Abstract
Background.
The evolution of primary glioblastoma (GBM) is poorly understood. Multifocal GBM (ie, multiple synchronous lesions in one patient) could elucidate GBM development.
Methods.
We present the first comprehensive study of 12 GBM foci from 6 patients using array-CGH, spectral karyotyping, gene expression arrays, and next-generation sequencing.
Results.
Multifocal GBMs genetically resemble primary GBMs. Comparing foci from the same patient proved their monoclonal origin. All tumors harbored alterations in the 3 GBM core pathways: RTK/PI3K, p53, and RB regulatory pathways with aberrations of EGFR and CDKN2A/B in all (100%) patients. This unexpected high frequency reflects a distinct genetic signature of multifocal GBMs and might account for their highly malignant and invasive phenotype. Surprisingly, the types of mutations in these genes/pathways were different in tumor foci from the same patients. For example, we found distinct mutations/aberrations in PTEN, TP53, EGFR, and CDKN2A/B, which therefore must have occurred independently and late during tumor development. We also identified chromothripsis as a late event and in tumors with wild-type TP53. Only 2 events were found to be early in all patients: single copy loss of PTEN and TERT promoter point mutations.
Conclusions.
Multifocal GBMs develop through parallel genetic evolution. The high frequency of alterations in 3 main pathways suggests that these are essential steps in GBM evolution; however, their late occurrence indicates that they are not founder events but rather subclonal drivers. This might account for the marked genetic heterogeneity seen in primary GBM and therefore has important implications for GBM therapy.
Keywords: monoclonal origin, multifocal glioblastoma, tumor evolution, tumor genetics, tumor heterogeneity
Glioblastoma (GBM) is the most frequent primary malignant brain tumor in adults, with an average survival time of less than 12 months despite multimodal therapeutic strategies. GBMs can be broadly classified into primary if they are thought to arise de novo and secondary if they progress from lower-grade gliomas.1 The extent of their heterogeneity has recently come to light through the efforts of The Cancer Genome Atlas (TCGA).2–7 However, the evolution of primary GBMs has not been fully understood.8
The incidence of multiple synchronous GBM lesions in the same patient is reported between 0.5% and 35% of diagnosed GBM cases with an average of ~10%.8–11 Multifocal GBMs are associated with worse outcome and poorer overall survival times compared with single focus GBMs.9 There are 2 theories about the nature of these multiple GBMs: they could either arise as independent tumors (also referred to as multicentric gliomas by some authors) or originate from a single tumor (termed multifocal gliomas).12,13 Up to now, multiple GBMs have been grouped into one of the 2 categories based on whether a physical connection was presumed or not. However, due to the highly invasive nature of GBMs, it is hard to exclude the existence of a connection in cases deemed to be multicentric. We therefore use the term “multifocal GBM” throughout this manuscript to mean any case of multiple GBM lesions in a single patient.
While there is a plethora of genetic data on GBM in general, genetic studies on multifocal GBMs are very rare. In an earlier study, we analyzed 3 isolated GBM lesions from the same patient using spectral karyotyping (SKY), microsatellite analysis, and sequencing of PTEN and TP53.14
This study confirmed the monoclonal origin of one case of multifocal GBM. In the current study, we included the aforementioned patient and added 5 more patients with multifocal GBM. Altogether, we analyzed 12 tumors from these 6 patients using a genome-wide and comprehensive approach. We confirmed the monoclonal nature of all investigated multifocal GBMs, identified a characteristic genetic signature, and further differentiated between early and late events in gliomagenesis, thus demonstrating parallel genetic evolution in these tumors. To the best of our knowledge, this is the largest study that has comprehensively compared GBM foci from the same patient.
Materials and Methods
Patients and Ethics Statement
Tissue samples were obtained from 6 patients with newly diagnosed multiple synchronous lesions of GBM without evidence of a connection by MRI undergoing surgery at the Klinik und Poliklinik für Neurochirurgie, Universitätsklinikum Carl Gustav Carus (Dresden, Germany). Details about the patients and samples are provided in the Supplementary materials and methods (Supplementary File S2) and Supplementary Table S1 in File S1. Patients had given prior informed written consent for use of their material for research purposes (EK 179082004). The procedures described here followed German legal regulations for research using human material and are in compliance with the ethical standards laid down by the 1964 Declaration of Helsinki.
