Electrical signals initiate all rapid physiological events that take place on the millisecond time scale, from long-range electrical communication within bacterial biofilm communities1, to coordinated swimming of protozoa like Paramecium2, and hitting a fastball in major league baseball3. Initiation of electrical signaling requires voltage-gated sodium and/or calcium (NaV and/or CaV) channels, which are transmembrane proteins that open their pore in response to membrane depolarization. They conduct Na+ and Ca2+ into cells to rapidly depolarize the cell membrane and initiate conducted action potentials (Fig. 1a)3,4. Immediately afterwards, the voltage-gated potassium (KV) channels open and mediate efflux of K+ to terminate the electrical signal and restore the membrane to its resting potential in preparation for upcoming action potentials (Fig. 1a)3,4. The essential functions of NaV and CaV channels required for initiating action potentials are (i) steeply voltage-dependent activation, (ii) rapid pore opening, (iii) highly selective ion permeability, and (iv) voltage-dependent inactivation to terminate the increase in cation influx3. These steps in channel function are considered in sequence below.
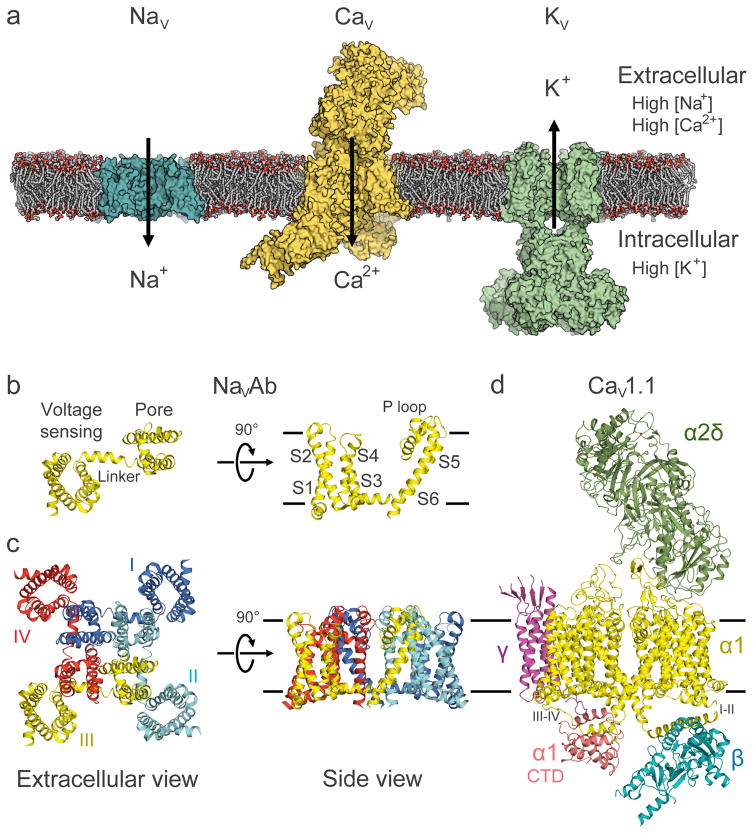
Figure 1. Overall architecture of voltage-gated sodium, calcium, and potassium channels.
a, Model of representative NaV, CaV, and KV channels in a lipid membrane. From left to right: bacterial NaVAb (cyan), mammalian CaV1.1 complex with auxiliary subunits (yellow), and mammalian KV1.2/2.1 chimera complex with cytoplasmic β subunits (green). b, Structure of the bacterial sodium channel NaVAb single subunit. The structure comprises the voltage-sensing module (S1–S4) connected to the pore module (S5–S6) via the S4–S5 linker. The P loop connects the S5 and S6 segments and contains the ion selectivity filter motif (Fig. 5). c, NaVAb homotetramer with domain swapping illustrated by different color for each subunit. The pore is located at the center of the tetramer with the voltage-sensing module interacting with the pore module of the neighboring subunit. d, Single particle cryo-EM reconstruction of CaV1.1 with NaVAb-like core α1 subunit highlighted in yellow and the α1 C-terminal domain in pink. Auxiliary α2δ, β, and γ subunits are colored in green, cyan, and magenta, respectively. Black lines depict membrane boundaries. The C-terminal domain of CaV1 channels is exceptionally large, and only partially resolved in the cryo-EM structure (Fig. 1d). The structure reveals an unexpected interaction between the intracellular domain III–IV linker and the intracellular C-terminal domain of the α1 subunit. The domain III–IV linker serves as the fast-inactivation gate in eukaryotic sodium channels (see below)11, and structure-function studies suggest that it also interacts with the C-terminal domain87,88. The functional significance of this conserved interaction in CaV1 channels is unknown.
Subunit architecture of voltage-gated sodium and calcium channels
The NaV and CaV channel proteins were first identified by photoaffinity labeling, purification, and reconstitution5,6, and their amino acid sequences were determined by cDNA cloning and sequencing7,8. Analysis of cDNA sequences revealed that they have a common evolutionary ancestor within a superfamily of ion channel proteins9,10. The fundamental structural building block of NaV and CaV channels is a noncovalently or covalently linked tetramer of six-transmembrane (TM) subunits or domains that surround a central pore11,12. Prokaryotic NaV channels are composed of four 6-TM subunits in a homotetramer13,14. The crystal structure of the bacterial sodium channel NaVAb revealed the fundamental fold and TM architecture for these channels (Fig. 1)15, which is shared with KV channels (Box 1). The first four TM helices (S1–S4) form the voltage-sensing module that responds to changes in membrane potential. The S5 and S6 TM segments and the connecting P loop form the pore module that mediates selective ion permeation. These two functional modules are connected by the S4–S5 linker helix, which is oriented approximately parallel to the membrane surface. The four subunits assemble in a domain-swapped manner, in which each voltage-sensing module interacts most closely with the pore-forming module of the neighboring subunit (Fig. 1c).
Box 1. Voltage-Gated Potassium Channels.
In the yin and yang of electrical signaling, NaV and CaV channels initiate electrical signals and KV channels terminate them (Fig. 1)1. Here we provide an outline and references to reviews on KV channels, which are not covered in this article. KV channels are homo- or heterotetramers of four homologous subunits with structures similar to one subunit or domain of an NaV or CaV channel 2. The structure of their voltage-sensing module is similar to NaVAb3. Their voltage-dependent gating is now thought to resemble that of NaV and CaV channels, with outward movement of the S4 segment through a “charge-transfer center” within the hydrophobic constriction site in the center of the voltage-sensing module4–6. The open pore of KV1.2 resembles the open pore of NaVMs with a substantial orifice formed by the intracellular ends of the S6 segments3. Selectivity and conductance of K+ proceeds via a very different mechanism from NaV and CaV channels. K+ is conducted as a completely dehydrated cation and interacts only with backbone carbonyls, which form a characteristic, highly conserved ion selectivity filter7. KV channels inactivate by two mechanisms8. N-type inactivation involves an inactivation ball tethered at the amino terminus of the pore-forming α subunit or the auxiliary β subunit, which is thought to enter and block the pore from the intracellular side9. C-type (or P-type) inactivation involves conformational changes in the ion selectivity filter and S6 segments, and it generally resembles slow inactivation of NaV channels10. Detailed presentations of this work on KV channels can be found in the reviews and articles cited below10.
- 1.Hille B. Ionic Channels of Excitable Membranes. 3. Sinauer Associates Inc; 2001. [Google Scholar]
- 2.Jan LY, Jan YN. Voltage-gated potassium channels and the diversity of electrical signalling. J Physiol. 2012;590:2591–2599. doi: 10.1113/jphysiol.2011.224212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 4.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vargas E, et al. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol. 2012;140:587–594. doi: 10.1085/jgp.201210873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen MO, et al. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a potassium channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 8.Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 9.Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 10.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jan LY. Voltage-gated potassium channels. Ion Channel Database, IUPHAR/BPS Guide to Pharmacology. http://www.guidetopharmacology.org/
Eukaryotic NaV and CaV channels are composed of four homologous 6-TM ion channel domains covalently linked to form a 24-TM protein7,8. Early biochemical studies showed that NaV and CaV channels are expressed as multisubunit complexes with auxiliary subunits5,6, but their pore-forming α subunits are sufficient for function as voltage-gated ion channel16. Cryo-electron microscopy (cryo-EM) provided a low resolution structure of a NaV channel17. Recent studies with modern state-of-the-art equipment revealed the molecular architecture of the CaV1.1 channel of skeletal muscle with its complete set of auxiliary subunits at a resolution of 3.6 Å (Fig. 1d), a major breakthrough in structural biology of eukaryotic ion channels18,19. The central α1 subunit of CaV1.1 has a core structure analogous to NaVAb. As expected from extensive biochemical studies6, the α1 subunit is associated with an extracellular α2δ subunit, an intracellular β subunit having a structure resembling the isolated β subunits studied previously20–22, and a 4-TM γ subunit (Fig. 1d).
