Arabidopsis mutants with negligible DHAR activity can maintain wild-type ascorbate status, but decreased oxidation of glutathione alters downstream responses to intracellular H2O2.
Abstract
The complexity of plant antioxidative systems gives rise to many unresolved questions. One relates to the functional importance of dehydroascorbate reductases (DHARs) in interactions between ascorbate and glutathione. To investigate this issue, we produced a complete set of loss-of-function mutants for the three annotated Arabidopsis (Arabidopsis thaliana) DHARs. The combined loss of DHAR1 and DHAR3 expression decreased extractable activity to very low levels but had little effect on phenotype or ascorbate and glutathione pools in standard conditions. An analysis of the subcellular localization of the DHARs in Arabidopsis lines stably transformed with GFP fusion proteins revealed that DHAR1 and DHAR2 are cytosolic while DHAR3 is chloroplastic, with no evidence for peroxisomal or mitochondrial localizations. When the mutations were introduced into an oxidative stress genetic background (cat2), the dhar1 dhar2 combination decreased glutathione oxidation and inhibited cat2-triggered induction of the salicylic acid pathway. These effects were reversed in cat2 dhar1 dhar2 dhar3 complemented with any of the three DHARs. The data suggest that (1) DHAR can be decreased to negligible levels without marked effects on ascorbate pools, (2) the cytosolic isoforms are particularly important in coupling intracellular hydrogen peroxide metabolism to glutathione oxidation, and (3) DHAR-dependent glutathione oxidation influences redox-driven salicylic acid accumulation.
Recent years have witnessed an ever-growing focus on the roles of reactive oxygen species (ROS) in plants, with molecules such as hydrogen peroxide (H2O2) implicated in numerous developmental processes and environmental responses (Dietz et al., 2016). Key questions remain concerning exactly how the signaling functions of these reactive species are mediated (Foyer and Noctor, 2016). When produced inside the cell, the accumulation of ROS is limited by their reactivity with multiple antioxidant systems found in plants. In the aqueous phase, such systems include catalases, ascorbate, glutathione, and peroxiredoxins (Rouhier et al., 2002; Tripathi et al., 2009; Mhamdi et al., 2010b; Foyer and Noctor, 2011; Smirnoff, 2011; Awad et al., 2015). At least some of the signaling effects of ROS may involve secondary changes in such antioxidative systems (Han et al., 2013a).
Ascorbate and glutathione often are considered to work together in ROS removal. Following oxidation, ascorbate can be regenerated through several reactions, only one of which, reduction of dehydroascorbate (DHA), is known to be strongly dependent on glutathione (Foyer and Noctor, 2011; Smirnoff, 2011). When the ascorbate-glutathione pathway was originally formalized as a chloroplast route for ROS processing, DHA reduction was proposed to be nonenzymatic (Foyer and Halliwell, 1976). Although the chemical reaction between reduced (thiol) glutathione (GSH)5 and DHA is rapid, proteins that catalyzed GSH-dependent ascorbate formation from DHA have been partially purified and characterized from several plants, including spinach (Spinacia oleracea), rice (Oryza sativa), and poplar (Populus spp.; Foyer and Halliwell, 1977; Hossain and Asada 1984; Kato et al., 1997; Shimaoka et al., 2000; Lallement et al., 2016). A recent survey of taxonomically diverse species suggests that plants contain between two and four genes encoding dehydroascorbate reductase (DHAR; Zhang et al., 2015). In Arabidopsis (Arabidopsis thaliana), three genes that belong to the glutathione S-transferase (GST) superfamily were annotated to encode DHARs, and all three proteins were shown to have DHAR activity in vitro (Dixon et al., 2002; Dixon and Edwards, 2010).
Studies using tobacco (Nicotiana tabacum) lines overexpressing or underexpressing DHAR revealed roles for the enzyme in determining stomatal opening, plant performance, and ascorbate contents (Chen et al., 2003; Chen and Gallie, 2004, 2006). Other studies have reported positive effects of overexpressing various DHARs (Kwon et al., 2001, 2003; Wang et al., 2010; Yin et al., 2010; Le Martret et al., 2011; Chang et al., 2017). Nevertheless, because plants contain various proteins able to catalyze GSH-dependent DHA reduction and because the reaction can occur nonenzymatically at quite high rates, questions remain surrounding the exact significance of DHARs. Indeed, kinetic modeling of the chloroplast ascorbate-glutathione pathway suggested that the chemical reaction could be largely predominant (Polle, 2001). For these reasons, the physiological importance of DHARs in oxidative stress has been subject to some debate (Morell et al., 1997; Foyer and Mullineaux, 1998; Smirnoff, 2011), and the biological roles of the endogenous genes, and the relationships between them, remain to be elucidated.
While DHA reduction is considered to be important to regenerate ascorbate, and therefore to prevent its irreversible breakdown (Parsons and Fry, 2012), proteins that can oxidize GSH may be important in regulating redox signaling pathways (Han et al., 2013a, 2013b). Glutathione accumulation, largely in the form of GSSG, has long been known to occur in response to increased intracellular H2O2 (Smith et al., 1984; May and Leaver, 1993), but the extent to which such effects are DHA dependent is not known (Rahantaniaina et al., 2013). Other systems, such as GSTs or glutaredoxin-peroxiredoxins, may couple ROS removal to GSH oxidation (Rouhier et al., 2002; Dixon et al., 2009; Tripathi et al., 2009). Although the ascorbate-glutathione pathway has become accepted as an important route for ROS metabolism in plants, the extent to which the two antioxidants interact in vivo remains to be established (Noctor et al., 2000; Foyer and Noctor, 2011).
In Arabidopsis, three genes are annotated to encode DHAR (Dixon et al., 2002). While the encoded proteins may allow DHAR activity in both the chloroplast and the cytosol, some studies point to DHAR1 localization in the peroxisomes (Dixon et al., 2002; Reumann et al., 2009; Grefen et al., 2010; Tang and Yang, 2013). To date, three studies have appeared using single loss-of-function mutants. The earliest reported a substantial effect on the extractable activity (about 30% of the wild-type value) and effects on ozone resistance in dhar2 (Yoshida et al., 2006). Very recently, a much smaller decrease was reported in a line lacking DHAR2 function, with most of the activity associated with DHAR1. Single mutants for both genes were reported to show modified ascorbate and glutathione redox states following photooxidative stress (Noshi et al., 2017). A similar study of DHAR3 also reported decreased activity and altered responses to photooxidative stress (Noshi et al., 2016).
In this work, we aimed to investigate the following distinct but related questions. What are the reactions that are directly responsible for glutathione oxidation during oxidative stress? To what extent are ascorbate and glutathione redox coupled in optimal and oxidative stress conditions? How important are the different genes encoding Arabidopsis DHAR? To answer these questions, we generated a complete set of single, double, and triple loss-of-function mutants for the three Arabidopsis DHARs and analyzed the localization of the three proteins in parallel. We also introduced all mutant combinations into a catalase-deficient background (cat2) to produce double, triple, and quadruple mutant lines. This approach was chosen because decreased catalase activity forces H2O2 metabolism through alternative systems that are dependent on cellular reductants (Mhamdi et al., 2010b). Our analysis of the dhar mutant lines, grown in parallel in identical conditions, shows that DHAR activity can be decreased to negligible levels without marked effects on plant growth or the ascorbate pool. However, the three DHARs have partly redundant roles in ensuring glutathione oxidation when H2O2 metabolism is enhanced, and their combined absence in these conditions impacts plant phenotypes and pathogenesis-related signaling.
RESULTS
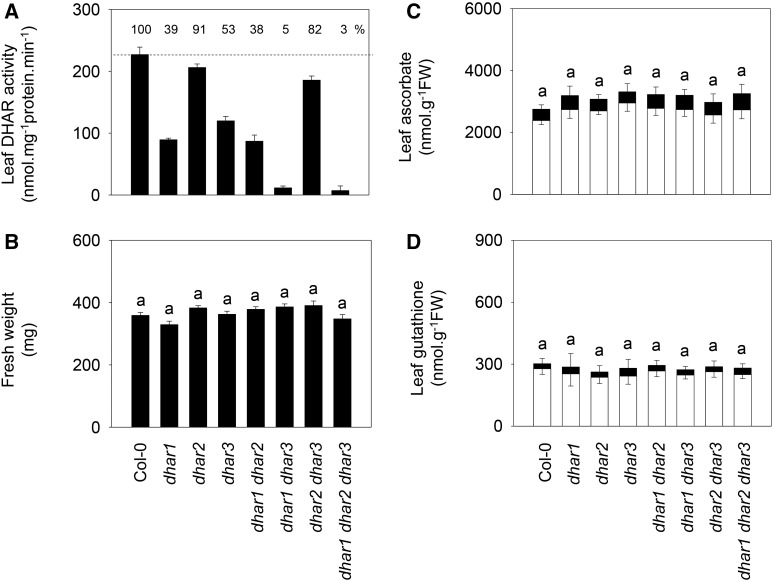
To analyze the roles of the three DHAR genes, we obtained Arabidopsis T-DNA mutants with insertions in the corresponding coding sequences. Homozygous lines were readily obtained for dhar mutations, and reverse transcription-PCR confirmed that they were all knockouts or severe knockdowns (Supplemental Fig. S1, left). Loss of DHAR1 or DHAR3 function decreased extractable leaf activity by about 50%, while the effect of the dhar2 mutation was much less marked (Fig. 1A). From these lines, three double mutants were produced. The dhar1 dhar2 double mutant showed similar activity to dhar1, while the dhar1 dhar3 combination decreased DHAR activity to 5% of the wild-type level (Fig. 1A). Combining the dhar2 and dhar3 mutations led to an increase in DHAR activity compared with the dhar3 single mutant, pointing to a compensatory effect that was presumably linked to DHAR1, because a triple mutant, in which all three genes were mutated, showed negligible activity (Fig. 1A).
Figure 1.
Functional characterization of single, double, and triple dhar mutants. A, Extractable DHAR activity in leaf extracts. Plants were grown 3 weeks under 16 h of light/8 h of dark at an irradiance of 200 µmol m−2 s−1 and 65% relative humidity. Percentage DHAR activity relative to Columbia-0 (Col-0; dotted line) is indicated at the top of the frame. Data are means ± se of six different extracts. B, Fresh weight (FW) of Col-0 and mutants. Plants were sampled in the same conditions as in A. Values are means ± se of 15 plants. C and D, Leaf contents of ascorbate (C) and glutathione (D) in Col-0 and dhar mutants. White bars, Reduced forms; black bars, oxidized forms. Values are means ± se of three plants, sampled in the same conditions as in A and B. The same letter indicates no significant difference by Student’s t test at P < 0.05 (for total leaf pools in C and D; redox states are shown in Supplemental Table S1).
Functional Impact of dhar Mutations in Optimal and Oxidative Stress Conditions
When grown in standard conditions, none of the single, double, or triple mutants showed any difference in phenotype compared with Col-0 (Fig. 1B; Supplemental Fig. S2). Similarly, leaf ascorbate and glutathione pools remained at wild-type levels and reduction states (Fig. 1, C and D; Supplemental Table S1).
As a first test of the role of each DHAR in stress responses, mutants were grown in vitro in agar containing stressful concentrations of cadmium, mannitol, salt, or paraquat, and root growth was measured (Supplemental Fig. S3). Loss of DHAR function caused a slight increase in cadmium sensitivity, which was more evident at low concentrations and which was apparent even in the three single mutants. Growth on mannitol was not substantially affected by the mutations, but dhar1 dhar2, dhar1 dhar3, and the triple mutant showed somewhat enhanced sensitivity to NaCl (Supplemental Fig. S3). All three single mutations enhanced sensitivity to the oxidative stress caused by paraquat, although combination of the mutations did not reinforce this effect (Supplemental Fig. S3).
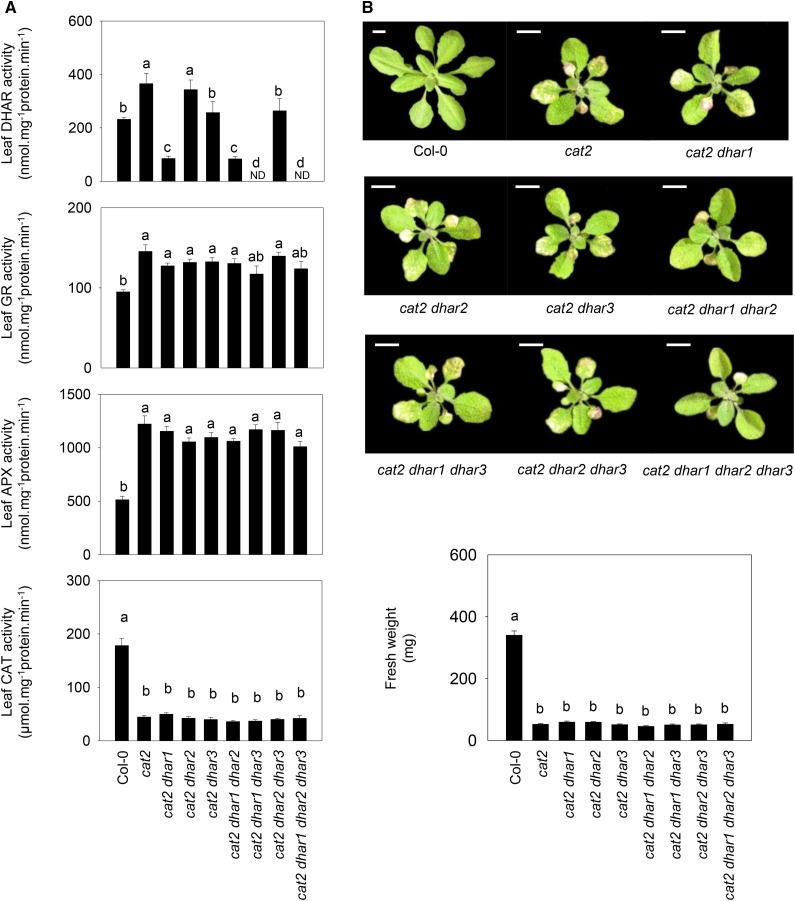
Taken together, the above results suggest that none of the three DHARs is required for plant development but that they may interact in a complex manner to determine stress resistance. To explore this point further, the three mutations were introduced into a catalase-deficient background in which enhanced intracellular H2O2 availability increases the demand on reductants such as ascorbate and glutathione (Mhamdi et al., 2010b). Thus, we produced double cat2 dhar1, cat2 dhar2, and cat2 dhar3 mutants, and from these lines, we produced three triple and one quadruple mutant (Supplemental Fig. S1, right). Analysis of extractable leaf DHAR in cat2 revealed that the activity was increased in response to the oxidative stress caused by catalase deficiency (Fig. 2A, top). Analysis of double cat2 dhar mutants suggested that DHAR1 was responsible for most of the DHAR activity in the cat2 background and that the remainder was largely due to DHAR3. Activities in cat2 and cat2 dhar2 were similar (Fig. 2A, top), suggesting that, as in the Col-0 background, DHAR2 makes a minor contribution to the extractable leaf activity. Consistent with this, no activity was detected in cat2 dhar1 dhar3 or the quadruple cat2 dhar1 dhar2 dhar3 mutant (Fig. 2A, top).
Figure 2.
Major antioxidative enzyme activities (A) and phenotypes (B) in the cat2 mutant carrying T-DNA insertions in DHAR1, DHAR2, and DHAR3 singly or in combination. A, Extractable leaf activities of DHAR, GR, APX, and CAT after 3 weeks of growth at an irradiance of 200 µmol m−2 s−1 in a 16-h photoperiod. Data are means ± se of nine biological replicates for DHAR leaf activity and six for other enzymes. Different letters indicate significant differences at P < 0.05. ND, Not detected. B, Photographs of mutants and rosette fresh weight, determined in the same conditions as in A. For fresh weight, values are means ± se of 15 to 24 plants. Bars = 1 cm.
As reported previously, the cat2 mutant showed only about 20% of wild-type leaf catalase activity (Queval et al., 2009), and this was not affected by any of the combinations of dhar mutations (Fig. 2A, bottom). The activities of two other enzymes of the ascorbate-glutathione pathway, glutathione reductase (GR) and ascorbate peroxidase (APX), were increased alongside DHAR in cat2, but the presence of the dhar mutations had little effect on either (Fig. 2A, middle). The dhar mutations did not affect rosette mass in the cat2 background (Fig. 2B, bottom), but two mutant combinations significantly impacted the cat2-triggered phenotype (Fig. 2B, top). While cat2 and most of the derived cat2 dhar lines showed clearly visible hypersensitive response-like lesions on specific leaves, these were much less apparent in cat2 dhar1 dhar2 and cat2 dhar1 dhar2 dhar3 (Fig. 2B, top).
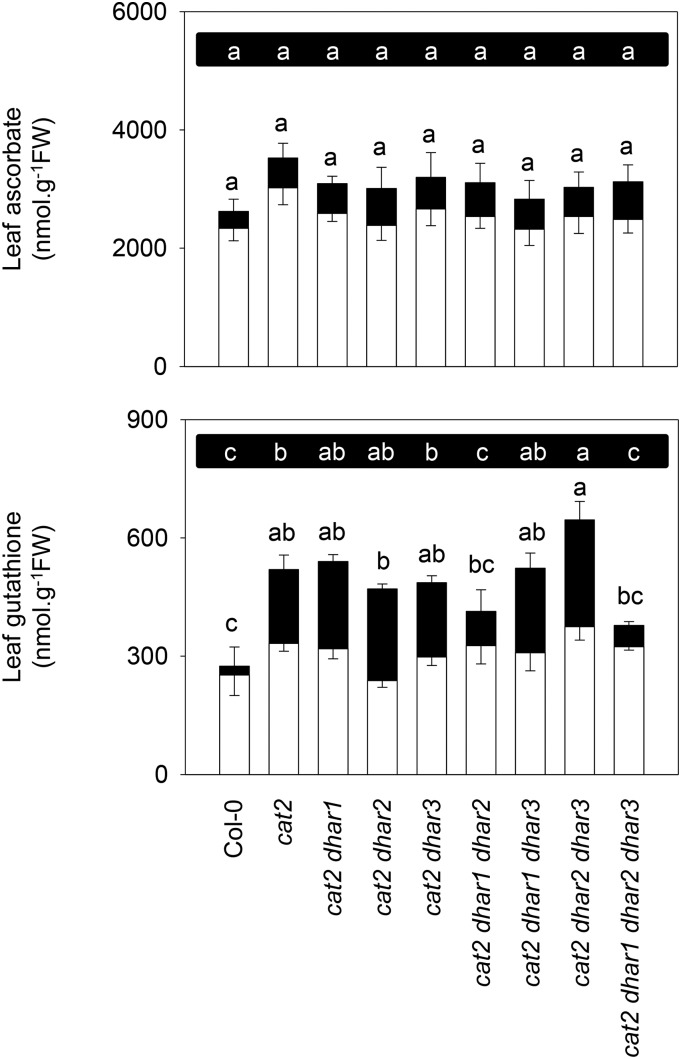
Genetically or pharmacologically induced catalase deficiency causes the accumulation of glutathione (Smith et al., 1984; May and Leaver, 1993; Willekens et al., 1997; Mhamdi et al., 2010b; Queval et al., 2011). Accordingly, leaf glutathione was about 2-fold higher in cat2 than in Col-0, with the difference being almost entirely due to GSSG (Fig. 3, bottom). As a result, the glutathione reduction state, which was above 90% in Col-0, was only 64% in cat2 (Supplemental Table S2). Glutathione was significantly less oxidized in the triple cat2 dhar1 dhar2 and quadruple cat2 dhar1 dhar2 dhar3 mutants (Supplemental Table S2), and this was accompanied by decreased total glutathione in these two lines (Fig. 3, bottom). All other cat2 dhar lines showed similar glutathione status to cat2. As reported previously, ascorbate is less affected in cat2 than is glutathione (Mhamdi et al., 2010a, 2010b; Han et al., 2013a). No difference in total ascorbate contents was observed in any of the mutants relative to Col-0 (Fig. 3, top). However, several lines in the cat2 background showed a slightly but significantly more oxidized ascorbate pool as a result of the presence of additional mutations in dhar (Supplemental Table S2).
Figure 3.
Effects of dhar mutations on oxidative stress-triggered changes in ascorbate and glutathione. White bars, Reduced forms; black bars, oxidized forms; FW, fresh weight. Values are means ± se of three plants, sampled at 3 weeks at an irradiance of 200 µmol m−2 s−1 in a 16-h photoperiod. Different letters indicate significant differences in oxidized forms (top row of letters on black background) or total contents (bottom row of letters) at P < 0.05 by one-way ANOVA with posthoc Tukey’s honestly significant difference, a test calculator for comparing multiple treatments.
Subcellular Localization of the Three DHARs
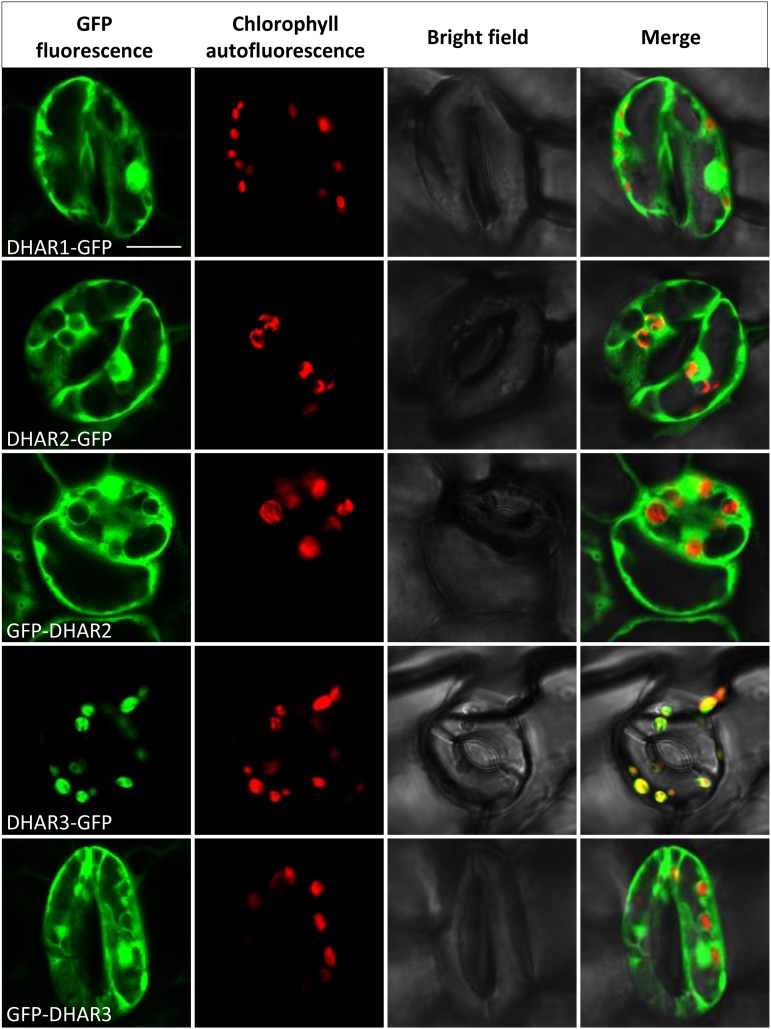
Fusion proteins were constructed for all three DHARs with the GFP tag fused C or N terminally (DHAR-GFP or GFP-DHAR, respectively). The six constructs were transformed into the quadruple cat2 dhar1 dhar2 dhar3 and the triple dhar1 dhar2 dhar3 mutant backgrounds, and T1 plants were analyzed. Despite repeated attempts, we were not able to regenerate plants expressing GFP-DHAR1 in either the cat2 dhar1 dhar2 dhar3 or dhar1 dhar2 dhar3 background. DHAR1-GFP showed a cytosolic localization in the leaves of the quadruple mutant background (Fig. 4). A similar localization also was observed for DHAR1-GFP in the roots of this line (Supplemental Fig. S4) and in the leaf cells of the dhar1 dhar2 dhar3 triple mutant background (Supplemental Fig. S5). Fluorescence patterning did not overlap with the positive control for peroxisomal localization (Supplemental Fig. S6).
Figure 4.
Subcellular localization of DHAR-GFP fusion proteins. Full-length sequences of AtDHAR1, AtDHAR2, and AtDHAR3 were tagged with GFP in the N-terminal (GFP-DHAR) or C-terminal (DHAR-GFP) position and introduced into the cat2 dhar1 dhar2 dhar3 quadruple mutant. No transformants for GFP-DHAR1 were recovered. The images correspond to epidermal cells of 10-d-old seedlings. From left to right are GFP fluorescence (green), chlorophyll autofluorescence (red), bright-field, and merged images. Bar = 10 µm.
DHAR2-GFP and GFP-DHAR2 fusions showed similar fluorescence patterns to the negative GFP control, confirming a cytosolic localization in the leaves of both genetic backgrounds (Fig. 4; Supplemental Fig. S5). Both DHAR2 fusions also gave a cytosolic signal in the roots (data not shown). In contrast to DHAR1 and DHAR2, DHAR3-GFP was associated with the chloroplast. The DHAR3-GFP signal showed close overlap with chlorophyll autofluorescence in the leaves of both the complemented quadruple and triple mutants (Fig. 4; Supplemental Fig. S5). To establish whether DHAR3 also might be targeted to the mitochondria, we compared GFP fluorescence with MitoTracker signals and with a positive control for dual chloroplast-mitochondrion targeting (PDF1B-GFP) in roots. MitoTracker fluorescence revealed no overlap with the DHAR3-GFP signal (Supplemental Fig. S4). Whereas PDF1B-GFP fluorescence patterns were observed in both the larger plastids and the smaller punctate mitochondria, DHAR3-GFP signals were observed only in plastids (Supplemental Fig. S4). When the GFP was fused to the DHAR3 N terminus (GFP-DHAR3), no association with the chloroplast was observed; instead, a diffuse pattern of staining appeared (Fig. 4; Supplemental Fig. S5), consistent with targeting of the native DHAR3 to the chloroplast through an N-terminal signal peptide sequence. In conclusion, these data suggest that DHAR1 and DHAR2 are cytosolic while DHAR3 is localized in plastids. This distribution was not affected by the presence of the cat2 mutation.
Functional Complementation of the cat2 dhar1 dhar2 dhar3 Quadruple Mutant
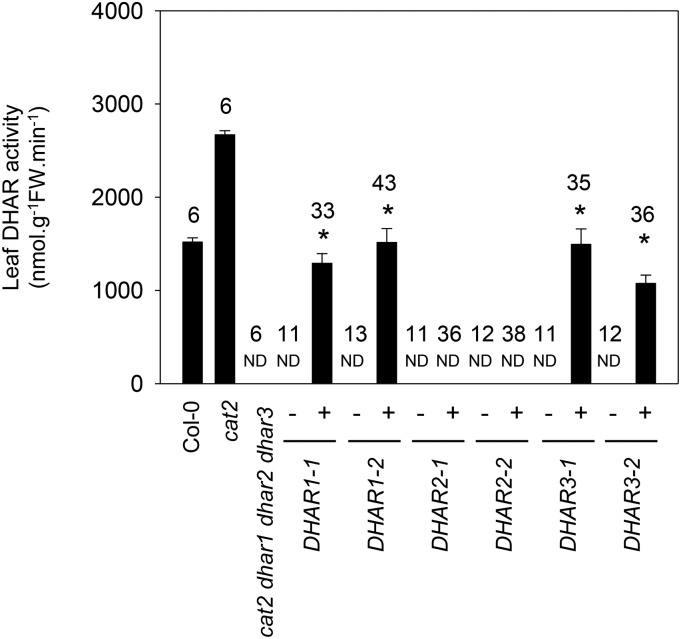
To further evaluate the importance of the three DHARs in oxidative stress responses, we analyzed the ability of DHAR-GFP fusion proteins for each of the three DHARs to complement extractable activity, glutathione contents, and phenotypes in T2 individuals of the cat2 dhar1 dhar2 dhar3 mutant. For each DHAR, two independent lines were obtained that showed a 3:1 segregation for antibiotic resistance in T2 (Supplemental Table S3). In resistant DHAR1-GFP and DHAR3-GFP plants, DHAR activity was restored to about 50% of the value in cat2 (Fig. 5). No increase in extractable DHAR activity was detected in antibiotic-resistant individuals from the quadruple mutant complemented with DHAR2-GFP (Fig. 5).
Figure 5.
DHAR activities in the cat2 dhar1 dhar2 dhar3 line complemented with DHAR1, DHAR2, or DHAR3 (two independent lines each). Enzyme activities were determined in leaf extracts from 15-d-old T2 plants. Plus or minus indicates the presence or absence of the transgene in the plants in which activity was measured. The number of plants assayed is given above the columns for each genotype. Asterisks indicate significant differences at P < 0.05 from the activity in the uncomplemented cat2 dhar1 dhar2 dhar3 mutant. FW, Fresh weight; ND, not detected.
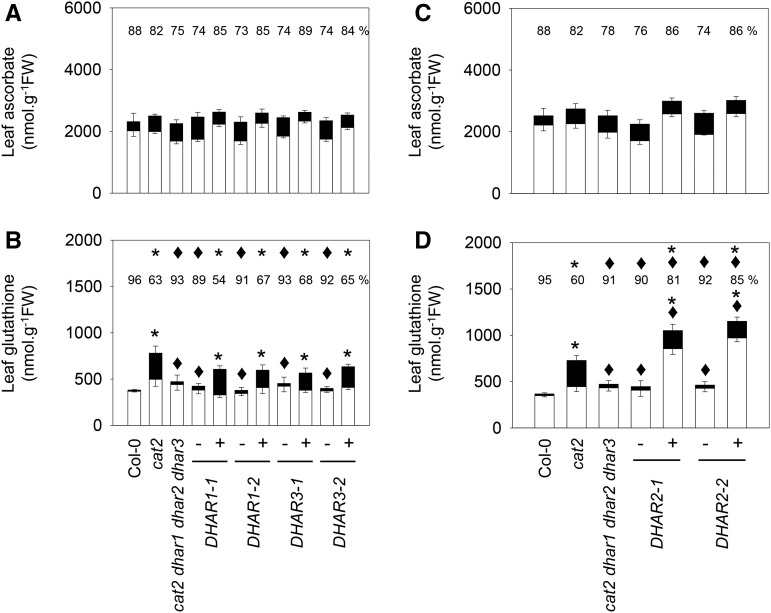
Complementation of cat2 dhar1 dhar2 dhar3 plants with DHAR1 or DHAR3 caused significant increases in GSSG (Fig. 6B). As a result, the glutathione status of DHAR1-GFP+ and DHAR3-GFP+ plants was more similar to that of the single cat2 mutant than to that of the parent quadruple mutant line (Fig. 6B). No effect on ascorbate was observed (Fig. 6A). Despite the lack of a detectable increase in extractable activity in DHAR2-GFP+ complemented lines, introduction of this protein also increased glutathione contents and decreased the glutathione reduction state (Fig. 6D). However, the effect was somewhat different from that produced by the introduction of DHAR1 or DHAR3. Complementation with DHAR2 caused glutathione to increase above cat2 levels and to remain more reduced than in cat2, albeit less reduced than in the uncomplemented quadruple mutant line (Fig. 6D). Again, no effect on ascorbate was observed (Fig. 6C).