DNA and RNA Extraction
DNA and RNA were extracted from fresh-frozen tumor tissue and primary cell cultures by phenol:chloroform extraction using standard protocols and QIAGEN miRNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions, respectively. Tumor content was evaluated on hematoxylin-and-eosin sections by an experienced neuropathologist and had to be at least 80%.
Array Comparative Genome Hybridization
Array comparative genome hybridization (array-CGH) was performed on Agilent’s SurePrint G3 Human CGH Microarray Kit 2x400K (Design ID 021850) according to the manufacturer’s instructions, with the exception that the labeling of reference and test DNA was reversed. Details are available in the Supplementary materials and methods (Supplementary File S2).
Spectral Karyotyping
For preparation of metaphase chromosomes, patient-derived primary tumor cell cultures were treated with Colcemid (0.035 µg/ml) for 60 minutes, followed by incubation in 0.075M KCl for 20 minutes at 37°C and fixation in methanol/acetic acid (3:1) at room temperature. Cell suspensions were dropped onto glass slides and hybridized for 3 days with a self-made SKY hybridization probe cocktail, as previously described.15 For details see Supplementary materials and methods (Supplementary File S2).
Sanger Sequencing
All coding exons of TP53 (NM_000546.4) and PTEN (NM_000314.4) as well as exon 4 of IDH1 (NM_005896.3) and IDH2 (NM_002168.3) and the TERT (NM_001193376.1) promoter were PCR-amplified and sequenced using primers given in Supplementary Table S2 in S1 File.
Panel Next-generation Sequencing
Tumor DNA and DNA from matched blood was enriched using the Illumina TruSight Cancer Panel protocol (Illumina Inc., San Diego) and sequenced on an Illumina MiSeq Desktop sequencer to an average coverage of 300–400x. Variant calling was done using the CLC Genomic Workbench (Qiagen, Aarhus, Denmark) with a threshold of 1% supporting reads. Only rare variants (<1% in the general population) with potential effect on the protein level were considered. Details are provided in the Supplementary materials and methods (Supplementary File S2).
Gene Expression Analysis
Expression analysis was performed on RNA from fresh-frozen tumor tissues and on 3 commercially available normal brain samples (see Supplementary materials and methods in S2 File). Only RNAs with RIN>8 were included (n = 6, both foci from patients 1 and 2 and focus 1 from patients 4 and 6). RNA was treated according to manufacturer’s protocol and hybridized onto Agilent’s SurePrint G3 Human Gene Expression 8x60K microarrays, Design ID028004. Analysis of differentially expressed genes, pathway enrichment, and subtype classification was done as previously described16,17 and was elaborated in Supplementary materials and methods (Supplementary File S2).
