New evidence for ABA-induced stomatal closure in fern and known evidence in earlier diverging lineages does not support the hypothesis that stomatal responsiveness to ABA evolved first in seed plants.
Abstract
Abscisic acid (ABA)-driven stomatal regulation reportedly evolved after the divergence of ferns, during the early evolution of seed plants approximately 360 million years ago. This hypothesis is based on the observation that the stomata of certain fern species are unresponsive to ABA, but exhibit passive hydraulic control. However, ABA-induced stomatal closure was detected in some mosses and lycophytes. Here, we observed that a number of ABA signaling and membrane transporter protein families diversified over the evolutionary history of land plants. The aquatic ferns Azolla filiculoides and Salvinia cucullata have representatives of 23 families of proteins orthologous to those of Arabidopsis (Arabidopsis thaliana) and all other land plant species studied. Phylogenetic analysis of the key ABA signaling proteins indicates an evolutionarily conserved stomatal response to ABA. Moreover, comparative transcriptomic analysis has identified a suite of ABA-responsive genes that differentially expressed in a terrestrial fern species, Polystichum proliferum. These genes encode proteins associated with ABA biosynthesis, transport, reception, transcription, signaling, and ion and sugar transport, which fit the general ABA signaling pathway constructed from Arabidopsis and Hordeum vulgare. The retention of these key ABA-responsive genes could have had a profound effect on the adaptation of ferns to dry conditions. Furthermore, stomatal assays have shown the primary evidence for ABA-induced closure of stomata in two terrestrial fern species P. proliferum and Nephrolepis exaltata. In summary, we report, to our knowledge, new molecular and physiological evidence for the presence of active stomatal control in ferns.
Fossil plants have provided insights into the broader physiological and ecological adaptations that enable land plants to reduce water loss via specialized morphological structures (Edwards et al., 1998; Raven, 2002). Early land plants were exposed to high levels of UV radiation, elevated temperatures and extremely dry soils. Thus, adaptations to regulate gas exchange via active control mechanisms would have helped avoid catastrophic dehydration and allow early land plants to survive long enough to reproduce (Ruszala et al., 2011). Stomata were a key innovation in the earliest phases of the colonization of land by terrestrial plants, an event that fundamentally altered the global landscape and climate. Stomata have been observed in Silurian fossils of sporophytes and gametophytes of early land plants (Raven, 2002). The presence of structurally defined stomata in the epidermis of above-ground organs of sporophytes appears to be the ancestral condition for terrestrial plants (Qiu et al., 1998; Raven, 2002). That these structures appear more or less unchanged for over 400 million years (Edwards et al., 1998; Beerling and Franks, 2009) suggests that they are an essential adaptation to terrestrial plant life. Land plants acquired stomata to regulate gas exchange by opening and closing of the stomatal pore (Hetherington and Woodward, 2003; Berry et al., 2010; Chater et al., 2011). While liverworts lack stomata, mosses and most other land plants have a typical stomatal pore surrounded by at least a pair of guard cells (Franks and Farquhar, 2007; Chen et al., 2017).
The response of stomatal guard cells to hydraulic and nonhydraulic signaling supports a fundamental role of abscisic acid (ABA) in plant signaling in a changing environment (Brodribb and McAdam, 2011; Pantin et al., 2013). ABA induces stomatal closure in many seed plant species (Willmer and Fricker, 1996; Wolf et al., 2006; Chen et al., 2012b, 2016; Wang et al., 2013). ABA-induced stomatal closure has also been reported in a lycophyte, Selaginella uncinata (Ruszala et al., 2011), and the mosses Physcomitrella patens and Funaria hygrometrica, but not in all species of mosses and lycophytes (Brodribb and McAdam, 2011; Merced and Renzaglia, 2014; Lind et al., 2015). However, the role of ABA in stomatal regulation in the other major clades of land plants is still under debate. A major shift in the stomatal control process between lycophytes and ferns versus seed plants (gymnosperms and angiosperms) has been proposed; fern and lycophyte guard cells lack responsiveness to endogenous ABA, which evolved subsequently in seed plants (Brodribb and McAdam, 2011; McAdam and Brodribb, 2012a, 2012b; McAdam et al., 2016). The ecological significance of this change is that seed plants may have a greater capacity than lycophytes and ferns to actively control water loss during drought (McElwain, 2011). However, molecular evidence supporting this shift is still lacking (Chater et al., 2011, 2013; Ruszala et al., 2011; Lind et al., 2015; Chen et al., 2017).
Investigations of ABA signaling and membrane transport to date have focused largely on angiosperms (Blatt, 2000). ABA metabolism and transport, ABA perception and signal transduction, and ABA signal response and modulation are parts of the ABA signaling pathway in plants (Cutler et al., 2010; Hauser et al., 2011). ABA metabolism includes genes encoding zeaxanthin epoxidases (ZEPs; Marin et al., 1996), 9-cis-epoxycarotenoid dioxygenase (NCEDs; North et al., 2007), abscisic aldehyde oxidases (Seo et al., 2000) that are crucial for ABA biosynthesis, and CYP707As encoding ABA 8′-hydroxylases in the ABA catabolic pathway (Saito et al., 2004). ABA transport is mediated by the ATP binding cassette transporters ABCG25 and ABCG40 (Kang et al., 2010; Kuromori et al., 2010) and nitrate transporter NRT1.2 (Kanno et al., 2012). Cytosolic ABA perception consists of Pyrabactin resistance (PYR)/PYR Like (PYL)/regulatory component of ABA receptor (RCAR; Ma et al., 2009; Park et al., 2009), protein phosphatase 2Cs (PP2Cs; Schweighofer et al., 2004) and snf1-related protein kinase 2 (SnRK2; Umezawa et al., 2009). RCAR-PP2C complex formation leads to inhibition of PP2C activity, thereby allowing activation of SnRK2s. Two G protein-coupled receptors-Type G proteins GTG1 and GTG2 were identified as plasma membrane ABA receptors (Pandey et al., 2009). ABA-induced reactive oxygen species (ROS) and nitric oxide (NO) production down-regulates the activity of the PP2C phosphatases and activates Ca2+-permeable channels and anion channels (Grabov and Blatt, 1998; Hamilton, et al., 2000; Köhler and Blatt 2002; Garcia-Mata et al., 2003; Wang et al., 2013; Chen et al., 2016). Interactions with the H2S gasotransmitter have also been implicated to overlap with ABA regulation of guard cell ion transport (Papanatsiou et al., 2015). During ABA-induced stomatal closure, elevated cytosolic Ca2+ activates Ca2+-dependent protein kinases that activate Ca2+ channel (Harmon et al., 2000) and directly phosphorylate PP2Cs and targets like slow anion channels (SLAC1/SLAHs; Vahisalu et al., 2008; Geiger et al., 2009, 2010). While the ABA pathways are firmly resolved in angiosperms, a comprehensive comparative study in ferns, sister group to the seed plants, is needed to address the conflicting results (Brodribb and McAdam, 2011; Hanada et al., 2011; Ruszala et al., 2011).
The evolutionary timing of the acquisition of active stomatal control has emerged as a key question because of its importance for understanding the influence on the rise of global vegetation. In this study we addressed fundamental questions regarding stomatal regulation in ferns, a group that to date has been understudied, so as to better evaluate the evolution of this key adaptation across land plants. Was active stomatal regulation retained in ferns, following its origin in early land plants? We utilized genomic, transcriptomic and physiological tools to test the hypothesis that ABA-induced stomatal closure governed by membrane transporters and ABA signaling components is ancestral in ferns. Our results suggest that genes of ABA reception and signaling components are found in all stomata-bearing terrestrial plants including ferns. Our molecular and physiological evidence indicates that active stomatal control is present not only in seed plants but also in early diverged extant vascular plants and ferns. These findings discount previous claims (McAdam et al., 2016) that ferns lack the necessary signaling components for active, ABA-mediated stomatal closure.
