The hypomorphic mutant speechless-5 reveals a brassinosteroid-dependent mechanism for stomata formation and the involvement of the bHLH domain of SPEECHLESS in proper expression of some target genes.
Abstract
The asymmetric cell divisions necessary for stomatal lineage initiation and progression in Arabidopsis (Arabidopsis thaliana) require the function of the basic helix-loop-helix (bHLH) transcription factor SPEECHLESS (SPCH). Mutants lacking SPCH do not produce stomata or lineages. Here, we isolated a new spch-5 allele carrying a point mutation in the bHLH domain that displayed normal growth, but had an extremely low number of sometimes clustered stomata in the leaves, whereas the hypocotyls did not have any stomata. In vivo tracking of leaf epidermal cell divisions, combined with marker lines and genetic analysis, showed that the spch-5 leaf phenotype is dosage dependent and results from the decreased ability to initiate and amplify lineages, defects in asymmetric cell fate allocation, and misorientation of asymmetric division planes. Notably, application of brassinosteroids (BRs) partly rescued the stomatal leaf phenotype of spch-5. Transcriptomic analysis combining spch-5 with BR treatments revealed that the expression of a set of SPCH target genes was restored by BRs. Our results also show that BR-dependent stomata formation and expression of some, but not all, SPCH target genes require the integrity of the bHLH domain of SPCH.
Stomata, which capture carbon dioxide for photosynthesis while controlling excessive water loss, are set through a genetic program connected to environmental and physiological factors (Raven, 2002; Casson and Hetherington, 2010; Dow et al., 2014). Stomatal development in Arabidopsis (Arabidopsis thaliana) takes place through a stereotyped cell division and differentiation pathway that also ensures proper patterning (Supplemental Fig. S1). Current understanding of the process involves the signaling peptides EPIDERMAL PATTERNING FACTORs (EPFs) with both positive and negative downstream effects (Torii, 2015). The peptides are perceived by receptor complexes that contain several combinations of receptor-like kinases (RLKs) and TOO MANY MOUTHS (TMM), a receptor-like protein that lacks the kinase domain (Meng et al., 2015; Han and Torii, 2016). The signal is sent through a mitogen-activated protein kinase (MAPK) phosphorylation cascade led by the MAPKKK YODA that finally affects the activity of three related basic helix-loop-helix (bHLH)-type transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA (Lampard et al., 2009; Meng et al., 2015; Simmons and Bergmann, 2016). These bHLH proteins act as positive drivers of cell divisions and fate decisions from the initiation of stomatal lineages to their completion with the formation of two guard cells (Pillitteri and Torii, 2007), cooperating with the paralogous proteins INDUCER OF CBF EXPRESSION1 (ICE1), which is also known as SCREAM (ICE1/SCRM), and SCRM2, probably through heterodimerization (Kanaoka et al., 2008). SPCH is crucial for the entry asymmetric cell divisions (ACDs) that create the stomatal lineages and is also required for the subsequent amplification ACDs, leading to an increased number of cells in the lineages (and, thus, in the leaf epidermis) and for the spacing ACDs that prevent the differentiation of stomatal clusters during satellite lineage formation. SPCH activity is negatively regulated by phosphorylation through the MAPK cascade led by YODA (MacAlister et al., 2007; Lampard et al., 2008). SPCH activity is also influenced by brassinosteroids (BRs) via BIN2-mediated phosphorylation and inactivation of both SPCH (Gudesblat et al., 2012) and the YODA-MAPK pathway (Kim et al., 2012; Khan et al., 2013). Whereas SPCH phosphorylation by BIN2 results in stomata repression, the inactivation of the YODA cascade releases repression; thus, it promotes stomata formation. Thus, BRs have opposite effects on the SPCH activity, and the output may depend on how prominent each of the two pathways is, as both promotion and repression have been reported in hypocotyls (Gudesblat et al., 2012; Wang et al., 2015) and in cotyledons and leaves (Kim et al., 2012; Khan et al., 2013).
By means of chromatin immunoprecipitation sequencing (ChIP-seq) combined with transcriptomics, various genes directly regulated by SPCH have been described (Lau et al., 2014), but, to date, no direct DNA binding for SPCH, alone or with other partners, has been demonstrated. The central role of SPCH in stomata development has been underlined by recent models proposing that SPCH creates a transcriptionally positive feedback loop with ICE1, up-regulating the expression of the signaling peptide EPF2 and TMM, whereas the RLK ERECTA is outside the loop (Horst et al., 2015). In this pathway, EPF2 would buffer the positive feedback loop of SPCH/ICE1 heterodimers and its high diffusivity would spread lateral inhibitory effects to neighbor cells around stomatal precursors, guaranteeing strictly localized SPCH accumulation (Horst et al., 2015). The detailed developmental processes and mechanisms involving SPCH are still not totally understood, partly because loss-of-function SPCH mutants do not initiate lineages, because they are defective in entry ACDs. The involvement of SPCH in amplification and spacing divisions was based on the weak allele spch-2 (MacAlister et al., 2007), highlighting the potential of non-null mutants to discover gene functions.
Here, we isolated and characterized a hypomorphic SPCH allele, spch-5. This mutant carries a mutation within the bHLH domain of the protein and displays several stomatal pattern defects, such as strongly decreased stomatal and epidermal cell production, stomatal clustering, increased pavement cell size, lack of physical and fate asymmetry in the stomatal lineage divisions, misorientation of the division planes, and widespread transcriptional changes. Notably, BR treatment reverted the low stomatal numbers of spch-5 and, partly, the clustering phenotype and transcriptomic signature of the mutant. Combining genetic, biochemical, and transcriptomic approaches with transactivation assays, we found possible molecular components and mechanisms involved in SPCH functions that are dependent or independent on the integrity of its bHLH domain, suggesting that direct DNA binding might be needed only for some of them.
RESULTS
Identification of the spch-5 Mutant
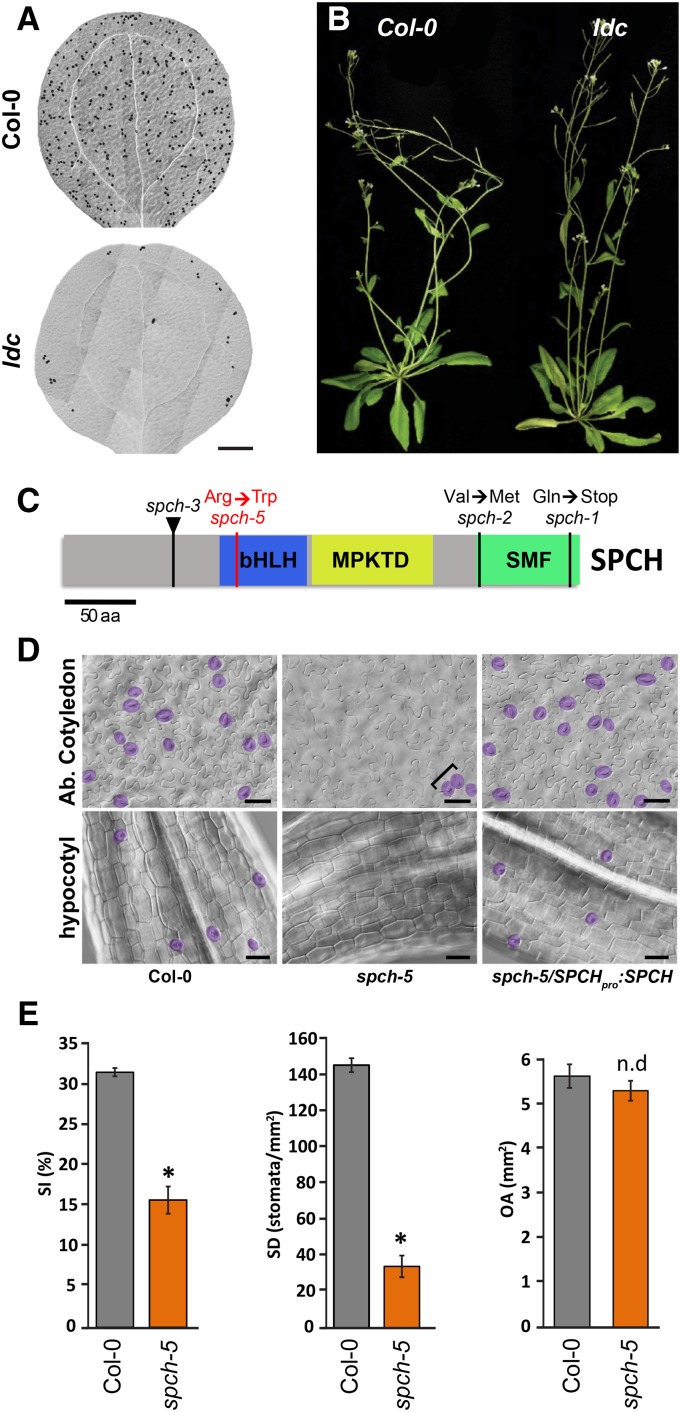
To identify regulators of stomatal patterning, we screened EMS-mutagenized Arabidopsis Columbia-0 (Col-0) seeds for aberrant epidermal phenotypes in the cotyledons of M2 seedlings. We isolated the low density and clustering (ldc) mutant that had a strikingly low stomatal density in both adaxial and abaxial cotyledon epidermis and occasional stomatal clusters (Fig. 1A; Supplemental Fig. S2). Homozygous ldc plants were fertile, grew normally, and looked morphologically similar to the wild-type Col-0 plants (Fig. 1B). Genetic analysis indicated that ldc is a recessive single-gene mutant because the epidermis of the F1 plants from a cross between ldc and Col-0 was normal and the F2 progeny segregated wild type to ldc phenotypes according to the expected 3-to-1 ratio (n = 40; χ2 significance level of 5%). Genetic mapping revealed a complete linkage between the gene responsible for the ldc phenotype and the marker MNB8 in chromosome 5 (Supplemental Fig. S3A), located at 58 kb of SPCH. We sequenced the SPCH coding region in ldc plants and identified a C/T change at position 331 that generated an Arg/Trp substitution at position 111, in the basic region of the bHLH domain (Fig. 1C; Supplemental Fig. S3B). Transformation of ldc with a SPCHpro:SPCH construct fully rescued the mutant phenotype (Fig. 1D). In addition, crossing homozygous ldc plants with plants heterozygous for the spch-3 null allele (MacAlister et al., 2007) created heterozygous ldc/spch-3 individuals that presented a very low stomatal abundance. Altogether, these results confirmed that the mutation in the SPCH locus is the cause of the ldc phenotype. Therefore, we will hereafter refer to this ldc mutation as spch-5.
Figure 1.
Phenotype of ldc mutants and molecular identification of the spch-5 mutation. A, DIC images of entire 15-d-old abaxial cotyledons of Col-0 and ldc. B, Growth phenotypes of 40-d-old Col-0 and ldc plants. C, Diagram of SPCH showing the spch-5 mutation (red) causing an Arg-to-Trp change within the bHLH domain (blue box). Other alleles have point mutations outside the bHLH domain (spch-1 and spch-2) or T-DNA insertions (spch-3; black triangle). D, DIC micrographs showing the abaxial epidermis of 23-d-old cotyledons and 10-d-old hypocotyls in Col-0, spch-5, and the complemented spch-5/SPCHpro:SPCH-GFP line. Stomata are false colored in purple for easier identification. Stomatal clusters are marked by brackets. E, Graphs representing stomatal index, stomatal density, and organ area in the abaxial epidermis of 23-d-old cotyledons. Gray and orange bars denote Col-0 and spch-5, respectively. Asterisks indicate P < 0.05 (Student’s t test; n = 10), and n.d indicates P > 0.05. Error bars represent se. Bars = 0.5 mm in A and 50 µm in D.
The spch-5 Epidermal Phenotype Differs among Organs
Next, stomatal abundance and patterning were examined in different plant organs. As measured by the stomatal index (SI; number of stomata/total number of epidermal cells × 100) and the stomatal density (SD; number of stomata per mm2), the number of stomata produced in the abaxial cotyledon epidermis of spch-5 was largely reduced, with 50% and 23% lower SI and SD than those of the Col-0 (Fig. 1E). Despite the stomatal scarcity, 29% of the stomata in spch-5 appeared in clusters, most of them formed by two stomata and rarely by three or more (29% ± 5% in spch-5 compared to 0.6% ± 0.3% in Col-0; n = 10; mean ± se).