Results and Discussion
Multifocal GBMs Genetically Resembled Typical Primary GBMs but With a Particular Mutation Profile
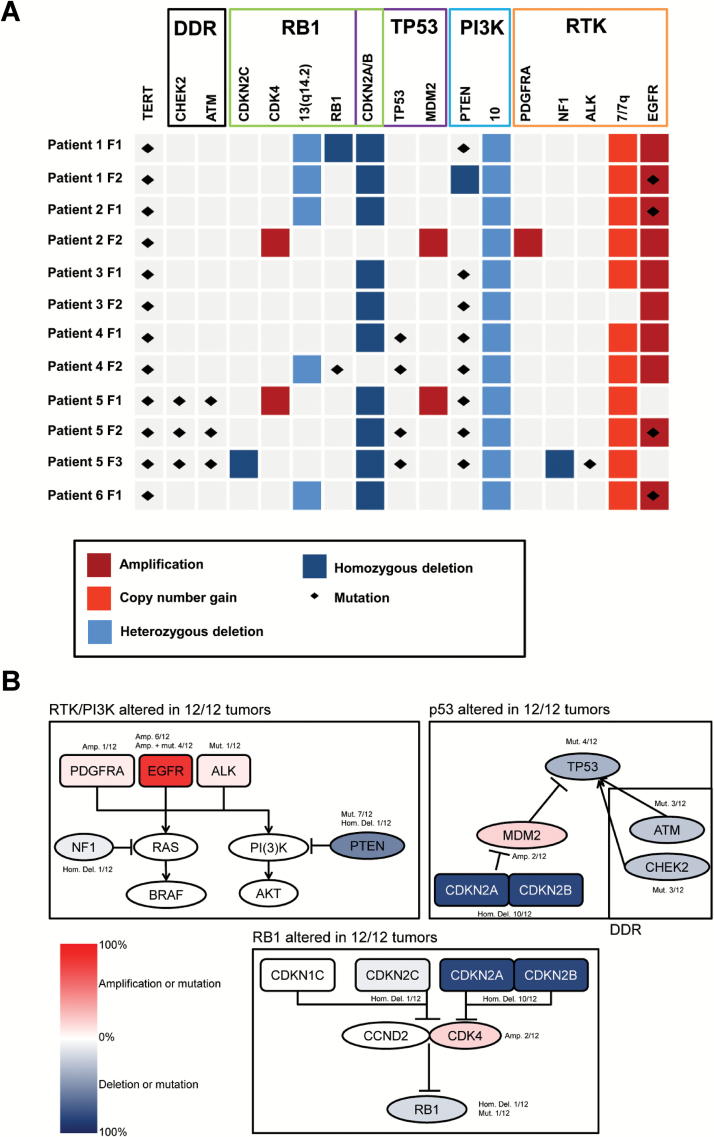
The multifocal GBMs analyzed in our study match the typical genetic profile of primary GBM: combined loss of chromosome 10 and gain of chromosome 7, partial loss of 9p (including CDKN2A/B), EGFR amplification, and lack of IDH1/2 mutations.1,3,18 Aberrations enriched in our tumors are shown in Fig. 1A, and the complete results are summarized in Table 1 and Supplementary Table S4.
Fig. 1.
Most common aberrations and affected pathways in 12 multifocal glioblastomas (GBMs) from 6 patients: (A) Most common copy number variations (CNVs) and mutations in our cohort per tumor focus (F) and patient (Y-axis) and pathway (X-axis). (B) Frequencies of copy number variations (CNVs) and mutations mapped to 3 core signaling pathways (framed in black). Abbreviations: Amp, amplification; DDR, DNA damage response; Hom. Del., homozygous deletion; Mut., mutation; RTK, receptor tyrosine kinase.
Table 1.
Overview of results from array-comparative genome hybridization array-CGH, SKY, and expression analyses of 12 tumor foci from 6 patients with multifocal GBM
| Array-CGHa | SKY | Expression | ||
|---|---|---|---|---|
| Copy number loss | Copy number gain | Subtypeb | ||
| Patient 1 F1 | -1p36.31p36.23,-1p35.1p34.4,-9p23p13.2, --9p22.1p21.3[CDKN2A/B]*,-10,-13,-14,-16p12.2p12.1, -16q,-18p,-21q21.2,-22 | +7p21.3q11.21,++7p11.2[EGFR]*,+7q21.11qter | n.d. | Mesenchymal |
| Patient 1 F2 | -1p36.32p36.22,-9p22.1p21.3,--9p21.3[CDKN2A/B]*, -9p21.1p13.2,-10,--10q23.2q23.31[PTEN], (-12pterp12.3),-13,--13q14.2[RB1], -14,-18p,-21q21.2,-22 | +7,++7p11.2[EGFR]*,(+9q31.1qter) | n.d. | Mesenchymal |
| Patient 2 F1 | -9p23p21.1,--9p21.3[CDKN2A/B],-10,-13,-14q11q21.1, -21q11.2q21.1,-21q22.2qter | +2p21,+7,++7p11.2[EGFR]*,+15q23.31,+19,+20pterp12. 3,+20p11.22qter | n.d. | Classical |
| Patient 2 F2 | -6q16.3,-10,-12q13.13qter,-14q11.2q21.1, -21q11.2q21.1,-22q11.23qter | +1p36.12,+1q32.1,++2p25.3p24.1[MYCN]cth,+3p25.3, +3q28,++4q12q13.1[KIT,PDGFRA],+7,++7p11.2[EGFR]*, +9q34.12q34.13,+11p15.4,+11p13,+11q13.4,+12p,++12 q14.1q24.13[CDK4,MDM2]cth,+14q22.2,+14q32.2,+14q3 2.31,+15q24.3,+17q11.2,+17q21.1q21.2,+17q23.1q23.2 ,+19,+20pterp12.3,+20p11.22qter,++22q11.22,+22q11.2 3,++22q11.23 | n.d. | Classical |
| Patient 3 F1 | -6q25.3,-7p21.1,-8q22.3q23.1,-8q23.3q24.12, --9p21.3[CDKN2A/B],-9q34.13q34.3,-10,-11q13.1q13.2, -11q13.2q13.3 | +3q, +7,++7p11.2[EGFRvIII]* | n.d. | n.d. |