RESULTS
Genomic Evidence of an Essential Set of ABA Signaling Genes and Proteins in Ferns
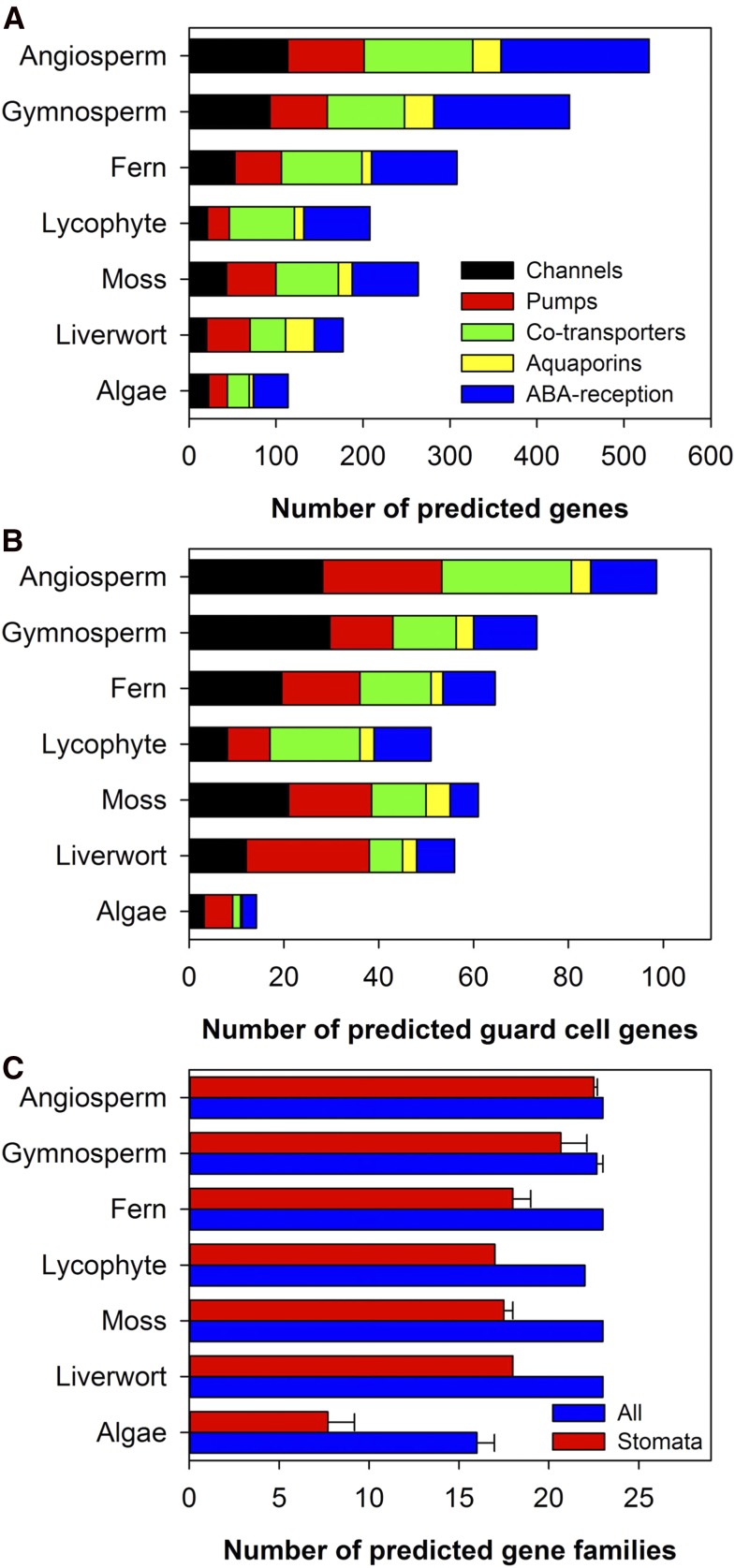
To identify potential orthologs of 63 ABA reception and membrane transport genes known to be involved in stomatal regulation in Arabidopsis (Arabidopsis thaliana), we performed bioinformatics analyses of predicted stomatal ABA receptor and membrane transport genes in 23 gene families across 36 species (including chlorophytic and streptophytic green algae, read algae and plants; Figs. 1 and 2). There was an overall increase in the number of these genes and gene families from algae to seed plants (Fig. 1). To avoid potential bias when using a single selection criterion for the comparative analysis of candidate genes, we tested a range of selection criteria (e.g. E-value less than 10−10 and E-value less than 10−5 with or without query coverage greater than 50%) that all give comparable results (data not shown). Using a simple selection criterion of E-value < 10−5, we found that the water fern species Azolla filiculoides and Salvinia cucullata have orthologs of all 23 gene families (Fig. 2). Among those 23 gene families, we found orthologs of 109 and 71 putative stomatal guard cell genes and 364 and 233 genes in the 23 families of A. filiculoides and S. cucullata, respectively (Fig. 2, A and B; Supplemental Tables S1–S4). The corresponding numbers are 336 and 138 genes in all 23 tested families for Arabidopsis and a charophyte alga, Klebsormidium flaccidum, respectively (Fig. 2A; Supplemental Tables S1 and S2). Even with the strict selection criteria (E-value < 10−10 and query coverage > 50%), A. filiculoides and S. cucullata have at least one putative guard cell gene in 19 and 17 of the 23 ABA reception and membrane transport gene families, respectively (Fig. 2B; Supplemental Tables S3 and S4). The number of genes for water ferns were similar to those of the other nonseed plants such as the lycophyte Selaginella moellendorffii and moss P. patens, but much higher than those for the representative algal species (Figs. 1 and 2).
Figure 1.
Number of predicted membrane transporters and ABA reception complex proteins and families in different taxa. A and B, The number of predicted transporters and ABA reception families (A) and guard cell genes (B) are grouped into seven taxa. The GENESIS (http://genesis-sim.org/) simulation environment was used to estimate the similarity among protein sequences (A) and family (B). Candidate protein sequences were selected by BLASTP (NCBI) software searches that satisfied the criteria of E-value < 10−10 and query coverage > 50% for guard cell proteins and E-value < 10−5 for all proteins. C, The number of gene families in different taxa. Data are mean ± se for those with error bars.
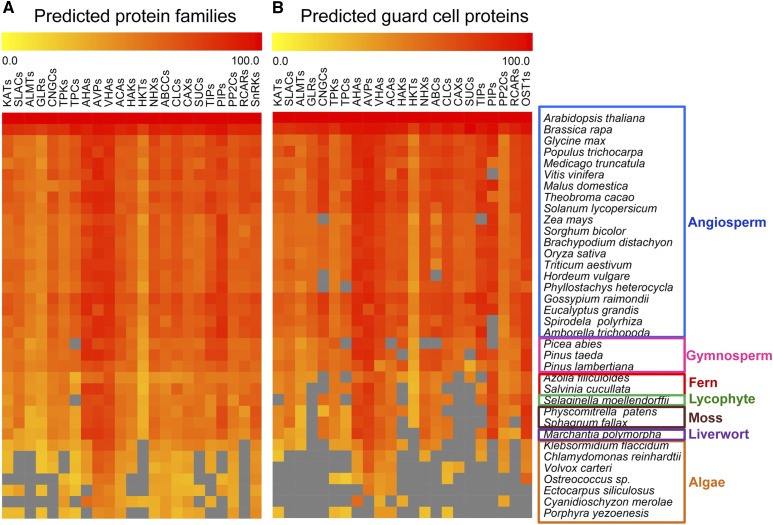
Figure 2.
Similarity heat map for the evolution of membrane transporters and ABA reception complex proteins in different species. A and B, the GENESIS simulation environment was used to estimate the similarities among protein family (A) and sequences (B). Candidate protein sequences were selected by BLASTP software searches that satisfied the criteria of E-value < 10−5 only (A) and E-value < 10−10 and query coverage > 50% (B). Colored squares indicate protein sequence similarity from zero (yellow) to 100% (red). Clades are indicated by different colors on the right: angiosperm (blue), gymnosperm (pink), fern (red), lycophyte (green), moss (brown), liverwort (purple), and algae (orange). Gray squares indicate that no proteins were found that satisfied the selection criteria. KATs represent the AKTs/KATs/GORKs proteins. ABCC, ATP-binding cassette C transporter; ACA, autoinhibited Ca2+-ATPase; AHA, Arabidopsis plasma membrane H+-ATPase; AKT, Arabidopsis inwardly rectifying K+ channel; ALMT, aluminum-activated malate transporter; AVP, Arabidopsis vacuolar H+-pyrophosphatase; CAX, cation proton exchanger; CLC, chloride channel; CNGC, cyclic nucleotide gated channel; GORK, guard cell outwardly rectifying K+ channel; GLR, Glu receptor-like Ca2+ channel; GORK, gated outwardly rectifying K+ channel; HAK, high-affinity K+ transporter; HKT, high-affinity K+/Na+ transporter; KAT, guard cell inwardly rectifying K+ channel; NHX, Na+/H+ antiporter; TPK, tonoplast K+ channel; OST1, open stomatal1; PIP, plasma membrane intrinsic protein; PP2C, Protein Phosphatase 2C; RCAR, regulatory component of ABA receptor; SLAC, slow anion channel; SnRK2, SNF1-related protein kinase2; SUC, Suc transporter; TIP, tonoplast intrinsic protein; TPC, two-pore channel; TPK, two-pore channel, a guard cell membrane transporter; VHA, vacuolar H+-ATPase.
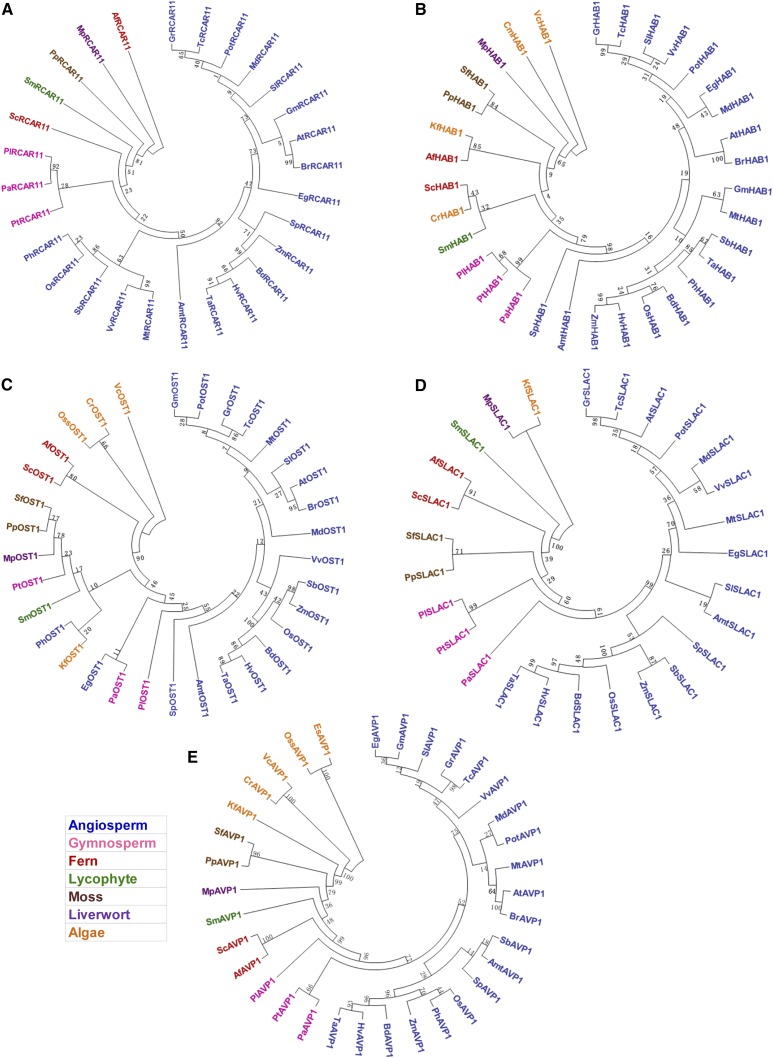
We next used the predicted protein sequences of RCAR11s, PP2Cs, SnRK2s, SLAC1s, and vacuolar H+-pyrophosphatase1 in 36 species for phylogenetic analysis. Core ABA signaling has been evolutionarily present in land plants including ferns (Figs. 2 and 3). All the PP2C and SnRK2s protein families have been identified across all tested land plant and algal species, but RCARs are not found in the seven algae examined (Figs. 2 and 3). Importantly, it appears that the two key ion transporters SLAC1 and AVP1 evolved in concert with green plant evolution (Fig. 3, D and E) as plants adapted to land.