The spch-5 mutant did not form ectopic stomata in normally stomataless organs, such as petals and stamen filaments. In sepals, pedicels, siliques, and stamens, stomata were present, with occasional patterning mistakes in sepals (Supplemental Fig. S4). Cylindrical organs were severely affected in the mutant; notably, spch-5 hypocotyls lacked stomata (Fig. 1D; no stomata found in n = 10), and spch-5 stems barely produced stomata compared to the wild type (Supplemental Fig. S4). In the abaxial epidermis of the third leaf, the SD of spch-5 was ∼54% lower than that of the wild type, although the SI was the same in the two genotypes (Supplemental Fig. S5, A and B). Interestingly, the size of the cotyledons and the third leaf was unaffected in spch-5 plants (Fig. 1E; Supplemental Fig. S5B). We calculated the pavement cell density (PCD) for the abaxial epidermis of the third leaf. In spch-5, the PCD value was half that in Col-0 (Supplemental Fig. S5B). The size of individual pavement cells was also measured. The mutant epidermis had an overrepresentation of large cells compared to the wild type (Supplemental Fig. S5, C and D). We estimated the number of total stomata and pavement cells from previous parameters (SD, PCD, and organ area) in the abaxial epidermis of the third leaf and the cotyledon. In both organs, the pavement cell number was much higher in Col-0 than in spch-5 (33,336 versus 18,046 in the leaf and 1,791 versus 927 in cotyledons, respectively). The total number of stomata in the leaf was 10,216 and 5,565 in the wild type and mutant plants, respectively. In mutant cotyledons, the number dramatically diminished to 183 stomata compared to 828 in the wild type. Thus, the spch-5 mutation reduces the number of stomata and pavement cells in the epidermis without impairing the mechanisms that compensate pavement cell number and size (Hisanaga et al., 2015).
Developmental Bases of the spch-5 Phenotype
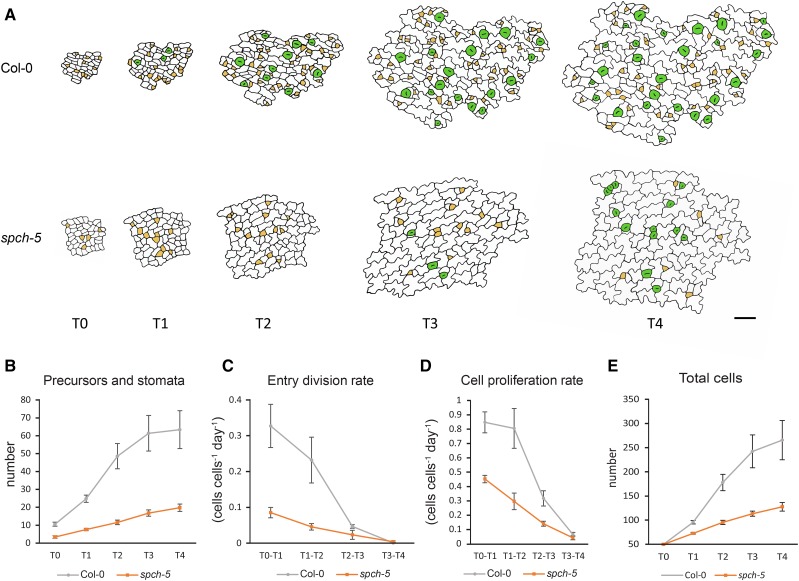
Because SPCH directs the entry, spacing, and amplifying ACDs during stomatal formation (MacAlister et al., 2007), we assessed whether the spch-5 mutant had alterations in any of these divisions. To this end, we studied the kinetics of cell division and differentiation during stomatal lineage progression in the abaxial epidermis of the third leaf of spch-5 and Col-0. Epidermal imprints were obtained from the same primordium at 24-h intervals (T0 through T4), registering, for each time point, the number of stomata and stomata precursors (meristemoids plus guard mother cells), the number of total epidermal cells, and the number of entry divisions (Fig. 2). The spch-5 mutation did not affect the persistence of protodermal cells competent to initiate stomatal lineages in the developing epidermis because at T0, both genotypes presented some stomatal precursors that augmented in the following 4 d. Stomata were first detected in Col-0 at T1, but did not appear consistently until T3 in spch-5 (Fig. 2A). The number of stomatal lineages (precursors plus stomata) increased gradually over time in both genotypes, albeit it was much lower in the mutant at all the time points examined (Fig. 2B). The mutant exhibited a markedly reduced frequency of entry divisions at early time points (T0 through T2), when most of the stomatal lineages were initiated in Col-0 (Fig. 2C). In Col-0, 85% of the T0 cells initiated stomatal lineages, whereas only 30% did so in spch-5 (Supplemental Fig. S6A). A similar evolution was observed for the cell proliferation rate and, consequently, the total cell number increased only limitedly in spch-5, particularly at late time points (Fig. 2, D and E). The number of stomatal lineage ground cells (SLGCs), which originate from amplifying divisions of the meristemoid, was also significantly lower in spch-5 than in Col-0 (Supplemental Fig. S6B). At the latest time point examined, spch-5 had produced only 31% of the total stomata plus precursors and 48% of the cells produced by Col-0. Surprisingly, both Col-0 and spch-5 displayed a similar proportion of primary lineages undergoing satellization (35% and 37%, respectively; Supplemental Fig. S6C), indicating that spch-5 had seemingly no impact on spacing divisions. In addition, the differentiation of guard cells in spch-5 was delayed compared to that of Col-0 (Supplemental Fig. S6D).
Figure 2.
In vivo tracking of epidermal cell divisions and stomatal differentiation in leaf primordia. A, Abaxial epidermis of third-leaf primordia in Col-0 and spch-5 plants followed in vivo with serial resin imprints. Epidermal replicas were inspected at 24-h intervals for 4 d (T0–T4). Drawings are reproductions from representative micrographs of the Col-0 and spch-5 serial imprints at the times indicated. Stomatal precursors are marked in yellow and stomata in green. B to E, Graphs of the number of stomata and stomata precursor cells (B), entry division rate (C), cell proliferation rate (D), and number of total cells (E) from leaves (n = 5) per genotype and with 50 cells at the initial field (T0) of 50 cells. Error bars represent se. Bars = 100 µm in A.
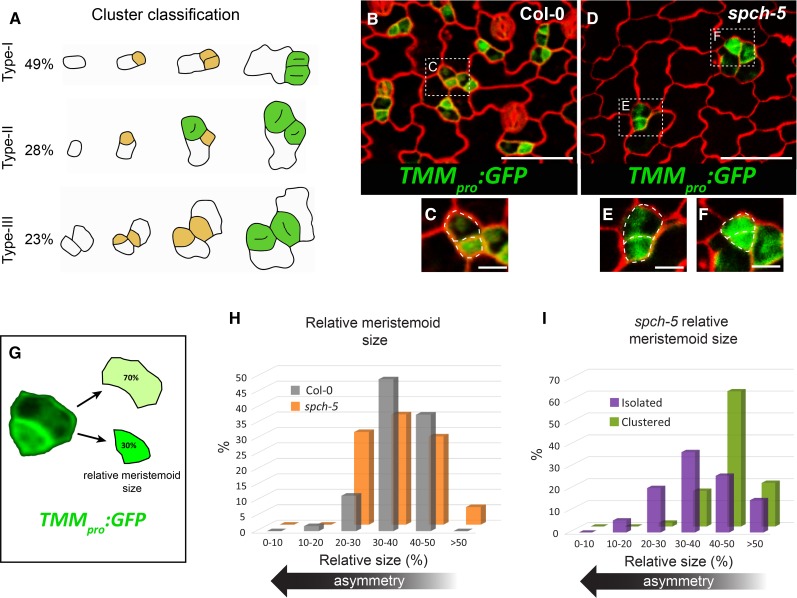
One of the most striking features of spch-5 plants was the combination of a low stomatal abundance with stomatal clustering (affecting 18% of the stomata). Thus, we determined the specific alterations in the developmental pathway by reconstructing stomatal lineage histories of 128 clusters in the same leaf imprint series. Clusters were classified and quantified based on their origin (Fig. 3A). Most clusters (49%; type-I) originated from meristemoid divisions that produced two daughter cells with the same stomatal fate. Type-II clusters (28%) arose from misoriented SLGC spacing divisions, leading to differentiation of satellite stomata adjacent to preexisting primary stomata or precursors. In type-III (23%), stomata pairs derived from different, but neighboring, primary lineages. Hence, spch-5 showed a reduced ability to assign fate asymmetry to the two meristemoid daughter cells and was defective in signaling mechanisms between stomata and/or stomatal precursors.
Figure 3.
Defective ACD in spch-5. A, Stomatal 2-mer cluster ontogeny followed through serial resin imprints as in Figure 2. The relative percentage of the three different cluster ontogenies found in spch-5 was calculated with data collected for 4 d at 24-h intervals from 128 clusters in seven plants. Drawings are representative examples reproduced from micrographs of the serial imprints. Stomatal precursors are marked in yellow and stomata in green. B and D, Confocal images of the adaxial epidermis of 3-d-old cotyledons in Col-0 (B) and spch-5 (D), carrying the marker TMMpro:GFP (green) and stained with propidium iodide (red). C, E, and F, Magnified fields from B and D. Cell outlines are indicated as dotted lines. G, Procedure to estimate relative meristemoid size as the percentage of the mother cell area that corresponds to each of the two daughter cell products of an asymmetric division. H, Graph representing the relative meristemoid size, as determined in G after lineage cell divisions in Col-0 (gray) and spch-5 (orange). I, Relative meristemoid size in spch-5 type-I clusters (green) and isolated stomata (purple) by means of serial resin imprints (see “Materials and Methods” for details). Bars = 100 µm in B and D and 20 µm in C, E, and F.
To investigate the possible causes of this lack of asymmetric fate in the most abundant type-I clusters, we used TMMpro:GFP as a marker of stomatal lineage cells (Nadeau and Sack, 2002; Fig. 3, B–F). As reported (Nadeau and Sack, 2002) in Col-0, TMMpro:GFP was highly expressed in meristemoids, at a much lower level in the youngest SLGCs, and only rarely in other lineage cells (Fig. 3, B and C). In spch-5, sometimes two adjacent small lineage cells showed an equally high GFP signal (Fig. 3, D and F), although other lineages had a proper asymmetric marker allocation (Fig. 3E). These small GFP-expressing cells appear to be the products of a recent cell division and one of them frequently exhibited a characteristically meristemoid morphology.
To assess whether spch-5 also affected the physical asymmetry of meristemoid divisions, we used Col-0 and spch-5 lines with the TMMpro:GFP marker (Fig. 3G). The mean and median relative cell area of meristemoids was very similar in both genotypes: 37% and 36% of the combined area of the two daughters (the meristemoid and its sister SLGC) in Col-0 and spch-5, respectively (Fig. 3H). However, we found differences in spch-5 between the meristemoid relative area in lineages producing either one isolated stoma or two stomata in contact (type-I clusters). When meristemoids generated daughter cells with an unequal fate (leading to isolated stomata), the size of the daughter cells was also dissimilar and the new meristemoid had a mean relative area of 38% (Fig. 3I) as previously described for Col-0 (Dong et al., 2009). In contrast, in lineages leading to type-I clusters, the smallest daughter cell had a mean relative area of 45% (Fig. 3I). These results indicate that in spch-5 the two cell products of some meristemoid divisions have a similar size and lack fate asymmetry.
To analyze whether the stomatal cluster development in spch-5 involved failures in the mechanisms that ensure the correct orientation of the division plane in ACDs, we took advantage of the regular rosette-like arrangement of lineage cells developed by mute mutants (Pillitteri et al., 2007; Triviño et al., 2013; Supplemental Fig. S7). In the mute-3 spch-5 double mutant, disorganized groups of small cells replaced the typical rosette pattern of mute-3 lineages. These small cells were stomatal lineage cells executing ACDs with misoriented division planes because their TMMpro:GFP expression varied to various extents.
Together, these data evidence that the very low stomatal numbers and moderate clustering in spch-5 result from the defective capacity for entry and amplifying divisions in the stomatal lineages, dysfunctions in the physically asymmetric divisions of meristemoids, difficulties to assign an unequal fate to the two daughter cells, and improper orientation of these divisions between and within lineages.