Figure 6.
Ascorbate and glutathione in the cat2 dhar1 dhar2 dhar3 line complemented with DHAR1, DHAR2, or DHAR3. White bars, Reduced forms; black bars, oxidized forms; FW, fresh weight. A and B, DHAR1 and DHAR3 complemented lines. C and D, DHAR2 complemented lines. Plants were grown on soil from germination in a 16-h/8-h day/night regime for 3 weeks. Plus or minus indicates the presence of absence of the transgene in the plants in which activity was measured. Data are means ± se of between three and nine biological repeats. Asterisks indicate significant differences in total ascorbate or glutathione contents in mutants and complemented lines compared with Col-0, and diamonds indicate significant differences compared with cat2, at P < 0.05. Numbers above the columns indicate percentage reduced (100 × reduced form/total content). Above these numbers, asterisks indicate significant differences compared with Col-0, and diamonds indicate significant differences compared with cat2, at P < 0.05.
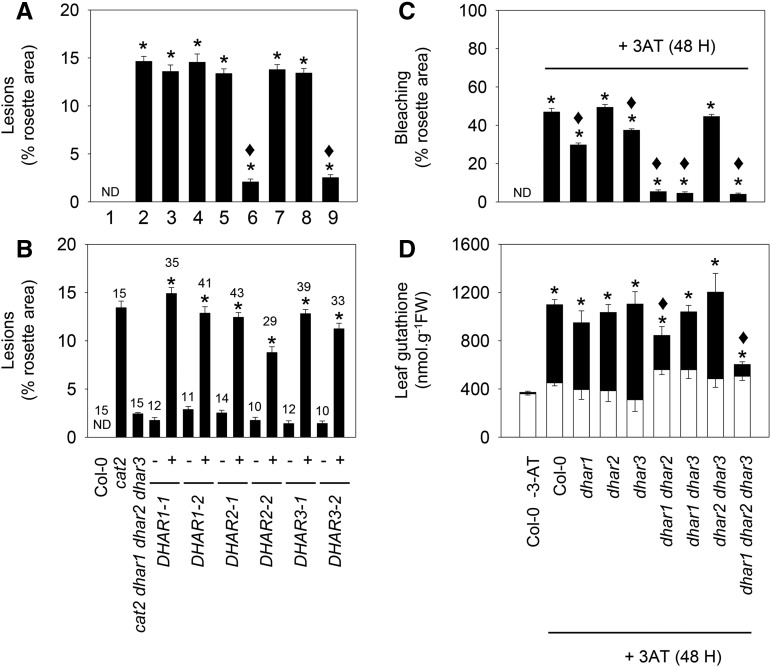
Complementation with DHAR1, DHAR2, or DHAR3 restored the cat2 lesion phenotype in the cat2 dhar1 dhar2 dhar3 quadruple mutant. Two experiments with different batches of T2 seed revealed a 3:1 phenotype segregation (Supplemental Table S4). In a first step to analyze this effect more closely, lesions were quantified using imaging software. Quantification in Col-0 and the eight cat2 dhar loss-of-function lines showed that all lines carrying the cat2 mutation presented lesions on about 15% of the total rosette surface, except the two lines in which DHAR1 and DHAR2 functions were both lost (cat2 dhar1 dhar2 and cat2 dhar1 dhar2 dhar3; Fig. 7A). The cat2-dependent lesions could be fully restored by complementation of the quadruple mutant with any of the three DHARs (Fig. 7B). The role of the different DHARs in oxidative stress linked to catalase deficiency was further tested using 3-AT. Like the cat2 mutation, catalase inhibition by this herbicide engages GSH-dependent processes, which become apparent as the accumulation of GSSG (May and Leaver, 1993). Spraying with 3-AT caused a bleaching phenotype within 2 d in the wild type (Supplemental Fig. S7). The appearance of this phenotype was inhibited in dhar1 and, more clearly, in dhar1 dhar2 and dhar1 dhar3 double mutants as well as in the triple dhar1 dhar2 dhar3 mutant line (Fig. 7C; Supplemental Fig. S7). Similar to the effects observed in the cat2 background (Fig. 3), 3-AT-induced accumulation of GSSG was decreased significantly in dhar1 dhar2 and, especially, in dhar1 dhar2 dhar3 (Fig. 7D). Consequently, glutathione remained significantly more reduced in these lines than in the wild type treated with 3-AT (Supplemental Table S5).
Figure 7.
Influence of dhar mutations on lesions triggered by the cat2 mutation (left) or by the catalase inhibitor 3-amino-1,2,4-triazole (3-AT; right). A, Effects of different dhar mutant combinations on cat2-triggered lesions. Numbers indicate genotypes as follows: 1, Col-0; 2, cat2; 3, cat2 dhar1; 4, cat2 dhar2; 5, cat2 dhar3; 6, cat2 dhar1 dhar2; 7, cat2 dhar1 dhar3; 8, cat2 dhar2 dhar3; and 9, cat2 dhar1 dhar2 dhar3. Asterisks indicate significant differences in percentage of lesions on cat2 background mutant leaves compared with Col-0, and diamonds indicate significant differences when they were compared with cat2, at P < 0.05. ND, Not detected. Values are means ± se of 15 plants. B, Quantification of lesions in the cat2 dhar1 dhar2 dhar3 quadruple mutant retransformed (+) or not (−) with DHAR1, DHAR2, or DHAR3. The number of plants analyzed in complemented lines is indicated above each bar. Asterisks indicate significant differences in percentage of lesions on the complemented line compared with the untransformed quadruple mutant. C, Extent of bleaching in Col-0 and different dhar mutant lines caused by exposure to 2 mm 3-AT. 3-AT was sprayed onto the leaf surface of 3-week-old plants, and lesions were quantified and samples taken 48 h later. Asterisks indicate significant effects of 3-AT treatment compared with nontreated Col-0, and diamonds indicate significant differences between treated dhar mutants and Col-0, at P < 0.05. Values are means ± se of 30 plants. D, Leaf glutathione content after 3-AT treatment. White bars, reduced forms; black bars, oxidized forms; FW, fresh weight. Data are means ± se of three plants. For total glutathione, asterisks indicate significant effects of 3-AT treatment compared with nontreated Col-0, and diamonds indicate significant differences between dhar mutants and Col-0, at P < 0.05.
Roles of DHARs in Oxidative Stress-Triggered Activation of the Salicylic Acid Pathway and Cell Death
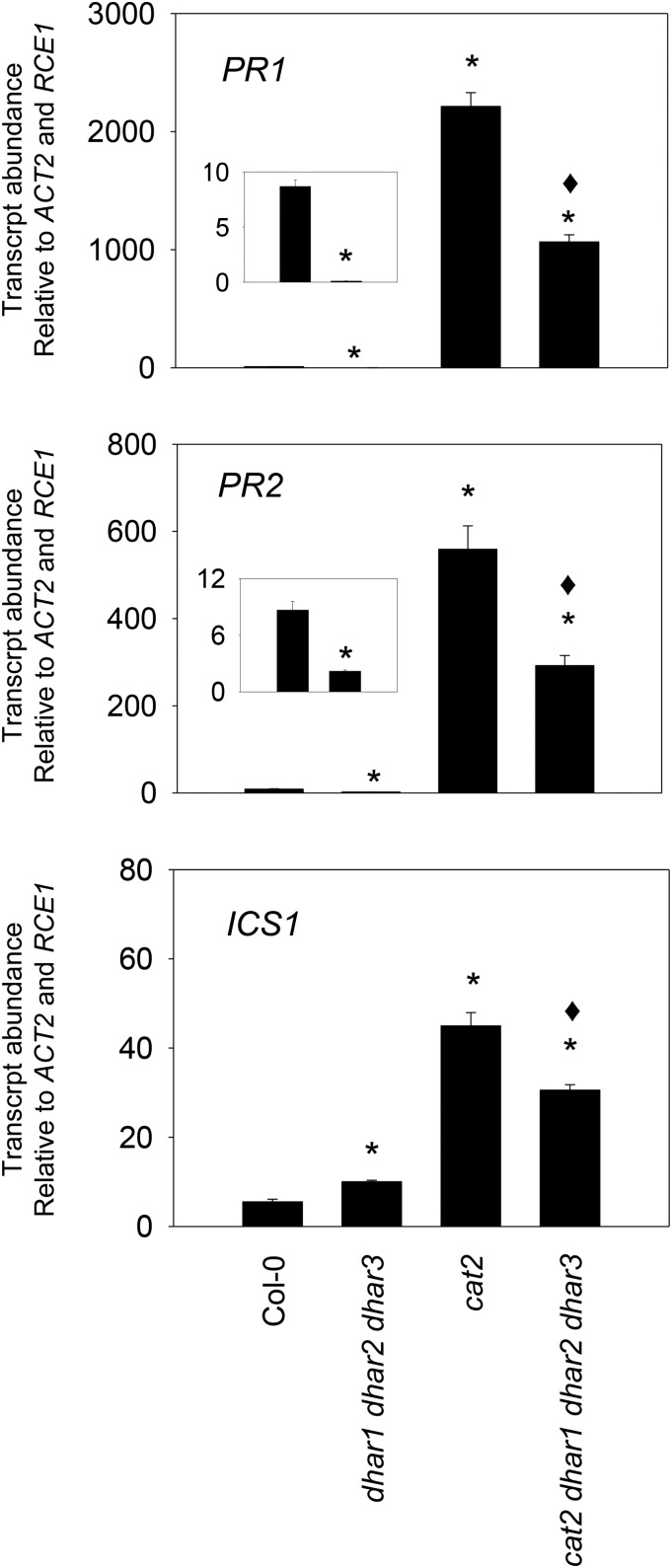
To explore the processes underlying the phenotypic effects described above, we analyzed the effects of the three dhar mutations on transcripts of a key gene involved in salicylic acid (SA) synthesis (ICS1) and two SA-dependent pathogenesis-related genes (PR1 and PR2). All three genes were markedly induced in response to oxidative stress in cat2 but less so in the quadruple cat2 dhar1 dhar2 dhar3 line (Fig. 8). In the Col-0 background, the triple loss of DHAR function also decreased basal levels of PR1 and PR2 transcripts, although ICS1 transcripts were increased slightly (Fig. 8).
Figure 8.
SA-linked gene expression in Col-0, cat2, and derived lines carrying all three dhar mutations. PR1, PR2, and ICS1 transcripts were quantified by quantitative reverse transcription-PCR in 3-week-old plants using two reference genes (ACTIN2 and RCE1). Inset graphs show Col-0 (left) and dhar1 dhar2 dhar3 (right). Data are means ± se of three biological replicates and have been multiplied by 100 for ease of expression. Significant differences between mutants and Col-0 at P < 0.05 are indicated by asterisks, while diamonds indicate significant differences between cat2 and the quadruple mutant.
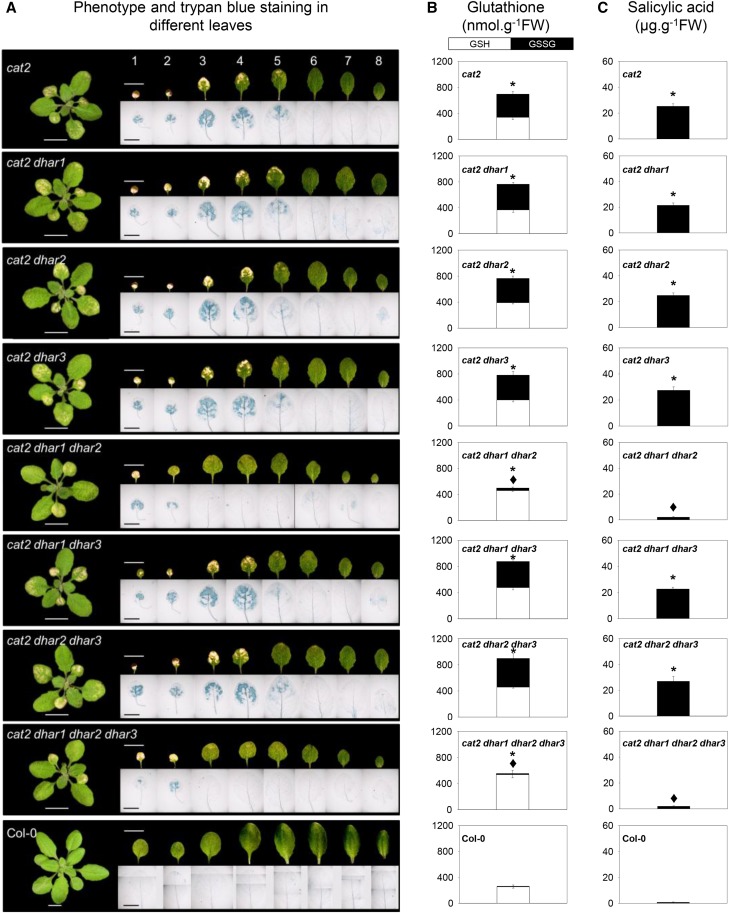
In a second approach, we profiled the lesion phenotype of the leaves from Col-0 and all eight lines containing cat2 and dhar mutations in combination. In the cat2 single mutant, cell death was apparent on the oldest leaves, both as whitish lesions and Trypan Blue-positive staining (Fig. 9A). Similar phenotypes to cat2 were observed in cat2 dhar1, cat2 dhar2, cat2 dhar3, cat2 dhar1 dhar3, and cat2 dhar2 dhar3 (Fig. 9A). By contrast, lesions were restricted to one or two of the very oldest leaves in cat2 dhar1 dhar2 and the quadruple mutant (Fig. 9A). Hence, a clear phenotypic difference was apparent between these two lines and the others carrying the cat2 mutation, particularly on leaves 4 and 5 (Fig. 9A). Measurements of glutathione specifically in these leaves confirmed the effect of the combined dhar1 and dhar2 mutations on cat2-triggered effects on glutathione (Fig. 9B).
Figure 9.
Detailed analysis of the effects of dhar mutations on the cat2 phenotype and their relationship to localized glutathione status and SA contents. A, Photographs of rosette leaves and Trypan Blue staining for dead cells in the different mutants. Bars = 1 cm in leaf photographs and 5 mm in Trypan Blue photographs. B, Leaf glutathione contents in leaves 4 and 5. White bars, GSH; black bars, GSSG; FW, fresh weight. Values are means ± se of three biological replicates. C, SA contents in leaves 4 and 5. Values are means ± se of six biological replicates. In B and C, asterisks indicate significant differences in total glutathione and SA contents in mutants compared with Col-0, and diamonds indicate significant differences between cat2 dhar mutants and cat2, at P < 0.05. For all these experiments, plants were grown for 3 weeks under 16 h of light/8 h of dark at an irradiance of 200 µmol m−2 s−1 and 65% relative humidity.
Analysis of SA in the same leaves revealed that, while all other cat2-carrying genotypes accumulated SA to around 20 µg g−1 fresh weight in leaves 4 and 5, SA remained at close to wild-type levels in cat2 dhar1 dhar2 and in the quadruple mutant (Fig. 9C). To assess the significance of this effect, the quadruple mutant was selected for an analysis of resistance to virulent bacteria. While bacterial growth was substantially lower in cat2 than in the wild type, the simultaneous presence of the three dhar mutations caused partial loss of this induced resistance (Supplemental Fig. S8). Although no difference in bacterial growth was observed between Col-0 and dhar1 dhar2 dhar3, SA accumulated somewhat less in response to infection in the triple mutant (Supplemental Fig. S8). Similarly, the combined loss of the three DHAR functions decreased SA accumulation following infection of the quadruple mutant compared with cat2 (Supplemental Fig. S8).
DISCUSSION
This study was conducted with the aim of resolving several outstanding questions related to the importance of genes encoding DHAR and the functioning of the ascorbate-glutathione pathway in H2O2 processing. From a parallel analysis of a complete set of Arabidopsis mutants for the three DHAR-encoding genes, together with transformed lines complemented with each DHAR, we are led to the following conclusions.
Almost All of the Extractable DHAR Activity in Arabidopsis Leaves Is Attributable to Cytosolic DHAR1 and Chloroplastic DHAR3
In addition to the chloroplast and cytosol, DHAR activity has been reported in purified pea (Pisum sativum) peroxisomes and mitochondria (Jiménez et al., 1997). Targeting experiments in Arabidopsis have established that these organelles contain APX, MDHAR, and GR, in some cases as a result of dual targeting with the chloroplast or cytosol (Chew et al., 2003; Kataya and Reumann, 2010). Although DHAR1 does not contain a known peroxisomal targeting sequence, it was assigned a localization in this organelle on the basis of proteomics analyses and experiments with yellow fluorescent protein (YFP) fused to the DHAR1 C terminus (Reumann et al., 2009). For unknown reasons, we were unable to recover GFP-DHAR1 plants. However, we could not find any evidence that the protein is found in organelles from analysis of lines transformed with constructs with GFP fused to the DHAR1 C terminus. Our data suggest that DHAR1 is a cytosolic protein, in agreement with another study using YFP (Grefen et al., 2010). If so, two of the Arabidopsis DHARs would be cytosolic, as DHAR2 was clearly located in this compartment, whether the GFP was fused N or C terminally.
A pea gene encoding GR was the first to be shown to encode targeting sequences directing dual localization in the chloroplast and mitochondria (Creissen et al., 1995). This observation was confirmed in Arabidopsis, where APX, MDHAR, and DHAR activities also were detected in mitochondria (Chew et al., 2003). In our study here, DHAR3-GFP signals were clearly associated with the chloroplast but not the mitochondria. Our study, therefore, suggests that the leaf DHAR activity in Arabidopsis is distributed approximately equally between the cytosol and the chloroplast, linked mainly to the activities of DHAR1 and DHAR3, respectively. A minor contribution to the cytosolic activity would come from DHAR2. This is consistent with two recent reports on single dhar mutants (Noshi et al., 2016, 2017) but in contrast to an earlier analysis of dhar2 (Yoshida et al., 2006).
The subcellular distribution we report for the Arabidopsis DHARs is consistent with earlier predictions (Dixon et al., 2002) as well as with information available for the three poplar genes (Tang and Yang, 2013). Localization of DHARs was recently compared in maize (Zea mays), Picea abies, and Selaginella moellendorffii (Zhang et al., 2015). All three species contained one gene encoding chloroplastic DHAR and either one or two genes encoding cytosolic isoforms, with a fourth gene in maize coding for a protein localized in the vacuole (Zhang et al., 2015). Interestingly, both of the DHARs we found to be cytosolic were identified in a proteomics study of proteins associating with the plasma membrane, along with several other GSTs and peroxidases (Marmagne et al., 2007). Proteins other than the three DHARs, such as certain glutaredoxins or GSTs (Chew et al., 2003; Dixon et al., 2009), may be responsible for DHAR activities reported previously to be associated with the peroxisomes and mitochondria. However, very low residual activities were detected in both the dhar1 dhar2 dhar3 triple and cat2 dhar1 dhar2 dhar3 quadruple mutants (Figs. 1 and 2), indicating that other proteins probably have relatively low capacities.