Figure 3.
Phylogenetic trees of key ABA signaling proteins in species of plants and algae. A to E, The regulatory component of ABA receptor 11 (RCAR11; A), ABA insensitive1 (ABI1; B), open stomatal 1 (OST1; C), slow anion channel1 (SLAC1; D), and vacuolar H+-pyrophosphatase1 (AVP1; E) were estimated. The maximum-likelihood method was used to construct the trees and evolutionary distances were computed in the software MEGA 6. Bootstrap values are shown next to each branch of the trees. Clades are indicated by different colors: angiosperm (blue), gymnosperm (pink), fern (red), lycophyte (green), moss (brown), liverwort (purple), and algae (orange). Af, Azolla filiculoides; Amt, Amborella trichopoda; At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Br, Brassica rapa; Cm, Cyanidioschyzon merolae; Cr, Chlamydomonas reinhardtii; Eg, Eucalyptus grandis; Es, Ectocarpus siliculosus; Gm, Glycine max; Gr, Gossypium raimondii; Hv, Hordeum vulgare; Kf, Klebsormidium flaccidum; Md, Malus domestica; Mp, Marchantia polymorpha; Mt, Medicago truncatula; Os, Oryza sativa; Oss, Ostreococcus sp.; Pa, Picea abies; Ph, Phyllostachys heterocycla; Pl, Pinus lambertiana; Pot, Populus trichocarpa; Pp, Physcomitrella patens; Pt, Pinus taeda; Py, Porphyra yezoenessi; Sb, Sorghum bicolor; Sc, Salvinia cucullata; Sf, Sphagnum fallax; Sl, Solanum lycopersicum; Sm, Selaginella moellendorffii; Sp, Spirodela polyrhiza; Ta, Triticum aestivum; Th, Theobroma cacao; Vc, Volvox carteri; Vv, Vitis vinifera; Zm, Zea mays.
Fern Retains ABA-Responsive Genes for Active Stomatal Control
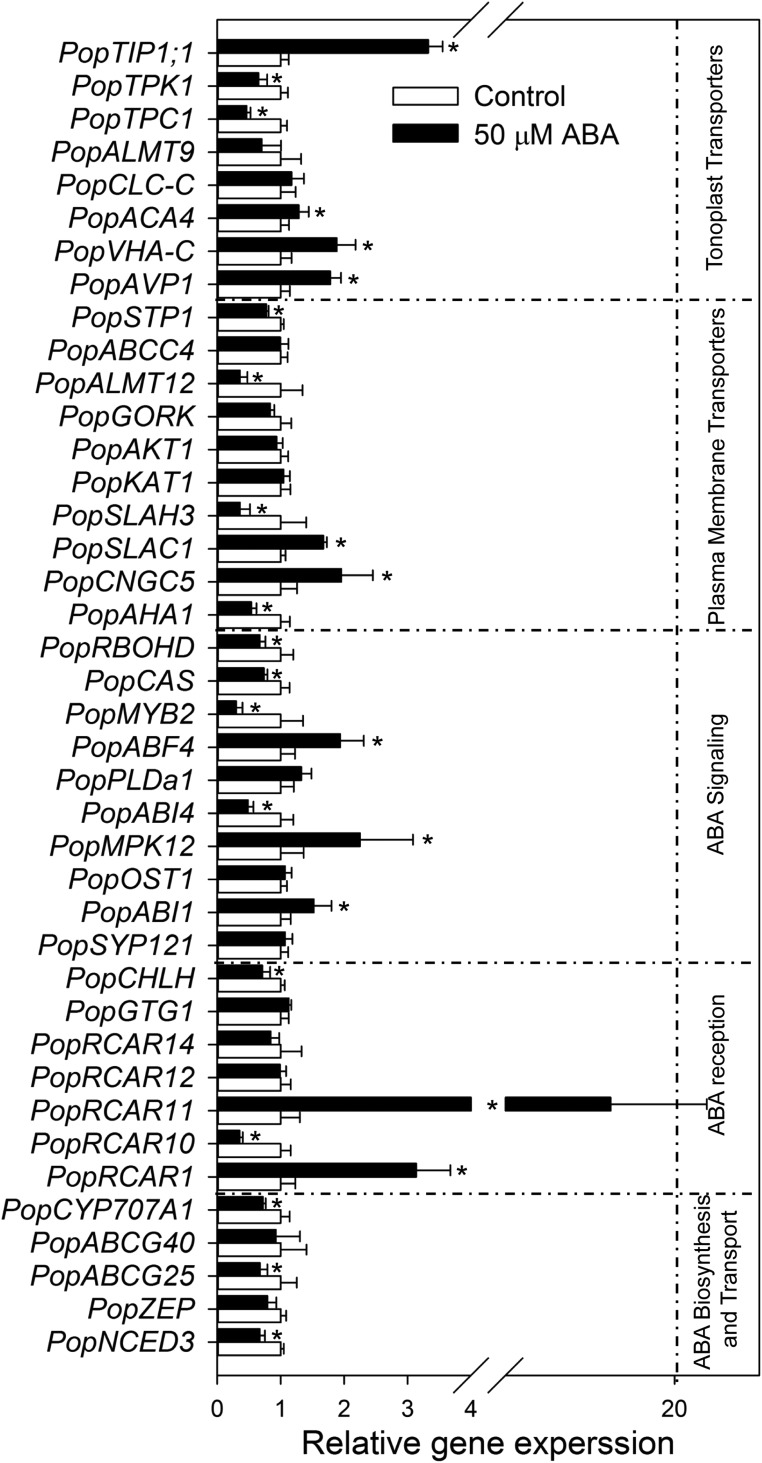
The water fern species examined may not respond to ABA due to their unusual guard cell and pore development (Busby and Gunning, 1984). We therefore used two terrestrial fern species Polystichum proliferum and Nephrolepis exaltata (both are more typical of ferns in terms of morphology and habitat) in the following experiments. Well-established ABA responsive genes and signaling pathway in Arabidopsis (Supplemental Table S5) were used as a control for the ABA-induced gene expression in epidermal layers of P. proliferum. The results showed that ferns share core ABA metabolism, reception, signaling, and membrane transporter genes (Figs. 4 and 5) with Arabidopsis, Hordeum vulgare, Oryza sativa, P. patens, and K. flaccidum (Supplemental Table S5). In quantitative real-time PCR (qPCR) experiments, all 40 selected key genes (Supplemental Table S6) were successfully amplified from the ABA-treated lower epidermis of P. proliferum and many orthologous genes in Arabidopsis guard cells were significantly up-regulated or down-regulated in P. proliferum compared to the control (Fig. 4). We detected at least one gene family member in P. proliferum representing each core node of the ABA signaling pathways (Figs. 4 and 5).
Figure 4.
Relative expression of ABA signaling genes in epidermal peels of P. proliferum. Forty genes involved in the ABA signaling pathway were classified into five groups by function. Relative expression levels were calculated and normalized. Data are mean ± se. Significant up- or down-regulation is indicated with an asterisk at P < 0.05.
Figure 5.
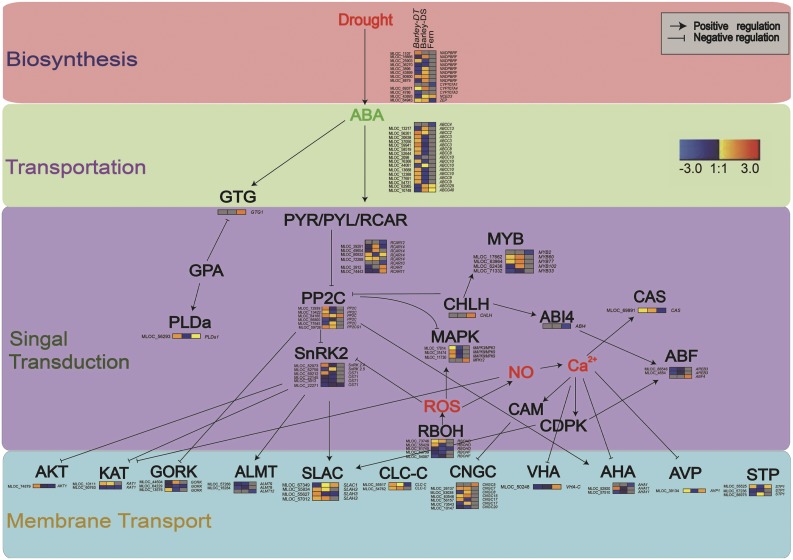
Conserved ABA signaling pathway in epidermis of H. vulgare and P. proliferum. The signaling network is formed by four main functional categories: ABA biosynthesis (pink), ABA transportation (green), signal transduction (purple), and the downstream ion channels and transporters (light blue). Heatmap of transcriptome of H. vulgare epidermis in drought stress (left and middle) and qPCR of P. proliferum epidermis in ABA treatment (right) are shown. ABCC, ATP-binding cassette C transporter; ABCG, ATP-binding cassette G; ABF, ABA responsive elements-binding factor; ABI4, ABA insensitive4; AHA, Arabidopsis plasma membrane H+-ATPase; AKT1, Ser/Thr kinase1; ALMT, aluminum-activated malate transporter; AREB, AREB-like protein; AVP, Arabidopsis vacuolar H+-pyrophosphatase; CAS, calcium sensing receptor; CHLH, protoporphyrin IX magnesium chelatase, subunit H; CLC-C, chloride channel C; CNGC, cyclic nucleotide gated channel; CYP707A, cytochromeP450 family 707 superfamily A; GORK, gated outwardly rectifying K+ channel; GTG, GPCR-type g protein1; KAT, guard cell inwardly rectifying K+ channel; MAPK, mitogen activated kinase-like protein; MYB, MYB domain protein; NADPBRF, NAD(P)-binding Rossmann-fold superfamily protein; NCED3, 9-cis-epoxycarotenoid dioxygenase6; OST1, open stomata1; PLDa1, phospholipase Dα1; PP2C, Protein Phosphatase 2C; RBOH, respiratory burst oxidase homolog protein; RCAR, regulatory component of ABA receptor; SLAC, slow anion channel; STP1, sugar transporter1; VHA, vacuolar H+-ATPase; ZEP, zeaxanthin epoxidase.