Molecular Basis of the spch-5 Dysfunction
The spch-5 point mutation produced a substitution of Arg R111 by a Trp in the bHLH domain (Fig. 1C; Supplemental Fig. S3B) predicted to bind the canonical G-box (CACGTG) through the conserved residues H-E-R (H104, E108, and R112 in SPCH; Supplemental Fig. S3B). Although the mutated Arg (R111) residue in SPCH-5 is not a G-box-binding residue, it is highly conserved among species (Carretero-Paulet et al., 2010), and in the E47 transcription factor of Drosophila melanogaster, it forms hydrogen bonds with the DNA backbone to stabilize the interaction of the Glu that contacts the G-box (E108 in SPCH) (Ellenberger et al., 1994). In our model of the SPCH bHLH domain, created with the human protein Myc-Max (PDB ID = 1 nkp) as template, a similar hydrogen bond between R111 and E108, the critical position for DNA binding, could be detected (Supplemental Fig. S8). Accordingly, when R111 was replaced by Trp in the SPCH-5 protein, the resulting W111 did not establish this bond with E108, theoretically compromising the DNA-binding capabilities of SPCH-5. Although ChIP-seq or ChIP-PCR with SPCH have been carried out (Lau et al., 2014; Horst et al., 2015), a direct DNA binding of SPCH could not be demonstrated. A yeast one-hybrid assay was used to test whether SPCH and SPCH-5 bind the canonical G-box motif with PHYTOCHROME-INTERACTING FACTOR4 (PIF4) as control. The results were negative (Supplemental Fig. S9), similarly as those reported for the maize (Zea mays) factor R that shares the presence of bHLH and ACT domains with SPCH (MacAlister and Bergmann, 2011; Kong et al., 2012). Deletion of the ACT domain of the R factor, but not of SPCH, allowed G-box binding (Kong et al., 2012; Supplemental Fig. S9). No interaction with the G-box was detected in yeast producing both SPCH and ICE1 proteins (Supplemental Fig. S9), suggesting that SPCH-ICE1 heterodimers do not bind the canonical G-box motif in this assay.
Mutations in the bHLH domain might impair dimerization with other transcription factors (Massari and Murre, 2000). To test dimerization capabilities in planta, we used the bimolecular fluorescence complementation (BiFC) assay in Nicotiana benthamiana leaves. GFP expression indicated heterodimer assembly with both SCRMs for SPCH and for SPCH-5 (Supplemental Fig. S10A). In contrast, no homodimer was formed for SPCH in BiFC (Supplemental Fig. S10B), as reported previously (Kanaoka et al., 2008).
Subsequently, we chose to evaluate whether SPCH-5 might have altered DNA-binding capacities. We constructed SPCHPPP, a SPCH variant with the critical DNA-contacting residues (H104, E108, and R112) substituted by prolines, a modification that impairs DNA binding in other bHLH proteins (Pace and Scholtz, 1998; Maerkl and Quake, 2009; De Masi et al., 2011). BiFC assays determined that the SPCHPPP variant was able to interact with both SCRMs in planta (Supplemental Fig. S10C). To check the capacity of this variant to sustain stomatal development, we introduced the GFP-tagged SPCH-5 or SPCHPPP driven by the SPCH promoter into the null spch-3 background (designated SPCH-5/spch-3 and SPCHPPP/spch-3, respectively) and compared several independent homozygous lines with the previously described spch-3 line fully complemented with SPCHpro:SPCH-GFP (designated SPCH/spch-3; Gudesblat et al., 2012). Like the spch-5 mutant, lines producing the SPCH-5 or SPCHPPP proteins were fertile and had a normal growth, but they formed few stomata, some of them clustered, in cotyledons and leaves, and lacked stomata in hypocotyls (Supplemental Fig. S11, A–C). The expression pattern of SPCH-GFP and the two mutant versions did not differ qualitatively in young abaxial cotyledons (Supplemental Fig. S11, D–F). Taken together, these results show that SPCH-5 and SPCHPPP lead to very similar phenotypes, indicating that these lesions in the bHLH domain of SPCH, putatively affecting the DNA binding, have functional consequences.
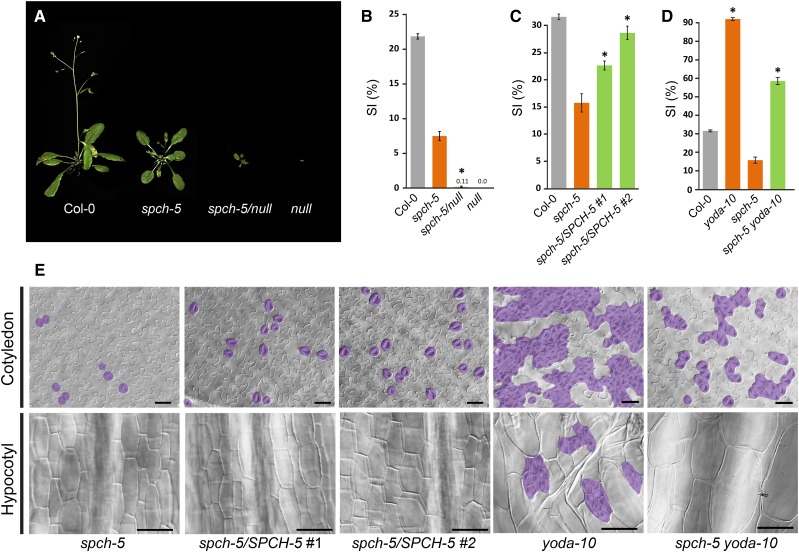
SPCH-5 Dosage Affects the Stomatal Phenotype
According to its phenotype and recessive behavior, we hypothesized that spch-5 is a partial loss-of-function (hypomorphic) allele and, as such, might have a dose effect. Hence, we crossed spch-5 plants with plants heterozygous for the null spch-3 mutant to produce plants with only one copy of the spch-5 allele (spch-5/null). Plants with only one SPCH-5 copy had a very limited growth (Fig. 4A), although they produced some seeds after prolonged cultivation, and their adaxial cotyledon epidermis barely produced stomata (Fig. 4B), implying a dosage sensitivity and corroborating the hypomorphic nature of the spch-5 allele. We also introduced extra copies of SPCH-5 by transforming spch-5 plants with the transgene SPCHpro:SPCH-5-GFP. Two double homozygous independent lines, each carrying four SPCH-5 doses, produced statistically significantly more stomata than spch-5 (Fig. 4C). Although the alleviation of the mutant phenotype was evident in abaxial cotyledons with extra copies of SPCHpro:SPCH-5, hypocotyls remained unable to produce stomata (Fig. 4E). These observations reveal that spch-5 showed a dose-dependent phenotype in cotyledons, but, in hypocotyls, that it behaves as a null mutant.
Figure 4.
SPCH-5 dosage effects on the stomatal phenotype. A, Growth phenotype of the different genotypes tested for gene dosage effects 33 d after sowing in soil. B, Adaxial stomatal index of soil-grown fully expanded cotyledons (25 d for all the genotypes, except for spch-5/null that needed 35 d because of its delayed growth). C, Abaxial SI of 23-d-old cotyledons in Col-0 (gray), spch-5 (orange), and two independent spch-5 lines carrying extra copies of the transgene SPCHpro:SPCH-5 (green). D, Abaxial SI of 23-d-old cotyledons in Col-0 (gray), spch-5 and yoda-10 single mutants (orange), and spch-5 yoda-10 double mutants (green). Asterisks in B to D indicate P < 0.05 (Student’s t test) when compared to spch-5. Error bars represent se. E, Representative DIC images of the cotyledon epidermis quantified in C and D and the 10-d-old hypocotyl epidermis of the same genotypes. Hypocotyls show clustered stomata in yoda-10 and complete absence of stomata in spch-5, two independent lines of spch-5 with two extra copies of SPCHpro:SPCH-5, and spch-5 yoda-10 double mutant. Stomata are colored in purple. Bars = 50 µm.
The amount of active SPCH-5 protein was genetically increased by eliminating YODA, a known SPCH repressor that decreases SPCH activity via phosphorylation (Bergmann et al., 2004; Lampard et al., 2008). The double spch-5 yoda-10 mutant still produced more stomata than Col-0 in cotyledons, but both SI and clustering were significantly reduced compared to those of yoda-10 (Fig. 4, D and E). Interestingly, the loss of the YODA function in a spch-5 background did not allow stomata formation in hypocotyls, in which the stomatal production remained blocked in the double mutant (Fig. 4E).
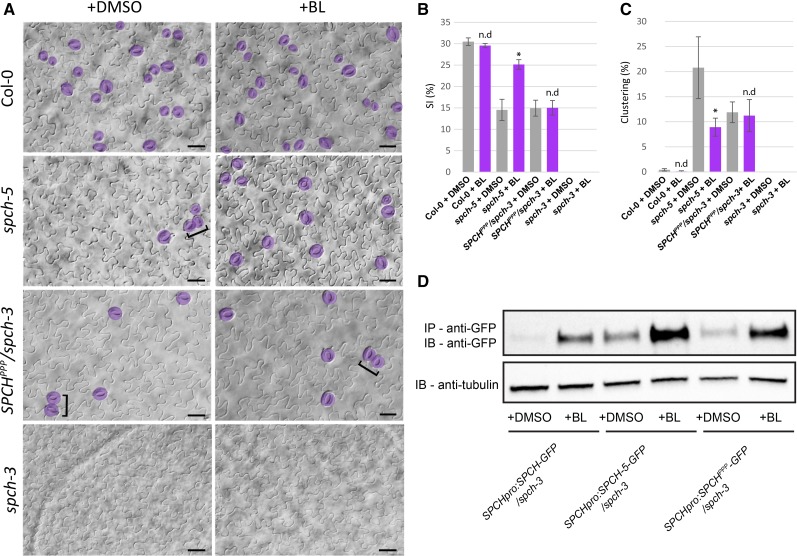
The SPCH protein can be stabilized by application of brassinolide (BL), the most active BR (Gudesblat et al., 2012). In terms of stomata production, its effect is mostly evident in hypocotyls. As reported before (Gudesblat et al., 2012), 50 nm BL had no distinctive effect on the stomatal phenotype of Col-0 cotyledons compared to plants grown in control medium (Fig. 5, A–C). In contrast, when spch-5 plants were grown on BL, the SI increased significantly and the stomatal clustering diminished (from 21% ± 6% in mock to 9% ± 2% in BL medium; Fig. 5, A–C). Remarkably, BL did not affect the epidermal phenotypes of either the spch-3 lines expressing SPCHPPP or the spch-2 mutant (Fig. 5; Supplemental Fig. S12). The spch-3 cotyledons did not produce stomata either in control or in BL-containing medium (Fig. 5, A and B), as previously reported for hypocotyls (Gudesblat et al., 2012). To determine whether the BL treatment affected the SPCH-5 accumulation in spch-5 as reported for SPCH in Col-0 (Gudesblat et al., 2012), we generated homozygous plants carrying the SPCHpro:SPCH-5-GFP fusions in a spch-3 null background. Immunoprecipitation of SPCH-5-GFP revealed that a short BL treatment remarkably increased protein accumulation compared to the quasi-undetectable protein found in nontreated plants (Fig. 5D). Hence, BL mediated the SPCH-5 stabilization, providing a possible mechanism for the partial rescue of the spch-5 phenotypes by the BL treatment. Similarly, BL treatment of SPCHpro:SPCHPPP-GFP led to protein stabilization (Fig. 5D); in contrast, the stomatal phenotype of this line was unaffected by BL (Fig. 5, A–C). Altogether, our results support the notion that, at least in cotyledons, the phenotypes associated with the dysfunctional SPCH-5 protein are highly dependent on protein dosage and that increasing the SPCH-5 amounts or activity can overcome the low stomatal production of spch-5.