Arabidopsis Can Maintain Leaf Ascorbate Pools and Redox States When DHAR Activity Is Negligible, Even in Conditions of Oxidative Stress
In agreement with our previous analyses, leaf ascorbate was highly reduced in Arabidopsis Col-0 (about 90%). A high ascorbate reduction state is predicted if most of the pool is found in compartments that contain nonlimiting regenerating systems dependent on more powerful reductants, such as ferredoxin, NAD(P)H, and glutathione (Foyer and Noctor, 2011, 2016). Previous studies in tobacco have implicated DHAR as a major player determining ascorbate redox state and contents. A DHAR-silenced tobacco line with a decrease of about 70% in leaf activity had a more oxidized ascorbate pool, while ascorbate was more reduced in strong overexpressors (Chen et al., 2003; Chen and Gallie, 2004, 2006). The Arabidopsis genotypes reported here show decreases in DHAR activity from 10% to 100% as a result of gene-specific loss of function. In stark contrast to the studies in tobacco, we observed no marked effects on ascorbate contents or redox state. Significantly enhanced oxidation, relative to cat2, was observed in some of the cat2 dhar lines but the effects were slight, with ascorbate remaining about 80% reduced (Supplemental Table S2). We cannot discount a role for DHARs in maintaining ascorbate in other conditions. For example, changes in ascorbate redox state were reported in dhar1 and dhar2 single mutants subjected to high light (Noshi et al., 2016, 2017). However, these effects were relatively slight. Our analysis, in which all combinations of the three dhar mutations were examined, provide little evidence for a major role for DHARs in maintaining ascorbate pools in Arabidopsis grown in moderate light with or without intracellular oxidative stress. The ability to maintain a largely reduced leaf ascorbate pool when DHAR activity is negligible may be explained by the presence of other routes that can either reduce DHA or else avoid its formation by reducing MDHA. Such mechanisms notably include nonenzymatic reduction of DHA by GSH and reduction of MDHA by chloroplast ferredoxin or by NAD(P)H-dependent reductases (Polle, 2001; Smirnoff, 2011; Gest et al., 2013; Johnston et al., 2015; Noctor, 2015).
DHARs Cooperate in Glutathione Oxidation Triggered by Oxidative Stress
Although complete loss of DHAR function in dhar1 dhar2 dhar3 did not greatly affect ascorbate, it clearly impacted the responses to oxidative stress triggered by catalase deficiency. Accumulation of GSSG is the clearest and most readily quantifiable biochemical marker of altered redox state caused by a decreased capacity for the removal of intracellular H2O2 (Willekens et al., 1997; Queval et al., 2009, 2011). By comparison, reproducible increases in H2O2 itself are much more difficult to detect in cat2 (Mhamdi et al., 2010a; Noctor et al., 2015). This suggests that, when catalase capacity is decreased, alternative H2O2-reducing reactions are readily elicited, dampening increases in H2O2 and instead leading to the oxidation of key redox buffers such as glutathione. Exactly how this occurs remained unclear: DHARs are only one of several classes of GSH-dependent enzymes that could potentially be involved in H2O2 processing, and genes encoding several of these enzymes can be up-regulated alongside GSSG accumulation (Mhamdi et al., 2010a; Rahantaniaina et al., 2013). Given the differential oxidation of glutathione compared with ascorbate, one simple interpretation would be that GSH is coupled to H2O2 metabolism via pathways that are ascorbate independent, such as GSH-dependent peroxidases. Our data provide in planta evidence against this interpretation, because they show that at least one DHAR must be functional in order to observe a significant oxidation of glutathione when H2O2 availability is increased. If all three DHARs are knocked out, GSSG accumulation, whether induced pharmacologically or genetically, is almost completely inhibited (Figs. 6 and 7).
Of the three double loss-of-function combinations, the clearest effect on glutathione status was observed for dhar1 dhar2. This underscores the potential importance of the cytosolic isoforms in coupling ascorbate and glutathione pools and is consistent with previous studies demonstrating the importance of cytosolic APX1 or GR1 in cat2-triggered responses (Mhamdi et al., 2010a; Vanderauwera et al., 2011) The importance of the cytosol in this context may be related to the peroxisomal location of the H2O2 that becomes available when catalase is deficient, although decreased glutathione oxidation also was reported for single dhar1 and dhar2 mutants after exposure to high light (Noshi et al., 2017). DHAR3 may be more significant in chloroplastic processing, although our data provide little evidence that this isoform plays a specific role in response to paraquat-induced oxidative stress, which largely originates in this organelle (Supplemental Fig. S3). Intriguingly, although glutathione was only marginally more oxidized in cat2 dhar1 dhar2 than in cat2 dhar1 dhar2 dhar3, the chloroplastic DHAR3 was just as effective as the cytosolic DHAR1 in restoring cat2 glutathione status (Fig. 6). Differences between cat2 dhar1 dhar2 dhar3 DHAR3+, in which glutathione status was quite similar to cat2 (Fig. 6), and cat2 dhar1 dhar2, in which GSSG accumulation was somewhat lower (Fig. 3), may reflect differences in DHAR3 expression levels. In any case, the ability of chloroplastic DHAR3 to complement the quadruple mutant raises questions about the potential importance of redox exchange across the inner envelope membrane. Chloroplasts have long been known to be competent in GSSG and DHA uptake (Anderson et al., 1983), and more recently, chloroplast envelope glutathione and ascorbate transporters have been identified in Arabidopsis (Maughan et al., 2010; Miyaji et al., 2015).
While our data provide in planta evidence that DHARs are important in coupling ascorbate and glutathione pools during oxidative stress, they also identify important questions for further study. The first relates to why ascorbate stays largely reduced even when glutathione is markedly oxidized and why DHAR function can affect glutathione but not ascorbate. Several factors may explain these observations. As discussed above, GSH-independent mechanisms exist for regenerating ascorbate, notably from MDHA, implying that a relatively minor part of ascorbate regeneration is linked to glutathione oxidation (Polle, 2001). Another important factor relates to differences in the compartmentation of the two antioxidants. In many cell compartments, glutathione and ascorbate are highly reduced under most conditions (Meyer et al., 2007; Noctor et al., 2016), with DHA and GSSG accumulating at specific locations. Much of the GSSG accumulated during oxidative stress is found in the vacuole, thus allowing it to escape reduction by GR (Queval et al., 2011). We are currently investigating the processes responsible for this distribution, which may be significant in at least two respects. First, it could be crucial to allow GSSG generated in compartments such as the cytosol to be useful as a stable tissue marker of oxidative stress. Second, it may contribute to the homeostasis of the NADP(H)-glutathione redox couples and, therefore, influence cellular redox signaling.
The second question relates to the exact biochemical function of DHAR2. We found little genetic evidence that this protein is able to catalyze the GSH-dependent conversion of DHA to ascorbate. Extractable DHAR assays revealed little effect of either the dhar2 mutation or of complementation with DHAR2. This is surprising because the recombinant DHAR2 has been shown to be competent in GSH-dependent reduction of DHA, albeit with lower specific activities compared with DHAR3 and, particularly, the other cytosolic isoform, DHAR1 (Dixon et al., 2002). Our study suggests that DHAR2 is active in GSH oxidation in planta, although we note that complementation of the cat2 dhar1 dhar2 dhar3 mutant with this protein produced a different glutathione signature from that observed when the mutant was complemented with DHAR1 or DHAR3. Further work will be required to elucidate this question, although we tentatively suggest that DHA is not the only physiological acceptor substrate for the DHAR2-dependent oxidation of GSH. This might explain why DHAR2 can contribute to glutathione oxidation in vivo even though the associated DHAR activity, measured in vitro, appears to be negligible.
Compromised DHAR Activity Uncouples Oxidative Stress from Activation of the SA Pathway
Loss of CAT2 function induces SA accumulation in the absence of pathogens and associated pathogenesis-related responses, including resistance to bacteria (Chaouch et al., 2010, 2012). The induction of these responses is influenced by glutathione status (Mhamdi et al., 2010a; Han et al., 2013a). Here, we show that when glutathione remains close to wild-type status (in cat2 dhar1 dhar2 or the quadruple mutant), much of the SA response is abolished (Figs. 8 and 9). Thus, cooperation between the DHARs, and particularly the cytosolic forms, is required to couple oxidative stress to the activation of downstream signaling. The importance of the cytosolic isoforms could be linked to their partial recruitment to the plasma membrane (Marmagne et al., 2007), possibly to set appropriate conditions for redox signaling at this location. However, the close correlation between glutathione and SA contents (Fig. 9) suggests that the status of this key cellular thiol/disulfide compound also may be an important factor. If so, this would add to data showing that SA-related signaling is compromised in mutants for chloroplast-cytosol glutathione exchange (Maughan et al., 2010) and in cat2 lines in which glutathione synthesis is partially blocked (Han et al., 2013a). It also provides further evidence that the activation of H2O2-triggered signaling inside the cell depends on a secondary modulation of antioxidant status, notably that of glutathione.
In conclusion, our study found no evidence for an essential role for any single DHAR but provided direct in planta evidence that coupling between ascorbate and glutathione is functionally important in H2O2 metabolism. DHARs seem to be dispensable for the maintenance of leaf ascorbate, even in stressful conditions. Rather, one function of these proteins may be to adjust intracellular glutathione status in oxidative stress conditions to ensure the appropriate activation of signaling through downstream pathways such as those involving SA. If so, these enzymes might more appropriately be named glutathione dehydrogenases rather than dehydroascorbate reductases.
MATERIALS AND METHODS
Plant Materials and Mutant Characterization
All the Arabidopsis (Arabidopsis thaliana) mutants used in this study were in the Col-0 genetic background, and all seeds were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info). Lines carrying T-DNA insertions in the DHAR1 (At1g19570), DHAR2 (At1g75270), and DHAR3 (At5g16710) genes were identified using insertion mutant information obtained from the SIGnAL Web site (http://signal.salk.edu). While T-DNA lines for DHAR1 and DHAR2 were obtained from the SALK collection (SALK_005238 and SALK_026089, respectively), the T-DNA line for DHAR3 was from the SAIL collection (SAIL_435_A09). The cat2 mutant was cat2-1 (Queval et al., 2009). From these lines, the following resources were produced by crossing: dhar1 dhar2, dhar1 dhar3, dhar2 dhar3, dhar1 dhar2 dhar3, cat2 dhar1, cat2 dhar2, cat2 dhar3, cat2 dhar1 dhar2, cat2 dhar1 dhar3, cat2 dhar2 dhar3, and cat2 dhar1 dhar2 dhar3. After verification of heterozygotes in F1 plants by PCR, double, triple, and quadruple homozygotes were identified similarly in the F2 generation and allowed to produce F3 seeds, which were used for experiments. Expression analyses were performed by semiquantitative reverse transcription-PCR. Total RNA was extracted with TRIzol reagent (Invitrogen) following the manufacturer’s instructions. RT and first-strand cDNA synthesis were performed using the SuperScript III First-Strand Synthesis System (Invitrogen). ACTIN2 transcripts were measured as a loading control. Primer sequences are listed in Supplemental Table S6.
Plasmid Constructs and Plant Transformation
Full-length cDNA clones of Arabidopsis DHARs were obtained from the RIKEN BioResource Center Arabidopsis full-length cDNA collection (https://www.brc.riken.jp). The cDNA clones encoding DHAR1 (RAFL19-03-N19), DHAR2 (RAFL09-83-E23), and DHAR3 (RAFL19-84-K20) were moved as SalI-NotI fragments into the vector pENTR2b (Invitrogen) and sequenced, using primer pairs listed in Supplemental Table S6. An LR reaction was then used to transfer cDNAs from the resulting entry vectors to the appropriate Gateway binary destination vectors fused according to the manufacturer’s instructions (Invitrogen). The pB7WGF2 vector was used for N-terminal GFP fusion, and the pH7FWG2 vector was used for C-terminal GFP fusion (Karimi et al., 2002). In these constructs, the expression of DHAR1, DHAR2, or DHAR3 was driven by the 35S promoter of Cauliflower mosaic virus. Purified plasmids were analyzed and sequenced to confirm successful fusion constructs. Agrobacterium tumefaciens strain GV3101 pMP90 (Koncz and Schell, 1986) was transformed with confirmed binary vector constructs (Höfgen and Willmitzer, 1988) and used to transform developing floral tissues of 4-week-old Arabidopsis triple dhar1 dhar2 dhar3 and quadruple cat2 dhar1 dhar2 dhar3 mutant plants using the floral dip method (Clough and Bent, 1998).
Subcellular Localization and Genetic Complementation
Ten-day-old seedlings of the triple dhar1 dhar2 dhar3 and quadruple cat2 dhar1 dhar2 dhar3 mutants were stably transformed with DHAR1-GFP or GFP-DHAR1, DHAR2-GFP or GFP-DHAR2, and DHAR3-GFP or GFP-DHAR3. Transformed T1 seeds were germinated on agar and identified based on their resistance to hygromycin or were screened using a Leica dissecting microscope equipped with a mercury lamp and epifluorescence filter set (Karimi et al., 2002). Resistant and PCR-positive transgenic plants were transferred to a growth chamber and maintained up to the T2 generation, when segregation analysis was performed on complemented cat2 dhar1 dhar2 dhar3 lines. Two independent lines were selected and used for the analysis of enzyme activities, glutathione, and ascorbate.
To determine subcellular localization, leaf sections from T1 transformed plants were mounted onto a slide in water and imaging was performed with a Leica SP8 scanning confocal microscope using a 63× PLAN APO oil-immersion objective (numerical aperture, 1.4). GFP was excited with a 488-nm laser, and fluorescence was detected in a 493- to 530-nm detection window. As a chloroplast marker, chlorophyll autofluorescence was excited at 633 nm and detected at 698 to 748 nm. As peroxisome and dual chloroplast-mitochondrion markers, lines expressing GFP-MFP2 (A5 line; Cutler et al., 2000) or PDF1B-GFP (Giglione et al., 2000) were used. To check for possible mitochondrial localization, transformants were stained with the mitochondrion-selective probe MitoTracker Red (fluorescent dye; CMTMRos; Invitrogen) following the instruction manual. Fluorescence was excited with a 578-nm laser and detected in a 582- to 609-nm detection window. Both detection channels were recorded using hybrid detectors (Hamamatsu) with sequential acquisition to avoid any cross talk.
Plant Growth and Stress Treatments
For plants grown on soil, seeds were first incubated for 2 d at 4°C and then transferred to a controlled-environment growth chamber in a 16-h photoperiod and an irradiance of 200 μmol m−2 s−1 at leaf level, 20°C/18°C, 65% humidity, and given nutrient solution twice per week. Unless stated otherwise, plants were sampled after 3 weeks of growth. Samples were rapidly frozen in liquid nitrogen and stored at −80°C until analysis. All data are means ± se of at least three biological replicates obtained from different plants, and experiments were repeated at least twice.
For in vitro stress treatments, seeds were sown on commercially available 0.5× Murashige and Skoog medium (basal salt mixture; M0221; Duchefa) solidified with 0.8% agar (Phyto-Agar HP696; Kalys) and grown in a 16-h-light/8-h-dark, 20°C growth chamber without sugar. Plants were grown vertically for 10 d on medium supplemented with 0, 10, or 50 μm CdCl2, 50 or 100 mm NaCl, 100 or 200 mm mannitol, and 0.01 or 0.1 µm paraquat. For 3-AT treatment, seeds were sown on soil as described above. After 3 weeks, plant leaves were sprayed once with 2 mm 3-AT, then leaf samples were collected after 48 h and immediately frozen with liquid nitrogen for further analysis. To assess resistance to a virulent bacterium, plants grown in soil as above were inoculated with Pseudomonas syringae pv tomato strain DC3000 as described by Chaouch et al. (2012) using a bacterial titer of 106 colony-forming units mL−1. Samples were taken for counting immediately after infiltration (0 h) or 48 h later. Quadruplicate biological samples, each of two leaf discs, were used.
Lesion Quantification, Trypan Blue Staining, and Pathogen Tests
Percentage lesion area in cat2, or bleaching induced by 3-AT treatment, was quantified using IQmaterials software. For Trypan Blue staining, rosette leaves were infiltrated three times under vacuum with lactophenol trypan solution (2.5 mg mL−1 Trypan Blue, 25% lactic acid, 23% water-saturated phenol, 25% glycerol, and water). Samples were then washed with distilled water and heated over boiling water. After cooling, samples were cleared with a chloral hydrate solution (8 g of chloral hydrate dissolved in 2 mL of 50% glycerol and 1 mL of water). The solution was replaced several times. Samples were mounted in chloral hydrate solution and observed with a macroscope (Nikon AZ100 Multizoom).
Transcript Quantification
Total RNA was extracted with TRIzol (Invitrogen) following the manufacturer’s instructions. RNA quality was determined by gel electrophoresis, and concentration was estimated using a Nanodrop spectrophotometer at 260 nm. RT and first-strand cDNA synthesis were performed using the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR was performed according to Queval et al. (2009). Primer sequences are listed in Supplemental Table S6.
Enzyme Assays and Metabolite Analysis
Extractable enzyme activities were measured according to protocols detailed by Noctor et al. (2016). Briefly, catalase was measured by the removal of H2O2 monitored at 240 nm, DHAR as GSH-dependent formation of ascorbate from DHA at 265 nm, APX as H2O2-dependent ascorbate oxidation at 290 nm, and GR as GSSG-dependent NADPH oxidation at 340 nm. Oxidized and reduced forms of glutathione and ascorbate were measured by plate-reader assay as described by Queval and Noctor (2007). Total SA was measured according to the protocol of Chaouch et al. (2010).
Accession Numbers
Sequences of genes studied in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following gene identifier codes: At1g19570 (DHAR1), At1g75270 (DHAR2), At5g16710 (DHAR3), At4g35090 (CAT2), At2g14610 (PR1), At3g57260 (PR2), At1g74710 (ICS1), At3g18780 (ACT2), and At4g36800 (RCE1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genotyping and analysis of transcript abundance in dhar mutant lines.
Supplemental Figure S2. Photographs of dhar mutants after 3 weeks of growth.
Supplemental Figure S3. Effects of various stresses on root growth in dhar mutants.
Supplemental Figure S4. Subcellular localization of DHAR1 and DHAR3 in roots.
Supplemental Figure S5. Subcellular localization of DHARs in the absence of the cat2 mutation.
Supplemental Figure S6. Comparison of DHAR1-GFP signals in leaves with a positive control for peroxisomal targeting.
Supplemental Figure S7. Phenotypes of Col-0 and dhar mutants 2 d after spraying with 3-AT.
Supplemental Figure S8. Effect of dhar1 dhar2 dhar3 triple mutation on resistance to P. syringae pv tomato DC3000 in the Col-0 and cat2 backgrounds.
Supplemental Table S1. Ascorbate and glutathione redox states in Col-0 and dhar mutants.
Supplemental Table S2. Ascorbate and glutathione redox states in Col-0, cat2, and cat2 dhar mutants.
Supplemental Table S3. T2 segregation of DHAR-complemented lines growing in vitro on agar.
Supplemental Table S4. T2 segregation of lesion phenotypes in complemented lines growing in soil.
Supplemental Table S5. Glutathione redox states in Col-0 and dhar mutants after treatment with 3-AT.
Supplemental Table S6. DNA primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, the Nottingham Arabidopsis Stock Centre for supplying seed stocks, and RIKEN for cDNA clones; Alison Baker (University of Leeds) and Cécile Raynaud (Institute of Plant Sciences Paris-Saclay) for supplying reporter lines for control GFP experiments; Myroslawa Miginiac-Maslow for discussions and comments on the article in preparation; and Jean-Claude Pasquet for valuable practical advice and help.
Glossary
- ROS
reactive oxygen species
- DHA
dehydroascorbate
- GSH
reduced (thiol) glutathione
- GSSG
glutathione disulfide
- Col-0
Columbia-0
- 3-AT
3-amino-1,2,4-triazole
- SA
salicylic acid
Footnotes
This work was supported by the Agence Nationale de la Recherche (grant Cynthiol to G.N.), by the Doctoral School ED129 Sciences de l’Environnement d’Ile de France (Bourse Ministérielle Ph.D. studentship to M-S.R.), and by Michel Dron (Institute of Plant Sciences Paris-Saclay) and the Université de Paris Sud (funding toward Ph.D. studies for S.L.).
Articles can be viewed without a subscription.
References
- Anderson JW, Foyer CH, Walker DA (1983) Light-dependent reduction of dehydroascorbate and uptake of exogenous ascorbate by spinach chloroplasts. Planta 158: 442–450 [DOI] [PubMed] [Google Scholar]
- Awad J, Stotz HU, Fekete A, Krischke M, Engert C, Havaux M, Berger S, Mueller MJ (2015) 2-Cysteine peroxiredoxins and thylakoid ascorbate peroxidase create a water-water cycle that is essential to protect the photosynthetic apparatus under high light stress conditions. Plant Physiol 167: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Sun H, Yang H, Wang X, Su Z, Chen F, Wei W (2017) Over-expression of dehydroascorbate reductase enhances oxidative stress tolerance in tobacco. Electron J Biotechnol 25: 1–8 [Google Scholar]
- Chaouch S, Queval G, Noctor G (2012) AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J 69: 613–627 [DOI] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Vanderauwera S, Mhamdi A, Vandorpe M, Langlois-Meurinne M, Van Breusegem F, Saindrenan P, Noctor G (2010) Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol 153: 1692–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gallie DR (2004) The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 16: 1143–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gallie DR (2006) Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol 142: 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Young TE, Ling J, Chang SC, Gallie DR (2003) Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc Natl Acad Sci USA 100: 3525–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O, Whelan J, Millar AH (2003) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278: 46869–46877 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Creissen G, Reynolds H, Xue Y, Mullineaux P (1995) Simultaneous targeting of pea glutathione reductase and of a bacterial fusion protein to chloroplasts and mitochondria in transgenic tobacco. Plant J 8: 167–175 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Mittler R, Noctor G (2016) Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol 171: 1535–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Davis BG, Edwards R (2002) Functional divergence in the glutathione transferase superfamily in plants: identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem 277: 30859–30869 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Edwards R (2010) Glutathione transferases. The Arabidopsis Book 8: e0131, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Hawkins T, Hussey PJ, Edwards R (2009) Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot 60: 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Halliwell B (1976) The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta 133: 21–25 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Halliwell B (1977) Purification and properties of dehydroascorbate reductase from spinach leaves. Phytochemistry 16: 1347–1350 [Google Scholar]
- Foyer CH, Mullineaux PM (1998) The presence of dehydroascorbate and dehydroascorbate reductase in plant tissues. FEBS Lett 425: 528–529 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2016) Stress-triggered redox signalling: what’s in pROSpect? Plant Cell Environ 39: 951–964 [DOI] [PubMed] [Google Scholar]
- Gest N, Garchery C, Gautier H, Jiménez A, Stevens R (2013) Light-dependent regulation of ascorbate in tomato by a monodehydroascorbate reductase localized in peroxisomes and the cytosol. Plant Biotechnol J 11: 344–354 [DOI] [PubMed] [Google Scholar]
- Giglione C, Serero A, Pierre M, Boisson B, Meinnel T (2000) Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J 19: 5916–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G (2013a) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid Redox Signal 18: 2106–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Mhamdi A, Chaouch S, Noctor G (2013b) Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ 36: 1135–1146 [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Asada K (1984) Purification of dehydroascorbate reductase from spinach and its characterisation as a thiol enzyme. Plant Cell Physiol 25: 85–92 [Google Scholar]
- Jiménez A, Hernández JA, Del Río LA, Sevilla F (1997) Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol 114: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston EJ, Rylott EL, Beynon E, Lorenz A, Chechik V, Bruce NC (2015) Monodehydroascorbate reductase mediates TNT toxicity in plants. Science 349: 1072–1075 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kataya AR, Reumann S (2010) Arabidopsis glutathione reductase 1 is dually targeted to peroxisomes and the cytosol. Plant Signal Behav 5: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Urano J, Maki Y, Ushimaru T (1997) Purification and characterisation of dehydroascorbate reductase from rice. Plant Cell Physiol 38: 173–178 [Google Scholar]
- Koncz C, Schell J (1986) The promoter of Ti-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kwon SY, Ahn YO, Lee HS, Kwak SS (2001) Biochemical characterization of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J Biochem Mol Biol 34: 316–321 [Google Scholar]
- Kwon SY, Choi SM, Ahn YO, Lee HS, Lee HB, Park YM, Kwak SS (2003) Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J Plant Physiol 160: 347–353 [DOI] [PubMed] [Google Scholar]
- Lallement PA, Roret T, Tsan P, Gualberto JM, Girardet JM, Didierjean C, Rouhier N, Hecker A (2016) Insights into ascorbate regeneration in plants: investigating the redox and structural properties of dehydroascorbate reductases from Populus trichocarpa. Biochem J 473: 717–731 [DOI] [PubMed] [Google Scholar]
- Le Martret B, Poage M, Shiel K, Nugent GD, Dix PJ (2011) Tobacco chloroplast transformants expressing genes encoding dehydroascorbate reductase, glutathione reductase, and glutathione-S-transferase, exhibit altered anti-oxidant metabolism and improved abiotic stress tolerance. Plant Biotechnol J 9: 661–673 [DOI] [PubMed] [Google Scholar]
- Marmagne A, Ferro M, Meinnel T, Bruley C, Kuhn L, Garin J, Barbier-Brygoo H, Ephritikhine G (2007) A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol Cell Proteomics 6: 1980–1996 [DOI] [PubMed] [Google Scholar]
- Maughan SC, Pasternak M, Cairns N, Kiddle G, Brach T, Jarvis R, Haas F, Nieuwland J, Lim B, Müller C, et al. (2010) Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc Natl Acad Sci USA 107: 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ, Leaver CJ (1993) Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol 103: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat Y, Saindrenan P, Issakidis-Bourguet E, Gouia H, Renou JP, et al. (2010a) Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G (2010b) Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61: 4197–4220 [DOI] [PubMed] [Google Scholar]
- Miyaji T, Kuromori T, Takeuchi Y, Yamaji N, Yokosho K, Shimazawa A, Sugimoto E, Omote H, Ma JF, Shinozaki K, et al. (2015) AtPHT4;4 is a chloroplast-localized ascorbate transporter in Arabidopsis. Nat Commun 6: 5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell S, Follmann H, De Tullio M, Häberlein I (1997) Dehydroascorbate and dehydroascorbate reductase are phantom indicators of oxidative stress in plants. FEBS Lett 414: 567–570 [DOI] [PubMed] [Google Scholar]
- Noctor G. (2015) Lighting the fuse on toxic TNT. Science 349: 1052–1053 [DOI] [PubMed] [Google Scholar]
- Noctor G, Lelarge-Trouverie C, Mhamdi A (2015) The metabolomics of oxidative stress. Phytochemistry 112: 33–53 [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Foyer CH (2016) Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant Cell Environ 39: 1140–1160 [DOI] [PubMed] [Google Scholar]
- Noctor G, Veljovic-Jovanovic S, Foyer CH (2000) Peroxide processing in photosynthesis: antioxidant coupling and redox signalling. Philos Trans R Soc Lond B Biol Sci 355: 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noshi M, Hatanaka R, Tanabe N, Terai Y, Maruta T, Shigeoka S (2016) Redox regulation of ascorbate and glutathione by a chloroplastic dehydroascorbate reductase is required for high-light stress tolerance in Arabidopsis. Biosci Biotechnol Biochem 80: 870–877 [DOI] [PubMed] [Google Scholar]
- Noshi M, Yamada H, Hatanaka R, Tanabe N, Tamoi M, Shigeoka S (2017) Arabidopsis dehydroascorbate reductase 1 and 2 modulate redox states of ascorbate-glutathione cycle in the cytosol in response to photooxidative stress. Biosci Biotechnol Biochem 81: 523–533 [DOI] [PubMed] [Google Scholar]
- Parsons HT, Fry SC (2012) Oxidation of dehydroascorbic acid and 2,3-diketogulonate under plant apoplastic conditions. Phytochemistry 75: 41–49 [DOI] [PubMed] [Google Scholar]
- Polle A. (2001) Dissecting the superoxide dismutase-ascorbate-glutathione-pathway in chloroplasts by metabolic modeling: computer simulations as a step towards flux analysis. Plant Physiol 126: 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Jaillard D, Zechmann B, Noctor G (2011) Increased intracellular H2O2 availability preferentially drives glutathione accumulation in vacuoles and chloroplasts. Plant Cell Environ 34: 21–32 [DOI] [PubMed] [Google Scholar]
- Queval G, Noctor G (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Queval G, Thominet D, Vanacker H, Miginiac-Maslow M, Gakière B, Noctor G (2009) H2O2-activated up-regulation of glutathione in Arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol Plant 2: 344–356 [DOI] [PubMed] [Google Scholar]
- Rahantaniaina MS, Tuzet A, Mhamdi A, Noctor G (2013) Missing links in understanding redox signaling via thiol/disulfide modulation: how is glutathione oxidized in plants? Front Plant Sci 4: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, Linka N, Switzenberg R, Wilkerson CG, Weber AP, et al. (2009) In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 150: 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP (2002) Glutaredoxin-dependent peroxiredoxin from poplar: protein-protein interaction and catalytic mechanism. J Biol Chem 277: 13609–13614 [DOI] [PubMed] [Google Scholar]
- Shimaoka T, Yokota A, Miyake C (2000) Purification and characterization of chloroplast dehydroascorbate reductase from spinach leaves. Plant Cell Physiol 41: 1110–1118 [DOI] [PubMed] [Google Scholar]
- Smirnoff N. (2011) Vitamin C: the metabolism and functions of ascorbic acid in plants. Adv Bot Res 59: 107–177 [Google Scholar]
- Smith IK, Kendall AC, Keys AJ, Turner JC, Lea PJ (1984) Increased levels of glutathione in a catalase-deficient mutant of barley (Hordeum vulgare L.). Plant Sci Lett 37: 29–33 [Google Scholar]
- Tang ZX, Yang HL (2013) Functional divergence and catalytic properties of dehydroascorbate reductase family proteins from Populus tomentosa. Mol Biol Rep 40: 5105–5114 [DOI] [PubMed] [Google Scholar]
- Tripathi BN, Bhatt I, Dietz KJ (2009) Peroxiredoxins: a less studied component of hydrogen peroxide detoxification in photosynthetic organisms. Protoplasma 235: 3–15 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Suzuki N, Miller G, van de Cotte B, Morsa S, Ravanat JL, Hegie A, Triantaphylidès C, Shulaev V, Van Montagu MCE, et al. (2011) Extranuclear protection of chromosomal DNA from oxidative stress. Proc Natl Acad Sci USA 108: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xiao Y, Chen W, Tang K, Zhang L (2010) Increased vitamin C content accompanied by an enhanced recycling pathway confers oxidative stress tolerance in Arabidopsis. J Integr Plant Biol 52: 400–409 [DOI] [PubMed] [Google Scholar]
- Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W (1997) Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J 16: 4806–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang S, Eltayeb AE, Uddin MI, Yamamoto Y, Tsuji W, Takeuchi Y, Tanaka K (2010) Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 231: 609–621 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Tamaoki M, Shikano T, Nakajima N, Ogawa D, Ioki M, Aono M, Kubo A, Kamada H, Inoue Y, et al. (2006) Cytosolic dehydroascorbate reductase is important for ozone tolerance in Arabidopsis thaliana. Plant Cell Physiol 47: 304–308 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Wang W, Yang HL, Li Y, Kang XY, Wang XR, Yang ZL (2015) Molecular properties and functional divergence of the dehydroascorbate reductase gene family in lower and higher plants. PLoS ONE 10: e0145038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.