PopNCED3 and PopZEP, two key genes of ABA biosynthesis, were significantly down-regulated when ABA was applied to P. proliferum. The expression levels of P. proliferum orthologs PopABCG25 and PopABCG40, which transport ABA, were slightly reduced by ABA treatment. The expression of ABA catabolic gene, PopCYP707A1 was significantly reduced upon ABA treatment (Fig. 4). ABA treatment resulted in up-regulation of ABA receptor genes PopRCAR1 and PopRCAR11 up to 18-fold while the expression of PopRCAR10 was significantly reduced. The ortholog of the plasma membrane ABA receptor gene PopGTG1 showed no response to ABA. Moreover, the ortholog of ABA insensitive1 was significantly upregulated and open stomata1 was little affected. Surprisingly, the transcripts of MAPK 12 (Jammes et al., 2009) PopMPK12 in P. proliferum was doubled by ABA treatment. Among other signaling components for ABA response, transcription factors ABA insensitive4 and myb domain protein2 were down-regulated while ABRE binding factor4 was significantly up-regulated by ABA. The Calcium sensor (Han et al., 2003) PopCAS and the ROS homeostasis gene PopRBOHD were both down-regulated in ABA-treated epidermis of P. proliferum (Fig. 4).
Most of the transporter genes were differentially regulated in leaf epidermis of P. proliferum after ABA treatment. Expressed orthologs of Arabidopsis guard cell S-type anion channel homolog3, PopALMT12, Ca2+ activated vacuolar K+ channel, and slow vacuolar Ca2+ channel were significantly down-regulated while PopSLAC1 and cyclic nucleotide gated channel5 was up-regulated upon ABA treatment. Importantly, the plasma membrane H+-ATPase 1 and Suc/H+ cotransporter1 were significantly down-regulated, but the expression of vacuolar H+-pyrophosphatase1 and vacuolar H+-ATPase C was up-regulated by ABA. Furthermore, a key ABA responsive water channel encoding tonoplast intrinsic protein1;1 gene was up-regulated by >300% (Fig. 4). These observations indicate a set of signaling pathways that regulate gene expression in ways similar to that of angiosperms. However, the fact that these genes may be regulated similarly does not necessarily mean that the responsiveness of guard cells to ABA is the same in ferns.
Comparative Transcriptomic Analysis Reveals Conserved Genes for Active Stomatal Regulation across Land Plant Lineages
To identify genes potentially involved in stomatal regulation along the green plant tree of life, we first interrogated a collection of microarray and RNA-sequencing (RNA-seq) data sets from the angiosperms Arabidopsis (Leonhardt et al., 2004; Wang et al., 2011) and O. sativa (Lenka et al., 2011), the moss P. patens (Stevenson et al., 2016) and the green algae K. flaccidum (Holzinger et al., 2014). We also conducted qPCR on the fern P. proliferum (Fig. 4) and RNA-seq on the angiosperm H. vulgare (Fig. 5). The combined results further supported our hypothesis that the ABA signaling pathway is conserved throughout the examined plants despite only a limited number of genes tested with qPCR in P. proliferum (Figs. 4 and 5; Supplemental Table S6). The ABA- or drought-induced transcriptomic data showed that there are a few thousand genes up-regulated or down-regulated and that these genes vary among species. However, key genes are conserved across the studied species (Supplemental Table S5).
The presence of a gene in an epidermal preparation does not mean that it is expressed in guard cells or that it participates in guard cell ABA signaling. However, our recent work on comparative analysis of RNA-sequencing of epidermal peels and guard cell protoplasts has shown similar transcriptome profiles in A. thaliana (C. Zhao, Y. Wang, and Z.-H. Chen, unpublished data). Here, we isolated RNA from lower leaf epidermal peels of P. proliferum and H. vulgare. The use of H. vulgare genotypes has yielded a total of 6,676 differentially expressed genes (DEGs) from epidermal peels (Supplemental Table S5). The data provided clear transcriptome profiles of stomatal-specific transcripts. RNA-seq on whole leaves is not likely to have good representation of guard cell DEGs due to masking effects from the low ratio (1:100) of guard cells to all other leaf cells (C. Zhao, Y. Wang, and Z.-H. Chen, unpublished data). Epidermal peels reduce the ratio of guard cells to other cells ratio to approximately 1:5, which significantly increases the detection of guard cell transporter transcripts that are usually in low abundance (Chen et al., 2016). In our RNA-seq experiments on H. vulgare epidermal peels, 579 ABA signaling, 55 ABA biosynthesis and transport, 87 ABA reception components, and 117 membrane transport DEGs were detected (Fig. 5). Based on the RNA-seq of H. vulgare leaf epidermal peels, we were able to construct an ABA signaling pathway containing most of the key ABA-responsive genes identified in drought-tolerant and drought-sensitive H. vulgare genotypes and in P. proliferum (Fig. 5). Interestingly, qPCR data collected from leaf epidermal peels of P. proliferum were able to fit into most of the important nodes of the ABA signaling pathways (Fig. 5). Therefore, these data provided transcriptomic evidence that stomata in P. proliferum may respond to ABA.
ABA Induces Stomatal Closure in Two Fern Species
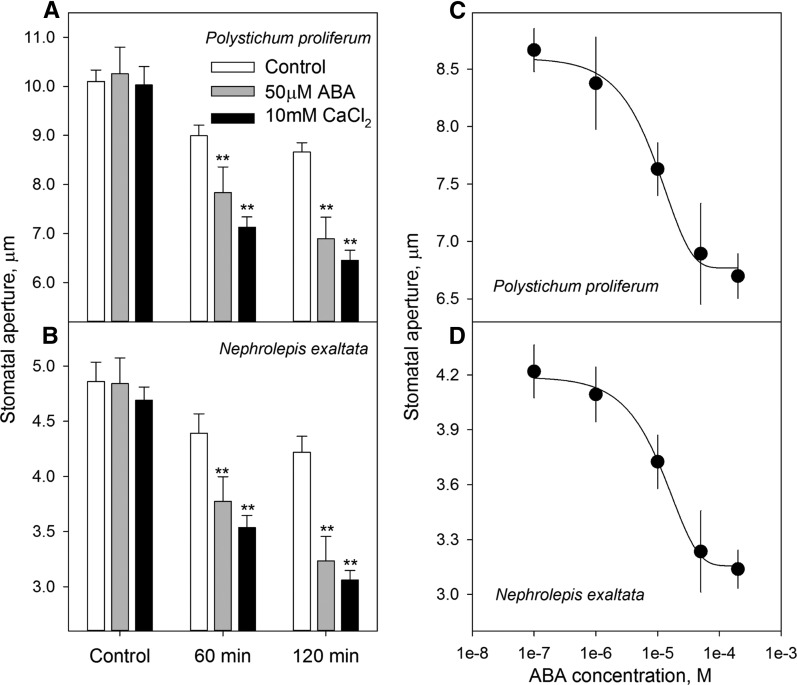
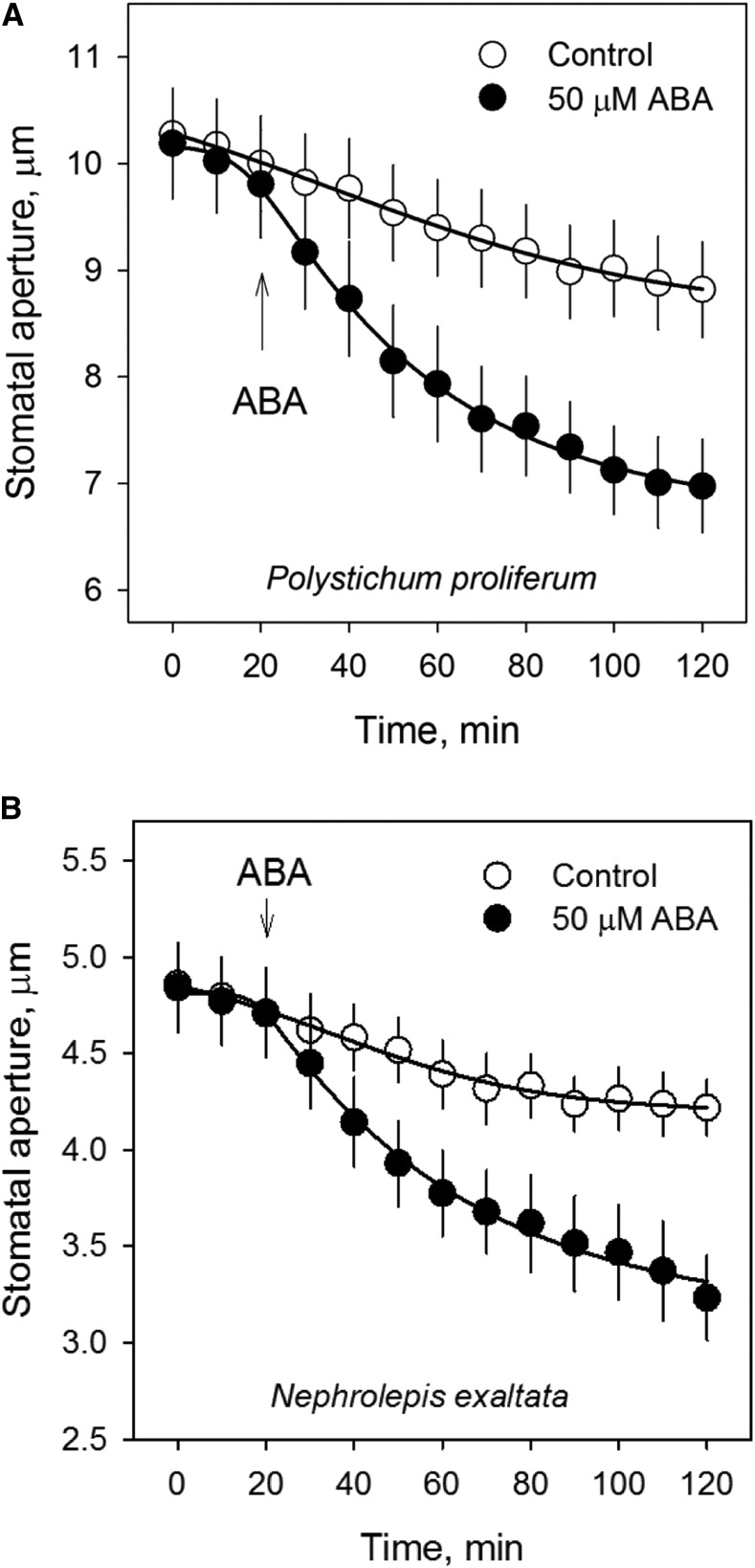
Despite the genomic and transcriptomic findings on the conservation of ABA-responsive genes and signaling pathways, this evidence alone does not prove that the stomata of ferns respond to ABA. Therefore, we explored the physiological impact of ABA on stomatal closure in two fern species. We used a well-established methodology adapted from stomatal assay and electrophysiology (Blatt et al., 1990; Chater et al., 2011; Chen et al., 2012a), which was different from the previously published work in ferns (Brodribb and McAdam, 2011; McAdam and Brodribb, 2012a, 2012b). In epidermal peels of the two fern species P. proliferum and N. exaltata, most of the stomata were open during the day in a growth chamber (Figs. 6–8). Stomata of P. proliferum and N. exaltata showed only a slight decrease in the measuring buffer for a period of 120 min under the microscopic light at 100 μmol m−2 s−1 photosynthetically active radiation (PAR; Fig. 6, A and B). Stomatal sensitivity was also tested with 10 mm CaCl2 (commonly used to trigger Ca2+-induced stomatal closure), which led to enhanced stomatal closure (Fig. 6, A and B). Most importantly, stomatal aperture was significantly decreased (P < 0.01) by 30.8% and 44.9% at 60 min and 120 min, respectively, in 50 μm ABA treatment in P. proliferum (Fig. 6A) and did so in a dose-dependent manner (Fig. 6, C and D) in both fern species. Moreover, ABA-induced stomatal closure was consistent in epidermal peels of both P. proliferum and N. exaltata (Fig. 7).