Figure 5.
spch-5 epidermal phenotype alleviated by BRs through protein stabilization. A, Representative images of some plants scored in B and C, with stomata marked in purple and clusters denoted by brackets. Bars = 50 µm. B, Stomatal index of abaxial epidermis of 23-d-old cotyledons from plants grown on control medium (with DMSO as mock treatment; gray) or with 50 nm BL (purple). C, Clustering percentage in 23-d-old abaxial cotyledons of spch-5 under control medium (gray) or BL (purple). Error bars represent se. Asterisks indicate P < 0.05, and n.d indicates P > 0.05 (Student’s t test) compared to the control medium. D, Protein immunoprecipitation with anti-GFP antibody (top) in protein extracts from spch-3 plants transformed with SPCHpro:SPCH-5-GFP or SPCHpro: SPCHPPP-GFP. Antitubulin antibody used as loading control (bottom).
spch-5 and SPCHPPP/spch-3 Partially Share Transcriptomic Signatures
To better understand the molecular bases for the spch-5 phenotype, we carried out a microarray-based transcriptomic analysis with 3-d-old seedlings from wild-type (Col-0), spch-5, and SPCHPPP-complemented spch-3 plants (SPCHPPP/spch-3). Cotyledons of these young seedlings had abundant developing stomatal lineages that highly express SPCH variants (Supplemental Fig. S11, D and E). In this analysis, spch-3 was not included because at this developmental time the mutant seedlings cannot be accurately identified in segregating populations. In addition, it is well known that spch-3 lacks stomatal lineages (MacAlister et al., 2007) and that BL treatment does not change this phenotype (this work). It is also established that spch-3 does not express lineage marker genes, both through observation of promoter-reporter fusions and transcriptomic analysis (Horst et al., 2015; MacAlister et al., 2007). Given that most stomatal regulators are low-expressing genes, hereafter we defined differentially expressed genes (DEGs) as those with at least an 1.5-fold change in transcript abundance and with a P value lower than 0.05. The analysis identified 3,035 and 4,401 DEGs in the spch-5 and SPCHPPP/spch-3 lines, respectively, compared to Col-0 (Fig. 6; Supplemental Data Sets S1 and S2). The relative expression levels for a panel of genes was further tested by qPCR, and the results showed similar expression tendencies for all 10 genes as estimated by the microarray analysis (Supplemental Fig. S13). The Gene Ontology analysis with ClueGO (Bindea et al., 2009) revealed that the functional categories overrepresented in the spch-5 down-regulated genes were cell division and expansion, microtubule-associated processes, hormone responses, and His kinases (Supplemental Fig. S14). No categories for metabolic processes were overrepresented, supporting the impression that the general physiology of spch-5 is not significantly different from the wild type at this early seedling stage. Of all spch-5 DEGs, 1,476 were up- and 1,559 were down-regulated with fold changes ranging between −13.4 and 16.2 (Supplemental Data Set S1). Among the down-regulated genes were the known stomatal regulators EPF2, TMM, POLAR, and FAMA (fold changes between −8.4 and −5.2) as well as EPF1, SPCH, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), STOMATAL DENSITY AND DISTRIBUTION1 (SDD1), MUTE, ICE1, and SCRM2 (fold changes between −3.7 and −1.6). Several other genes previously reported to be expressed in stomatal lineages, but with still unknown functions, were down-regulated in spch-5, including AT2G40670, AT1G33930, AT3G17640, AT1G26600, AT5G07280, and AT5G62210 (Pillitteri et al., 2011). The guard cell-expressed genes AT1G03440 and AT3G57600 (Hachez et al., 2011) and the putative positive regulators of stomatal development HOTHEAD (HTH), AT4G34000, and AT5G60890 (de Marcos et al., 2015) were also down-regulated. Interestingly, no known stomatal regulators appeared among the genes up-regulated in spch-5. However, some transcription factors of which the overexpression triggers epidermal and/or stomatal aberrations (AT1G29160, AT3G16770, AT4G39070, and AT5G25810; de Marcos et al., 2015) were up-regulated in spch-5. Interestingly, in the SPCHPPP/spch-3 line, EPF2, TMM, POLAR, and FAMA were down-regulated as well, but genes, such as BASL, ICE1, SCRM2, or SDD1 remained unaltered (Supplemental Data Set S2).
Figure 6.
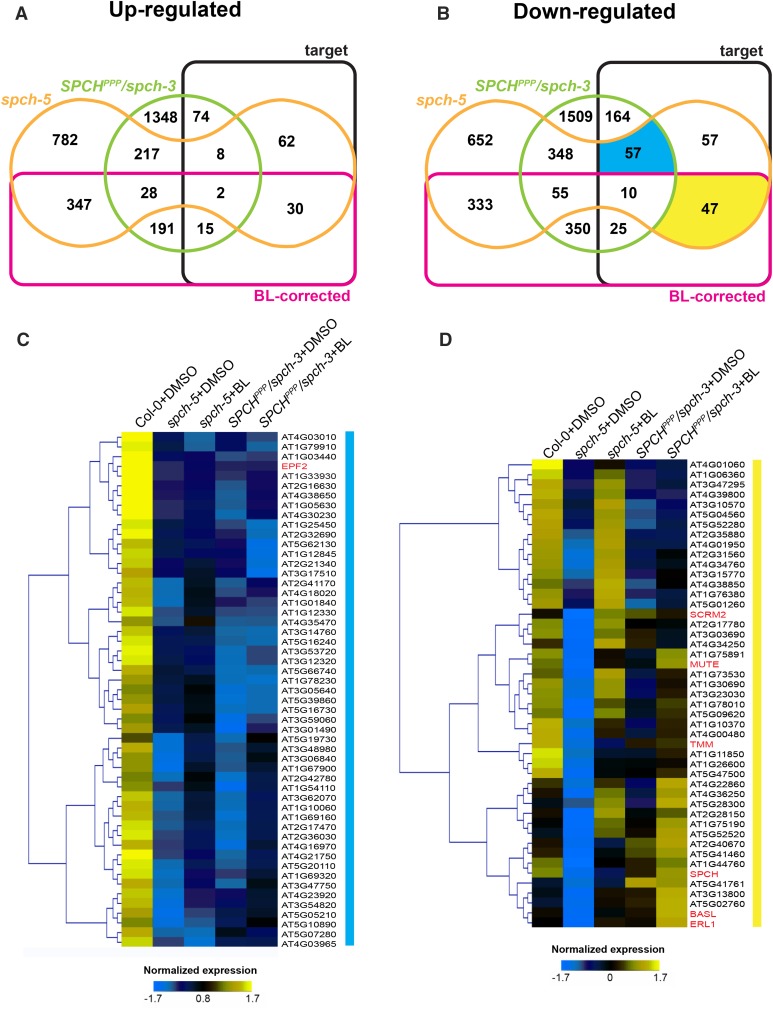
Comparative expression profiles for the BL response of SPCH target genes differentially expressed in spch-5 and/or SPCHPPP. A and B, Edwards’ Venn diagram for DEGs in spch-5 and/or SPCHPPP compared to SPCH target and BL-corrected genes. Up-regulated (A) and down-regulated (B) in spch-5 and/or SPCHPPP. C and D, Z-score normalized log2 gene expression for nontreated Col-0, spch-5, and SPCHPPP plants (DMSO controls) and for BL-treated spch-5 and SPCHPPP plants, corresponding to the yellow and blue sector in B. Highlighted sectors represent spch-5 and SPCHPPP common down-regulated target genes not regulated by BL (blue) and spch-5 exclusive target genes corrected by BL addition (yellow). Color key is given below the heat maps.
Previously, SPCH had been shown to regulate the expression of BR biosynthesis and signaling genes (Lau et al., 2014). However, with the exception of the BRASSINOSTEROID INSENSITIVE1 (BRI1) receptor, which was slightly down-regulated (−1.54 fold change; P value 0.05), other key components of the BR biosynthesis and signaling pathway were not differentially expressed in spch-5 (Supplemental Data Sets S1 and S5), suggesting that this mutation impacts BR-related gene networks in a very specific manner, rather than producing a general effect. The same results were elicited by the SPCHPPP mutation because only AT1G69010 and AT3G30180 were moderately down-regulated and AT1G19350 up-regulated (Supplemental Data Sets 2 and 5).
Recent ChIP-seq studies provided a list of 1,517 high-confidence target genes of SPCH (Lau et al., 2014). Because the lesion in spch-5 suggests an altered DNA-binding potential, we sorted the spch-5 DEGs on the basis of being, or not, potentially high-confidence targets of SPCH. Within the spch-5 DEG set, we found 273 high-confidence SPCH targets, of which 102 were up- and 171 down-regulated (Fig. 6, A and B; Supplemental Data Sets S6 and S7), some of which shared with SPCHPPP/spch-3, particularly down-regulated ones (Fig. 6B). Therefore, nearly 18% of the SPCH targets were differentially expressed in spch-5, a proportion much higher than the 4.2% expected by random hits.
As BRs partially rescued the stomatal phenotype of the spch-5 mutant, but not that of the SPCHPPP/spch-3 transgenic lines, we also investigated by means of a transcriptomic analysis whether treatments with BL would recover the wild-type expression of DEGs in spch-5, in contrast to SPCHPPP/spch-3 (Fig. 6, A and B; Supplemental Data Sets S3 and S4). The BL treatment was effective because the elicited changes in Col-0 matched those previously described (Nemhauser et al., 2006), such as a more than 2-fold change increase in transcripts for AT5G50335, AT5G25190, AT5G37770, and AT2G43290, and, conversely, a more than 2-fold change decrease in the AT1G76240, AT4G16670, and AT5G57785 transcripts (Supplemental Data Set S5).
By combining all mentioned data above, we obtained 12 intersections in each Venn diagram for up- and down-regulated genes in spch-5 and SPCHPPP/spch-3 plants (Fig. 6, A and B; Supplemental Data Sets S6 and S7). For further analysis, we focused on the SPCH target genes with an expression unaltered by the BR treatment in both mutant SPCH versions (blue sector in Fig. 6B) and on the SPCH target genes that responded to BRs in spch-5, but not in SPCHPPP/spch-3 (yellow sector in Fig. 6B). We hypothesized that the blue intersection would include genes of which the proper expression strictly depended on the integrity of the SPCH DNA-binding domain, regardless of the BR application. Indeed, the heat map representation clearly supported this behavior (Fig. 6C). The only known stomatal gene found in this list was EPF2 (Supplemental Data Set S6). The EPF2 transcripts were dramatically reduced in spch-5 and in the plants expressing SPCHPPP and were not restored by the BR treatment (Supplemental Data Sets S1 and S2; Supplemental Fig. S13). Consistent with the fact that BRs mediate the increase of stomatal numbers only in spch-5, the yellow sector would include putative BR-responsive genes with a function in stomatal development (Fig. 6B). Indeed, the heat map representation of the normalized expression values (Fig. 6D) showed that the 47 genes within this sector included a notable group of stomatal regulators, including SPCH, MUTE, SCRM2, TMM, ERECTA-LIKE1 (ERL1), and BASL. Interestingly, whereas BASL was down-regulated (fold change −2.99) and later recovered by application of BRs (2.5-fold increase) in spch-5, it had wild-type levels in the SPCHPPP line (Supplemental Data Sets S1, S2 and S7; Supplemental Fig. S13).
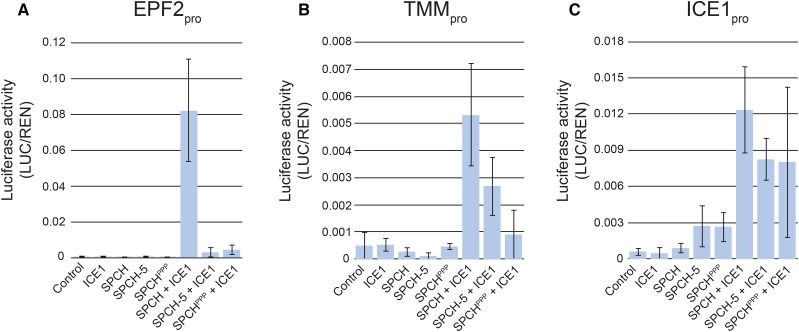
To test whether the transcriptional signatures of spch-5 and SPCHPPP/spch-3 correlated with the functionality of the corresponding SPCH variants, we performed transactivation assays in N. benthamiana. As effectors, we used ICE1 and the three SPCH versions driven by the cauliflower mosaic virus 35S promoter, and as reporters, luciferase fusions to the promoters of three SPCH targets (EPF2, TMM, and ICE1) with differential expression behavior in the three genotypes used for transcriptomics (Fig. 7). As previously shown (Horst et al., 2015), the EPF2 promoter expression was only activated by coexpressing ICE1 and SPCH, but coexpression of ICE1 with either SPCH-5 or SPCHPPP did not activate this promoter. The TMM promoter reporter activity was induced by the cotransfection of ICE1 with the wild-type SPCH and, to a lower level, by SPCH-5 and SPCHPPP, whereas the ICE1 promoter reporter was similarly activated by coexpressing ICE1 with any of the three SPCH versions. As spch-5 is nearly null in the activation of specific SPCH targets (notably EPF2, but not TMM and ICE1), these results provide a functional mechanism for its misfunction and is fully consistent with its stomatal phenotype. Given that SPCH-5 and SPCHPPP showed very similar transactivation abilities, these results also support the notion that proper regulation of selected SPCH target genes requires the integrity of its bHLH domain.