| Patient 3 F2 | -5q35.2,-7p21.1,-7q21.11,--9p21.3[CDKN2A/B],-10 | +3q, ++7p11.2[EGFR]*,++7q36.3 | n.d. | n.d. |
| Patient 4 F1 | -Yq11.221qter,-9p24.3,-9p24.2,-9p24.1p21.1, --9p21.3[CDKN2A/B],-10,-12pterq12,-13q34 | +Ypterq11.221,++7p21.2p21.1,++7p21.1, ++7p15.3,++7p15.3p14.3,++7p14.3,++7p14.3, ++7p11.2[EGFR],++7p11.2,+7q,++12p11.22p11.21, +12p11.21p11.1 | 61~79,XX,-Y,der(1)t(1;12)(q11;p?),+der(7) t(7;?Y)(p11;?p),+der(7)t(7;12)(p11;?p)ins(7;3) (p11;?),+der(7)t(7;12)(p11;q11),del(9)(?p21p24) x2,-12,-21 | Classical |
| Patient 4 F2 | -3pterp12.3,-10,-13q12.13q12.3,-13q14.2q21.1,-22 | +7pterp14.2,++7p11.2[EGFR],+7q,+9q31.3qter | 84~91,XXYY,der(3)t(3;9)(p?12.3;?q31) x2,+7,+der(7)t(7;12)(q36;?),-10,-22,-22,der(22) t(2;22)(?;p11) | n.d. |
| Patient 5 F1 | -1p35.1p34.3,-1p33p32.3,-6p22.3,-9p22.3p13.3, --9p21.3[CDKN2A/B]*,-10q21.2qter,-12q12, -12q13.2q13.3,-12q14.1,-12q14.3,-12q15,-15q26.1 | +7,++12q13.3q14.1[CDK4],++12q14,++12q14. 3q15,++12q15[MDM2],+19,+20,+21 | n.d. | n.d. |
| Patient 5 F2 | -1p36.32p36.22,-1p35.1p34.3,-1p33p32.3, -9p23p22.3,-9p22.2p21.2,--9p21.3[CDKN2A/B]*, -10q21.2qter,-15q26.1 | +7p22.3p13,++7p12.3,++7p12.2,++7p12.1, ++7p11.2[EGFR],+7q11.23q21.11,+7q21. 11q36.3,+19,+20,+21 | 80~90,XXYY,-1,der(1)t(1;15)(p3?3;q2?6.1),der(3)ins(3;5)(q2?1;?q2?q2),+del(7)(p?)x2,del(10)(q2?1.2)x2,-13,-13,der(15)t(1;15)(p3?4.3;q2?6.1)ins(15;1)(q2?4;p3?4.3p3?3) x2,+19,+19,+20,+20,+21 | n.d. |
| Patient 5 F3 | -1p35.1p34.3,-1p33p32.3,--1p32.3[CDKN2C], -1q,-3p,-5q11.2q13.3,-9p23p22.3,-9p22.2p21.2, --9p21.3[CDKN2A/B]*,-10p11.2p11.1,-10q21.3qter, -11q23.3q25,-13q14.4q31.1,-14,-15q26.1,-15q26.1q26.3, -17,--17q11.2[NF1],-19q13.43,-22 | +5p,+5q12.1,+7,+10p15.3p11.21,+10q11.2q21.2, +19p13.2p13.11,+20 | 60~90,XXYY,-1,der(1)t(1;15)(p3?3;q2?6.1), -3,idic(3)(p11.2),-4,der(5)del(5)(p12)del(5)(q?11.2q?13)x2,+i(5)(p10)x2,+7,+9,+10,del(10)(q2?1.2)x2,del(11)(q2?3),-12,del(13)(q?14q?31), -14,-14,der(15)t(1;15)(p3?4.3;q2?6.1)ins(15;1)(q2?4;p3?4.3p3?3)x2[11],der(15)ins(15;1)(q2?4;p3?4.3p3?3)t(15;15)(q2?5;q2?5)[11],der(15) t(1;15)(p3?4.3;q25)[11],-17,+20,-22[cp22] | n.d. |
|
Patient 6
F1 |
-6,-9,--9p21.3p21.1[CDKN2A/B],-10q11.23, -10q21.2q22.2cth,-10q23.2qtercth,-13q13.3q22.2, -14q31.3 | +X,(+2),+2p22.1,(+3),+4q12,(+5),+7,++7p11.2[EGFR], +8q24.13,(+12),+12q15,+15q22.31,(+17),(+19), +19p13.2,(+20),(+21),(+22) | n.d. | Classical / Mesenchymal |
Aberrations shared between the foci from the same patient (eg, same breakpoints in array-CGH) are marked in bold. Common aberrations that are found in both foci but have different breakpoints between foci from the same patient and therefore must have occurred independently are marked with an*.
Array-CGH: Only a short notation of aberrations is used instead of the recommended International System for Human Cytogenetic Nomenclature (ISCN) for CNVs due to lack of space. Important genes involved in amplifications or homozygous deletions in array-CGH have been added in square brackets. Aberrations in parentheses were found in < 50% of the cells.
Expression subtypes are given according to the Verhaak classification. Abbrevations: -, loss; --, homozygous deletion; +, gain; ++, amplification; array-CGH, array comparative hybridization; cp, composite karyotype; cth, chromothripsis; del, deletion; der, derivative chromosome; dup, duplication; idic, isodicentric chromosome; ins, insertion; p, short arm; q, long arm; SKY, spectral karyotyping; t, translocation; ter, terminal.
We detected single copy loss of chromosome 10 including the PTEN locus in all (12/12) tumors. All 12 tumors also carried a TERT promoter mutation (Fig. 1A, Supplementary Table S4). The 2 mutations in our cohort (chr5:1,295,228C>T [C228T] and chr5:1,295,250C>T [C250T]) are the most common TERT mutations in cancer and cause its transcriptional overexpression.19 We also identified protein-disrupting PTEN mutations in all foci of 4 patients (9/12 tumors, 75%). Two patients carried protein-disrupting mutations in TP53 (4/12 tumor foci, 33%) (Supplementary Table S4). Other aberrations typical of GBM included high-level amplification of chromosome 12 including CDK4 and MDM2 loci (2/12), homozygous NF1 deletion (1/12), PDGFRA amplification (1/12), CDKN2C homozygous deletions (1/12), and RB1 mutation or homozygous deletion (2/12) (Fig. 1A). Additionally, we identified mutations in DNA damage sensing and repair genes in ATM (3/12) and CHEK2 (3/12). In a recent study that retrospectively analyzed the TCGA dataset to identify genetic and epigenetic changes associated with multifocality in GBM, the authors also found that these tumors are very similar in their copy number variation (CNV) profile to solitary GBM.20 The authors identified higher expression of CYB5R2 associated with multifocality and poorer survival in GBM and proposed CYB5R2 as a potential prognostic and diagnostic marker. We also found this gene to be downregulated compared with normal brain (Table 3 in S1 File). However, expression levels of CYB5R2 varied substantially between patients and also between tumor foci within one patient, indicating that CYB5R2 might not represent a suitable biomarker (at least in our cohort).