Figure 6.
Stomatal responsiveness to ABA and CaCl2 in ferns. A to D, Stomatal aperture of P. proliferum (A) and N. exaltata (B) in the control, 50 μm ABA, and 10 mm CaCl2 treatments at 0, 60, and 120 min. B, ABA dose-dependent (0.1, 1, 10, 50, and 200 μm ABA) stomatal closure in P. proliferum (C) and N. exaltata (D). Data are mean ± se (n = 30–40 stomata from five biological replicates). Asterisks indicate significant difference at P < 0.01 level.
Figure 8.
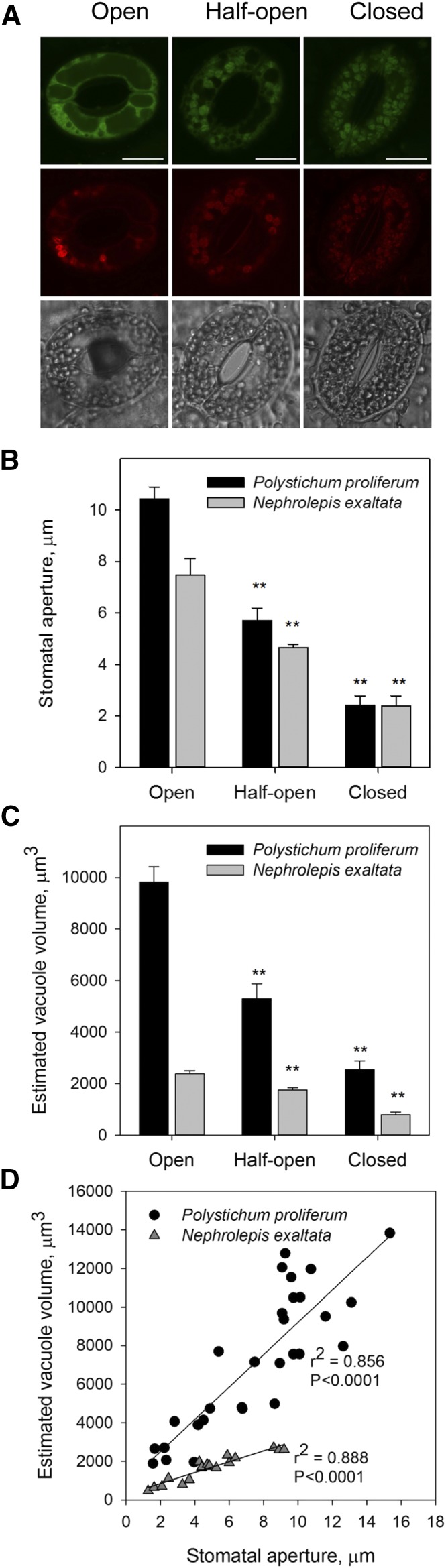
ABA-induced stomatal guard cell volume changes in ferns. A, Representative confocal images of open, half-open, and closed stomata in P. proliferum. Green and red fluorescence indicate the ROS and endomembrane stain, respectively. Bars = 20 μm. B and C, Stomatal aperture (B) and estimated vacuole volume (C) of open, half-open, and closed stomata in P. proliferum and N. exaltata. Data are mean ± se (n = 6–10 stomata from three biological replicates). D, Correlation between stomatal aperture and estimated vacuole volume. Data are plotted from all the measured stomata in confocal imaging.
Figure 7.
ABA-induced stomatal closure in ferns. A and B, Stomatal aperture of P. proliferum (A) and N. exaltata (B) in the control (0–20 min) and 50 μm ABA (20–120 min) treatment. Blank (control) stomatal aperture measurements were also conducted for 120 min in both species. Data are mean ± se (n = 30–40 stomata from five biological replicates).
Vacuoles are critical for controlling guard cell volume, which regulates the opening and closure of stomata (Blatt, 2000; Franks et al., 2001; Shope et al., 2003; Chen et al., 2012b). We tested whether stomatal closure is related to vacuole changes (Gao et al., 2005; Meckel et al., 2007) of guard cells in P. proliferum and N. exaltata. ROS staining using confocal microscopy showed that large vacuoles are found in guard cells of open stomata of P. proliferum and N. exaltata (Fig. 8A). After ABA treatment, vacuoles shrank significantly in guard cells of half-open and closed stomata (Fig. 8, B and C). Interestingly, the estimated total volume of vacuoles of each stomata was significantly correlated with the stomatal aperture (P < 0.0001) in both fern species (Fig. 8D).
DISCUSSION
Evolutionary Genomic Analysis Reveals the Genetics of Active Stomatal Regulation in Ferns
Molecular biological and genomic analyses provide evidence that a core ABA signaling pathway (Hauser et al., 2011) consisting of PYR/PYL/RCAR, PP2Cs, and OST1-like kinases and their target genes were established during the transition from an aquatic to a terrestrial environment over 400 million years ago during the origin of land plants (Munemasa et al., 2015). Nonetheless, the origin of the active response of stomata to endogenous and environmental cues is still unclear. Stomata of the moss P. patens and in the lycophyte S. uncinata respond to environmental signals in a manner similar to those observed in seed plants. The orthologs of the key ABA signaling components in drought response are involved in stomatal control in early land plants (Raven, 2002; Chater et al., 2011, 2013; Ruszala et al., 2011). ABA-responsive PpLEA-1::GUS lines of P. patens revealed a pattern of localized expression around the stomatal ring of sporophytes, which further strengthens previous observations of the ABA response of stomata in mosses (Garner and Paolillo, 1973; Chater et al., 2011; Stevenson et al., 2016). The stomatal response of the Arabidopsis ost1 mutant to ABA is rescued by substitution with the P. patens ortholog PpOST1. Also, the targeted knockout of the Ppost1-1 exhibits a significantly attenuated ABA response of stomata. It indicates important features for guard cell ABA signaling during land plant evolution (Cuming et al., 2007; Lind et al., 2015). ABA-related genes from Arabidopsis generate 11 orthologous clusters of ABA-related genes from A. thaliana, Arabidopsis lyrata, Populus trichocarpa, O. sativa, S. moellendorffii, and P. patens. Phylogenetic analyses indicated that the common ancestor of these six species possessed most of the key protein functions of ABA-related genes, suggesting that the expansion of the gene families related to ABA signaling pathways may have contributed to the sophisticated stress tolerance mechanisms of seed plants (Hanada et al., 2011).
Here, we filled a crucial gap in our understanding of ABA signaling by including genomic and transcriptomic evidence for ABA signaling of ferns. The essential orthologs to Arabidopsis core ABA reception components (RCARs, OST1s, and PP2Cs) and guard cell membrane transporters (e.g. SLAC1/SLAHs, AHAs, TPKs, ALMTs, and AVPs) are found in the two water fern (Figs. 2 and 3) and a land fern species (Fig. 4). These highly conserved ABA signaling components reinforce the pivotal role that stomata have played in the evolution of terrestrial plants (Raven, 2002; O’Donoghue et al., 2013). Although the functions of those key genes in ferns need to be determined in future experiments, our results suggest that ferns are equipped with a set of essential ABA signaling genes necessary for ABA-induced stomatal closure. These findings discount previous claims based on a more limited analysis centered on a single kinase pathway leading to control of a subset of anion channels (McAdam et al., 2016).
Stomatal Membrane Transport and ABA Signaling Genes Are Conserved in a Terrestrial Fern
Comparative transcriptomic analysis among P. proliferum, H. vulgare, O. sativa, Arabidopsis, P. patens, and K. flaccidum (Supplemental Table S5; Fig. 5) has generated some important insights into the ABA responsiveness in fern stomata. Our results for ABA-induced gene expression in P. proliferum are consistent with those of the model plant Arabidopsis (Leonhardt et al., 2004; Wang et al., 2011), O. sativa (Lenka et al., 2011), H. vulgare (Fig. 5), and P. patens (Stevenson et al., 2016). Further evidence for establishing the evolutionary timeline for the appearance of functional ABA signaling gene expression in guard cells could be helpful to refine evolutionary models (Munemasa et al., 2015).