Voltage Sensing
The most unique feature of NaV and CaV channels is their exquisite sensitivity to transmembrane potential. Hundreds of transmembrane proteins are subject to the same plasma membrane potential but are unaffected by it. What structural and chemical mechanisms are responsible for this unique voltage sensitivity? In their classic work on voltage clamp analysis of Na+ currents in the squid giant axon, Hodgkin and Huxley predicted that voltage sensing must involve movement of charged “particles” across the membrane electrical field in the process of channel activation4. The capacitative “gating currents” resulting from the transmembrane movement of these gating charges were measured about 20 years later 23. The initial amino acid sequence of a NaV channel revealed a striking concentration of positive charges repeated at intervals of three positions in the S4 alpha-helical segmen7. It was subsequently proposed that the S4 segment has a TM position in both resting and activated states and bears the gating charges that drive voltage-dependent activation24,25. These ‘sliding-helix’ or ‘helical-screw’ models posited that: (i) the positive gating charges were neutralized in their TM positions by formation of ion pairs with negatively charged amino acid side chains in surrounding TM segments; (ii) the negative internal membrane potential exerted an electrostatic force to pull those gating charges inward toward the cytosol; and (iii) depolarization released the gating charges to move outward along a spiral pathway, exchanging ion pair partners, carrying the capacitative gating current outward, and initiating conformational changes that result in pore opening24,25. Extensive work on sodium channels has tested this hypothesis, and strong support has emerged from several approaches (reviewed in 26). Highlights of this previous work include: (i) Neutralization of the S4 gating charges by site-directed mutagenesis shifts the voltage dependence of activation and reduces its steepness27. (ii) Neurotoxin binding studies revealed that the S4 segment is in a TM orientation in both resting and activated states28,29. (iii) Chemical labeling studies showed that the S4 segment moves outward through a short TM pathway in the voltage-sensing module during activation30–32. (iv) Disulfide-locking studies showed that the S4 gating charges exchange ion pair partners during outward motion of the S4 segment33–36. These findings support all of the essential features of the sliding-helix/helical-screw model of gating.
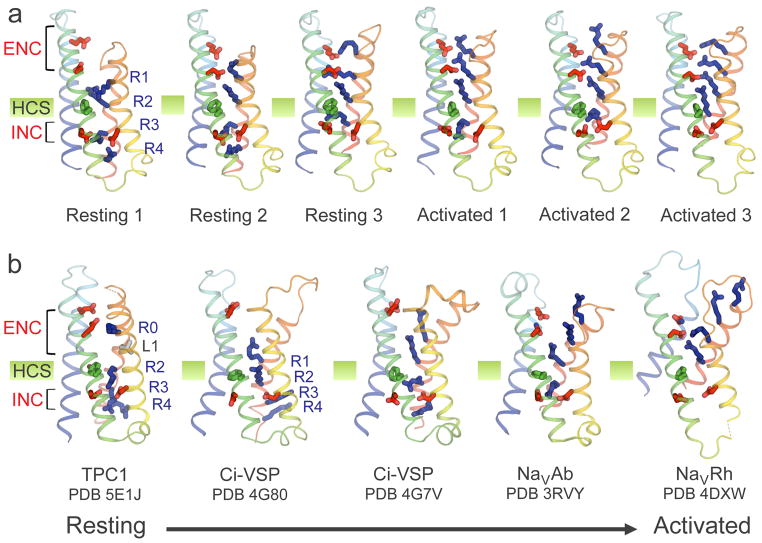
Despite these extensive biophysical studies, the structural basis for voltage sensor function and the chemical mechanisms that support gating charge movement have remained unclear. Recent studies using structural modeling and X-ray crystallographic analysis of multiple classes of ion channels have given new insight into these fundamental questions. The structure of the voltage sensor of the bacterial sodium channel NaVAb has a conformation (Fig. 215) almost identical to the voltage sensor of the KV1.2/2.1 channel chimera (see Box 137). The S1–S2 and S3–S4 helical hairpins form a V-shaped aqueous cleft between them (Fig. 2). A hydrophobic constriction site (HCS, Fig. 2, green) seals the voltage sensor at its center, preventing TM movement of water or ions through the cleft15. The four Arg gating charges in the S4 segment (R1–R4, Fig. 2, blue) are arrayed across the membrane at an angle of ~25° from perpendicular. The R1–R3 gating charges are on the extracellular side of the hydrophobic constriction site (Fig. 2), where they interact with the negatively charged side chains of amino acid residues in the extracellular negative cluster (ENC, Fig. 2, red), as well as other hydrophilic side chains15, backbone carbonyls15, and water in the aqueous cleft. The R4 gating charge is on the intracellular side of the hydrophobic constriction site, where it interacts with negatively charged amino acid side chains in the intracellular negative cluster (INC, Fig. 2, red). The narrow hydrophobic constriction site creates a focused electric field between the aqueous cleft on the extracellular side of the voltage sensor and the cytosol on the intracellular side, as suggested from previous studies32,38. This snapshot of NaVAb structure suggests that the S4 segment could move inward or outward, translocating gating charges through the hydrophobic constriction site in response to changes in electrical field15.
Figure 2. Structure of the voltage-sensing module of NavAb.
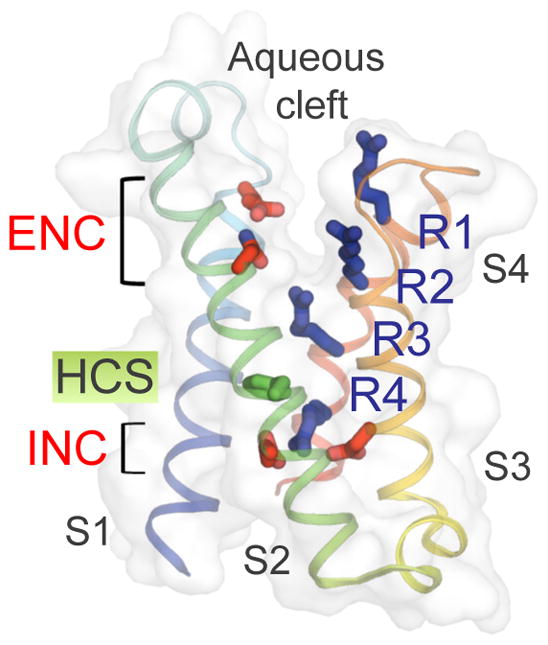
The TM helices are colored from S1 to S4 segments in blue to red spectrum. Side chains of the extracellular and intracellular negative charge cluster (ENC and INC) amino acids are highlighted in red and side chains of the positive gating charge Arg residues (R1–R4) in blue. The hydrophobic constriction site (HCS) is shown in green. The aqueous cleft can be seen between the S1–S2 and S3–S4 hairpins from the overlaying semi-transparent surface.
Molecular modeling and recent X-ray crystallographic studies support the sliding helix/helical screw mechanism of voltage sensor function. Using the Rosetta Membrane ab initio computational modeling system and molecular constraints from disulfide-locking experiments, a series of three resting and three activated states of the voltage sensor were modeled at high resolution for the bacterial NaV channel NaChBa36. The positions of the S4 segments illustrate progressive outward movement of the gating charges and exchange of ion-pair and hydrophilic interactions as they move from Resting States 1–3 outward to Activated States 1–3 (Fig. 3a). Experimental support for these conformational transitions comes from X-ray crystal structures of multiple ion channels and related proteins captured in a series of distinct conformations (Fig. 3b). The crystal structure of NaVAb shows the voltage sensor in a conformation similar to Activated State 2 (compare Fig. 2 vs. 3a). Disulfide locking the voltage sensor in that conformation caused activation followed by inactivation of NaChBac34. Gating charges R1–R3 are external to the hydrophobic constriction site in this state. KV1.2/2.1 (Box 137) and the voltage-gated calcium channel CaV1.118 were also captured in this state, which must be a highly stable state for voltage sensors at 0 mV as in a protein crystal. In contrast, the voltage sensor of the bacterial sodium channel NaVRh was captured in a further activated state in which the R4 gating charge has moved more completely across the hydrophobic constriction site than in Activated State 3 of NaChBac; (Fig. 3b39). Because the R4 gating charge is on the outer edge of the hydrophobic constriction site, this structure likely reflects full activation because the focused electric field can no longer exert electrostatic force on the gating charges. In contrast to these activated states, the structures of the resting states of voltage-gated ion channels have been particularly difficult to study because they exist in cells at the resting membrane potential of approximately −90 mV, and there is no membrane potential in protein crystals.
Figure 3. Structural models of resting and activated states of voltage-sensing modules.
a, Structural models of voltage-sensing module of NaChBac from Rosetta Membrane computational modeling. The Arg gating charges on S4 moves outward from the most resting state (Resting 1) to the most activated state (Activated 3) via several intermediates, passing through the HCS and exchanging their interactions with different INC and ENC side chains. b, Structures of voltage-sensing modules from available X-ray crystallographic structures of voltage-sensitive ion channels and enzyme. From left to right: domain 2 (resting state) of plant TPC1; resting state of Ci-VSP; activated state of Ci-VSP; activated state of NaVAb; inactivated state of NaVRh. Same color scheme as Fig. 2 is used.
The voltage-sensitive phosphatase Ci-VSP has a voltage sensor linked to a lipid phosphatase catalytic domain40. Its voltage dependence is shallow compared to voltage-gated ion channels, consistent with movement of a single gating charge during activation41. In a landmark study, Perozo and colleagues captured this voltage sensor in two functional states, resting and activated, the first time that structures of multiple states of the same voltage sensor have been determined41 (Fig. 3b). The structures are similar to Resting State 1 and Resting State 2 predicted from structural modeling studies of NaChBac (Fig. 3a)36. In another landmark advance, the research groups of Youxing Jiang42 and Robert Stroud43 determined the crystal structure of a Two Pore Channel (TPC1), which is composed of two 6-TM voltage-gated ion channel domains in a single polypeptide and functions in the membrane as a noncovalent dimer. In their structures, the first voltage sensor adopts an activated conformation, as usual for voltage-gated ion channels, while the second voltage sensor is locked in a resting state (Fig. 3b) by binding of calcium, a potent channel regulator. In these structures, a deeper resting state than in Ci-VSP is captured, with the R1–R3 gating charges all located on the intracellular side of the hydrophobic constriction site. The S4 segment is retracted even further toward the intracellular side than in Resting State 1 of NaChBac (Fig. 3a). This series of conformational states of voltage sensors from widely different voltage-sensitive proteins (Fig. 3b) fits surprisingly closely with the predicted series of resting and activated states of the voltage sensors of bacterial NaV channels and KV channels (Fig. 3a, Box 1)36.
What chemical interactions stabilize the gating charges in the lipid membrane and catalyze their movement in response to changes in membrane potential? All of the structures in Fig. 3b show the gating charges interacting with hydrophilic and negatively charged side chains in the extracellular aqueous cleft and the intracellular negative cluster. When gating charges move through the hydrophobic constriction site, they leave interactions with hydrophilic and negatively charged moieties on one side of the barrier and make new interactions with hydrophilic and negatively charged groups on the other side. This exchange of ion-pair and hydrophilic interactions follows precisely the prediction of the sliding-helix model of voltage-sensor function, which posited that the gating charges would move across the membrane by exchange of ion-pair partners24. It seems likely that this sliding-helix mechanism, or a variation on this theme, is responsible for voltage-dependent activation of all voltage-gated ion channels and therefore represents the chemical basis for electromechanical coupling in voltage sensors.
Pore Opening
Classic studies of potassium channels showed that they open at the intracellular end44, and the first ion channel crystal structures of the pore-only bacterial KcsA channels revealed tight closure by crossing of the four pore-lining (S6 equivalent) segments that form the activation gate (Box 145). Since then, crystal structures of pore modules with and without voltage sensors from several bacterial NaV channels have been determined15,39,46–49. The pre-open state structure of NaVAb (with I217C mutation) (Fig. 1b and Box 2) and structures of a number of other pore-only constructs revealed similar closed-pore conformations15. In the NaVAb structure, all four voltage sensors are activated but the pore is tightly closed with average diameters of 7.6 Å and 4.4 Å from the centers of diagonally opposing nearest atoms of I217C and Met 221, respectively (Fig. 4a). This pre-open state is prepared to spring open with all four subunits participating in a simultaneous conformational change to the open state (Box 2). The domain-swapped organization of the subunits of bacterial sodium channels argues for concerted opening, because structural clashes would occur if the subunits undergo the pore-opening conformational change individually.
Box 2. Structures and States.
Structures determined by X-ray crystallography or cryo-EM are essentially fixed snapshots of one functional state of the protein. However, because function cannot usually be measured in the crystal or frozen cryo-EM sample, assigning the probable functional state requires use of indirect information. So far, X-ray crystallography and cryo-EM studies of intact voltage-gated Na+ and Ca2+ channels have revealed two probable functional states—pre-open and inactivated1–3. The initial structure of NaVAb (with I217C mutation) was proposed to be in the pre-open state, one of the required functional states for a tetrameric ion channel that opens by a concerted mechanism. In NaVAb/I217C, all four voltage sensors are activated, the selectivity filter is open and symmetric, but the activation gate is tightly closed1. This fits the expectation of a pre-open state that is poised to spring open on the msec time scale via a conformational change of the activation gate. NaVAb/I217C could conceivably also represent an inactivated state; however, extensive studies of slow inactivation of mammalian and bacterial sodium channels by mutagenesis and molecular modeling support the idea that a change in the conformation of the ion selectivity filter to a nonconductive state is a critical part of the slow-inactivation process4,5. Since molecular dynamics studies indicate that the ion selectivity filter is in an open conformation in NaVAb/I217C6–8, it is not likely to be in a slow-inactivated state. On the other hand, the structures of NaVAb wild-type and NaVRh fit the criteria for slow-inactivated states2,3. In both structures, there are substantial rearrangements of the ion selectivity filter, the central cavity, and the activation gate, which result in pores that are partially collapsed throughout their length. The rectangular or parallelogram shapes of the ion selectivity filters in these channel structures are unlikely to be effective in Na+ conductance, because Na+ prefers four planar waters of hydration in square array, exactly the size and shape of the ion selectivity filter of NaVAb/I217C, which is distorted in the putative inactivated states of NaVAb/WT and NaVRh. A critical need in understanding the full series of conformational changes that NaV channels can undergo is to determine the structures of resting and open states, which will fill in the gaps between these putative pre-open and inactivated conformations.
The available structures of NaV and CaV channels do allow further conclusions about the structures of functionally important states of their domains. The voltage sensors have been captured in activated states multiple times1–3, and both the TPC channel from Arabidopsis thaliana9 and the voltage-sensitive phosphatase Ci-VSP from Ciona intestinalis10 provide the first glimpses of the resting-state structures of voltage sensors. Similarly, the pore-only constructs of NaVMs and NaVAe offer initial views of the structure of the open activation gate11,12. Nevertheless, the field eagerly awaits the structures of complete NaV and CaV channels in resting and open states, arguably the two most important functional states of those channel families.
- 1.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, Clapham DE, Yan N. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vilin YY, Ruben PC. Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem Biophys. 2001;35:171–190. doi: 10.1385/CBB:35:2:171. [DOI] [PubMed] [Google Scholar]
- 5.Pavlov E, Bladen C, Winkfein R, Diao C, Dhaliwal P, French RJ. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys J. 2005;89:232–242. doi: 10.1529/biophysj.104.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti N, Ing C, Payandeh J, Zheng N, Catterall WA, Pomes R. Catalysis of Na+ permeation in the bacterial sodium channel NaVAb. Proc Natl Acad Sci U S A. 2013;110:11331–11336. doi: 10.1073/pnas.1309452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiteux C, Vorobyov I, Allen TW. Ion conduction and conformational flexibility of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A. 2014;111:3454–3459. doi: 10.1073/pnas.1320907111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finol-Urdaneta RK, Wang Y, Al-Sabi A, Zhao C, Noskov SY, French RJ. Sodium channel selectivity and conduction: prokaryotes have devised their own molecular strategy. J Gen Physiol. 2014;143:157–171. doi: 10.1085/jgp.201311037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kintzer AF, Stroud RM. Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature. 2016;531:258–262. doi: 10.1038/nature17194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, McGreevy R, Villalba-Galea CA, Hulse RE, Roux B, Schulten K, Kossiakoff A, Perozo E. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagneris C, Naylor CE, McCusker EC, Wallace BA. Structural model of the open-closed-inactivated cycle of prokaryotic voltage-gated sodium channels. J Gen Physiol. 2015;145:5–16. doi: 10.1085/jgp.201411242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, Loussouarn G, Minor DL., Jr Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol. 2014;426:467–483. doi: 10.1016/j.jmb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
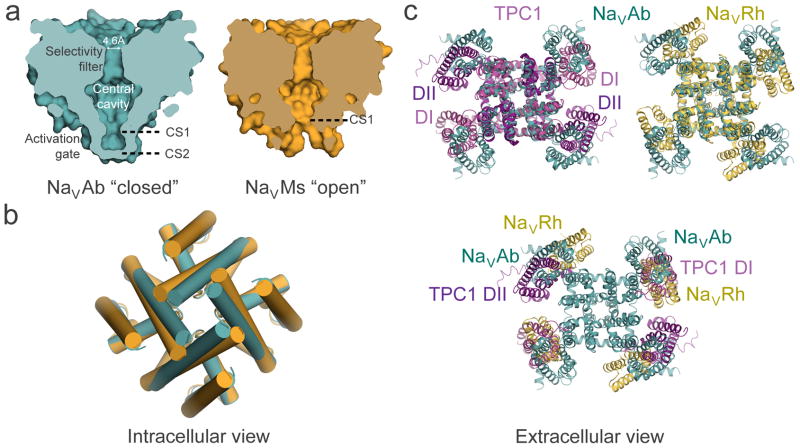
Figure 4. Structural model of conformational changes during channel activation and pore opening.
a, Pore-opening conformational change between the closed pore of NaVAb and the open pore of NaVMs. NaVAb structure (left, cyan) contains two pore constriction sites (CS1 and CS2) with CS2 site completely sealing the pore. NaVMs structure (right, orange) contains one pore constriction site (CS1) that remains open to allow hydrated sodium ion to pass through. b, Superposition of NaVAb and NaVMs pores viewed from the intracellular side. A counterclockwise twisting motion of the S6 segment from the closed pore of NaVAb to the open pore of NaVMs shifts the end of S6 helix outward to dilate the pore diameter. c, Channel activation involves clockwise rotation of the voltage sensor around the pore. Top left: Superposition of TPC1 (light purple for activated state domain I (DI) and dark purple for resting state domain II (DII)) and NaVAb (cyan) structures. Top right: Superposition of NaVRh (yellow) and NaVAb (cyan) structures. Bottom: Overlay of voltage-sensing modules in TPC1, NaVAb, and NaVRh as in the top panel but with the pore modules of TPC1 and NaVRh omitted for clarity. The voltage-sensing module progressively rotates around the pore from the most resting state in TPC1 DII to increasingly activated states in NaVAb, TPC1 DI, and NaVRh.
What conformational changes are involved in pore opening? A structure of the pore-only construct of NaVMs revealed an open activation gate, dilated to a diameter of ~8.4 Å (Fig. 4a)47,50,51. This diameter would accommodate passage of hydrated Na+ 50. Therefore, this structure provides the first experimental evidence for the conformation of an open sodium channel activation gate. Analysis of the closed (NaVAb) and open (NaVMs) pore structures indicates movement of the S6 segments in a twisting manner (Fig. 4b), analogous to the iris of a camera.
Although the S4 segment moves outward through the voltage-sensing module during voltage-dependent activation, recent structures suggest that repositioning of the voltage-sensing module with respect to the pore module drives pore opening. Overlay of the resting and activated voltage-sensing modules in domains II and I of TPC1 (Fig. 4b, left), as well as the activated and further-activated voltage-sensing modules of NaVAb and NaVRh (Fig. 4b, right), reveals a striking clockwise rotation of the voltage-sensing modules with respect to the pore. The rotation observed in these four voltage sensor structures follows the sequence TPC1/DII>NaVAb>TPC1/Di>NaVRh (Fig. 4c), consistent with the relative extents of outward movement of S4 segments in these structures (Fig. 3a). This comparison indicates that the voltage-sensing module rotates in the plane of the membrane around the pore module, exerting a torque on the S4–S5 linker along the inner membrane surface rather than pulling it outward (Fig. 4c15,50). That rigid-body motion of the S4-S5 linker drives rotation and subtle bending of the pore-lining S6 segments, which open the activation gate in an iris-like transition15,50,51 (Fig. 4b).
The structure of NaVAb with four voltage sensors activated but the pore still closed provides evidence for a concerted mechanism of pore opening. This proposed concerted opening mechanism raises an important thermodynamic question. How is the electrostatic energy of outward movement of each S4 segment through the membrane electric field captured and summated to drive the subsequent concerted opening of the pore? Conformational transition of the S4 segment from low-energy alpha helix (i to i+4 H-bond) to higher-energy 310 helix (i to i+3) as it moves through the gating pore may provide a mechanism to capture electrostatic energy in the activated conformation of the voltage sensor26,36. Disulfide-locking studies of the movement of the S4 segment support outward translocation as a 310 helix, which allows each gating charge to interact with the same set of neutralizing residues during transit through the gating pore36. In this model, the electrostatic energy of the electric field would be captured by the energy-requiring transition from alpha helix to 310 helix26. Consistent with this idea, the gating charges outside of the hydrophobic constriction site in NaVAb are in a 310 helix15, whereas KV1.2 has an open pore and its gating charges are in alpha-helical conformation52. Transformation of this 310 helix back to alpha helix would provide energy on the order of 7 kcal/mol53 to drive the pore-opening transition. Structures of closed and open pores in a complete voltage-gated channel are required to test this hypothesis for capturing the energy of the electric field to drive concerted opening.
Concerted pore opening is likely to be physiologically important for NaV channels, which must depolarize excitable cells within less than one millisecond with a smoothly and steeply rising change in membrane potential. A stepwise rising phase reflecting individual conformational changes of four S6 segments would not give optimal electrical stimulation for rapid conduction in axons and or neurotransmitter release in nerve terminals. The four-domain mammalian NaV channels have additional specializations that control kinetics and voltage dependence of activation, such that their four domains activate in a specific voltage/time sequence: III>I>II≫IV54.
Surprisingly in light of these ideas on NaV channel gating, recent structures of KV10.1 (or Eag1)55 and TRPV656 revealed homotetrameric ion channels with structurally distinct S4-S5 linkers and no domain-swapping. However, these channels activate more slowly in their physiological context, so nonconcerted, independent pore-opening movements may be sufficient for their function.
Ion Conductance and Selectivity
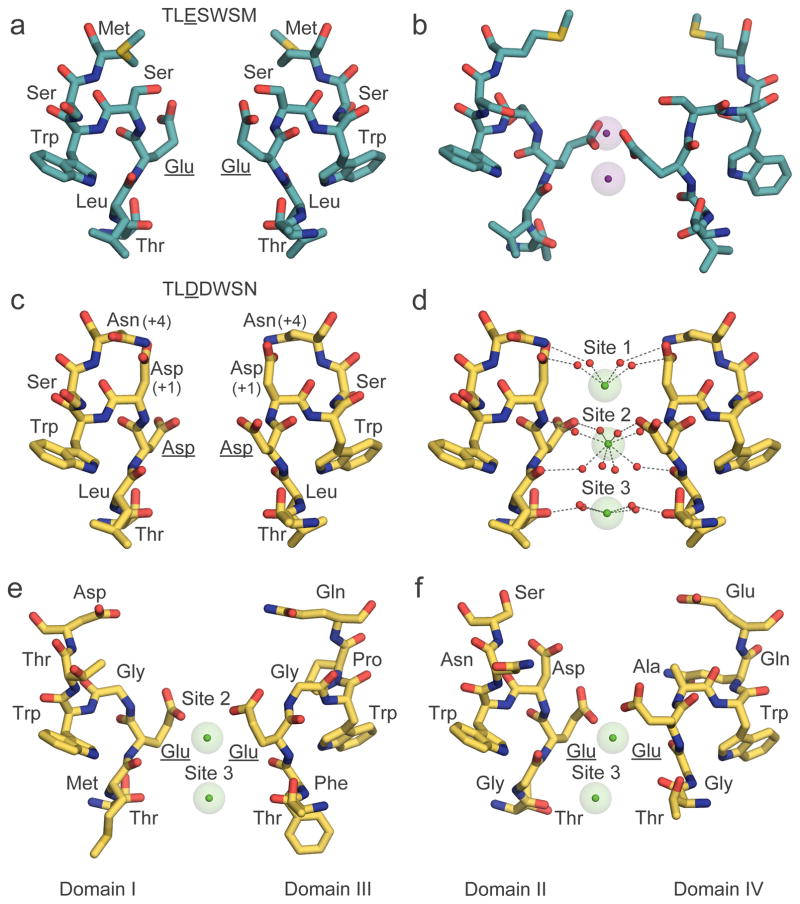
In order to effectively depolarize excitable cells, sodium and calcium channels must be highly selective. Sodium channels have a selectivity sequence of Na+>K+>Ca2+≫Cl− (~1.00:0.08:0.02:<0.01)57,58. Because Na+ and Ca2+ have similar ionic diameters, ion selectivity must be based on chemical interactions rather than molecular sieving. The pore of NaVAb is composed of a wide outer vestibule, a narrow ion selectivity filter, a large water-filled central cavity, and an intracellular activation gate (Fig. 4a). The selectivity filter formed by Thr-Leu-Glu (TLESWSM (Fig. 5a)) is highly conserved among bacterial sodium channels. Its extracellular edge contains negatively charged side chains of four Glu residues that form a high field-strength site with an internal orifice of ~4.6 Å × 4.6 Å. The high field-strength site is followed by central and inner coordination sites of similar internal dimensions composed of backbone carbonyls of the Leu and Thr residues (Fig. 5a). As the ionic diameter of Na+ is ~2.1 Å59, the space between the ion and the coordinating carboxyls and carbonyls must be filled with water, indicating that Na+ is conducted as a hydrated cation. Hydration of Na+ by an inner shell of water molecules in tetragonal bipyramidal or octahedral geometry is a common configuration in protein crystals containing Na+ 60, and it would fit the square geometry of the NaVAb pore very well. The structure of NaVAb15 revealed two key differences in the mechanism of ion conductance between NaV and KV channels: the negatively charged high field-strength site meets the incoming Na+, in contrast to the ion selectivity filter of KV channels which is made completely of backbone carbonyls; and Na+ is conducted as a hydrated cation, whereas K+ is completely dehydrated (Box 161). Thus, the fundamental chemistry of ion permeation in NaV and CaV (see below) channels differs from KV channels. Analysis of amino acid sequences in the pore domain indicated that KV channels form a different clade of the voltage-gated ion channel superfamily than NaV and CaV channels9, and the structural work on bacterial NaV channels shows that this difference in channel structure and function was already well-established in prokaryotes12.
Figure 5. Chemical mechanism of ion permeation and selectivity of NaV and CaV channels with structural models of their ion selectivity filters with ions bound.
a, Na+ selectivity filter (TLESWSM) in NaVAb. b, Representative conformations of Na+ selectivity filter from molecular dynamic simulations of sodium permeation in NaVAb. Conformational dunking of Glu side chain of the high-field strength site allows direct coordination of Na+ ions. c, Ca2+ selectivity filter (TLDDWSN) in CaVAb. d, Hydrated Ca2+ bound in the CaVAb selectivity filter. e, Ca2+ selectivity filter of CaV1.1 from Domains I (TMEGWTD) and III (TFEGWPQ). f, Ca2+ selectivity filter of CaV1.1 from Domains II (TGEDWNS) and IV (TGEAWQE). Na+ (purple) and Ca2+ (green) ions are shown with semi-transparent ionic sphere. Dash lines indicate network of interactions among coordinated water molecules with the ions and protein atoms from high-filed strength site (Glu in NaVAb and CaV1.1, and Asp in CaVAb, underlined) and backbone carbonyls of Leu and Thr. For clarity, only two opposing subunits in the tetramer are shown. Of note, a distantly related non-voltage-gated Ca2+ channel has a different architecture of its outer pore with Asp residues from each subunit directly binding dehydrated Ca2+ in a closed state structure56. The significance of this binding mode in ion conductance is unknown.
The chemistry of Na+ permeation
Molecular dynamics studies revealed complex chemical interactions of conducted Na+ ions with the selectivity filter62–66. Molecular dynamics simulations at a physiological concentration of extracellular Na+ and a membrane potential of 0 mV, where cellular Na+ current is maximal during an action potential, indicate that the Glu side chains of the high field-strength site move in a ‘dunking’ motion with each permeating cation (Fig. 5b)62. This finding was a surprise because the classical model of the ion selectivity filter suggests that it remains rigid as ions move through it3. Na+ moves inward with different ratios of bound water and coordinating Glu carboxyls, resulting in a degenerate network of chemical pathways that may increase ion conductance by increasing entropy in the permeation process62.
Permeating Na+ occupies multiple sites in the conductance pathway. The primary site of occupancy has eight points of coordination formed by the four Glu carboxyls of the high field-strength site and the four backbone carbonyls of the central Leu site62. This finding agrees with crystallographic studies of NaVMs, which reveal Na+ bound in the selectivity filter coordinated by the four conserved Glu carboxyls and four Leu carbonyls49. Na+ also binds in the extracellular vestibule and in the internal site formed by backbone carbonyls of Thr in molecular dynamics simulations62, which show rapid movement through the selectivity filter at rates >107 ions per second, consistent with the high conductance of sodium channels.
A major remaining challenge is understanding sodium permeation in vertebrate NaV channels, which have a different ion selectivity filter motif67,68. Instead of four Glu residues at the high field-strength site, mammalian NaV channels typically have Asp/Glu/Lys/Ala in their four homologous domains67, and the positively charged Lys residue in domain III is crucial for Na+ selectivity68. Evolution of invertebrate NaV and CaV channels has led to a wide array of combinations of amino acid residues in the selectivity filter, which always include at least two negative charges at this high field-strength site69.
The chemistry of Ca2+ permeation
Calcium channels face a difficult task, as they are required to conduct Ca2+ rapidly and selectively in the face of a 70-fold higher concentration of Na+ in the extracellular fluid. How do calcium channels prevent monovalent cation permeation and at the same time conduct Ca2+ rapidly and selectively? Biophysical studies suggested a multi-site knock-off mechanism in which high affinity binding of Ca2+ prevents permeation of monovalent cations, while closely spaced Ca2+ binding sites allow high throughput as entering Ca2+ knocks off bound Ca2+ ions by electrostatic repulsion70–72. NaVAb can be converted to a Ca2+-selective form (CaVAb) for high-resolution structural studies by three Asp mutations to yield TLDDWSD: two of these mutations add negative charge in the vestibule at positions +1 and +4 with respect to the high field-strength site and one widens the lumen and alters the hydrogen-bonding pattern at the high field-strength site by substitution of Asp for Glu (Fig. 5c73,74). Substitution of Asn instead of Asp at the +4 position also gives a highly Ca2+-selective channel. Analysis of the crystal structure of CaVAb with Ca2+ bound revealed a ladder of four Ca2+ binding sites: one in the outer vestibule at the +4 position and three in the selectivity filter — one at the +1 position, a second positioned between the high field-strength carboxyls and the Leu backbone carbonyls at the central site, and a third at the inner Thr backbone carbonyl site (Fig. 5d73). Because these sites are spaced at distances of ~4 Å, strong repulsion would occur between bound Ca2+ ions if all sites were occupied simultaneously. Therefore, we believe that ion conductance operates through alternating occupancy of these sites. When Sites 1 and 3 are occupied, approach of an extracellular Ca2+ to the outer vestibule site causes electrostatic repulsion, pushing the Ca2+ occupying Site 1 inward to Site 2 and the Ca2+ occupying Site 3 into the cytosol. These two Ca2+ ions in the vestibule and Site 2 quickly move inward to Sites 1 and 3, respectively, to complete the catalytic cycle. The high concentration of Ca2+ on the extracellular side assures that Ca2+ ions always move inward. In addition to this knock-off effect, the CaVAb structure also shows that rapid conductance is supported by stepwise movement of Ca2+ up and down a staircase of energy barriers from the outer vestibule, to Site 1 formed by the substituted Asp at the +1 position, on to Site 2, which is the high field-strength site, then down again via Site 3 formed by Thr at the −1 position, and finally into the central cavity and cytosol. According to Eyring rate theory, this stepwise movement over smaller energy barriers will allow much more rapid ion conductance75.
The recent 3.6 Å-resolution cryo-EM structure of CaV1.1 revealed the chemical basis for Ca2+ selectivity in mammalian channels bearing an asymmetric selectivity filter formed from four homologous, but not identical, domains. Two Ca2+ ions are bound in this structure at positions similar to Sites 2 and 3 in CaVAb (Fig. 5e–f vs. 5d). Ca2+ coordination in the selectivity filter involves the high-field strength Glu residues from all four domains that contribute side chain carboxyl groups to Site 2, and the two preceding residues (Thr and Met/Gly/Phe/Gly in the four domains) that provide backbone carbonyls to Site 3, as observed in CaVAb. Ca2+ in the mammalian channel is arranged in a non-symmetric configuration at Site 3, off the central axis and closer to domain II (Fig. 5f).
Like Na+, Ca2+ is too small in diameter to interact directly with its coordinating ligands in the ion selectivity filter and therefore must be conducted as a hydrated ion. Consistent with this model, high-resolution X-ray crystallography structures of Ca2+ bound to CaVAb reveal electron density attributable to water molecules in the first hydration shell positioned between Ca2+ and its coordinating ligands (Fig. 5d)73. These densities are strongest at the high field-strength site, where two quartets of water molecules can be observed above and below bound Ca2+ in favorable cases. These results provide direct evidence that CaV channels select Ca2+ by interaction with its inner hydration shell rather than with the ion itself.
Voltage-Dependent Inactivation
Virtually all NaV and CaV channels inactivate during prolonged depolarization to prevent continuous Na+ or Ca2+ influx. Two types of inactivation are well-known: fast inactivation in 0.5 to 10 msec4 and voltage-dependent slow inactivation in tens to hundreds of milliseconds or longer76. In vertebrate nerve and muscle, fast inactivation causes decay of the Na+ current with a half-life of 1–3 msec3, whereas slow inactivation accumulates during repetitive action potentials and reduces the number of NaV channels that can be activated by a depolarizing stimulus77. The molecular and structural mechanisms of these two forms of sodium channel inactivation are distinct78.
Slow inactivation of NaV and CaV channels
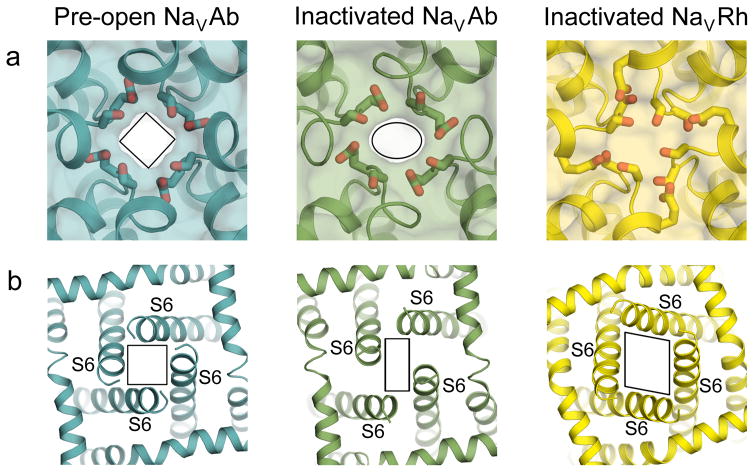
Slow, voltage-dependent inactivation is a common feature of all NaV channels77,79. This process is thought to involve amino acid residues in the pore-lining S6 segments77,79 Conformational changes underlying this slow form of inactivation have been captured in crystal structures of bacterial NaV channels. For NaVAb, physiological studies revealed a multi-step slow inactivation process with time constants from 7–10 msec up to seconds46. Structures of wild-type NaVAb and NaVRh portrayed the channel in slow-inactivated states39,46 characterized by asymmetric collapse of the pore (Fig. 6 and Box 2). At the selectivity filter, the arrangement of the high field-strength site is distorted from nearly square to oval or parallelogram (Fig. 6a)39,46. At the central cavity, the arrangement of amino acid residues is distorted to a parallelogram configuration39,46. Similarly, at the intracellular activation gate, the ends of the four S6 segments describe a parallelogram rather than a square (Fig. 6b)39,46. These changes are caused by coordinated movements of two S6 segments toward the central axis and two S6 segments away from it. The intracellular inactivation gate is closed in these structures, but it is likely that the distortion of the selectivity filter would also make the channels nonconductive or poorly conductive because its shape would no longer fit the optimum geometry for a hydrated Na+ ion.
Figure 6. Conformational changes in the pore associated with slow inactivation.
a, An extracellular view of the pore through the selectivity filter. The selectivity filter collapses from a four-fold symmetric shape in pre-open NaVAb (left) to an oval shape in inactivated NaVAb (middle) to a completely closed pore in inactivated NaVRh (right). b, An intracellular view of the pore at the C-termini of the S6 segments. The pore distorts from a square shape in the pre-open NaVAb structure to a parallelogram shape in the inactivated NaVAb and NaVRh structures.
What drives asymmetric pore collapse during slow inactivation? Molecular dynamics simulations of NaVAb suggested that the Glu residues at the high field-strength site and the pore domain become highly flexible during ion conduction62. As hydrogen bonds from Glu side chains in the high field-strength site in the selectivity filter break away from the adjacent Ser side chains in concert with Na+ translocation62,80, fluctuations in the structure of the selectivity filter are coupled to bending motions of the S6 helices, which result in breaking backbone hydrogen bonds at key conserved residues that are important for slow inactivation80. These fluctuations on the microsecond time scale may eventually lead to transition from a conducting state to a nonconducting state via partial pore collapse. Because slow inactivation is a conserved function of sodium channels, it is likely that this structural transition underlies the slow-inactivation process across phylogeny.
CaV channels also share a slow voltage-dependent inactivation process from protozoa to man81–84. Voltage-dependent slow inactivation of CaV channels is usually measured using Ba2+ as the permeant ion, which prevents more rapid Ca2+-dependent inactivation. Under these conditions, voltage-dependent inactivation half-lives for CaV channels range from 20 msec to hundreds of msec81–84, similar to slow inactivation of NaV channels. Because bacterial NaV channels are the likely evolutionary precursors to CaV channels, we speculate that this voltage-dependent inactivation process of CaV channels has the same underlying pore-collapse mechanism as NaVAb and NaVRh.
Fast inactivation of sodium channels
Eukaryotic NaV channels have evolved an additional fast inactivation process that terminates the Na+ current with a half-life of 1–3 msec, originally described by Hodgkin and Huxley4. Early biophysical studies showed that this process is separate from slow inactivation78. Extensive structure-function studies show that fast inactivation of NaV channels requires a conserved hydrophobic motif in the short intracellular loop connecting domains III and IV, which is thought to fold into the structure and occlude the intracellular mouth of the pore11. The structure of the fast inactivation gate itself has been determined in solution by NMR to be an alpha helix preceded by two beta turns that array the key hydrophobic motif (typically Ile-Phe-Met) ready for interaction with the pore85. However, because bacterial NaV channels do not have an equivalent intracellular loop, further structural understanding of fast inactivation awaits future high-resolution structural studies of eukaryotic NaV channels.
A Consensus Model for the Chemical Basis for Electrical Excitability
We have focused on the emerging chemical and structural basis for electrical excitability, with emphasis on the NaV and CaV channels. These channels have a common ancestor9, which is likely to be a bacterial NaV channel13,14. This evolutionary perspective leads to important inferences about the fundamental chemical basis of electrical signaling. Voltage sensing and voltage-dependent activation are conserved properties of all voltage-gated ion channels. A broad consensus supports a sliding-helix model of voltage sensor function86. Conformational changes in the voltage-sensing module couple voltage-dependent activation to pore opening via an iris-like widening of the bundle of the S6 segments in a concerted fashion to allow for all-or-none increase in ion permeation. Both NaV and CaV channels utilize a similar mechanism for ion conduction12. Their selectivity filters interact with the hydrated cations, and they conduct ions across the membrane through strong interaction and partial dehydration at a high field-strength site followed by sequential interactions with sites formed by backbone carbonyls. Motions of Glu side chains in the high field-strength site provide a knock-off mechanism in which an approaching ion on the extracellular side moves down its electrochemical gradient, knocks off a bound ion in the selectivity filter, and drives it into the cytosol. A primordial slow-inactivation mechanism terminates the inward movement of sodium and calcium ions in the time frame of tens to hundreds of milliseconds by mediating a coordinated asymmetric collapse of the pore12,77. This ordered series of molecular and chemical events is sufficient to generate conducted electrical signals that coordinate cell movement and function in even the simplest protozoans and are highly conserved in form and function in all eukaryotes. The fast-inactivation mechanism is an evolutionary innovation that distinguishes eukaryotes from prokaryotes and provides much more rapid inactivation that is needed for complex repetitive firing of electrical signals. The electrical signals generated by NaV channels, and the resulting intracellular Ca2+ transients generated by CaV channels, serve to coordinate cell function on the millisecond time scale, and they become increasingly sophisticated and refined from protozoa to humans. This fundamental electrical signaling mechanism is essential to complex forms of life as we know them.
References*
*We have annotated articles that have made major contributions to the advances in structural biology of NaV and CaV channels that are the focus of this review.
- 1.Prindle A, et al. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527:59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckert R, Brehm P. Ionic mechanisms of excitation in Paramecium. Annu Rev Biophys Bioeng. 1979;8:353–383. doi: 10.1146/annurev.bb.08.060179.002033. [DOI] [PubMed] [Google Scholar]
- 3.Hille B. Ionic Channels of Excitable Membranes. 3. Sinauer Associates Inc; 2001. [Google Scholar]
- 4.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catterall WA. The molecular basis of neuronal excitability. Science. 1984;223:653–661. doi: 10.1126/science.6320365. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi M, Seagar MJ, Jones JF, Reber BF, Catterall WA. Subunit structure of dihydropyridine-sensitive calcium channels from skeletal muscle. Proc Natl Acad Sci U S A. 1987;84:5478–5482. doi: 10.1073/pnas.84.15.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noda M, et al. Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature. 1984;312:121–127. doi: 10.1038/312121a0. [DOI] [PubMed] [Google Scholar]
- 8.Tanabe T, et al. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature. 1987;328:313–318. doi: 10.1038/328313a0. [DOI] [PubMed] [Google Scholar]
- 9.Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 10.Zakon HH. Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proc Natl Acad Sci U S A. 2012;109(Suppl 1):10619–10625. doi: 10.1073/pnas.1201884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catterall WA. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 12.Catterall WA, Zheng N. Deciphering voltage-gated Na+ and Ca2+ channels by studying prokaryotic ancestors. Trends Biochem Sci. 2015;40:526–534. doi: 10.1016/j.tibs.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. This article opened up a new era in structure-function studies of sodium and calcium channels by revealing a family bacterial ancestors of simple structure that can be easily manipulated by molecular biological methods and can be studied by x-ray crystallography. [DOI] [PubMed] [Google Scholar]
- 14.Koishi R, et al. A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 15.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. This article presented the first crystal structure of a sodium channel and revealed new structura insights into the voltage sensor, ion selectivity filter and pore, and and the drug receptor sites and fenestrations leading to them. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noda M, et al. Expression of functional sodium channels from cloned cDNA. Nature. 1986;322:826–828. doi: 10.1038/322826a0. [DOI] [PubMed] [Google Scholar]
- 17.Sato C, et al. The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature. 2001;409:1047–1051. doi: 10.1038/35059098. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, et al. Structure of the voltage-gated calcium channel CaV1.1 complex. Science. 2015;350:aad2395. doi: 10.1126/science.aad2395. This article (and Ref. 19) gave the first structural views of a eukaryotic calcium channel. It revealed the subunit architecture of the calcium channel complex and the structure of the central α1 subunit, including the transmembrane fold of the voltage sensor and pore and the ion selectivity filter. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, et al. Structure of the voltage-gated calcium channel CaV1.1 at 3.6 A resolution. Nature. 2016;537:191–196. doi: 10.1038/nature19321. This article (and Ref. 18) gave the first structural views of a eukaryotic calcium channel. It revealed the subunit architecture of the calcium channel complex and the structure of the central α1 subunit, including the transmembrane fold of the voltage sensor and pore and the ion selectivity filter. [DOI] [PubMed] [Google Scholar]
- 20.Van Petegem F, Clark KA, Chatelain FC, Minor DL., Jr Structure of a complex between a voltage-gated calcium channel β-subunit and an α-subunit domain. Nature. 2004;429:671–675. doi: 10.1038/nature02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YH, et al. Structural basis of the α1-β subunit interaction of voltage-gated Ca2+ channels. Nature. 2004;429:675–680. doi: 10.1038/nature02641. [DOI] [PubMed] [Google Scholar]
- 22.Opatowsky Y, Chen CC, Campbell KP, Hirsch JA. Structural analysis of the voltage-dependent calcium channel β subunit functional core and its complex with the α1 interaction domain. Neuron. 2004;42:387–399. doi: 10.1016/s0896-6273(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 24.Catterall WA. Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 25.Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci U S A. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuhmer W, et al. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 28.Rogers JC, Qu Y, Tanada TN, Scheuer T, Catterall WA. Molecular determinants of high affinity binding of alpha-scorpion toxin and sea anemone toxin in the S3-S4 extracellular loop in domain IV of the Na+ channel alpha subunit. J Biol Chem. 1996;271:15950–15962. doi: 10.1074/jbc.271.27.15950. [DOI] [PubMed] [Google Scholar]
- 29.Cestèle S, et al. Voltage sensor-trapping: Enhanced activation of sodium channels by beta-scorpion toxin bound to the S3–S4 loop in domain II. Neuron. 1998;21:919–931. doi: 10.1016/s0896-6273(00)80606-6. [DOI] [PubMed] [Google Scholar]
- 30.Yang N, Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15:213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 31.Yang N, George AL, Jr, Horn R. Probing the outer vestibule of a sodium channel voltage sensor. Biophys J. 1997;73:2260–2268. doi: 10.1016/S0006-3495(97)78258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 33.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci U S A. 2008;105:15142–15147. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci U S A. 2009;106:22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeCaen PG, Yarov-Yarovoy V, Scheuer T, Catterall WA. Gating charge interactions with the S1 segment during activation of a Na+ channel voltage sensor. Proc Natl Acad Sci U S A. 2011;108:18825–18830. doi: 10.1073/pnas.1116449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. This article presented the first complete, high-resolution model of the resting and activated states of a voltage sensor based on Rosetta Membrane modeling and disulfide locking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 38.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. This article revealed a highly activated conformation of the voltage sensor and, with Ref. 46, revealed the structural basis for the pore-collapse mechanism of slow inactivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 41.Li Q, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. This article on an invertebrate voltage-sensitive phosphatase revealed the structural change upon activation of a voltage sensor for the first time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo J, et al. Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature. 2016;531:196–201. doi: 10.1038/nature16446. This article (with Ref. 42) revealed the structure of a two-pore channel (TPC) for the first time, including the first structure of an ion channel voltage sensor in the resting state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kintzer AF, Stroud RM. Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature. 2016;531:258–262. doi: 10.1038/nature17194. This article (with Ref. 41) revealed the structure of a two-pore channel (TPC) for the first time, including the first structure of an ion channel voltage sensor in the resting state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armstrong CM. Interaction of tetraethylammonium ion derivatives with the potassium channels of giant axon. J Gen Physiol. 1971;58:413–437. doi: 10.1085/jgp.58.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 46.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. This article, with Ref. 39, revealed the structural basis for the pore-collapse mechanism of slow inactivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCusker EC, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. This article gave the first view of an open activation gate in a pore-only structure of a sodium channel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaya D, et al. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol. 2014;426:467–483. doi: 10.1016/j.jmb.2013.10.010. This article revealed the high-resolution structure of the four-helix bundle formed by the C-terminal tail of a bacterial sodium channel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naylor CE, et al. Molecular basis of ion permeability in a voltage-gated sodium channel. EMBO J. 2016;35:820–830. doi: 10.15252/embj.201593285. This article showed binding of spdium ions in the selectivity filter of a sodium channel for the first time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bagneris C, Naylor CE, McCusker EC, Wallace BA. Structural model of the open-closed-inactivated cycle of prokaryotic voltage-gated sodium channels. J Gen Physiol. 2015;145:5–16. doi: 10.1085/jgp.201411242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bagneris C, et al. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat Commun. 2013;4:2465. doi: 10.1038/ncomms3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 53.Smythe ML, Huston SE, Marshall GR. The molten helix: effects of solvation on the α to 310 helical ttransition. J Am Chem Soc. 1995;117:5445–5452. [Google Scholar]
- 54.Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol. 2002;120:629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whicher JR, MacKinnon R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saotome K, Singh AK, Yelshanskaya MV, Sobolevsky AI. Crystal structure of the epithelial calcium channel TRPV6. Nature. 2016;534:506–511. doi: 10.1038/nature17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hille B. The permeability of the sodium channel to metal cations in myelinated nerve. J Gen Physiol. 1972;59:637–658. doi: 10.1085/jgp.59.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finol-Urdaneta RK, et al. Sodium channel selectivity and conduction: prokaryotes have devised their own molecular strategy. J Gen Physiol. 2014;143:157–171. doi: 10.1085/jgp.201311037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahler J, Persson I. A study of the hydration of the alkali metal ions in aqueous solution. Inorg Chem. 2012;51:425–438. doi: 10.1021/ic2018693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a potassium channel- Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 62.Chakrabarti N, et al. Catalysis of Na+ permeation in the bacterial sodium channel NaVAb. Proc Natl Acad Sci U S A. 2013;110:11331–11336. doi: 10.1073/pnas.1309452110. This article presented molecular dynamic simulations that provide the first description of sodium selectivity and conductance under physiological conditions of ionic composition and membrane potential, and revealed that Glu side chains in the high field-strength site move inward with each round of sodium transport, creating degenerate modes of sodium conductance coordinated by different combinations of waters of hydration and carboxylate ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulmschneider MB, et al. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A. 2013;110:6364–6369. doi: 10.1073/pnas.1214667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barber AF, et al. Hinge-bending motions in the pore domain of a bacterial voltage-gated sodium channel. Biochim Biophys Acta. 2012;1818:2120–2125. doi: 10.1016/j.bbamem.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corry B, Thomas M. Mechanism of ion permeation and selectivity in a voltage gated sodium channel. J Am Chem Soc. 2012;134:1840–1846. doi: 10.1021/ja210020h. [DOI] [PubMed] [Google Scholar]
- 66.Stock L, Delemotte L, Carnevale V, Treptow W, Klein ML. Conduction in a biological sodium selective channel. J Phys Chem B. 2013;117:3782–3789. doi: 10.1021/jp401403b. [DOI] [PubMed] [Google Scholar]
- 67.Heinemann SH, Terlau H, Stühmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 68.Favre I, Moczydlowski E, Schild L. On the structural basis for ionic selectivity among Na+, K+, and Ca2+ in the voltage-gated sodium channel. Biophysical Journal. 1996;71:3110–3125. doi: 10.1016/S0006-3495(96)79505-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stephens RF, Guan W, Zhorov BS, Spafford JD. Selectivity filters and cysteine-rich extracellular loops in voltage-gated sodium, calcium, and NALCN channels. Front Physiol. 2015;6:153. doi: 10.3389/fphys.2015.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almers W, McCleskey EW, Palade PT. A nonselective cation conductance in frog muscle membrane blocked by micromolar external Ca++ J Physiol. 1984;353:565–583. doi: 10.1113/jphysiol.1984.sp015351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Almers W, McCleskey EW. The nonselective conductance due to calcium channels in frog muscle: calcium-selectivity in a single file pore. J Physiol. 1984;353:585–608. doi: 10.1113/jphysiol.1984.sp015352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hess P, Tsien RW. Mechanism of ion permeation through calcium channels. Nature. 1984;309:453–456. doi: 10.1038/309453a0. [DOI] [PubMed] [Google Scholar]
- 73.Tang L, et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 2014;505:56–61. doi: 10.1038/nature12775. This article gave structural insights into the mechanism of calcium selectivity in a voltage-gated calcium channel, in which calcium ions interact with a series of four coordination sites and are conducted by a knock-off mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yue L, Navarro B, Ren D, Ramos A, Clapham DE. The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol. 2002;120:845–853. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dang TX, McCleskey EW. Ion channel selectivity through stepwise changes in binding affinity. J Gen Physiol. 1998;111:185–193. doi: 10.1085/jgp.111.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adelman WJ, Palti Y. The effects of external potassium and long duration voltage conditioning on the amplitude of sodium currents in the giant axon of the squid Loligo pealei. J Gen Physiol. 1968;54:589–606. doi: 10.1085/jgp.54.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vilin YY, Ruben PC. Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem Biophys. 2001;35:171–190. doi: 10.1385/CBB:35:2:171. [DOI] [PubMed] [Google Scholar]
- 78.Rudy B. Slow inactivation of the sodium conductance in squid giant axons. Pronase resistance. J Physiol. 1978;283:1–21. doi: 10.1113/jphysiol.1978.sp012485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pavlov E, et al. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys J. 2005;89:232–242. doi: 10.1529/biophysj.104.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boiteux C, Vorobyov I, Allen TW. Ion conduction and conformational flexibility of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A. 2014;111:3454–3459. doi: 10.1073/pnas.1320907111. This article provided evidence from molecular dynamics that dunking of the carboxylate side chains of the Glu residues in the high field-strength site is coupled to bending of the S6 segments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hennessey TM, Kung C. Slow inactivation of the calcium current of Paramecium is dependent on voltage and not internal calcium. J Physiol. 1985;365:165–179. doi: 10.1113/jphysiol.1985.sp015765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brehm P, Eckert R. Calcium entry leads to inactivation of calcium channel in Paramecium. Science. 1978;202:1203–1206. doi: 10.1126/science.103199. [DOI] [PubMed] [Google Scholar]
- 83.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 84.Nilius B, Benndorf K. Joint voltage and calcium dependent inactivation of Ca channels in frog atrial myocardium. Biomed Biochim Acta. 1986;45:795–811. [PubMed] [Google Scholar]
- 85.Rohl CA, et al. Solution structure of the sodium channel inactivation gate. Biochemistry. 1999;38:855–861. doi: 10.1021/bi9823380. [DOI] [PubMed] [Google Scholar]
- 86.Vargas E, et al. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol. 2012;140:587–594. doi: 10.1085/jgp.201210873. This article presented a consensus view of the voltage-dependent activation process of sodium and potassium channels that is consistent with the sliding helix model of voltage sensing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Motoike HK, et al. The Na+ channel inactivation gate is a molecular complex: a novel role of the COOH-terminal domain. J Gen Physiol. 2004;123:155–165. doi: 10.1085/jgp.200308929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kass RS. Sodium channel inactivation in heart: a novel role of the carboxy-terminal domain. J Cardiovasc Electrophysiol. 2006;17(Suppl 1):S21–S25. doi: 10.1111/j.1540-8167.2006.00381.x. [DOI] [PubMed] [Google Scholar]