Figure 7.
Transcriptional activation of SPCH targets by different SPCH versions. Transactivation assays in N. benthamiana leaves. Values represent the relative luciferase activity driven by EPF2 (A), TMM (B), and ICE1 promoters (C) when coinfiltrated with different combinations of effector constructs expressing ICE1, SPCH, SPCH-5, or SPCHPPP. An empty effector vector was used in control assays. Relative luciferase activities were calculated as firefly luciferase (LUC) activities normalized to Renilla luciferase (REN) activities. In the presence of ICE1, SPCH strongly transactivated the three reporters, whereas SPCH-5 and SPCHPPP had a transactivation ability null on EPF2pro, low on TMMpro, and similar to SPCH on ICEpro. Error bars represent se of three biological replicates.
Involvement of BASL and EPF2 in the BL Rescue of the spch-5 Phenotype
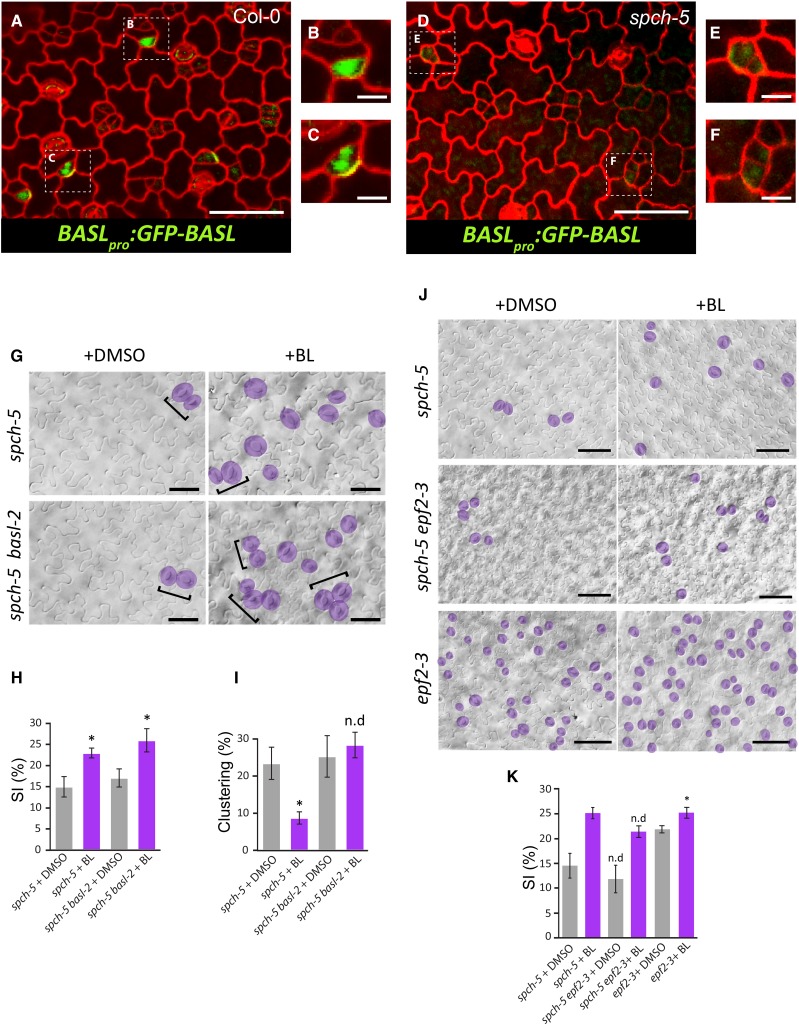
We hypothesized that the partial BR-dependent reduction of stomatal clustering in spch-5 is a consequence of the increase in BASL transcript levels (Fig. 6D; Supplemental Data Set S7), although we cannot exclude the contribution of the BR-dependent increase of TMM and EPF1 transcripts. As the asymmetry defects in spch-5 (Fig. 3) were similar to those reported for the basl mutant (Dong et al., 2009), we explored the role of BASL in the spch-5 phenotype. To this end, we introduced the BASLpro:GFP-BASL construct into spch-5 plants and followed the localization of the BASL protein in cotyledons (Fig. 8, A–F). In contrast to the wild type (Fig. 8, B and C), barely any GFP-BASL signal was detected (Fig. 8, E and F). Moreover, because spch-5 did not accumulate BASL, its stomatal patterning defects were expected to be similar to those of a spch-5 basl double null mutant. The spch-5 basl-2 double mutant was generated and, as hypothesized, behaved as the single spch-5 mutant in terms of stomatal clustering and SI (Fig. 8, G–I). However, the spch-5 phenotype was epistatic to that of basl-2 regarding both polarity defects in ACDs and hyperproliferation of stomatal lineage cells because the spch-5 basl-2 double mutant lacked the additional small epidermal cells that produce the typical lineage markers of the basl-2 mutants (Dong et al., 2009). BL addition to the medium induced an SI increase in the double mutant with the same magnitude as that of spch-5 (Fig. 8, G and H). In contrast, BL treatment did not reduce the stomatal clustering in the double mutant (Fig. 8I), suggesting that the partial alleviation elicited by BRs on the spch-5 stomatal clustering required the BASL function, whereas the BR-dependent increase in SI did not.
Figure 8.
Different involvement of BASL and EPF2 in the BL response of spch-5 plants. A and D, Confocal images of adaxial cotyledon epidermis from 5-d-old seedlings stained with propidium iodide (red) for visualization of cell outlines. GFP (green) marks the BASLpro:GFP-BASL localization. B, C, E, and F, Magnifications of A and D. G, DIC images representative of the epidermis scored in G and H. Stomata are highlighted in purple and clusters denoted by brackets. H and I, Stomatal index and clustering percentage in the abaxial cotyledon epidermis of 23-d-old plants grown in BL (purple) or DMSO (mock treatment; gray). Error bars represent se. Asterisks denote P < 0.05 (Student’s t test) in BL treatment compared to the control medium. n.d, no statistical difference. J, DIC images of abaxial cotyledon epidermis from 23-d-old spch-5 and spch-5 epf2-3 plants grown with BL or in control DMSO medium. Stomata are marked in purple. K, Quantification of the qualitative epidermal phenotypes shown in J, as stomatal index measurements. BL treatment (purple) and DMSO controls (gray). No differences at a 5% significance level (Student’s t test) are indicated as n.d when single and double mutants are compared in each treatment. Asterisk as in H and I. Error bars represent se. Bars = 100 µm in A and B, 50 µm in G and J, and 20 µm in B, C, E, and F.
One of the most striking findings of our microarray analysis was the strongly repressed expression of EPF2 in spch-5 (fold change of −13.4). We hypothesized that the EPF2 expression strictly depends on the integrity of the DNA-binding domain of the SPCH protein, compromised in spch-5. To understand the absence of the EPF2 function in spch-5, we generated the spch-5 epf2-3 double mutant. The genetic interaction analysis confirmed that spch-5 behaved as null mutant for the EPF2 function because single and double mutants had statistically undistinguishable SI values (Fig. 8K). Interestingly, spch-5 conferred an epistatic effect to the overproliferative epf2-3 phenotype. As observed for basl-2, the presence of the spch-5 mutation inhibited the formation of the abundantly arrested lineage cells typical of epf2-3 (Hara et al., 2009). When the plants were grown in BL-supplemented medium, both spch-5 and the spch-5 epf2-3 double mutant responded with an increased SI (Fig. 8K). A similar phenotype in response to BL was also observed in the epf2-3 mutant, which carried a wild-type SPCH allele (Fig. 8K). In contrast to BASL, BL treatment did not restore the EPF2 expression levels in spch-5 (Fig. 6C; Supplemental Data Set S7), but alleviated the mutant phenotype in the spch-5 epf2-3 double mutant (Fig. 8, J and K). Therefore, the impact of BL on spch-5 did not depend on the EPF2 function.
DISCUSSION
The Low Stomatal Abundance of spch-5 Results from Reduced ACDs in Stomatal Lineages
In spch-5 leaves, stomata are formed according to the normal ACD sequence, but the frequency of the various division types differs. The ability of spch-5 to execute entry divisions is reduced, although the expression of SPCH-5-GFP is detectable in many small epidermal cells at the early leaf developmental stages, indicating that SPCH-5 accumulates in protodermal cells, but often fails to trigger the transcriptional changes setting the meristemoid mother cell (MMC) stage. Entry divisions are particularly affected in cotyledons, where the adaxial epidermis has a severe phenotype with almost no stomata. Moreover, once meristemoids are formed, SPCH-5 does not promote efficiently their reiterated amplifying divisions and spch-5 meristemoids prematurely differentiate into guard mother cells (GMCs). As SPCH is essential for maintaining the self-renewal capacity of the meristemoid (Robinson et al., 2011), this function is evidently partly defective in spch-5. The meristemoid-to-GMC transition seems to be related to a decay in SPCH expression and presumably in SPCH activity in late meristemoids, concurrently with the expression of MUTE (Davies and Bergmann, 2014). Therefore, SPCH-5 may be inefficient to maintain the MUTE expression below its functional threshold, allowing it to be expressed prematurely and driving meristemoids to untimely exit ACD cycles and differentiation into stomata.
In contrast, the frequency of the spacing divisions that initiate satellite lineages is not affected in spch-5. Both entry and spacing divisions are specialized stomatal lineage-initiation events that involve the adoption of the MMC identity by distinct cell types, namely, protodermal cells and young SLGCs that probably differ in the negative context they impose to the SPCH-initiating function. These two processes are known to be under a distinct genetic control because some genes have been reported to specifically control satellization (Kutter et al., 2007; Yang et al., 2014) and present independent genetic variations in natural Arabidopsis accessions (Delgado et al., 2011). Such particular developmental characteristics may account for the observed differences in the lineage initiation activity of spch-5 that is sufficient in SLGCs, but limiting in the protoderm. In addition, the spch-5 transcriptional effects might favor the MMC fate in SLGCs because they often have high TMM expression levels that would attenuate repressive signals for spacing divisions. It remains to be tested whether late meristemoids in spch-5 produce low levels of the satellite inhibitory factors EPF1 and SDD1, as suggested by their low transcript abundance in spch-5 seedling samples (see below).
The reduction in stomatal lineage initiation and amplification divisions in spch-5 leads to a large decrease in the total epidermal cell number, but cotyledons and leaves reach a normal final area because of the larger size of the pavement cells than that of wild-type plants. This overall phenotype fits with the so-called phenomenon of “compensated cell enlargement” that results from the dynamic coordination between cell proliferation and expansion activities during leaf development (Hisanaga et al., 2015). The unchanged leaf area of spch-5 indicates that this mutation triggers a “perfect compensation,” whereas it is only partial in most studied genotypes (Ferjani et al., 2007). Interestingly, erecta mutants also exhibit perfect compensation; in this case, the overproduction of stomata and other epidermal cells by excessive SPCH activity is coupled to a reduction in cell sizes (Tisné et al., 2011). The young leaf apparently senses directly or indirectly the changes in SPCH activity, eliciting fully compensatory mechanisms among organ growth variables.
The Clustering Phenotype of spch-5 Results from Various Defects in ACDs
Although the formation of clustered stomata is an unexpected feature for a low SD mutant, several key pattern regulators appear among the SPCH high-confidence targets that are differentially regulated in spch-5; hence, the appearance of stomatal clusters in the mutant is not surprising. The most abundant were type-I clusters, characterized by the lack of asymmetry in the stomatal lineage ACDs. The establishment of physical and fate asymmetry in the cell products of ACDs were both affected to some extent in the mutant. In Arabidopsis, only BASL, which polarizes between the nucleus (meristemoid) and a discrete area at the cell membrane (SLGC; Dong et al., 2009), has been involved in asymmetrical stomatal divisions. The integrity of this protein is necessary for the uneven distribution of cell areas in ACD daughter cells products. Mutants in this locus behave similarly to spch-5 regarding lack of asymmetry and, consequently, cluster formation. In spch-5, BASL transcript levels are very low and the BASL-GFP protein is barely detectable. Type-II and type-III clusters are not related to polarity defects in ACDs, but to failures in cell-to-cell communication. The microarray data suggest that low expression levels of patterning genes, such as SDD1, might underlie this anatomical phenotype.
The spch-5 Phenotype Is Dosage Dependent
In spch-5 cotyledons, the stomatal production depends on gene dosage. As equal changes in the gene copy number of the wild-type SPCH allele do not alter stomatal production (Kanaoka et al., 2008), our results imply that the lineage initiation process is highly sensitive to the SPCH-5 protein amount. Indeed, the number of stomata also increased when the SPCH-5 protein was stabilized, either by elimination of the YODA signaling or by BR treatment. YODA and BR pathways result in phosphorylation of a number of SPCH amino acid residues, of which most are targets of both and some are BR specific (Lampard et al., 2008; Gudesblat et al., 2012). Although the biochemical consequences of the combinatorial phosphorylation remain unknown, the phosphorylation status of SPCH may plausibly be important in modulating protein conformation, DNA-binding capacity or specificity, or interacting partners, thus affecting qualitatively its behavior (Davies and Bergmann, 2014). Thus, not only the amount of SPCH-5, but also its phosphorylation status may underlie its positive response to BR.
Regardless of the presence or absence of BRs, the low stomatal production of spch-5 is related to an altered activity of the SPCH-5 protein. Recent work on the regulatory circuit operating at the initiation of stomatal lineages (Lau et al., 2014; Horst et al., 2015) has redefined the MMC as a cell that accumulates SPCH and its functional SCRMs partners above a certain threshold level (Han and Torii, 2016). To this end, SPCH is first widely and strongly expressed in the young protoderm, in which basal SCRMs levels are also present; then, probably as heterodimers, SPCH and SCRMs directly bind to the promoter and up-regulate the expression of the SCRM genes in the protoderm. According to this model, stochastic noise would account for some protodermal cells achieving the above-critical amounts of SPCH needed to activate SCRM proteins, thereby acquiring a stomatal-lineage character. Thus, the initiation activity conferred by SPCH-5 seems to be below the required threshold in most spch-5 protodermal cells. Such a limitation would be overcome by an increase in SPCH-5 amount or activity. In this hypothesis, we would expect a low expression of SCRM genes in spch-5 plants and an increased expression associated with the extra stomata formed by the BL treatment. Our transcriptomic analysis confirms both predictions: SCRM genes are down-regulated in spch-5 compared to Col-0, and they (particularly SCRM2) are up-regulated in BL-treated spch-5. In the line harboring the SPCHPPP variant, SCRMs transcripts do not respond to BRs, consistent with the inability of the BL treatment to increase the stomata production. It would be interesting to assess whether BR-mediated phosphorylation could also modulate the interaction between SPCH-5 and SCRMs.
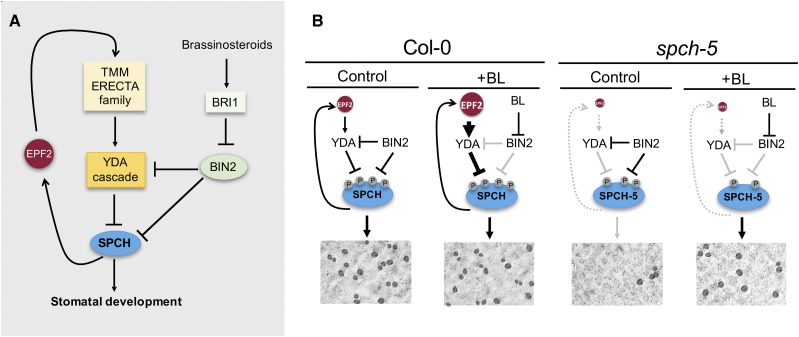
An intriguing question is why application of BRs promotes stomatal development in spch-5 cotyledons, whereas several studies have reported that BR restricts stomatal development in this organ (Kim et al., 2012; Khan et al., 2013). A tempting explanation stems from the absence of the EPF2 function, even after BR treatment, in spch-5 (Figs. 6C and 7K). Figure 9A summarizes stomatal promotion by SPCH, which also activates EPF2 transcription; EPF2 activates the YODA cascade after its perception by the membrane kinases complexes (TMM/ERECTA family), and the YODA cascade in turn inactivates SPCH by phosphorylation. The BIN2 kinase also phosphorylates components of the YODA cascade and SPCH (with opposite effects on stomata promotion), and the BIN2 is inactivated by BR. Figure 9B presents a model to interpret the spch-5 behavior. First, the absence of EPF2 signaling would generate a SPCH-5 protein hypophosphorylated in residues that are targets of the YODA pathway; this SPCH-5 phosphorylation status might be more sensitive to the BIN2-mediated hypophosphorylation in target residues of BRs established by the BL treatment, allowing quantitative and qualitative changes in the SPCH-5 functionality for transcriptional regulation. In addition, the absence of EPF2 in spch-5 will eliminate the negative feedback loop that inhibits SPCH-SCRMs accumulation, and even a modest increase in SPCH-5 activity would be highly amplified.
Figure 9.
Model for spch-5 phenotype and BR effects. A, Current simplified view of SPCH-mediated stomata promotion. SPCH is negatively regulated by the YODA cascade and by BIN2. In the initiation of the stomatal cell lineage, the YODA cascade is triggered by EPF2 through TMM and ERECTA family receptor complexes. SPCH activates EPF2 transcription, creating a feedback repression loop. BIN2 is a negative regulator of the repressive YODA cascade, and BIN2 activity is negatively regulated by BRs through the receptor BRI1. B, BR-dependent regulation of the SPCH-5 phosphorylation status. In Col-0, the negative feedback loop through EPF2 compensates for the negative and positive effects of BIN2 on SPCH activity; thus, the stomatal phenotype is independent of BL. In spch-5, because EPF2 and the negative loop are absent, the repressive YODA pathway operates very limitedly and most SPCH-5 phosphorylation is under BIN2 control. BL treatment inactivates BIN2, releasing the main phosphorylation-mediated repression on SPCH-5 and promoting protein accumulation and stomata development.
To validate our model in a SPCH wild-type context, we inspected the stomatal phenotype of the epf2-3 mutant in control and BL-containing medium. Consistently, we found that the epf2-3 mutant cotyledons (which are wild type for SPCH) responded to BL treatment with an increase in stomatal production, whereas Col-0 plants did not. Hence, our hypothesis that the enhanced BL response in spch-5 cotyledons could be due to the absence of EPF2 function is supported by the fact that in a wild-type SPCH background, lack of the EPF2 function produced a similar effect. It is worth mentioning that the phenotype of the spch-5 yoda-10 double mutant shows that, even in the absence of EPF2, YODA is at least partially active in spch-5, consistent with YODA and downstream MAPKs acting as a signaling hub for diverse SPCH-converging endogenous and exogenous stimuli (Wang et al., 2007; Colcombet and Hirt, 2008; Popescu et al., 2009).
Although the line expressing SPCHPPP has also very low EPF2 transcript levels, it does not respond phenotypically to BRs as spch-5, raising the possibility that SPCH-5 retains a partial bHLH activity that could be improved by BRs, whereas SPCHPPP would lack such property. Alternatively, the differences between the two lines might be due to variations between the regulation of the endogenous SPCH expression and of the sustained transgene expression of the SPCH promoter. SPCH has been proposed to regulate the expression of several genes involved in BR biosynthesis and signaling (Lau et al., 2014). We found no evidence that SPCH-5 is altered in such a regulation because the transcriptomic data indicate a similar signature for these genes in Col-0 and spch-5.
Molecular and Functional Basis of the spch-5 Phenotype
Recently, a ChIP-seq approach revealed that SPCH associates in vivo with nearly one-third of the Arabidopsis genes and that most of its binding sites are located in proximal gene promoters and are significantly enriched for the E-box motif variant CDCGTG (Lau et al., 2014). Nevertheless, the biochemical demonstration of the SPCH ability for direct interaction with specific DNA sequences remains elusive to conventional in vitro or heterologous system assays. Whether the SPCH transcriptional regulatory activity required direct DNA binding through its bHLH domain was questioned by the ability of transgenes expressing a SPCH variant lacking the critical bHLH residues for DNA binding (SPCHPGG) to direct stomata production in a spch-3 background (Davies and Bergmann, 2014). The spch-5 mutation unveils the implication of the SPCH bHLH domain in the regulation of a number of stomatal developmental processes. Moreover, the spch-5 phenotype strongly supports that SPCH is a DNA-binding transcription factor because, to our knowledge, no activity other than DNA binding has been reported for the bHLH basic region in which the spch-5 mutation locates.
Given that a fraction (∼18%) of the high-confidence SPCH targets are differentially expressed in spch-5, the mutation in the SPCH-5 protein may reduce the regulation efficiency of the expression of this subset of SPCH target genes. However, in spch-5, most SPCH target genes were expressed at normal levels, suggesting that SPCH does not require the complete bHLH function for some in vivo activities, possibly the reason for the formation of stomata in spch-5 plants as well as in the SPCHPPP/spch-3 line or in the previously reported SPCHPGG (Davies and Bergmann, 2014). Hence, SPCH seemingly regulates the transcription of its target genes by mechanisms differing from those used by MUTE that fully functions without its DNA-binding motif (Davies and Bergmann, 2014) or FAMA that strictly requires DNA-binding residues to function (Ohashi-Ito and Bergmann, 2006). These bHLH domain-independent activities have been proposed to act by interaction with specific partners, such as the SCRM proteins that may recruit SPCH and MUTE to regulatory regions of target genes (Han and Torii, 2016). As SPCH-5 retained wild-type dimerization abilities with SCRMs, this alternative mechanism might explain how SPCH-5 regulates most SPCH gene targets.
Our transcriptomic data provide molecular phenotypes that might support such different mechanisms in the SPCH functions. Among the genes deregulated in spch-5 that are SPCH targets, of which the transcription depends on the bHLH domain integrity, are BASL and EPF2, both implicated in the spch-5 phenotype, and probably other genes with the same behavior. Furthermore, these genes can be split into two groups: those with (such as BASL) and those without (such as EPF2) expression reversed by BRs. These differences are possibly rooted in distinct mechanisms for their SPCH-mediated regulation.
The transcriptional signatures of spch-5 and the SPCHPPP/spch-3 line only partially overlapped in the SPCH targets. As discussed above, SPCH-5 might retain a partial bHLH-dependent activity, whereas SPCHPPP might be null or the differences might stem from differences in the transgene expression. Another possibility relies on the reported MUTE-like activity of SPCH versions mutated in the bHLH domain (Davies and Bergmann, 2014). Although SPCH-5 does not display such MUTE-like activity because the double mutant spch-5 mute-3 does not form stomata, SPCHPPP, like SPCHPGG (Davies and Bergmann, 2014), might indeed, as reflected in the different transcriptome of this SPCHPPP line.
The SPCH-5 and the SPCHPPP proteins reported here have revealed a differential behavior of different SPCH target genes. Proper regulation of some of them requires integrity of the bHLH domain, whereas others seem to be unaffected by mutations in this domain that might compromise DNA binding. This observation was supported by the transactivation experiments where SPCH-5, together with ICE1, activated the promoters of TMM and ICE1, but not EPF2. SPCHPPP behaved similarly, but its capacity to activate both TMM and ICE1 promoters was lower and somehow erratic. The spch-5 mutation has also evidenced a BR-dependent mechanism that seems to compensate, at least partially, the lesion in the bHLH domain.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana), accession Col-0 (N1092), spch-3 (SAIL_36_B06), epf2-3 (SALK_047918), basl-2 (WiscDsLox264F02), and yoda-10 (SALK_105078) were obtained from the Nottingham Arabidopsis Stock Centre. The mutant mute-3 was isolated as described (Triviño et al., 2013). TMMpro:GUS-GFP and spch-2 were kind gifts of Fred Sack (The University of British Columbia; Nadeau and Sack, 2002), and Dominique Bergmann (Stanford University; MacAlister et al., 2007), respectively. The SPCHpro:SPCH-GFP (spch-3 background) line has been described previously (Gudesblat et al., 2012). spch-5 was isolated from a Col-0 EMS-mutagenized collection and carried a point mutation (see below). Lines carrying the translational fusions SPCHpro:SPCH-GFP, SPCHpro:SPCH-5-GFP, and SPCHpro:SPCHPPP-GFP were obtained by floral dip Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998) of plants heterozygous for the spch-3 allele or homozygous spch-5.
The following double mutants were generated: spch-5 yoda-10, spch-5 mute-3, spch-5 epf2-3, spch-5 basl-2, and the transheterozygous mutant spch-5/spch-3. The marker line carrying the fusion TMMpro:GUS-GFP in a Col-0 background was used to introduce the transgene into spch-5 and spch-5 mute-3 double mutant through sexual crosses.
All experiments were done under in vitro conditions, unless otherwise indicated. Seeds were surface-sterilized under overnight exposure to chlorine gas (Clough and Bent, 1998) and sown in Murashige and Skoog (MS) medium supplemented with 1% (w/v) Suc. After 2 to 5 d of stratification at 4°C in the dark, plates were placed at 21°C and a 16-h-light/8-h-dark photoperiod with 70 µmol m−2 s−1 of photosynthetically active radiation. When indicated, BL (Sigma-Aldrich) was added to the medium at a final concentration of 50 nm. For soil experiments, seeds were stratified at 4°C for 2 to 5 d in the dark and sown in peat pellets (Jiffy-7). Growth conditions were 21°C, 70% relative humidity, and 150 µmol m−2 s−1 photosynthetically active radiation.
Genotypes were determined by PCR with derived cleaved amplified polymorphic sequences or T-DNA genotyping approaches, with the primers listed in Supplemental Data Set S8.
Microscopy
For differential interference contrast (DIC) images, organs were hand-excised, fixed in ethanol:acetic acid 9:1 (v/v) for 16 h, replaced by 90% (v/v) ethanol, and rehydrated with ethanol dilutions with increasing water content, 70, 50, 30, and 10% ethanol, and pure distilled water as final step. All these incubations were done at room temperature and for 1 h each. Finally, a chloral hydrate:glycerol:water solution (8:1:2, w/v/v) was used to clear the tissues, whereafter the specimens were observed under a Nikon Eclipse 90i upright microscope with DIC optics and a DXM1200C camera for image acquisition.
For confocal images, a Leica TCS SP2 confocal inverted microscope was used for GFP and propidium iodide visualization. The propidium iodide solution (Sigma-Aldrich) was diluted in distilled water at a final concentration of 10 µg/mL and plants were submerged for 15 min for counterstaining of epidermal cell shapes.
Quantitative Analysis of Epidermal Phenotypes
SI (number of stomata/total number of epidermal cells × 100), SD (number of stomata per mm2), and PCD (number of pavement cells per mm2) were calculated by scoring two areas of 0.4 mm2 located on both sides of the median axis of the cotyledon or leaf (modified from Delgado et al., 2011). The ImageJ (Schneider et al., 2012) plug-ins “cell counter” and “grid” were used for counting the different cell types. Unless otherwise specified, 10 plants were examined (n = 10) for quantitative traits and the adaxial and/or the abaxial epidermis of 23-d-old cotyledons or 28-d-old third leaves were examined, corresponding to fully expanded stages of these organs under our conditions, respectively. Organ area and pavement cell size were measured at these fully expanded stages with ImageJ (Schneider et al., 2012).
The time-course experiment with serial imprints (Fig. 2) was as reported (de Marcos et al., 2016) without modifications. The term “stomatal precursors” was used for meristemoids and GMCs. The entry division percentage was calculated as the number of the initial 50 cells that enter the stomatal lineage during the T0-T4 period and the entry division rate corresponds to the increment in the number of entry divisions per day interval. The proportion of satellite lineages was calculated as the percentage of total lineages (including those leading to clustered stomata) that are satellite. SLGC/lineage estimates the number of nonstomatal lineage cells per stomata or stomata precursor at T4. In the total cell quantification, cells were counted at each time point, with stomata counted as two cells because they are formed by two guard cells. The cell proliferation rate was calculated as the increase in cell number over the different time intervals. For cluster classification, 128 clusters from seven plants were inspected. Clusters present at T4 were tracked backward in the previous imprints to reconstruct the cell divisions history that led to the development of contact stomata.
The relative meristemoid size (Fig. 3H) was calculated with 61 lineages from five different plants in Col-0 and 70 lineages from seven plants in spch-5. The cell area of daughter cells was determined in cells expressing the TMMpro:GFP marker. For spch-5, the relative meristemoid size (Fig. 3I) was measured in serial epidermal imprints tracking back 54 isolated stomata from four different plants and 55 type-I clustered stomata from seven spch-5 plants. Cell contours were drawn with the interactive display Cintiq 21UX (Wacom). Cell and organ areas were measured with ImageJ (Schneider et al., 2012). Clustering percentage was calculated as the proportion of stomata in clusters (two or more stomata in contact) to the total stomata.
Positional Cloning and Molecular Characterization of spch-5
The mutation was localized genomically by linkage analysis as described (Ponce et al., 2006) by means of the F2 progeny of a cross between the mutant (Col-0 background) and Ler (Landsberg erecta genetic background carrying the wild-type functional ERECTA allele). The SPCH genome was sequenced with total DNA extracted with the DNeasy Plant Mini Kit (Qiagen). The SPCH locus was selectively amplified by PCR with the HiFi PCR kit (KapaBiosystems) and specific primers (Supplemental Data Set S8). The PCR product was purified with the MinElute PCR purification kit (Qiagen) and sequenced with BigDye technology (Secugen).
Three-Dimensional SPCH Protein Modeling
The protein structure was modeled with the SWISS_MODEL workspace (Bordoli et al., 2009) and the Myc-Max protein as a template (PDB ID: 1 nkp). The structural consequences of the mutation present in spch-5 were predicted with the Swiss-PdbViewer 4.1 software (Guex and Peitsch, 1997).
DNA Engineering
Constructs used in this study are listed in Supplemental Data Set S9. The cloning strategies were the standard and the Multisite Gateway technologies. The primers are listed in Supplemental Data Set S8. The open reading frame of SPCHPPP was synthetically produced (GeneArt; Invitrogen) with the coding DNA sequence (CDS) of SPCH as template (GenBank accession number AY568670) with the following changes: substitutions of A→C at position 290 (290A→C), 322G→C, 323A→C, 334A→C, and 335G→C. At the protein level, these mutations led to the presence of prolines at amino acid positions 104, 108, and 112. The CDS of PIF4 in pDONR221 was a kind gift of Salomé Prat (CNB-CSIC, Madrid). SPCH and SPCH-5 were cloned from the cDNA of 10-d-old Col-0 seedlings with the same RNA extraction and cDNA synthesis approaches as indicated for the microarray hybridization. The CDS of ICE1 (GenBank accession number AY195621) and SCRM2 (GenBank accession number NM_101157) were purchased from the Arabidopsis Biological Research Center (U68804 and U60686, respectively) and subcloned into the pDONR221 vector (Invitrogen). The SPCH promoter sequence (−1 to −2,572) had been published previously (Gudesblat et al., 2012). Deletion of the SPCH-coding sequence (SPCHΔ273) was obtained by PCR-mediated amplification with the primers listed in Supplemental Data Set S8 and by subsequent cloning into pDONR221. Enhanced GFP was amplified from pK7WGF2 (Karimi et al., 2002) and subcloned into pDONR P2R-P3 (Invitrogen). All the entry clones used were sequenced prior to their recombination into the different destination vectors.
Yeast One-Hybrid Assays
The CDS-containing entry vectors were recombined into the Gateway-modified destination vectors pGADT7 (Clontech; Rombolá-Caldentey et al., 2014) and pDEST22 (Invitrogen) through LR reactions (LR clonase II mix; Invitrogen). These constructs and the empty pGADT7 vector were introduced into the MATa Leu auxotroph YM4271a (Liu et al., 1993). Bait sequences (wild-type G-box TGACACGTGGCATGACACGTGGCATGACACGTGGCA and mutated G-box TGACAATTGGCATGACAATTGGCATGACAATTGGCA) were synthesized with AttB4 and AttB1 Gateway recombination sites (Integrated DNA Technologies) and cloned into the pDONR P4-P1R vector with BP clonase (Invitrogen). A recombinational LR reaction (Invitrogen) was applied to introduce the bait sequences into the vector pMW#3 (Deplancke et al., 2006) that contains a LacZ reporter gene. The details of plasmid construction are described in Supplemental Data Set S9. The plasmid genome integrated at the ura3-52 locus by linearization of the constructs with NcoI or ApaI. For each DNA bait, a colony that exhibited a low self-activation was selected for subsequent yeast one-hybrid experiments. Transcription factor constructs were mobilized into the bait sequence-containing yeast strains by means of the low-efficiency transformation method (Walhout and Vidal, 2001). Selective media used were as follows: SD-Ura for bait sequences and SD-Leu for all transcription factors (pGADT7 vector), except for ICE1 (pDEST22) that was selected with SD-Trp. The yeast β-galactosidase assay kit (Thermo Scientific) was used for the binding experiments according to the manufacturer’s quantitative protocol. The absorbance data were obtained with a microplate reader (Epoch; BioTek). Normalized values (zi) of the β-galactosidase activity (xi) (Supplemental Fig. S11B) were calculated with the formula:
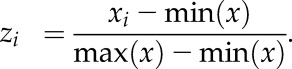 |
BiFC Assays in N. benthamiana Leaves
The BiFC constructs were obtained through the Multisite Gateway cloning technology (Invitrogen) with N- and C-terminal GFP derivatives (Boruc et al., 2010). Full-length open reading frames of the proteins of interest with and without STOP codons were recombined into the pDONR221 or pENTR/D-TOPO entry vectors (Invitrogen) with BP Clonase (Invitrogen) or the Directional TOPO cloning kit (Invitrogen). LR reactions with the LR Clonase II Plus enzyme (Invitrogen) produced translational fusions between the protein of interest and the GFP moieties, driven by the cauliflower mosaic virus 35S promoter. pH7m34GW and pK7m34GW were used as destination vector (Karimi et al., 2005). For detailed information on the plasmids, see Supplemental Data Set S9. Positive control full-length GFP derivatives were recombined with pK7FWG2 (Karimi et al., 2002). Constructions were transferred into A. tumefaciens GV3101 by electroporation, followed by identification in selective media. A. tumefaciens strains carrying the constructs of interest and a p19-harboring strain (Shamloul et al., 2014) in the abaxial side of N. benthamiana leaves were coinfiltrated as described (Boruc et al., 2010) with minor changes. At least five leaf segments per combination were examined under a confocal microscope (Leica TCS SP2) 3 to 5 d after infiltration. Interactions were considered positive when at least 10 cells emitted fluorescent GFP. As negative control, each split GFP constructs was individually infiltrated under the same experimental conditions without signal detection.
Dual Luciferase Transactivation Assay in N. benthamiana Leaves
The reporter and effector plasmids were transiently expressed in N. benthamiana leaves. Three to five days after infiltration, firefly luciferase (LUC) and Renilla luciferase (REN) were assayed with a dual-luciferase assay kit (Promega) and measured in a Synergy H1 plate reader (Biotek). Light emission was integrated over 15 s with 5 s of preread delay. The transactivation ability of the effectors was expressed as the ratio of LUC to REN. See Supplemental Data Set S8 and S9 for details about plasmid construction and primers used. Three biological replicates were assayed for each plasmid combination.
SPCH Protein Stability Assay
This assay was done according to Gudesblat et al. (2012) with minor modifications. SPCHpro:SPCH-GFP, SPCHpro:SPCH-5-GFP, and SPCHpro:SPCHPPP-GFP (spch-3 background) seedlings were grown in half-strength MS medium without Suc. Three days after sowing, 3 g of seedlings was incubated for 2 h in half-strength MS liquid medium supplemented with 100 nm BL (treatment) or the equivalent volume of DMSO (control) with gentle shaking. Protein extracts were prepared with ice-cold extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% [w/v] NP-40, and complete protease inhibitor tablet [Roche Diagnostics]). The protein extract was incubated with GFP-binding protein beads (GFP-Trap_A; Chromotek) for 4 h at 4°C and washed three times with washing buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.5% [w/v] NP-40). After centrifugation (2,500g) and discarding the washing buffer, beads were mixed with 2× SDS sample buffer and boiled for 5 min at 95°C. Samples were fractionated by SDS-PAGE (10% acrylamide) and analyzed by western blotting with an anti-GFP horseradish peroxidase-conjugated antibody (Monoclonal Antibody; Miltenyi Biotec) at a 1:10,000 dilution. Antitubulin antibody was used as loading control.
RNA Extraction and Microarray Hybridization
RNA was obtained from 50 seedlings, collected 3 d after sowing. Samples from three independent biological replicates were frozen in liquid nitrogen and RNA was extracted with TRIzol (Invitrogen), followed by column purification with the High Pure RNA extraction kit (Roche Diagnostics). RNA quality was determined by electrophoresis with a 2100 Bioanalyzer (Agilent). Samples were hybridized at the Centro Nacional de Biotecnología (Madrid) with the Agilent custom oligo microarrays 8×60K (reference GPL22511). This Arabidopsis microarray was designed by the Genomics Facility at the Centro Nacional de Biotecnología and covers 62,976 probes corresponding to 35,018 unique genes.
Total RNA (500 ng each) was amplified and labeled with cyanine 3 (Cy3) by means of the One-Color Low Input Quick Amp Labeling Kit (Agilent) according to the manufacturer’s instructions. Briefly, total RNA was converted into double-stranded cDNA with the oligo(dT)-T7 primer and AffinityScript Reverse Transcriptase (Agilent). cDNA was then used as template for in vitro transcription reaction with T7 RNA polymerase and incorporated into Cy3-CTP. Cy3-labeled cRNA was purified with RNeasy columns (Qiagen) and RNA yield and Cy3 incorporation were measured with a spectrophotometer (Nanodrop).
Probes were prepared and hybridized with the One-Color Microarray-Based Gene Expression Analysis (Agilent). Briefly, for each hybridization, 600 ng of Cy3-cRNA was added to 5 µL of 10× blocking agent, 1 µL of 25× fragmentation buffer in a 25-µL reaction, and incubated at 60°C for 30 min to fragment RNA, and stopped with 25 µL of 2× hybridization buffer. The samples were placed on ice and quickly loaded, hybridized at 65°C for 17 h, and then washed once in wash buffer 1 at room temperature (1 min) and once in wash buffer 2 at 37°C (1 min; GE Healthcare). Arrays were drained by centrifugation. Images for the Cy3 channel were captured with a DNA Microarray Scanner (Agilent) at a resolution of 2 µm, and spots were quantified with the Feature Extraction Software (Agilent).
Microarray Data Analysis
Background was corrected and quantile expression data were normalized with limma (Smyth and Speed, 2003; Smyth, 2004). Each probe was tested for changes in expression over replicates by using an empirical Bayes moderated t-statistic (Smyth, 2004). For further analysis, only one probe per gene was taken into account with the following criteria: the P values in the Col-0 were lower than those in spch-5 in control (DMSO) medium.
Genes were determined as differentially expressed when two criteria were met: P value < 0.05 and fold change > 1.5 or < −1.5, up- and down-regulated, respectively. For the Gene Ontogeny classification (Supplemental Fig. S16), the Cytoscape plug-in ClueGO (Bindea et al., 2009) was used with DEGs as input data. Heat maps (Fig. 6) were plotted by means of the TM4 MeV software (Saeed et al., 2003). The complete hierarchical clustering method was applied with average dot product as distance metrics, after Z score normalization of the log2 absolute expression values. Venn diagrams were obtained with the online application VENNY (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
qPCRs
The same RNA samples used for the microarray experiments were also utilized for qPCR. cDNA was synthesized with the High-Capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s instructions. The real-time amplification was monitored with the maxima SYBR green qPCR master mix (Thermo Scientific) on a LightCycler 480 II PCR amplification and detection instrument (Roche Diagnostics). For the specific primer sets used for amplification, see Supplemental Data Set S8. Each target gene was paired with two different reference genes (ACT2 [At3g18780] and UBQ10 [At4g05320]). Expression values were calculated with the efficiency method in the LightCycler 480 software version 1.5 (Roche Diagnostics).
Accession Numbers
Sequence data can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT5G53210 (SPCH), AT1G34245 (EPF2), AT3G06120 (MUTE), AT5G60880 (BASL), AT3G26744 (ICE1/SCRM), AT1G12860 (SCRM2), AT2G43010 (PIF4), AT3G18780 (ACT2), AT4G05320 (UBQ10), AT2G20875 (EPF1), AT1G80080 (TMM), AT5G05690 (CPD), AT4G18710 (BIN2), and AT1G04110 (SDD1). Microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE88950.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Stomatal development in Arabidopsis.
Supplemental Figure S2. Adaxial cotyledon phenotype exhibited by the ldc mutant.
Supplemental Figure S3. Genetic mapping of the ldc mutation and SPCH protein sequence indicating known domains and amino acid changes of the point mutants.
Supplemental Figure S4. Epidermal DIC images of several mature organs in Col-0 and spch-5.
Supplemental Figure S5. spch-5 third-leaf epidermal phenotype.
Supplemental Figure S6. Additional data derived from cell tracking of leaf primordia shown in Figure 2.
Supplemental Figure S7. Orientation of cell division planes in spch-5 and spch-5 mute-3 stomatal lineages.
Supplemental Figure S8. Homology-based three-dimensional structural prediction for the SPCH-5 protein.
Supplemental Figure S9. DNA-protein interactions involving SPCH-5.
Supplemental Figure S10. Protein-protein interactions involving SPCH-5.
Supplemental Figure S11. Phenotypic and protein expression analyses of spch-3 lines complemented with different SPCH versions.
Supplemental Figure S12. BL effect on spch-2 stomatal development.
Supplemental Figure S13. Quantitative PCR for microarray validation.
Supplemental Figure S14. Biological processes overrepresented in the spch-5 DEGs.
Supplemental Data Set S1. DEG in spch-5 (DMSO) versus Col-0 (DMSO) comparison.
Supplemental Data Set S2. DEG in SPCHPPP/spch-3 (DMSO) versus Col-0 (DMSO) comparison.
Supplemental Data Set S3. DEG in spch-5 (BL) versus spch-5 (DMSO) comparison.
Supplemental Data Set S4. DEG in SPCHPPP/spch-3 (BL) versus SPCHPPP/spch-3 (DMSO) comparison.
Supplemental Data Set S5. List for BR-related genes mentioned in the text.
Supplemental Data Set S6. Gene expression values of intersection sectors from Figure 6A.
Supplemental Data Set S7. Gene expression values of intersection sectors from Figure 6B.
Supplemental Data Set S8. List of primers used in this study.
Supplemental Data Set S9. List of plasmids used in the current study.
Supplementary Material
Acknowledgments
A.d.M. thanks the European Commission for an exchange fellowship of the ERASMUS Program. We thank Martine De Cock for help in preparing the manuscript.
Glossary
- MAPK
mitogen-activated protein kinase
- ACD
asymmetric cell division
- BR
brassinosteroid
- ChIP-seq
chromatin immunoprecipitation sequencing
- SI
stomatal index
- SD
stomatal density
- PCD
pavement cell density
- SLGC
stomatal lineage ground cell
- BiFC
bimolecular fluorescence complementation
- BL
brassinolide
- DEG
differentially expressed gene
- MMC
meristemoid mother cell
- GMC
guard mother cell
- MS
Murashige and Skoog
- DIC
differential interference contrast
- CDS
coding DNA sequence
References
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4: 1–13 [DOI] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Hetherington AM (2010) Environmental regulation of stomatal development. Curr Opin Plant Biol 13: 90–95 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Davies KA, Bergmann DC (2014) Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc Natl Acad Sci USA 111: 15585–15590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado D, Alonso-Blanco C, Fenoll C, Mena M (2011) Natural variation in stomatal abundance of Arabidopsis thaliana includes cryptic diversity for different developmental processes. Ann Bot (Lond) 107: 1247–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marcos A, Triviño M, Fenoll C, Mena M (2016) Too many faces for TOO MANY MOUTHS? New Phytol 210: 779–785 [DOI] [PubMed] [Google Scholar]
- de Marcos A, Triviño M, Pérez-Bueno ML, Ballesteros I, Barón M, Mena M, Fenoll C (2015) Transcriptional profiles of Arabidopsis stomataless mutants reveal developmental and physiological features of life in the absence of stomata. Front Plant Sci 6: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi F, Grove CA, Vedenko A, Alibés A, Gisselbrecht SS, Serrano L, Bulyk ML, Walhout AJM (2011) Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res 39: 4553–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA, et al. (2006) A gene-centered C. elegans protein-DNA interaction network. Cell 125: 1193–1205 [DOI] [PubMed] [Google Scholar]
- Dong J, MacAlister CA, Bergmann DC (2009) BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow GJ, Bergmann DC, Berry JA (2014) An integrated model of stomatal development and leaf physiology. New Phytol 201: 1218–1226 [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Fass D, Arnaud M, Harrison SC (1994) Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev 8: 970–980 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Schneider-Pizoń J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, Russinova E (2012) SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol 14: 548–554 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hachez C, Ohashi-Ito K, Dong J, Bergmann DC (2011) Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol 155: 1458–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S-K, Torii KU (2016) Lineage-specific stem cells, signals and asymmetries during stomatal development. Development 143: 1259–1270 [DOI] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T (2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hisanaga T, Kawade K, Tsukaya H (2015) Compensation: a key to clarifying the organ-level regulation of lateral organ size in plants. J Exp Bot 66: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Horst RJ, Fujita H, Lee JS, Rychel AL, Garrick JM, Kawaguchi M, Peterson KM, Torii KU (2015) Molecular framework of a regulatory circuit initiating two-dimensional spatial patterning of stomatal lineage. PLoS Genet 11: e1005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu J-K, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Khan M, Rozhon W, Bigeard J, Pflieger D, Husar S, Pitzschke A, Teige M, Jonak C, Hirt H, Poppenberger B (2013) Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J Biol Chem 288: 7519–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Michniewicz M, Bergmann DC, Wang Z-Y (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Pattanaik S, Feller A, Werkman JR, Chai C, Wang Y, Grotewold E, Yuan L (2012) Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc Natl Acad Sci USA 109: E2091–E2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Schöb H, Stadler M, Meins F Jr., Si-Ammour A (2007) MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19: 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, MacAlister CA, Bergmann DC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lampard GR, Lukowitz W, Ellis BE, Bergmann DC (2009) Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC (2014) Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wilson TE, Milbrandt J, Johnston M (1993) Identifying DNA-binding sites and analyzing DNA-binding domains using a yeast selection system. Methods 5: 125–137 [Google Scholar]
- MacAlister CA, Bergmann DC (2011) Sequence and function of basic helix–loop–helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol Dev 13: 182–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC (2007) Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- Maerkl SJ, Quake SR (2009) Experimental determination of the evolvability of a transcription factor. Proc Natl Acad Sci USA 106: 18650–18655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L (2015) Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Scholtz JM (1998) A helix propensity scale based on experimental studies of peptides and proteins. Biophys J 75: 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU (2007) Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. BioEssays 29: 861–870 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU (2007) Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Peterson KM, Horst RJ, Torii KU (2011) Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23: 3260–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce MR, Robles P, Lozano FM, Brotóns MA, Micol JL (2006) Low-resolution mapping of untagged mutations. Methods Mol Biol 323: 105–113 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. (2002) Selection pressures on stomatal evolution. New Phytol 153: 371–386 [DOI] [PubMed] [Google Scholar]
- Robinson S, Barbier de Reuille P, Chan J, Bergmann D, Prusinkiewicz P, Coen E (2011) Generation of spatial patterns through cell polarity switching. Science 333: 1436–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombolá-Caldentey B, Rueda-Romero P, Iglesias-Fernández R, Carbonero P, Oñate-Sánchez L (2014) Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element. Plant Cell 26: 2905–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamloul M, Trusa J, Mett V, Yusibov V (2014) Optimization and Utilization of Agrobacterium-mediated Transient Protein Production in Nicotiana. J Vis Exp 86: 51204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AR, Bergmann DC (2016) Transcriptional control of cell fate in the stomatal lineage. Curr Opin Plant Biol 29: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Tisné S, Barbier F, Granier C (2011) The ERECTA gene controls spatial and temporal patterns of epidermal cell number and size in successive developing leaves of Arabidopsis thaliana. Ann Bot (Lond) 108: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU. (2015) Stomatal differentiation: the beginning and the end. Curr Opin Plant Biol 28: 16–22 [DOI] [PubMed] [Google Scholar]
- Triviño M, Martín-Trillo M, Ballesteros I, Delgado D, de Marcos A, Desvoyes B, Gutiérrez C, Mena M, Fenoll C (2013) Timely expression of the Arabidopsis stoma-fate master regulator MUTE is required for specification of other epidermal cell types. Plant J 75: 808–822 [DOI] [PubMed] [Google Scholar]
- Walhout AJM, Vidal M (2001) High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297–306 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yang K, Le J (2015) Organ-specific effects of brassinosteroids on stomatal production coordinate with the action of TOO MANY MOUTHS. J Integr Plant Biol 57: 247–255 [DOI] [PubMed] [Google Scholar]
- Yang K, Jiang M, Le J (2014) A new loss-of-function allele 28y reveals a role of ARGONAUTE1 in limiting asymmetric division of stomatal lineage ground cell. J Integr Plant Biol 56: 539–549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.