The stomatal response to ABA in land plants may have been recruited from a preexisting transport and signaling network in their common ancestor and ferns appear to have these stomatal regulatory features. During drought stress, Arabidopsis AtNCEDs and AtZEPs (Xiong and Zhu, 2003) promote the biosynthesis of ABA, which is then transported to guard cells via AtABCG25 and AtABCG40 (Kang et al., 2010; Kuromori et al., 2010). At the same time, ABA catabolic gene AtCYP707As (Okamoto et al., 2006) is significantly reduced. For membrane transport, ABA inhibits AtAHA1 activity by reducing the phosphorylation of AtAHAs (Merlot et al., 2007) and AtKATs (Sato et al., 2009). ABA also triggers Ca2+ influx and Ca2+ release from internal stores to the cytosol that leads to elevated cytosolic Ca2+, which, in turn, suppresses AtKAT1 activity while promoting that of AtSLAC1 and depolarizing the membrane (Grabov and Blatt, 1998; Hamilton et al., 2000; Chen et al., 2010, 2012b). Membrane depolarization activates AtGORK, resulting in K+ efflux from guard cells (Hosy et al., 2003). AtSLAC1 is involved in anion efflux guard cells (Vahisalu et al., 2008) and is directly activated by OST1, which is involved in core ABA signaling (Geiger et al., 2009). AtALMT12 is a plasma membrane malate-induced, R-type anion channel in guard cells controlling stomatal response (Imes et al., 2013). The tonoplast chloride channel C AtCLC-C and AtALMT9 have roles in the anion fluxes that control stomatal movements (Jossier et al., 2010; De Angeli et al., 2013). The tonoplast AtTPK1 (Gobert et al., 2007) regulate stomatal closure via K+ release from vacuoles. Also, ABA-induced cytosolic pH change has a significant role for stomatal closing (Blatt and Armstrong, 1993; Grabov and Blatt, 1997, 1998; Chen et al., 2012b) and AtAVP1 regulated NO3− transport and cytosolic pH (Rea and Poole, 1993; Zancani et al., 2007). These genes along with others were found to be differentially expressed in the fern P. proliferum in the control and ABA treatment (Fig. 4). In addition, recruitment of the regulatory genes controlling root function evolved after stomata, providing plants with improved capacities for water and nutrient uptake from soils (Menand et al., 2007). The parallel acquisition of those two adaptive features allowed early land plants to access water and nutrients more readily.
Ferns Have Both Passive and Active Stomatal Regulation
The fern clade shows a remarkable diversity with 11,961 species, outnumbering the number of species of extant gymnosperms (Pryer and Schuettpelz, 2009; McElwain, 2011; Schuettpelz and Schneider, 2016) and only angiosperms have more species (>350,000; http://www.theplantlist.org/browse/A/). Passive control of stomata via regulation of xylem water supply presumably played a role in the success of the fern clade (Brodribb and McAdam, 2011; McAdam and Brodribb, 2012a, 2012b). The evolution of stomata and xylem are interconnected. Stomata primarily control transpirational water loss while the xylem modulates water supply. Their coordinated functions have to be fine-tuned in response to drought. A species with greater coordination of stomatal and xylem functions would have an advantage especially when comparing ferns to seed plants (Sperry, 2004; McElwain, 2011). However, leaky stomata in ferns have generally been considered a significant ecophysiological limitation (Testo and Watkins, 2012).
We cannot rule out the possibility that the stomata of some ferns do not respond to ABA, given the huge diversity of fern species and their habitats (Creese et al., 2014) and the fact that only few species were examined in this study. However, active control of stomatal opening and closure may be ancestral for land plants with stomata, as evidenced by stomatal ABA sensitivity in the moss P. patens and the lycophyte S. uncinata (Chater et al., 2011; Ruszala et al., 2011). Furthermore, addition of ABA caused dose-dependent stomatal closure and inhibition of stomatal opening in response to light in S. uncinata (Ruszala et al., 2011). The overall ABA dose-dependent stomatal closure in P. proliferum and N. exaltata (Fig. 6, C and D) was similar to that in S. uncinata. However, there is a notable difference in their stomatal sensitivity to ABA. ABA-induced significant stomatal closure in S. uncinata was observed in 1 μm ABA treatment (Ruszala et al., 2011), which was not found in ferns at this concentration (Fig. 6). Many taxa do not have ABA-induced stomatal closure, including some mosses and lycophytes (Brodribb and McAdam, 2011). Stomatal ABA sensitivity is present in lineages of vascular plants that diverged before ferns as well as in lineages that diverged after ferns. Will it be possible to have an evolutionary scenario in which stomatal ABA sensitivity is present in ferns?
Here, we showed that stomatal closure can be measured directly in excised epidermis in the presence of ABA (Figs. 6 and 7) in two terrestrial fern species P. proliferum and N. exaltata. We employed similar stomatal assay methods to those used in analyses of Arabidopsis, Vicia faba, and H. vulgare (Chen et al., 2010, 2012a; Liu et al., 2014). Our results are consistent with recent reports that stomatal response of ferns to CO2, vapor pressure deficit, and light is neither lost nor controlled only by hydraulic (i.e. passive) regulation of turgor pressure (Creese et al., 2014; Doi et al., 2015; Franks and Britton-Harper, 2016). A crucial point to consider is that the methodologies differ between this study and that by McAdam and Brodribb (2012a, 2012b), which introduced ABA to the transpiration stream to measure responses to endogenous ABA concentrations. The addition of exogenous ABA to epidermal peels and spraying of ABA used in this study could be largely responsible for the different results. Also, there is an overlapping fern species N. exaltata in our study and those of Brodribb and McAdam (2011). It appears that the sensitivity of N. exaltata responding to ABA under these two experimental conditions was very different and was not directly comparable. By contrast, the range of physiological and molecular techniques used in this study is more comparable to those used in moss and lycophyte species by Chater et al. (2011) and Ruszala et al. (2011). Clearly, it will be of interest to see whether, as a group, the fern might fall between two or more subsets with different stomatal characteristics.
Is SnRK2 Regulation of SLACs the Only Mechanism for Stomatal Closure?
OST1/SnRK2s have important roles in stomatal regulation via the activation of SLAC/SLAH anion channels in Arabidopsis (Geiger et al., 2010), as well as the species of moss (Chater et al., 2011) and lycophyte examined (Ruszala et al., 2011; Lind et al., 2015). However, McAdam et al. (2016) recently reported that GAIA1, a homolog of OST1 in the water fern Ceratopteris richardii, regulates ABA signaling for gametophyte sex determination, rather than stomatal regulation. Fern and lycophyte SnRK2s were found to be unable to activate native endogenous SLACs. Therefore, these authors concluded that the ABA-signaling pathway through SnRK2-mediated SLAC activation for stomatal closure may not appear to be operating in the fern C. richardii. Instead, they suggested that connection between ABA and stomatal control via the specific activation of SLAC/SLAH anions channels is a more recent innovation that did not evolve until after the divergence of ferns and seed plants (McAdam et al., 2016).
We call into question the conclusions of McAdam et al. (2016). Not only can we demonstrate ABA-induced stomatal closures in two fern species, but recent advances point to alterative pathways for ABA-induced stomatal closure. These findings imply that the focus on the OST1/SnRK2-SLAC1 pathway might not be universal in all land plants. Indeed, the suggestion is hardly surprising; in Arabidopsis, multiple calcium-dependent kinases CPK3, CPK5, CPK6, CPK21, and CPK23 (Brandt et al., 2012, 2015; Geiger et al., 2010; Scherzer et al., 2012) and our recent data on CPKs and CRKs (Pornsiriwong et al., 2017) are thought to modulate ABA-activated guard cell anion channels. Orthologous genes (e.g. OST1, CPKs, CRKs) can be found in the genome and transcriptome of C. richardii (D.B. Marchant, D.E. Soltis, and P.S. Soltis, unpublished data). Therefore, these data raise the question of whether CrSnRK2 in C. richardii is the only kinase essential for ABA-induced stomatal closure in ferns. We note, too, that McAdam et al. (2016) offer no stomatal aperture assays for C. richardii. Clearly, if alternative pathways for ABA signaling exist, then the lack of anion channel activation by CrSnRK2 cannot prove an ABA insensitivity of stomata of C. richardii. Further experiments should focus on the identification of CPKs and CRKs in C. richardii and their interaction with SLAC/SLAH anion channels in combination with stomatal physiological measurements to confirm whether there is a general ABA insensitivity of stomata in C. richardii. In conclusion, despite the advantages of passive hydraulic control for fern stomata, the molecular mechanisms of ABA-induced stomatal closure may exist in certain fern species.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Drought-tolerant (X5) and -sensitive (X54) Tibetan annual wild barley (Hordeum vulgare spp. spontaneum) genotypes were used in this study. Seeds were sown in 4-liter pots with potting mixture and five healthy and uniform seedlings were maintained per pot. Plants were grown in a greenhouse at a 22 ± 2°C and 60% relative humidity (RH) during the day, and 20 ± 2°C and 60% RH at night, in 12-h/12-h light/dark cycle. The average PAR was 400 μmol m−2 s−1. All plants were well watered before drought treatment, which was commenced 5 weeks after sowing. Fully expanded leaves were sampled when the water holding capacity decreased to 10% (v/v). Abaxial epidermal strips were peeled using a pair of fine forceps and frozen immediately in liquid N.
Small sporophytes of the ferns Polystichum proliferum and Nephrolepis exaltata were purchased from a local plant nursery (Bunnings) and grown in a growth chamber. Plants were grown under 12-h/12-h day/night, 20 ± 1°C, 100 µmol m−2 s−1 PAR, and 60% RH. Plants were irrigated weekly with half-strength Hoagland’s solution. Plants were grown for at least four weeks before stomatal assay and qPCR. Treatment was applied by spraying 50 μm ABA to the upper and lower epidermis of fern leaves for 1 h before epidermal peeling. We used fine forceps to peel abaxial epidermal strips from young fully expanded leaves.
Evolutionary Bioinformatics Analysis
All 63 query Arabidopsis (Arabidopsis thaliana) guard cell genes and 23 gene families are from Chen et al. (2017). Genome sequence data were downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) and Ensembl Plants (http://plants.ensembl.org/index.html). The genome sequences of Azolla filiculoides and Salvinia cucullata are from Assistant Professor Fay-Wei Li. Genesis software was used to estimate the similarity among protein sequences and families. Candidate protein sequences were selected by research using the software BLASTP (NCBI; https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) that satisfied the criteria of E-value and query coverage at different levels. Amino acid sequences were translated from nucleotide sequences using the software BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and the phylogenies were constructed with the software MEGA 6 (www.megasoftware.net) using the maximum-likelihood method of evolution (Tamura et al., 2013). Values from 1,000 bootstrap replicates are shown at the nodes.
