grinsven KW, van Den Abbeele J, van den Bossche P, et al. : Adaptations in the glucose metabolism of procyclic Trypanosoma brucei isolates from tsetse flies and during differentiation of bloodstream forms. Eukaryotic Cell. 2009;8(8):1307–11. 10.1128/EC.00091-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hovel-Miner G, Mugnier M, Papavasiliou FN, et al. : A Host-Pathogen Interaction Reduced to First Principles: Antigenic Variation in T. brucei. Results Probl Cell Differ. 2015;57:23–46. 10.1007/978-3-319-20819-0_2 [DOI] [PubMed] [Google Scholar]
- 44. Ferguson MA, Cross GA: Myristylation of the membrane form of a Trypanosoma brucei variant surface glycoprotein. J Biol Chem. 1984;259(5):3011–5. [PubMed] [Google Scholar]
- 45. Morita YS, Paul KS, Englund PT: Specialized fatty acid synthesis in African trypanosomes: myristate for GPI anchors. Science. 2000;288(5463):140–3. 10.1126/science.288.5463.140 [DOI] [PubMed] [Google Scholar]
- 46. Paul KS, Jiang D, Morita YS, et al. : Fatty acid synthesis in African trypanosomes: a solution to the myristate mystery. Trends Parasitol. 2001;17(8):381–7. 10.1016/S1471-4922(01)01984-5 [DOI] [PubMed] [Google Scholar]
- 47. Lee SH, Stephens JL, Paul KS, et al. : Fatty acid synthesis by elongases in trypanosomes. Cell. 2006;126(4):691–9. 10.1016/j.cell.2006.06.045 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Tanowitz HB, Scherer PE, Mota MM, et al. : Adipose Tissue: A Safe Haven for Parasites? Trends Parasitol. 2017;33(4):274–284. 10.1016/j.pt.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Palmer JJ, Surur EI, Goch GW, et al. : Syndromic algorithms for detection of gambiense human African trypanosomiasis in South Sudan. PLoS Negl Trop Dis. 2013;7(1):e2003. 10.1371/journal.pntd.0002003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Goodacre R, Vaidyanathan S, Dunn WB, et al. : Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22(5):245–52. 10.1016/j.tibtech.2004.03.007 [DOI] [PubMed] [Google Scholar]
- 51. Dettmer K, Aronov PA, Hammock BD: Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. 10.1002/mas.20108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Breitling R, Takano E: Synthetic Biology of Natural Products. Cold Spring Harb Perspect Biol. 2016;8(10): pii: a023994. 10.1101/cshperspect.a023994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Weelden SW, Fast B, Vogt A, et al. : Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J Biol Chem. 2003;278(15):12854–63. 10.1074/jbc.M213190200 [DOI] [PubMed] [Google Scholar]
- 54. Coustou V, Biran M, Breton M, et al. : Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J Biol Chem. 2008;283(24):16342–54. 10.1074/jbc.M709592200 [DOI] [PubMed] [Google Scholar]
- 55. Creek DJ, Anderson J, McConville MJ, et al. : Metabolomic analysis of trypanosomatid protozoa. Mol Biochem Parasitol. 2012;181(2):73–84. 10.1016/j.molbiopara.2011.10.003 [DOI] [PubMed] [Google Scholar]
- 56. Bringaud F, Biran M, Millerioux Y, et al. : Combining reverse genetics and nuclear magnetic resonance-based metabolomics unravels trypanosome-specific metabolic pathways. Mol Microbiol. 2015;96(5):917–26. 10.1111/mmi.12990 [DOI] [PubMed] [Google Scholar]
- 57. Saldivia M, Barquilla A, Bart JM, et al. : Target of rapamycin (TOR) kinase in Trypanosoma brucei: an extended family. Biochem Soc Trans. 2013;41(4):934–8. 10.1042/BST20130052 [DOI] [PubMed] [Google Scholar]
- 58. Barquilla A, Saldivia M, Diaz R, et al. : Third target of rapamycin complex negatively regulates development of quiescence in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2012;109(36):14399–404. 10.1073/pnas.1210465109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Saldivia M, Ceballos-Pérez G, Bart JM, et al. : The AMPKα1 Pathway Positively Regulates the Developmental Transition from Proliferation to Quiescence in Trypanosoma brucei. Cell Rep. 2016;17(3):660–70. 10.1016/j.celrep.2016.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Cestari I, Stuart K: Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc Natl Acad Sci U S A. 2015;112(21):E2803–12. 10.1073/pnas.1501206112 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Kolev NG, Ramey-Butler K, Cross GA, et al. : Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338(6112):1352–3. 10.1126/science.1229641 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Hall JE, Seed JR: Increased urinary excretion of aromatic amino acid catabolites by Microtus montanus chronically infected with Trypanosoma brucei gambiense. Comp Biochem Physiol B. 1984;77(4):755–60. 10.1016/0305-0491(84)90309-2 [DOI] [PubMed] [Google Scholar]
- 63. Rijo-Ferreira F, Pinto-Neves D, Barbosa-Morais NL, et al. : Trypanosoma brucei metabolism is under circadian control. Nat Microbiol. 2017;2: 17032. 10.1038/nmicrobiol.2017.32 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation