A PVX-based VIF assay was designed in order to identify amino acids in the FT protein that are essential for flowering and to examine floral induction by mono- and dicotyledonous FT genes.
Abstract
Virus-induced flowering (VIF) uses virus vectors to express Flowering Locus T (FT) to induce flowering in plants. This approach has recently attracted wide interest for its practical applications in accelerating breeding in crops and woody fruit trees. However, the insight into VIF and its potential as a powerful tool for dissecting florigenic proteins remained to be elucidated. Here, we describe the mechanism and further applications of Potato virus X (PVX)-based VIF in the short-day Nicotiana tabacum cultivar Maryland Mammoth. Ectopic delivery of Arabidopsis (Arabidopsis thaliana) AtFT by PVX/AtFT did not induce the expression of the endogenous FT ortholog NtFT4; however, it was sufficient to trigger flowering in Maryland Mammoth plants grown under noninductive long-day conditions. Infected tobacco plants developed no systemic symptoms, and the PVX-based VIF did not cause transgenerational flowering. We showed that the PVX-based VIF is a much more rapid method to examine the impacts of single amino acid mutations on AtFT for floral induction than making individual transgenic Arabidopsis lines for each mutation. We also used the PVX-based VIF to demonstrate that adding a His- or FLAG-tag to the N or C terminus of AtFT could affect its florigenic activity and that this system can be applied to assay the function of FT genes from heterologous species, including tomato (Solanum lycopersicum) SFT and rice (Oryza sativa) Hd3a. Thus, the PVX-based VIF represents a simple and efficient system to identify individual amino acids that are essential for FT-mediated floral induction and to test the ability of mono- and dicotyledonous FT genes and FT fusion proteins to induce flowering.
Modified plant viruses have emerged as powerful tools for dissecting gene function in plants. Such plant virus-based technology can be applied to facilitate or impede gene expression, resulting in gain- or loss-of-function phenotypes. Although virus-based technology was initially exploited for the purpose of high-level production of foreign proteins, such as recombinant subunit vaccines and pharmaceutical proteins for molecular pharming (Scholthof et al., 1996; Porta and Lomonossoff, 2002), plant RNA and DNA virus-based techniques such as small interfering RNA-mediated virus-induced posttranscriptional gene silencing (VIGS) has been extensively utilized to silence genes for functional genomic studies in dicots and monocots, including plants and crops recalcitrant to classical forward or reverse genetic manipulation (Lindbo et al., 1993; Kumagai et al., 1995; Ruiz et al., 1998; Liu et al., 2002, 2016; Becker and Lange, 2010; Senthil-Kumar and Mysore, 2011; Qin et al., 2015). Various virus-based technologies have been developed such as VIGS, microRNA-based VIGS (Tang et al., 2010; Chen et al., 2015a, 2015c), virus-based microRNA silencing (Sha et al., 2014), and virus-induced transcriptional gene silencing (Kanazawa et al., 2011; Chen et al., 2015b). More recently, virus-induced genome editing has been employed to introduce mutations to specific genes and produce null mutants in plants (Baltes et al., 2014; Ali et al., 2015; Yin et al., 2015). Apart from these gene knockdown or knockout techniques, virus expression vectors have become a valuable tool to examine the mobility of cellular RNAs and the role of mobile RNA signals in flowering (Li et al., 2009, 2011), potato tuberization (Cho et al., 2015), and RNA silencing (Ryabov et al., 2004; Zhou et al., 2008; Qin et al., 2012).
Virus-based technology is also a useful means to transiently express endogenous or exogenous genes for functional analysis in plant development and growth, plant response to biotic stresses, and viral DNA replication (Hong et al., 1997; van Wezel et al., 2002; Hong et al., 2003). For instance, expression of the tomato (Solanum lycopersicum) homeobox protein LeHB1 from a Potato virus X (PVX) vector converts floral organs into fruit-like structures, revealing a new function of LeHB1 in floral organogenesis in addition to its role in fruit ripening (Lin et al., 2008). Moreover, in tomato rin and Cnr mutants, viral expression of the MADS- or SBP-box transcription factor LeMADS-RIN or LeSPL-CNR leads to virus-induced gene complementation and causes nonripening mutant fruits to ripen (Zhou et al., 2012; Kong et al., 2013). Another example is the use of virus technology to study the roles of proteins and RNAs in the flowering process (Li et al., 2011; McGarry et al., 2017).
Flowering is induced by florigen, an endogenous signal whose production responds to environmental cues such as day length (Garner and Allard, 1922; Chailakhyan, 1936; Srikanth and Schmid, 2011). In Arabidopsis (Arabidopsis thaliana), the major component of florigen is a protein encoded by Flowering Locus T (AtFT; Koornneef et al., 1991; Kardailsky et al., 1999; Kobayashi et al., 1999; Turck et al., 2008; Pin and Nilsson, 2012). Many AtFT orthologs have been isolated from other species, for instance, rice (Oryza sativa) Hd3a, tomato SFT, tobacco (Nicotiana tabacum) NtFT4, and sugar beet (Beta vulgaris) BvFT2, and transgenic overexpression of these genes results in early flowering (Kojima et al., 2002; Lifschitz et al., 2006; Pin et al., 2010; Harig et al., 2012). Instead of generating transgenic plants, virus-based technique has also been used as a more rapid method to demonstrate the florigenic activity of FT proteins and RNA. Ectopic expression of FT proteins by Zucchini yellow mosaic virus (ZYMV) induces early flowering in cucurbits (Lin et al., 2007; Yoo et al., 2013). FT protein or mRNA introduced by PVX promotes flowering in short-day (SD) N. tabacum MD Mammoth (MM) under noninductive (i.e. nonflowering) long-day (LD) conditions (Li et al., 2009, 2011). These findings open up possibilities for further development and application of the virus-induced flowering (VIF) assay in plants (McGarry et al., 2017). Since then, several RNA and DNA viruses, including Apple latent spherical virus, Cotton leaf crumple virus, and Citrus leaf blotch virus, have been engineered to express FT for floral induction in soybean, apple, pear, gentian, and lisianthus plants (Yamagishi and Yoshikawa, 2011; Yamagishi et al., 2011, 2014, 2016; Fekih et al., 2016), cotton (McGarry and Ayre, 2012; McGarry et al., 2016), and citrus (Velázquez et al., 2016). These latest developments have generated broad interest in the application of VIF for the benefit of crop breeding (McGarry et al., 2017).
While AtFT is involved in floral induction in Arabidopsis, a closely related gene, TERMINAL FLOWER1 (AtTFL1), encodes a flowering inhibitor. Exchange of a single amino acid leads to functional conversion of AtFT (Y85H) to AtTFL1 (H88Y) and vice versa (Hanzawa et al., 2005). This raises an intriguing question about how and to what extent each individual amino acid residue contributes to AtFT functionality. A recent elegant study through PCR-based random mutagenesis coupled with large-scale Arabidopsis transformation identified 33 unique mutations that influence AtFT activity among approximately 36,000 mutated AtFT alleles. Specific point mutations of E109, Y138, Q140, or N152 can convert AtFT into a TFL1-like floral repressor (Ho and Weigel, 2014). Nonetheless, this is a very time-consuming approach, whereas VIF offers the potential of an alternate rapid, efficient, and less labor-intensive flowering assay to evaluate the influence of each amino acid residue, as well as the effect of epitope tags on florigenic activity.
In this article, we describe further characterization and development of the PVX-based VIF approach to assess FT protein function. Expression of Arabidopsis AtFT by PVX/AtFT led to no increase in the levels of endogenous NtFT4 mRNA, but it was sufficient to cause floral induction in MM tobacco in LD. Using the PVX-based VIF, we were able to demonstrate in various approaches to (1) study the impact of single amino acid mutations on the floral inducing function of the AtFT protein, (2) investigate the influence of His or FLAG tags on the AtFT activity, and (3) assess the function of tomato SFT and rice Hd3a to induce flowering in MM tobacco under noninductive conditions within a matter of weeks. Thus, this PVX-based VIF represents an efficient system for the functional analysis of proteins such as mono- and dicotyledonous FT and FT-like genes that are involved in flowering.
RESULTS
PVX-Based VIF
We have previously described the development of the PVX-based VIF assay in SD MM tobacco plants (Li et al., 2009); it involves three main steps. First, the FT (or any flowering gene) expression cassette needs to be cloned into the modified PVX-based gene expression vector (van Wezel et al., 2002) and verified by sequencing. Second, recombinant infectious PVX transcripts containing the FT or flowering gene mRNA are produced by in vitro transcription from the vector’s T7 promoter using a commercially available kit or self-assembled reaction mixtures, as described (van Wezel et al., 2002). Third, SD MM tobacco plants at the four- to six-leaf stage were mechanically inoculated with the in vitro transcribed infectious PVX RNA. Plants are grown under noninducing 16 h LD (or inducing 8 h SD) conditions and monitored for floral induction (Li et al., 2009).
In this article, we further characterized the PVX-based VIF by looking at the effect of virally expressed AtFT on the level of endogenous NtFT4 gene expression and also at whether the PVX or virally expressed AtFT RNA is seed transmissible. We then used this VIF in three different approaches to evaluate the influence of individual amino acids on AtFT activity, the impact of polypeptide tags on AtFT function, and the roles of monocot and dicot FT genes in flowering.
Viral Delivery of AtFT Induces Flowering But No Increase in Endogenous NtFT4 Expression in MM Tobacco under Noninductive Conditions
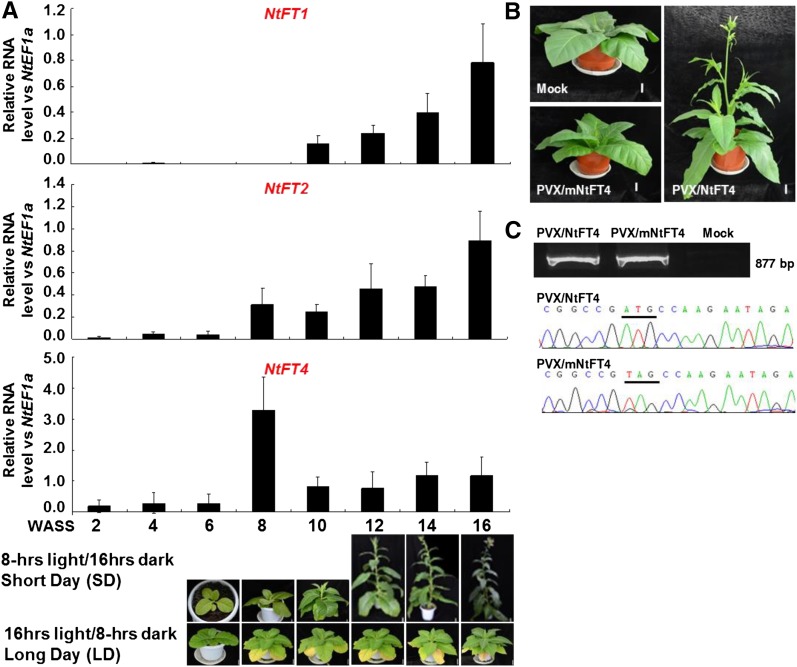
To investigate the mechanism through which VIF works, we analyzed the temporal expression profiles of the four known day-neutral tobacco FT-like genes NtFT1, NtFT2, NtFT3, and NtFT4 (Harig et al., 2012) in MM plants grown in SD (Fig. 1). Under SD of 8 h light/16 h dark (i.e. florally inductive conditions for MM tobacco), the plants bolted after 8 weeks and started to flower at 12 weeks after seed sowing (WASS), however, no bolting or flowering occurred when plants grown under noninductive LD conditions (16 h light/8 h dark; Fig. 1A). The relative expression levels of NtFT1 and NtFT2 were low until 16 WASS when flowers were already formed (Fig. 1A). The levels of NtFT3 expression were consistently too low to be reliably quantified by quantitative reverse transcription PCR (qRT-PCR).
Figure 1.
Floral induction by NtFT4 in SD N. tabacum MM. A, Expression of NtFTs in tobacco plants grown under inductive SD conditions was analyzed by qRT-PCR. Young leaf samples were taken at 2–16 WASS. Plants grown under SD had started to bolt after 8 WASS and flower after 12 WASS. No plants bolted or flowered under noninductive LD conditions. B, Viral expression of NtFT4 by PVX/NtFT4 induced SD tobacco to flower at 9 WASS in LD, 5 weeks postinoculation. Mock-inoculated controls and plants infected with PVX/mNtFT4 remained vegetative in LD. C, RT-PCR detection and sequencing confirmation of wild-type and mutant NtFT4 transcripts expressed in tobacco. Plants were photographed as indicated WASS in A or 9 WASS in B. Bar = 4 cm.
In contrast, NtFT4 was induced, and its expression was high at 8 WASS, i.e. around the time bolting was initiated. From 10 WASS onward, the expression of NtFT4 fell and was maintained at a constant level that was relatively higher than the other NbFTs until they increased at 16 WASS (Fig. 1A). Consistent with the fact that NtFT4 is known to be the true AtFT ortholog in day-neutral tobacco (Harig et al., 2012), viral expression of NtFT4 from PVX/NtFT4 in MM plants was able to induce flowering under noninductive LD (Fig. 1B; Supplemental Fig. S1; Supplemental Table S1). Control mock-inoculated tobacco and those plants infected with PVX/mNtFT4 containing a mutated nontranslatable NtFT4 mRNA did not flower (Fig. 1, B and C; Supplemental Fig. S1). Viral expression of wild-type or nonsense mutated NtFT4 in systemic tobacco leaf tissues was detected by RT-PCR and verified by direct sequencing (Fig. 1C; Supplemental Table S1). These data confirm that as in day-neutral N. tabacum, NtFT4 also promotes flowering in short-day MM tobacco.
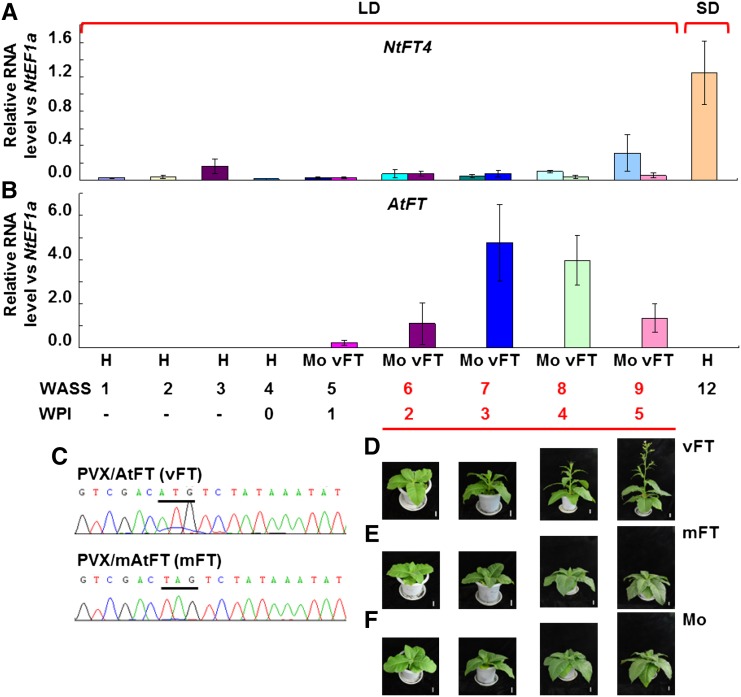
We previously showed that PVX-based expression of AtFT induces flowering in tobacco (Li et al., 2009, 2011). However, as the expression of the plant’s own endogenous FT gene(s) is still possible, the induction of flowering could either be a direct effect of the virally expressed FT gene or caused by an indirect induction of the expression of the endogenous FT gene. Such lack of understanding of the virus-induced flowering mechanism is also true for other published VIF systems (McGarry et al., 2017). To address this, we performed time-course experiments in PVX/AtFT-infected tobacco plants and examined the expression levels of AtFT and of the endogenous NbFT4 gene to investigate how they correlated with the flowering response (Fig. 2). Plants at 4 WASS (four to six leaves) were mock-inoculated or inoculated with PVX/AtFT or PVX/mAtFT (Supplemental Fig. S1A). As shown in Figure 2A, under LD conditions, endogenous NtFT4 was not induced in MM plants either before inoculation (WASS 1–4), or after mock-inoculation or inoculation with PVX/AtFT (WASS 5–9). However, wild-type or mutant AtFT mRNA was readily detected by qRT-PCR in viral infected plants at 5 WASS, i.e. 1 week postinoculation (WPI; Fig. 2B), and verified by direct sequencing (Fig. 1C). The level of AtFT transcripts reached a peak at 3 WPI (7 WASS) and then declined over the next 2 weeks (Fig. 1B). On the other hand, no AtFT was detectable in the control mock-inoculated plants (Fig. 1B). Consistent with the expression levels of AtFT mRNA, tobacco plants infected with PVX/AtFT started bolting at 2 WPI (6 WASS) onward and flowered at 4 to 5 WPI (8–9 WASS; Fig. 2D). Control plants that had been mock-inoculated or inoculated with PVX/mAtFT carrying a nonsense mutation in the AtFT mRNA start codon remained growing vegetatively in LD and did not flower (Fig. 2, E and F).
Figure 2.
Viral expression of AtFT induces flowering. A, Endogenous NtFT4 expression was not induced in systemic leaves of tobacco plants prior to inoculation (H), mock-inoculated (Mo), or infected with PVX/AtFT (vFT) under noninductive LD conditions. Levels of the NtFT4 transcripts were analyzed by qRT-PCR in noninoculated MM plants grown to flowering (12 WASS) under SD are shown for comparison. B, Ectopic expression of AtFT mRNA by PVX/AtFT. 0 indicates the inoculation week. C, RT-PCR detection and sequencing confirmation of virally expressed wild type and mutant AtFT mRNA (vFT and mFT, respectively). The start codon and the mutated start codon are underlined. D to F, Flowering phenotype of tobacco plants infected with PVX/AtFT (vFT), PVX/mAtFT (mFT), or mock (Mo) inoculation. Plants were photographed at 6, 7, 8, and 9 WASS. Bar = 4 cm.
Latent PVX-Based VIF Induces No Transgenerational Epiflowering
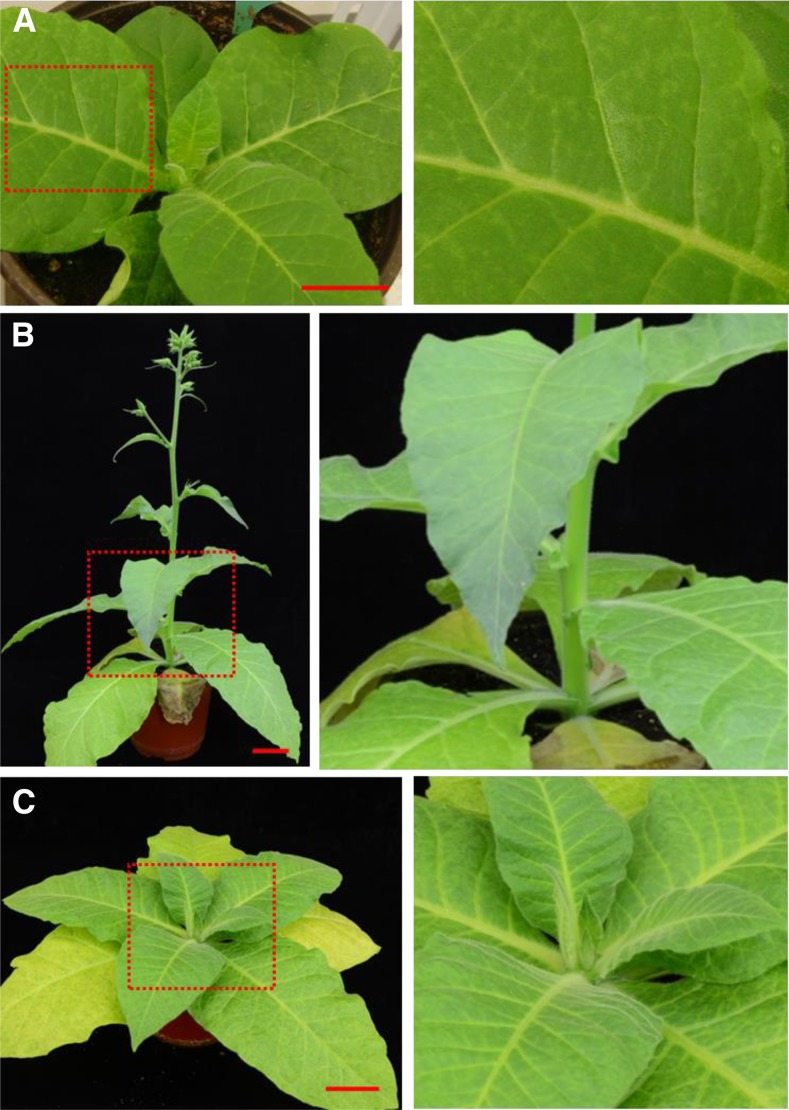
To assess whether the effects of virus infection on plant growth and development would affect the usefulness of the PVX-based VIF, we examined the development of systemic symptoms of PVX infection in MM tobacco grown in LD. Plants at 4 WASS (four to six leaves) were inoculated with PVX/AtFT or PVX/mAtFT (Fig. 3). Local infection occurred and chlorotic lesions appeared on inoculated leaves at approximately 7–10 d postinoculation (Fig. 3A). In contrast, in newly developing young leaves, no viral mosaic or chlorosis was observed, and systemic leaves were persistently symptomless (Fig. 3, B and C). Recombinant viral RNA was, however, present and readily detected in systemic leaf tissues (Figs. 1C and 2, B and C), suggesting PVX was able to establish latent infection in tobacco plants (Fig. 2, D and E). In all cases, tobacco plants infected with PVX virus vectors consistently appeared healthy (Figs. 1B and 2, D–F; Supplemental Fig. S1A; also see below), likely due to RNA silencing-mediated recovery of virus infection (Baulcombe, 2004).
Figure 3.
PVX latent infection of MM tobacco plants. A, A tobacco plant inoculated with PVX/AtFT developed local chlorotic lesions on the inoculated leaves. B, A tobacco plant infected with PVX/AtFT developed no systemic symptoms but was induced to flower. C, A tobacco plant inoculated with PVX/mAtFT did not show systemic symptoms and did not flower. Plants were photographed at 11 (A) or 34 (B and C) days postinoculation. Right panels in A to C show an enlarged section of the boxed region in the left. Bar = 5 cm.
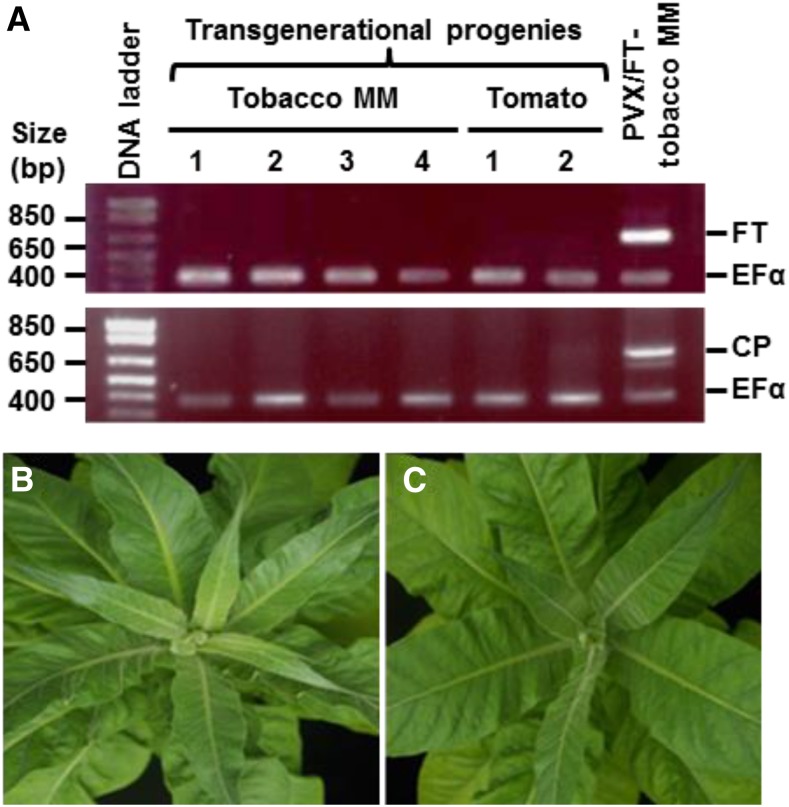
In order to investigate if the virally expressed AtFT and PVX RNA could be transmitted through seed to progeny, RT-PCR was performed on seedlings that were grown from seeds harvested from tobacco and tomato plants infected with PVX/AtFT (Fig. 4). Neither AtFT nor PVX coat protein RNA was detected in descendant seedlings (Fig. 4A). Moreover, once mature, these MM tobacco plants were unable to flower in LD (Fig. 4, B and C), in exactly the same as plants grown from seeds collected from noninfected MM tobacco plants grown to flowering in SD.
Figure 4.
PVX-based VIF induces no transgenerational flowering. A, RT-PCR detection of virally expressed Arabidopsis AtFT (FT) and PVX coat protein (CP) RNAs in seedlings grown from seeds collected from PVX/AtFT-infected tobacco (MM) and tomato plants. The positions and sizes of the 1 kb plus ladder marker are indicated. PVX-specific forward primer PP82 and gene-specific reverse primer PP356 were used for detection of the virally expressed AtFT (750 bp), TCPF and TCPR for PVX CP (714 bp), and EF1F and EF1R for the housekeeping gene NbEF1α (380 bp). Primer sequences are listed in Supplemental Table S1. The positive control was systemic leaf-tissue materials harvested from PVX/AtFT-infected MM tobacco. B, Tobacco plants grown from seeds harvested from PVX/AtFT-infected tobacco plants did not flower in LD. C, Tobacco grown from seeds collected from MM plants under SD remained in the vegetative phase in LD. Plants were photographed at 75 d after sowing seeds.
Application of PVX-Based VIF
Approach 1: VIF for Assessing the Role of Individual Amino Acids in AtFT Activity
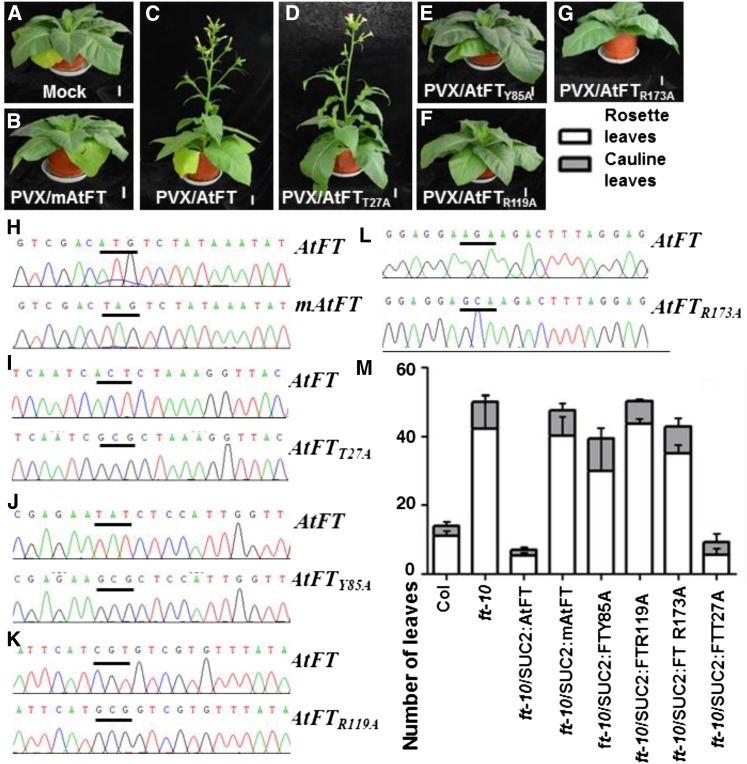
To test whether the PVX-based VIF could be used for assessing the influence of individual amino acids on AtFT activity, we substituted T27, Y85, R119, and R173, respectively, with Ala (A) and produced PVX/AtFTT27A, PVX/AtFTY85A, PVX/AtFTR119A, and PVX/AtFTR173A (Supplemental Figure S1A). We selected Y85 and R119 because their impacts on flowering have been shown in Arabidopsis (Bradley et al., 1997; Wickland and Hanzawa, 2015), while T27 and R173 of previously unknown contribution to AtFT activity were randomly selected for site-directed mutagenesis. Tobacco plants were inoculated with each of the recombinant PVX vectors and grown in LD (Fig. 5). Using PVX/AtFT as a positive control and PVX/mAtFT and mock-inoculation as negative controls, we found that viral expression of AtFTT27A, AtFTY85A, AtFTR119A, and AtFTR173A resulted in distinctive flowering phenotypes (Fig. 5, A–G). Similar to the mock-inoculated controls and to plants infected with PVX/mAtFT, those plants infected with PVX/AtFTY85A, PVX/AtFTR119A, or PVX/AtFTR173A were unable to flower (Fig. 5, A, B, and E–G). However, infection by PVX/AtFTT27A induced plants to flower at 5 WPI in a similar manner to the PVX/AtFT positive control (Fig. 5, C and D). Expression of AtFT, mAtFT, AtFTT27A, AtFTY85A, AtFTR119A, and AtFTR173A mRNAs were detected by RT-PCR and verified by direct sequencing (Fig. 5, H–L). These findings suggest that AtFTY85A, AtFTR119A, and AtFTR173A are defective in their ability to induce normal flowering compared to AtFT, while the T27A substitution did not affect the ability of AtFT to induce flowering.
Figure 5.
Impact of single amino acid on AtFT-mediated floral induction. A to F, Functional assessment of novel FT alleles using the PVX-based VIF. MM tobacco plants mock-inoculated (A), or infected with PVX/mAtFT (B), PVX/AtFT (C), PVX/AtFTT27A (D), PVX/AtFTY85A (E), PVX/AtFTR119A (F), and PVX/AtFTR173A (G) resulted in distinctive flowering phenotypes. Plants were photographed at 5 weeks postinoculation. Bar = 4 cm. H to L, RT-PCR detection and sequencing confirmation of virally expressed wild type and mutant AtFT mRNAs as indicated in each. The start codons and the mutated start codons are underlined. M, Flowering time of Arabidopsis Col, ft-10, and transgenic ft-10/SUC2:AtFT, ft-10/SUC2:mAtFT, ft-10/SUC2:AtFTY85A, ft-10/SUC2:FTR119A, ft-10/SUC2:AtFTR173A, and ft-10/SUC2:FTT27A plants under LD conditions. Flowering times were determined as the number of rosette (white bars) and cauline (gray bars) leaves in three to five independent single-copy transgenic lines for each gene. A representative line for each transgene is shown (also see Supplemental Table S2).
To make comparisons, we conducted transgenic complementation experiments in which the T-DNA insertion mutant ft-10 was transformed with a binary vector containing a SUC2:AtFT, SUC2:mAtFT, SUC2:AtFTT27A, SUC2:AtFTY85A, SUC2:AtFTR119A, or SUC2:AtFTR173A expression cassette under the control of the phloem-specific SUC TRANSPORTER2 (SUC2) promoter (Supplemental Fig. S1B; Clough and Bent, 1998). Three to five independent single-copy homozygous lines were generated for each transgene, and their flowering time in LD was analyzed. Similar to nontransformed ft-10 and transgenic control ft-10/SUC2:mAtFT plants, transgenic lines ft-10/SUC2:AtFTY85A, ft-10/SUC2:AtFTR119A, or ft-10/SUC2:AtFTR173A were late flowering when compared to wild-type Col plants. This is in agreement with our previous observation that MM tobacco plants inoculated with PVX vectors expressing AtFTY85A, FTR119A, or AtFTR173A did not flower at all (Fig. 5, E–G). However, transgenic expression of AtFT or AtFTT27A led to early flowering in ft-10/SUC2:AtFT and ft-10/SUC2:AtFTT27A plants (Fig. 5M; Supplemental Table S2), again consistent with the results of the PVX-based VIF (Fig. 5, A–G).
Approach 2: VIF for Assessing the Impact of Polypeptide Tags on AtFT Floral Induction
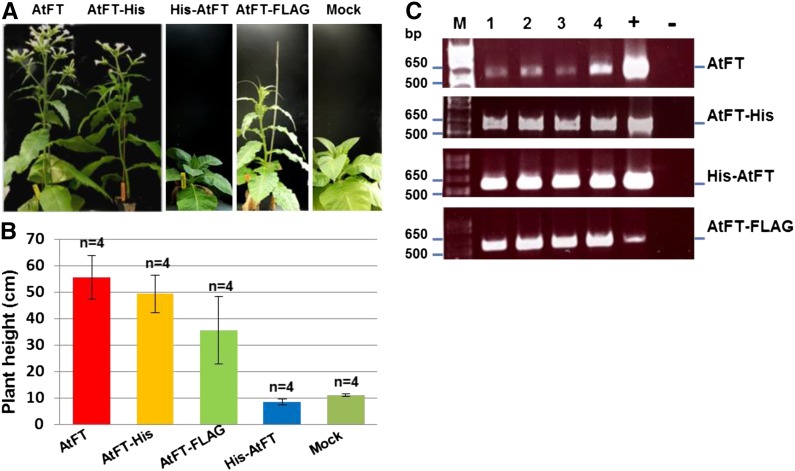
Polypeptide tags have been widely used to study correlations between function and spatial distribution of target proteins in tissues and cellular compartments. It has been documented that AtFT-GFP and AtFT-myc expressed in leaf companion cells are transported via the phloem over long-distance to the shoot apical meristem where these fusion proteins can initiate flowering in transgenic Arabidopsis (Abe et al., 2005; Wigge et al., 2005; Searle et al., 2006; Corbesier et al., 2007; Mathieu et al., 2007; Liu et al., 2012, Zhu et al., 2016). However, it remains to be clarified whether any free AtFT that may coexist with AtFT-GFP and AtFT-myc fusion proteins due to premature translational termination was responsible for floral induction in those transgenic plants. In contrast, viral expression of GFP-AtFT was functionally inactive in floral induction in tobacco under LD conditions (Li et al., 2009). This suggests peptide tags at the N or C terminus could affect AtFT florigenic activity. To test this, we generated PVX/His6-AtFT, PVX/AtFT-His6, and PVX/AtFT-FLAG (Supplemental Fig. S1A) and inoculated these viruses onto tobacco plants grown in LD (Fig. 6). At 1 WPI, chlorotic lesions were visible on inoculated leaves of all virus-infected, but not mock-inoculated plants. Similar to plants infected with PVX/AtFT, plants infected with PVX/AtFT-His6 started to bolt at 2 WPI and subsequently flowered at 5 to 6 WPI. Plants infected with PVX/FT-FLAG started bolting at 3 WPI and flowered at 8 to 9 WPI (Fig. 6A). The average height of the plants infected with PVX/AtFT, PVX/AtFT-His6, and PVX/AtFT-FLAG at 52 d postinoculation were 55, 49, and 35 cm, respectively (Fig. 6B). Plants infected with PVX/His6-AtFT, however, remained in their vegetative phase and behaved similarly to the mock controls with a mean height of 9 and 11 cm, respectively (Fig. 6, A and B). RT-PCR analysis showed that virally expressed free or tagged AtFT transcripts were present in systemic leaf tissues of all virus-infected plants (Fig. 6C). These data demonstrate that AtFT-His and free AtFT were equally efficient to induce flowering in tobacco. AtFT-FLAG was slightly less active than AtFT, and His-AtFT was completely inactive. These functional differences are likely to be due to the alternations in size, overall structure, solubility, and/or stability of the fusion protein.
Figure 6.
Influence of polypeptide tags on AtFT florigenic activity. A, MM plants infected with PVX expressing AtFT with and without different tags flowered differently under noninductive LD conditions. Plants were photographed at 52 d postinoculation. B, Effect of expression of AtFT with and without different tags, on bolting of tobacco plants. The average stem length (cm) at 52 d postinoculation is shown. C, RT-PCR detection of virally expressed Arabidopsis AtFT, AtFT-His, His-AtFT, and AtFT-FLAG mRNAs in systemic leaf tissues of four different tobacco plants (lanes 1–4) infected with PVX/AtFT (AtFT), PVX/AtFT-His (AtFT-His), PVX/His-FT (His-FT), or PVX/AtFT-FLAG (AtFT-FLAG). PVX-specific forward primer PP82 and a gene-specific reverse primer (Supplemental Table S1) were used for RT-PCR detection. Positive controls (+) were the respective recombinant plasmids, while no DNA was used in the negative control PCR. The positions and sizes of 1 kb plus ladder are indicated.
Approach 3: Functional Analyses of Mono- and Dicotyledonous FT Genes
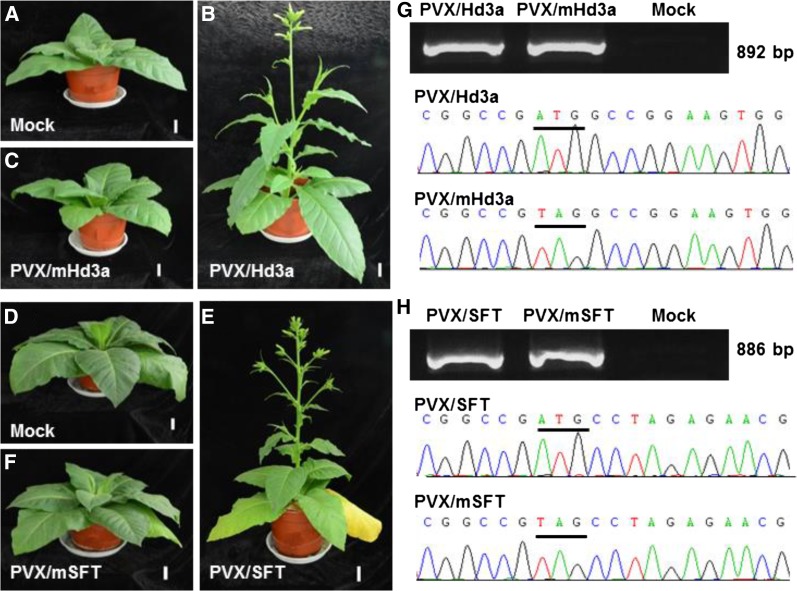
The FT gene belongs to a gene family that encodes closely related FT-like proteins, not all of which induce flowering. To identify which of the FT-like genes that have been identified in a plant species are the ones responsible for controlling floral induction, a rapid and efficient functional assay is necessary; previously this has been done through the generation of transgenic plant lines. We tested whether PVX-based ectopic expression of mono- and dicotyledonous FT genes was able to demonstrate their functional ability to induce flowering (Fig. 7). We cloned wild-type and nonsense mutated rice Hd3a and tomato SFT genes into the PVX vector to generate PVX/Hd3a, PVX/mHd3a, PVX/SFT, and PVX/mSFT (Supplemental Figure S1A; Supplemental Table S1). Tobacco plants were then inoculated with these viruses and grown in LD (Fig. 7, A–F). All plants infected with either PVX/Hd3a or PVX/SFT developed no systemic symptom but nevertheless bolted at 2 WPI and flowered at 5 to 6 WPI (Fig. 7, B and E). Mock controls and plants infected with PVX/mHd3a or PVX/mSFT did not flower (Fig. 7, A, C, D, and F). Expression of wild-type Hd3a or SFT mRNA from PVX/Hd3a or PVX/SFT, as well as mutated nontranslatable mHd3a or mSFT transcripts from PVX/mHd3a or PVX/mSFT were readily detectable by RT-PCR and were verified by direct sequencing (Fig. 7, G and H).
Figure 7.
Viral transient expression of mono- and dicot FT genes promotes SD tobacco flowering under noninducing LD conditions. A to C, MM tobacco plants were mock-inoculated or inoculated with PVX/Hd3a or PVX/mHd3a. D to F, Tobacco plants were mock-inoculated or inoculated with PVX/SFT or PVX/mSFT. In three separate experiments, ectopic expression of Hd3a or SFT caused tobacco plants to bolt at 2to 3 WPI and flower at 5 WPI. Plants mock-inoculated or infected with PVX/mHd3a or PVX/mSFT did not flower. Plants were photographed at 5 WPI. Bar = 4 cm. G and H, RT-PCR detection and sequencing confirmation of virally expressed wild-type and mutant rice Hd3a and SFT mRNA in systemic leaves of infected tobacco plants. The wild type and mutated start codons are underlined.
DISCUSSION
VIF has been developed in several plants and fruit trees (McGarry et al., 2017) since its early establishment in cucurbit (Lin et al., 2007) and tobacco (Li et al., 2009); however, the exact mechanism has not been fully investigated in each case. As the expression of the plant’s own endogenous FT gene(s) is still possible, it remains to be elucidated whether VIF was due to a direct or indirect effect of the virally expressed FT. Thus, further work is required to develop the full potential of VIF for the study of plant reproductive biology. Here, we demonstrate that viral expression of wild-type AtFT induces flowering of MM tobacco plants grown under noninductive LD conditions, without the involvement of the endogenous NtFT4 gene (Figs. 1 and 2).
There are two major concerns over pathogenic virus-induced flowering, which may restrict its usefulness in plant reproductive biology and crop breeding. First, viral infection often causes host plant developmental abnormalities and occasionally death. Plants respond to such biotic stresses by speeding up their reproductive process to flower earlier. This could complicate the interpretation of results of VIFs, such as those based on Zucchini yellow mosaic virus, Cotton leaf crumple virus, or Citrus leaf blotch virus, because all these viruses produced severe systemic symptoms in viral-infected plants (Lin et al., 2007; McGarry and Ayre, 2012; Velázquez et al., 2016). However, the PVX-infected tobacco plants exhibited normal vegetative and reproductive growth (Fig. 3), making PVX-VIF in MM tobacco a good system for studying the control of flowering. Secondly, VIFs based on seed-transmissible viruses are likely to hinder the technology from being used as a viable tool in breeding programs, whereas lack of seed transmission means progeny plants would be virus-free. Thus, if VIF is to be used commercially, the recombinant virus should not be transmitted through seed to the progeny, thus making it easier to contain and control. Indeed, we were unable to detect the presence of PVX RNA or virally expressed AtFT mRNA in next-generation seedlings of tobacco or tomato plants infected with PVX/AtFT, and these seedlings subsequently grew and flowered as wild-type plants (Fig. 4). This implies that neither PVX virus nor virally expressed AtFT mRNA is seed transmissible despite the fact that AtFT RNA is able to facilitate the entry of PVX to the apical shoot meristem (Li et al., 2011) and that PVX/AtFT infection cannot induce transgenerational epiflowering in progeny plants.
The transition from vegetative to reproductive growth is essential for most plants to complete a successful life cycle, and the tight regulation of when this transition occurs is important for their long-term evolutionary survival. In Arabidopsis, the key gene involved in this process is AtFT, which encodes a small protein consisting of 175 amino acid residues. Moreover, both AtFT and AtTFL1 (AtTFL1 is the TERMINAL FLOWER1 protein, an inhibitor of flowering in Arabidopsis) belong to the phosphatidylethanolamine-binding protein family, and only 39 nonconserved residues are found between the two functionally antagonizing proteins (Bradley et al., 1997; Ohshima et al., 1997; Wickland and Hanzawa, 2015). Genetic analyses revealed that late flowering in many Arabidopsis ft mutants often resulted from a dysfunctional AtFT with single amino acid substitution such as G171E in ft-1, R119H in ft-3, E84K in ft-4, and P94L in ft-6. A number of AtFT alleles with specific point mutations were further characterized through an intensive mutagenesis program (Ho and Weigel, 2014). While these studies were elegantly performed and extremely informative, a more rapid and less labor-intensive flowering assay would make it easier to efficiently evaluate the contribution of each of the 175 amino acids to the florigenic activity.
The efficacy of the PVX-VIF system was demonstrated in several approaches: (1) to assess the influence of individual amino acids on protein function—through this analysis we were rapidly able to show that, in addition to the known contributions of both Y85 and R119, residue R173 is essential for AtFT to induce flowering, whereas T27 is not (Fig. 5). (2) To assess the impact of various tags at the N or C terminus on protein function (Fig. 6)—in the case of the AtFT protein, we used this approach to show that N-terminal fusions are likely to disrupt AtFT function, consistent with previous findings (Li et al., 2009). (3) To analyze the floral inducing function of different mono- and dicotyledonous FT genes (Fig. 7)—the fact that virally expressed mono- and dicotyledonous FT genes can induce flowering in MM tobacco under noninductive conditions also provides further evidence for high conservation of the FT-mediated pathway in floral induction among many different plant species (Turck et al., 2008).
CONCLUSION
We have further developed our PVX-based VIF (Li et al., 2009) and demonstrated that it is a powerful tool for assessing the functions of FT and its mono- and dicotyledonous orthologs in flowering, as well as for evaluating the impact of specific amino acids or polypeptide tags on florigenic activity. Floral induction in this system is exclusively caused by the virally expressed FT genes independent of the endogenous tobacco FT. Compared with other virus-based flowering induction systems, the PVX-based VIF is (1) ideal for studying plant floral induction because symptomless PVX infection has no perceptible influence on plant growth and flowering. (2) Nonseed transmissibility of the virus also provides benefits in terms of biosafety and potential commercial application of the PVX-based VIF. (3) While our work has focused on the use of the PVX-based VIF for assessing the ability of modified, or different, FT proteins to induce flowering, the system can also be used more widely for the analysis of other proteins (or RNAs) involved in flowering, both in the floral induction and flower development processes. (4) The work described here can be easily translated to other viral expression vectors that could be used in other plant species besides tobacco. On the other hand, the current PVX-based VIF vector involves traditional cloning and production of infectious recombinant viral RNA transcripts by in vitro transcription for plant infection. To overcome these shortcomings, ongoing work in our laboratory aims to develop a more user-friendly version of the PVX-based VIF for systems biology and high-throughput analysis of FT and flowering-related genes, alleles, and mutants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tobacco plants (Nicotiana tabacum MD Mammoth) and Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col) and mutant ft-10 (seeds were obtained from the Nottingham Arabidopsis Stock Center) were grown under LD (16 h light/8 h dark) or SD (8 h light/16 h dark) conditions at 23°C. Tomato (Solanum lycopersicum var Ailsa Craig) was grown in glasshouse or growth room in LD at 23°C.
Construction of VIF and Plant Transformation Vectors
PVX/AtFT, PVX/mAtFT, PVX/Hd3a, PVX/mHd3a, PVX/SFT, PVX/mSFT, PVX/NtFT4, and PVX/mNtFT4 were constructed as previously described (Li et al., 2009). In brief, wild-type and mutant Arabidopsis AtFT were RT-PCR amplified using PrimeSTAR HS DNA polymerase and the primers RC0061/RC0062 or RC0066/RC0062 (Supplemental Table S1), digested with BspEI and SalI, and cloned into the BspEI/SalI sites of the PVX vector to produce PVX/AtFT and PVX/mAtFT, respectively (Supplemental Fig. S1A). Using a similar strategy, we constructed PVX/Hd3a, PVX/mHd3a, PVX/SFT, PVX/mSFT, PVX/NtFT4, and PVX/mNtFT4 but using different sets of primers: RC0536/RC0537 for PVX/Hd3a, RC0538/RC0537 for PVX/mHd3a, RC0541/RC0542 for PVX/SFT, RC0543/RC0542 for PVX/mSFT, RC0779/RC0780 for PVX/NtFT4, and RC1639/RC0780 for PVX/mNtFT4, respectively (Supplemental Fig. S1A; Supplemental Table S1). These genes were cloned into the EagI/SalI sites of the PVX vector.
Single amino acid mutated FT alleles were created by overlapping PCR and cloned into the PVX vector. Primers used for the construction of PVX/AtFTY85A, PVX/AtFTR119A, PVX/AtFTR173A, and PVX/AtFTT27A are listed in Supplemental Table S1.
To generate binary plant transformation vectors SUC2:AtFT, SUC2:AtFTY85A, SUC2:AtFTR119A, and SUC2:AtFTT27A, mutated AtFT genes were amplified using primers RC0783/RC0784 along with plasmid DNA temples PVX/AtFT, PVX/AtFTY85A, PVX/AtFTR119A, or PVX/AtFTT27A, digested with SalI and SacI, and cloned into the SalI/SacI sites of pBI101.3 (Supplemental Fig. S1B; Supplemental Table S1). To generate SUC2:mAtFT and SUC2:AtFTR173A, mutated genes were amplified using PVX/AtFT as templates along with primers RC0798/RC0784 or RC0783/RC0799, digested with SalI and SacI, and cloned into the SalI/SacI sites of pBI101.3 (Supplemental Figure S1B; Supplemental Table S1). All constructs were verified by nucleotide sequencing.
VIF in Tobacco and Transgenic Complementation in Arabidopsis ft-10
VIF was carried out in repeated experiments as previously described (Li et al., 2009). In each experiment, three to six young SD MM tobacco plants were mock-inoculated or inoculated with recombinant PVX and maintained in insect-free containment growth room at 23°C under LD. Transformation of Arabidopsis ft-10 plants was carried out using the floral-dip method, and transgenic plants were selected on 0.5× Murashige and Skoog medium supplemented with Kanamycin (Clough and Bent, 1998). Through selfing, three to five single-copy homozygous lines were generated for each transgene and used for analysis. Flowering time was calculated and represented by the average number of rosette and cauline leaves.
RT-PCR and qRT-PCR
Total RNA was extracted from Arabidopsis, MM, and tomato young-leaf tissues. First-strand cDNA was synthesized from DNase I-treated total RNAs by M-MLV Reverse Transcriptase according to the manufacturer’s instructions (Promega). RT-PCR was performed to detect virally expressed FT RNAs, and the resulting RT-PCR products were isolated and directly sequenced using primer PP82 as previously described (Li et al., 2009). qRT-PCR was performed in a CFX96 machine (Bio-Rad) using the iQ SYBR Green Supermix and gene-specific primers listed in Supplemental Table S1 following the manufacturer’s instructions. The amplification program for SYBR Green I was performed at 95°C for 10 s, 55°C for 30 s, and 72°C for 20 s. Triplicate quantitative assays were performed on each of triplicate cDNA samples. The relative quantification of each sample was determined by normalization to the amount of NtEF1α cDNA detected in the same sample. Relative expression level was calculated by the Equation 2−△△Ct as described (Qin et al., 2012).
Supplemental Data
The following materials are available.
Supplemental Figure S1. VIF vectors and Arabidopsis transformation constructs.
Supplemental Table S1. Primers used in this work.
Supplemental Table S2. Flowering time of Arabidopsis transgenic plants.
Supplementary Material
Acknowledgments
We thank David Baulcombe for providing the original PVX vector and Meiling He for technical support.
Glossary
- VIGS
virus-induced posttranscriptional gene silencing
- VIF
virus-induced flowering
- WASS
weeks after seed sowing
- WPI
weeks postinoculation
Footnotes
This work was supported by grants from the Ministry of Agriculture of China (National Transgenic Program 2016ZX08009-001-004 to Y.H.); the National Natural Science Foundation of China (NSFC 31370180 to Y.H., 31500251 to C.Q., and 31601765 to W.C.); Hangzhou Normal University (Pandeng Program 201108 to Y.H.); the Hangzhou City Education Bureau (Innovative Program for Science Excellence 20131028 to Y.H.); the UK Biotechnology and Biological Sciences Research Council (UK BBSRC–China Partnering Award BB/K021079/1 to S.J. and Y.H.); and the Zhejiang Provincial Natural Science Foundation (LY14C010005 to N.S.).
Articles can be viewed without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Ali Z, Abul-faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes NJ, Voytas DF, et al. (2015) Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant 8: 1288–1291 [DOI] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Becker A, Lange M (2010) VIGS—genomics goes functional. Trends Plant Sci 15: 1–4 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Chailakhyan MK. (1936) New facts in support of the hormonal theory of plant development. Dokl Akad Nauk SSSR (CR Acad Sci USSR) 4: 79–83 [Google Scholar]
- Chen W, Kong J, Lai T, Manning K, Wu C, Wang Y, Qin C, Li B, Yu Z, Zhang X, et al. (2015a) Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci Rep 5: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kong J, Qin C, Yu S, Tan J, Chen YR, Wu C, Wang H, Shi Y, Li C, et al. (2015b) Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci Rep 5: 9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang Q, Kong J, Hu F, Li B, Wu C, Qin C, Zhang P, Shi N, Hong Y (2015c) MR VIGS: microRNA-based virus-induced gene silencing in plants. Methods Mol Biol 1287: 147–157 [DOI] [PubMed] [Google Scholar]
- Cho SK, Sharma P, Butler NM, Kang IH, Shah S, Rao AG, Hannapel DJ (2015) Polypyrimidine tract-binding proteins of potato mediate tuberization through an interaction with StBEL5 RNA. J Exp Bot 66: 6835–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Fekih R, Yamagishi N, Yoshikawa N (2016) Apple latent spherical virus vector-induced flowering for shortening the juvenile phase in Japanese gentian and lisianthus plants. Planta 244: 203–214 [DOI] [PubMed] [Google Scholar]
- Garner WW, Allard HA (1922) Photoperiodism, the response of the plant to relative length of day and night. Science 55: 582–583 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102: 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harig L, Beinecke FA, Oltmanns J, Muth J, Müller O, Rüping B, Twyman RM, Fischer R, Prüfer D, Noll GA (2012) Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J 72: 908–921 [DOI] [PubMed] [Google Scholar]
- Ho WW, Weigel D (2014) Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26: 552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Saunders K, Stanley J (1997) Transactivation of dianthin transgene expression by African cassava mosaic virus AC2. Virology 228: 383–387 [DOI] [PubMed] [Google Scholar]
- Hong Y, Stanley J, van Wezel R (2003) Novel system for the simultaneous analysis of geminivirus DNA replication and plant interactions in Nicotiana benthamiana. J Virol 77: 13315–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa A, Inaba J, Shimura H, Otagaki S, Tsukahara S, Matsuzawa A, Kim BM, Goto K, Masuta C (2011) Virus-mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J 65: 156–168 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Kong J, Chen W, Shen J, Qin C, Lai T, Zhang P, Wang Y, Wu C, Yang X, Hong Y (2013) Virus-induced gene complementation in tomato. Plant Signal Behav 8: e27142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA 92: 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gu M, Shi N, Zhang H, Yang X, Osman T, Liu Y, Wang H, Vatish M, Jackson S, et al. (2011) Mobile FT mRNA contributes to the systemic florigen signalling in floral induction. Sci Rep 1: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang K, Zeng X, Jackson S, Zhou Y, Hong Y (2009) A cis element within flowering locus T mRNA determines its mobility and facilitates trafficking of heterologous viral RNA. J Virol 83: 3540–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka K, Miura E, Xoconostle-Cázares B, Gendler K, Jorgensen RA, Phinney B, et al. (2007) FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19: 1488–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG (1993) Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biol 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu N, Xie K, Jia Q, Zhao J, Chen T, Li H, Wei X, Diao X, Hong Y, Liu Y (2016) Foxtail mosaic virus-induced gene silencing in monocot plants. Plant Physiol 171: 1801–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- McGarry RC, Ayre BG (2012) Geminivirus-mediated delivery of florigen promotes determinate growth in aerial organs and uncouples flowering from photoperiod in cotton. PLoS One 7: e36746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry RC, Klocko AL, Pang M, Strauss SH, Ayre BG (2017) Virus-induced flowering: An application of reproductive biology to benefit plant research and breeding. Plant Physiol 173: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry RC, Prewitt SF, Culpepper S, Eshed Y, Lifschitz E, Ayre BG (2016) Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytol 212: 244–258 [DOI] [PubMed] [Google Scholar]
- Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F (1997) Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol Gen Genet 254: 186–194 [DOI] [PubMed] [Google Scholar]
- Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330: 1397–1400 [DOI] [PubMed] [Google Scholar]
- Pin PA, Nilsson O (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ 35: 1742–1755 [DOI] [PubMed] [Google Scholar]
- Porta C, Lomonossoff GP (2002) Viruses as vectors for the expression of foreign sequences in plants. Biotechnol Genet Eng Rev 19: 245–291 [DOI] [PubMed] [Google Scholar]
- Qin C, Shi N, Gu M, Zhang H, Li B, Shen J, Mohammed A, Ryabov E, Li C, Wang H, et al. (2012) Involvement of RDR6 in short-range intercellular RNA silencing in Nicotiana benthamiana. Sci Rep 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Zhang Q, He M, Kong J, Li B, Mohamed A, Chen W, Zhang P, Zhang X, Yu Z, et al. (2015) Virus technology for functional genomics in plants. In Poltronieri P and Hong Y, eds, Applied Plant Genomics and Biotechnology. Elsevier Woodhead Publishing, Cambridge, UK, pp 229–236 [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov EV, van Wezel R, Walsh J, Hong Y (2004) Cell-to-Cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. J Virol 78: 3149–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof HB, Scholthof KB, Jackson AO (1996) Plant virus gene vectors for transient expression of foreign proteins in plants. Annu Rev Phytopathol 34: 299–323 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16: 656–665 [DOI] [PubMed] [Google Scholar]
- Sha A, Zhao J, Yin K, Tang Y, Wang Y, Wei X, Hong Y, Liu Y (2014) Virus-based microRNA silencing in plants. Plant Physiol 164: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wang F, Zhao J, Xie K, Hong Y, Liu Y (2010) Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol 153: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- van Wezel R, Dong X, Liu H, Tien P, Stanley J, Hong Y (2002) Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol Plant Microbe Interact 15: 203–208 [DOI] [PubMed] [Google Scholar]
- Velázquez K, Agüero J, Vives MC, Aleza P, Pina JA, Moreno P, Navarro L, Guerri J (2016) Precocious flowering of juvenile citrus induced by a viral vector based on Citrus leaf blotch virus: A new tool for genetics and breeding. Plant Biotechnol J 14: 1976–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickland DP, Hanzawa Y (2015) The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol Plant 8: 983–997 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Kishigami R, Yoshikawa N (2014) Reduced generation time of apple seedlings to within a year by means of a plant virus vector: a new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnol J 12: 60–68 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Li C, Yoshikawa N (2016) Promotion of flowering by apple latent spherical virus vector and virus elimination at high temperature allow accelerated breeding of apple and pear. Front Plant Sci 7: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, Sasaki S, Yamagata K, Komori S, Nagase M, Wada M, Yamamoto T, Yoshikawa N (2011) Promotion of flowering and reduction of a generation time in apple seedlings by ectopical expression of the Arabidopsis thaliana FT gene using the Apple latent spherical virus vector. Plant Mol Biol 75: 193–204 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Yoshikawa N (2011) Expression of FLOWERING LOCUS T from Arabidopsis thaliana induces precocious flowering in soybean irrespective of maturity group and stem growth habit. Planta 233: 561–568 [DOI] [PubMed] [Google Scholar]
- Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y (2015) A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5: 14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SC, Chen C, Rojas M, Daimon Y, Ham BK, Araki T, Lucas WJ (2013) Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex. Plant J 75: 456–468 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ryabov E, Zhang X, Hong Y (2008) Influence of viral genes on the cell-to-cell spread of RNA silencing. J Exp Bot 59: 2803–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhang H, Lai T, Qin C, Shi N, Wang H, Jin M, Zhong S, Fan Z, Liu Y, et al. (2012) Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening. Sci Rep 2: 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu L, Shen L, Yu H (2016) NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat Plants 2: 16075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.