RNA-Seq
Total RNA was extracted from barley epidermal peels with the miRNeasy mini kit (Qiagen) following the manufacturer’s instructions. RNA quantity and quality check, library construction, and sequencing were performed as described by Dai et al. (2014) and Wang et al. (2016). The TruSeq RNA sample preparation kit (Illumina) was used to construct the library following the manufacturer’s instructions. Total RNA samples were purified by oligo(dT) beads and poly(A)-containing mRNA were then fragmented. The cDNA was generated by First/Second Strand Master Mix and Super Script II (Invitrogen) reverse transcription and adapters were ligated onto both ends to generate end-repaired DNA followed by amplification and enrichment. After purification, Ampure XP Beads (Agilent Genomics) were added to the PCR products and size-selected for sequencing using the 2100 Bioanalyzer (Agilent). The raw data obtained from the software HiSeq 2500 (Illumina) were cleaned by trimming and removing empty reads, low-quality bases (Q < 30 and length < 50 bp), and adaptor sequences at the 3′ end. We used the tool BWA (https://sourceforge.net/projects/bio-bwa/) to map clean reads to the genome reference of barley (http://plants.ensembl.org/) and the software Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) for gene reference (Langmead et al., 2009). The mapped reads assembling, abundance estimation and DEGs were done using the routine Cuffdiff of Cufflinks v2.1.1 (http://cole-trapnell-lab.github.io/cufflinks/; Roberts et al., 2011).
Quantitative RT-PCR
Quantitative PCR was carried out following the protocol of Chen et al. (2016) with some modification. RNA of epidermal peels of P. proliferum was extracted using Trizol reagent (Life Technologies) following the manufacturer’s procedure, and the residual genomic DNA was removed with amplification grade DNase I (Ambion). First-strand cDNA was synthesized with the SensiFAST Kit (Bioline). Transcript levels of the target genes were determined by the SensiFAST SYBR No-ROX Kit (Bioline) with Ceratopteris richardii-specific primers (Supplemental Table S6) for P. proliferum genes using a Rotor-Gene Q6000 (Qiagen). The primers design was based on the amplification of transcripts encoding conserved protein domains between C. richardii and Arabidopsis. qPCR conditions were: (1) polymerase activation at 95°C for 2 min; (2) 40 cycles of 5 s denaturation at 95°C, 10 s annealing at 63°C, and 15 s extension at 72°C; and (3) SYBR green signal data were acquired at the end. β-Tubulin (PopTUB) was used as the reference for normalization of relative gene expression. Final values were averaged from three independent biological replicates.
Stomatal Aperture Assay
Stomatal aperture assays were carried out using similar methods as described earlier (Chen et al., 2010, 2016; Eisenach et al., 2012). Peels from the lower epidermis of P. proliferum and N. exaltata plants were placed in glass-bottom petri dishes and were pretreated for 10 min in a measuring buffer [10 mm KCl and 5 mm 2-(N-morpholino)propanesulfonic acid at pH 6.1 with Ca(OH)2] under 100 µmol m−2 s−1 PAR light. The samples were imaged in the measuring buffer for 20 min as the control under a microscope (Nikon) with a model no. NIS-F1 CCD camera and a DS-U3 controller (both Nikon) attached. Treatments were applied as ABA (0.1, 1, 10, 50, and 200 μm) or CaCl2 (10 mm) and measured for another 100 min. Control experiments were also performed by measuring stomatal aperture in the measuring buffer for 120 min. All the stomatal aperture measurements were conducted under constant microscopy light of 100 µmol m−2 s−1 PAR to avoid dark-induced stomatal closure. Images were taken every 5 min and stomatal apertures were measured and analyzed with the software ImageJ (National Institutes of Health). For every data point, there were 30 to 40 stomata measured and the experiments were independently repeated five times.
Confocal Microscopy
ROS production in the cytoplasm of guard cells was determined using the fluorescent indicator 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA; Life Technologies). The styryl dye (N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide; cat. no. FM4-64; Life Technologies) has been reported to selectively stain tonoplast and endomembrane. Epidermal peels were pretreated with the opening buffer for 2 h before loading 20 µM H2DCFDA for 20 min in the dark, followed by a 5-min flush in the measuring buffer to remove excess dye. The strips were subsequently incubated in the measuring buffer containing 50 μM ABA for 120 min with a 10-min sampling interval under confocal microscopes (Zeiss and Leica). The fluorescence images were collected with excitation at 488 nm, and emission at 505 to 525 nm for H2DCFDA and emission at 610 to 640 nm for FM4-64 (Life Technologies). Vacuole volume of both guard cells was estimated using the formula: V = 4/3 × S × r, where S is the area of the vacuole and r is the radius of the guard cells. Guard cell volume was also calculated as a control to the vacuolar volume using the software Henry III (University of Glasgow).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Number of predicted membrane transporters and ABA reception complex proteins in 36 plant and algal species with E-value < 10−5.
Supplemental Table S2. Similarity analysis for the evolution of predicted membrane transporters and ABA reception complex proteins in 36 plant and algal species with E-value < 10−5.
Supplemental Table S3. Number of predicted proteins of guard cell transporters and ABA reception complex in 36 plant and algal species with E-value < 10−10 and query coverage > 50%.
Supplemental Table S4. Similarity analysis for the evolution of predicted guard cell transporters and ABA reception complex proteins in 36 plant and algal species with E-value < 10−10 and query coverage > 50%.
Supplemental Table S5. Comparative transcriptomic analysis of plant and algal species to ABA, drought, or desiccation stress.
Supplemental Table S6. Primers for quantitative RT-PCR of ABA signaling genes in the fern P. proliferum.
Note Added in Proof
A nonbiased approach to screen protein families across a wide range of species (see Fig. 2) inevitably will show a hit rate that does not cover 100% of the potential proteins, regardless of how well defined the criteria may be. We note the absence of two putative potassium channels that Gomez-Porras et al. (Gomez-Porras JL, Riaño-Pachón DM, Benito B, Haro R, Sklodowski K, Rodríguez-Navarro A, Dreyer I [2012] Phylogenetic analysis of K transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front Plant Sci 3: 167) identified by manual selection. We expect more details from these different species to come forward in the future, especially once functional information becomes available, and we present this analysis in contribution to the ongoing discussion from which a consensus will no doubt emerge.
Supplementary Material
Acknowledgments
We thank Assistant Professor Fay-Wei Li (Cornell University) and Professor Kathleen Pryer (Duke University) for the water fern genome sequences and David Randall, Gulei Jing, Xinze Zhao, Jiarun Zhang, and Tianyuan Li for their technical support.
Footnotes
Articles can be viewed without a subscription.
References
- Beerling DJ, Franks PJ (2009) Evolution of stomatal function in ‘lower’ land plants. New Phytol 183: 921–925 [DOI] [PubMed] [Google Scholar]
- Berry JA, Beerling DJ, Franks PJ (2010) Stomata: key players in the earth system, past and present. Curr Opin Plant Biol 13: 233–240 [DOI] [PubMed] [Google Scholar]
- Blatt MR. (2000) Cellular signaling and volume control in stomatal movements in plants. Annu Rev Cell Dev Biol 16: 221–241 [DOI] [PubMed] [Google Scholar]
- Blatt MR, Armstrong F (1993) K+ channels of stomatal guard cells: abscisic-acid-evoked control of the outward rectifier mediated by cytoplasmic pH. Planta 191: 330–341 [Google Scholar]
- Blatt MR, Thiel G, Trentham DR (1990) Reversible inactivation of K+ channels of Vicia stomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature 346: 766–769 [DOI] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA 109: 10593–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Munemasa S, Wang C, Nguyen D, Yong T, Yang PG, Schroeder JI (2015) Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 4: e03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, McAdam SA (2011) Passive origins of stomatal control in vascular plants. Science 331: 582–585 [DOI] [PubMed] [Google Scholar]
- Busby CH, Gunning BE (1984) Microtubules and morphogenesis in stomata of the water fern Azolla: an unusual mode of guard cell and pore development. Protoplasma 122: 108–119 [Google Scholar]
- Chater C, Gray JE, Beerling DJ (2013) Early evolutionary acquisition of stomatal control and development gene signalling networks. Curr Opin Plant Biol 16: 638–646 [DOI] [PubMed] [Google Scholar]
- Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, Gray JE, Beerling DJ (2011) Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr Biol 21: 1025–1029 [DOI] [PubMed] [Google Scholar]
- Chen ZH, Chen G, Dai F, Wang Y, Hills A, Ruan YL, Zhang G, Franks PJ, Nevo E, Blatt MR (2017) Molecular evolution of grass stomata. Trends Plant Sci 22: 124–139 [DOI] [PubMed] [Google Scholar]
- Chen ZH, Eisenach C, Xu XQ, Hills A, Blatt MR (2012a) Protocol: optimised electrophysiological analysis of intact guard cells from Arabidopsis. Plant Methods 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Hills A, Bätz U, Amtmann A, Lew VL, Blatt MR (2012b) Systems dynamic modeling of the stomatal guard cell predicts emergent behaviors in transport, signaling, and volume control. Plant Physiol 159: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Hills A, Lim CK, Blatt MR (2010) Dynamic regulation of guard cell anion channels by cytosolic free Ca2+ concentration and protein phosphorylation. Plant J 61: 816–825 [DOI] [PubMed] [Google Scholar]
- Chen ZH, Wang Y, Wang JW, Babla M, Zhao C, García-Mata C, Sani E, Differ C, Mak M, Hills A, Amtmann A, Blatt MR (2016) Nitrate reductase mutation alters potassium nutrition as well as nitric oxide-mediated control of guard cell ion channels in Arabidopsis. New Phytol 209: 1456–1469 [DOI] [PubMed] [Google Scholar]
- Creese C, Oberbauer S, Rundel P, Sack L (2014) Are fern stomatal responses to different stimuli coordinated? Testing responses to light, vapor pressure deficit, and CO2 for diverse species grown under contrasting irradiances. New Phytol 204: 92–104 [DOI] [PubMed] [Google Scholar]
- Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS (2007) Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol 176: 275–287 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dai F, Chen ZH, Wang X, Li Z, Jin G, Wu D, Cai S, Wang N, Wu F, Nevo E, Zhang G (2014) Transcriptome profiling reveals mosaic genomic origins of modern cultivated barley. Proc Natl Acad Sci USA 111: 13403–13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angeli A, Zhang J, Meyer S, Martinoia E (2013) AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat Commun 4: 1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Kitagawa Y, Shimazaki K (2015) Stomatal blue light response is present in early vascular plants. Plant Physiol 169: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D, Kerp H, Hass H (1998) Stomata in early land plants an anatomical and ecophysiological approach. J Exp Bot 49: 255–278 [Google Scholar]
- Eisenach C, Chen ZH, Grefen C, Blatt MR (2012) The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant J 69: 241–251 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Britton-Harper ZJ (2016) No evidence of general CO2 insensitivity in ferns: one stomatal control mechanism for all land plants? New Phytol 211: 819–827 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Buckley TN, Shope JC, Mott KA (2001) Guard cell volume and pressure measured concurrently by confocal microscopy and the cell pressure probe. Plant Physiol 125: 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJ, Farquhar GD (2007) The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol 143: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XQ, Li CG, Wei PC, Zhang XY, Chen J, Wang XC (2005) The dynamic changes of tonoplasts in guard cells are important for stomatal movement in Vicia faba. Plant Physiol 139: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata C, Gay R, Sokolovski S, Hills A, Lamattina L, Blatt MR (2003) Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc Natl Acad Sci USA 100: 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner D, Paolillo DJ (1973) On the functioning of stomates in Funaria. Bryologist 76: 423–427 [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, Romeis T, Hedrich R (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, Romeis T, Hedrich R (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Isayenkov S, Voelker C, Czempinski K, Maathuis FJ (2007) The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proc Natl Acad Sci USA 104: 10726–10731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Blatt MR (1998) Membrane voltage initiates Ca2+ waves and potentiates Ca2+ increases with abscisic acid in stomatal guard cells. Proc Natl Acad Sci USA 95: 4778–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Blatt MR (1997) Parallel control of the inward-rectifier K+ channel by cytosolic free Ca2+ and pH in Vicia guard cells. Planta 201: 84–95 [Google Scholar]
- Han S, Tang R, Anderson LK, Woerner TE, Pei ZM (2003) A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature 425: 196–200 [DOI] [PubMed] [Google Scholar]
- Hanada K, Hase T, Toyoda T, Shinozaki K, Okamoto M (2011) Origin and evolution of genes related to ABA metabolism and its signaling pathways. J Plant Res 124: 455–465 [DOI] [PubMed] [Google Scholar]
- Hamilton DW, Hills A, Köhler B, Blatt MR (2000) Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc Natl Acad Sci USA 97: 4967–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF (2000) CDPKs—a kinase for every Ca2+ signal? Trends Plant Sci 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Holzinger A, Kaplan F, Blaas K, Zechmann B, Komsic-Buchmann K, Becker B (2014) Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. PLoS One 9: e110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosy E, Vavasseur A, Mouline K, Dreyer I, Gaymard F, Porée F, Boucherez J, Lebaudy A, Bouchez D, Very AA, et al. (2003) The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc Natl Acad Sci USA 100: 5549–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imes D, Mumm P, Böhm J, Al-Rasheid KA, Marten I, Geiger D, Hedrich R (2013) Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J 74: 372–382 [DOI] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Kroniewicz L, Dalmas F, Le Thiec D, Ephritikhine G, Thomine S, Barbier-Brygoo H, Vavasseur A, Filleur S, Leonhardt N (2010) The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J 64: 563–576 [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler B, Blatt MR (2002) Protein phosphorylation activates the guard cell Ca2+ channel and is a prerequisite for gating by abscisic acid. Plant J 32: 185–194 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenka SK, Katiyar A, Chinnusamy V, Bansal KC (2011) Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind C, Dreyer I, López-Sanjurjo EJ, von Meyer K, Ishizaki K, Kohchi T, Lang D, Zhao Y, Kreuzer I, Al-Rasheid KA, et al. (2015) Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr Biol 25: 928–935 [DOI] [PubMed] [Google Scholar]
- Liu X, Mak M, Babla M, Wang F, Chen G, Veljanoski F, Wang G, Shabala S, Zhou M, Chen ZH (2014) Linking stomatal traits and expression of slow anion channel genes HvSLAH1 and HvSLAC1 with grain yield for increasing salinity tolerance in barley. Front Plant Sci 5: 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15: 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- McAdam SA, Brodribb TJ (2012a) Fern and lycophyte guard cells do not respond to endogenous abscisic acid. Plant Cell 24: 1510–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam SA, Brodribb TJ (2012b) Stomatal innovation and the rise of seed plants. Ecol Lett 15: 1–8 [DOI] [PubMed] [Google Scholar]
- McAdam SA, Brodribb TJ, Banks JA, Hedrich R, Atallah NM, Cai C, Geringer MA, Lind C, Nichols DS, Stachowski K, Geiger D, Sussmilch FC (2016) Abscisic acid controlled sex before transpiration in vascular plants. Proc Natl Acad Sci USA 113: 12862–12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwain JC. (2011) Ferns: a xylem success story. New Phytol 192: 307–310 [DOI] [PubMed] [Google Scholar]
- Meckel T, Gall L, Semrau S, Homann U, Thiel G (2007) Guard cells elongate: relationship of volume and surface area during stomatal movement. Biophys J 92: 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L (2007) An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Merced A, Renzaglia K (2014) Developmental changes in guard cell wall structure and pectin composition in the moss Funaria: implications for function and evolution of stomata. Ann Bot (Lond) 114: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, Giraudat J, Leung J (2007) Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26: 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North HM, De Almeida A, Boutin JP, Frey A, To A, Botran L, Sotta B, Marion-Poll A (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50: 810–824 [DOI] [PubMed] [Google Scholar]
- O’Donoghue MT, Chater C, Wallace S, Gray JE, Beerling DJ, Fleming AJ (2013) Genome-wide transcriptomic analysis of the sporophyte of the moss Physcomitrella patens. J Exp Bot 64: 3567–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Pantin F, Renaud J, Barbier F, Vavasseur A, Le Thiec D, Rose C, Bariac T, Casson S, McLachlan DH, Hetherington AM, Muller B, Simonneau T (2013) Developmental priming of stomatal sensitivity to abscisic acid by leaf microclimate. Curr Biol 23: 1805–1811 [DOI] [PubMed] [Google Scholar]
- Papanatsiou M, Scuffi D, Blatt MR, García-Mata C (2015) Hydrogen sulfide regulates inward-rectifying K+ channels in conjunction with stomatal closure. Plant Physiol 168: 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornsiriwong W, Estavillo GM, Chan KX, Tee EE, Ganguly D, Crisp PA, Phua SY, Zhao C, Qiu J, Park J, et al. (2017) A chloroplast retrograde signal, 3′-phosphoadenosine 5′-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer KM, Schuettpelz E (2009) Ferns (Monilophyta). In Hedges SB and Kumar S, eds, The Timetree of Life. Oxford University Press, Oxford, UK, pp 153–156 [Google Scholar]
- Qiu YL, Cho Y, Cox JC, Palmer JD (1998) The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394: 671–674 [DOI] [PubMed] [Google Scholar]
- Raven JA. (2002) Selection pressures on stomatal evolution. New Phytol 153: 371–386 [DOI] [PubMed] [Google Scholar]
- Rea PA, Poole RJ (1993) Vacuolar H+-translocating pyrophosphatase. Annu Rev Plant Biol 44: 157–180 [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Ruszala EM, Beerling DJ, Franks PJ, Chater C, Casson SA, Gray JE, Hetherington AM (2011) Land plants acquired active stomatal control early in their evolutionary history. Curr Biol 21: 1030–1035 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, Uozumi N (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Scherzer S, Maierhofer T, Al-Rasheid KAS, Geiger D, Hedrich R (2012) Multiple calcium-dependent kinases modulate ABA-activated guard cell anion channels. Mol Plant 5: 1409–1412 [DOI] [PubMed] [Google Scholar]
- Schuettpelz E, Schneider H (2016) A community-derived classification for extant lycophytes and ferns. J Syst Evol 54: 563–603 [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope JC, DeWald DB, Mott KA (2003) Changes in surface area of intact guard cells are correlated with membrane internalization. Plant Physiol 133: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry JS. (2004) Coordinating stomatal and xylem functioning—an evolutionary perspective. New Phytol 162: 568–570 [DOI] [PubMed] [Google Scholar]
- Stevenson SR, Kamisugi Y, Trinh CH, Schmutz J, Jenkins JW, Grimwood J, Muchero W, Tuskan GA, Rensing SA, Lang D, et al. (2016) Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE (ANR), a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 28: 1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testo WL, Watkins JE Jr (2012) Influence of plant size on the ecophysiology of the epiphytic fern Asplenium auritum (Aspleniaceae) from Costa Rica. Am J Bot 99: 1840–1846 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RS, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM (2011) Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu D, Yang Q, Zeng J, Jin G, Chen ZH, Zhang G, Dai F (2016) Identification of mild freezing shock response pathways in barley based on transcriptome profiling. Front Plant Sci 7: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen ZH, Zhang B, Hills A, Blatt MR (2013) PYR/PYL/RCAR abscisic acid receptors regulate K+ and Cl− channels through reactive oxygen species-mediated activation of Ca2+ channels at the plasma membrane of intact Arabidopsis guard cells. Plant Physiol 163: 566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer C, Fricker MD (1996) Stomata. Champman and Hall, London [Google Scholar]
- Wolf T, Heidelmann T, Marten I (2006) ABA regulation of K+-permeable channels in maize subsidiary cells. Plant Cell Physiol 47: 1372–1380 [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancani M, Skiera LA, Sanders D (2007) Roles of basic residues and salt-bridge interaction in a vacuolar H+-pumping pyrophosphatase (AVP1) from Arabidopsis thaliana. Biochim Biophys Acta 1768: 311–316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.