Noticeably, we observed EGFR amplifications and CDKN2A/B homozygous deletions in all patients and in 10/12 tumor foci (approximately 83%). This is more than the 30%–50% and 31%–52%, respectively, reported in GBMs in the literature.1,7 Co-occurrence of PTEN and TERT mutations with the former aberrations was found in 4/6 patients (67%) and in 6/12 tumor foci (50%)—more than one would expect from the literature (15% for combined PTEN mutations and EGFR amplification, 6% for the former with TERT promoter mutations).2,7,21–23 Notably, all tumors harbored at least one event in each of the 3 main signaling pathways in GBM – RTK/PI3K, p53, and RB12,7,16,24 (Fig. 1B). Although the number of samples in our study is too small to draw conclusions, EGFR amplification has been associated with an increased infiltrative and invasive phenotype and thus with multifocal appearance of GBM.25,26
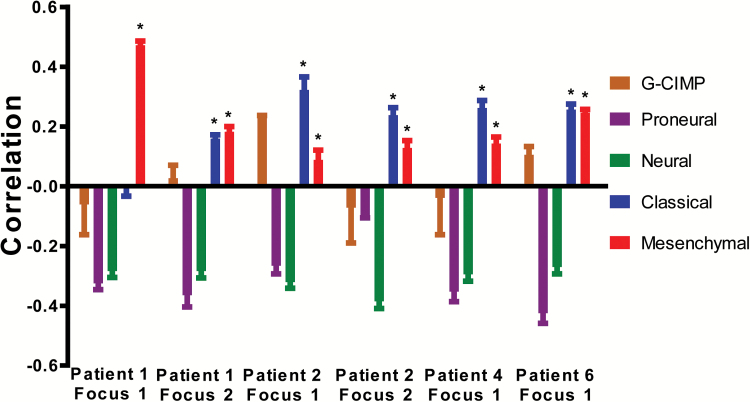
From the 6 tumors in which high-quality RNA was available for expression analysis, 5 tumors significantly correlated with both the Classical and Mesenchymal (and not the Proneural, Neural, or G-CIMP) expression subtypes according to the Verhaak and Brennan classification2,3 (P < 2 x 10–5 [Pearson’s product moment correlation test]); focus 1 from patient 1 only associated with the Mesenchymal subtype (Fig. 2, Supplementary Table S5). This matches the tumors’ mutation profile (EGFR amplification, lack of IDH1/2 mutation, etc.)2 We recently also identified a strong association with both Classical and Mesenchymal subtypes in primary glioblastoma samples.16 Possible explanations might be intratumor heterogeneity or biological similarity between both subtypes. Patel et al. showed that individual cells could cluster with different expression subtypes within one tumor.5 Based on the global gene expression profiles and the aberration spectrum, all multifocal GBMs in this study would classify under the “RTK II Classic” group according to Sturm et al.4
Fig. 2.
Association of gene expression profiles of 6 foci from 4 patients with the expression subtypes according to the Verhaak classification. Tumors were significantly associated with the Classical and Mesenchymal GBM subtypes (positive correlation) but not with the Proneural, G-CIMP, or Neural subtypes. Correlation scores (Y-axis) ranging from 1 (highly correlated) to −1 (highly anticorrelated) are obtained per sample for normalization with 3 different normal brain RNAs. Bars represent the median of correlation scores; error bars denote the interquartile range; * indicates significant correlations (P < .05).
SKY analysis of primary tumor cell lines derived from different tumor foci of patients 4 and 5 revealed that these cells were polyploid (hypertriploid to hypotetraploid) (see Table 1). In addition to identifying numerical and gross chromosomal aberrations found by array-CGH, we detected recurrent structural chromosome rearrangements such as translocations and formation of isochromosomes (Table 1, Supplementary Figure S1 in S1 File). For example, in the SKY analysis of cells derived from tumor focus 1 of patient 4, we found 3 different derivative chromosomes 7 due to complex translocations between chromosomes 7 and 12 in all metaphases analyzed, which is in line with array-CGH data showing multiple amplified regions on chromosome 7 and 12 (Table 1).
Multifocal GBMs Are of Monoclonal Origin
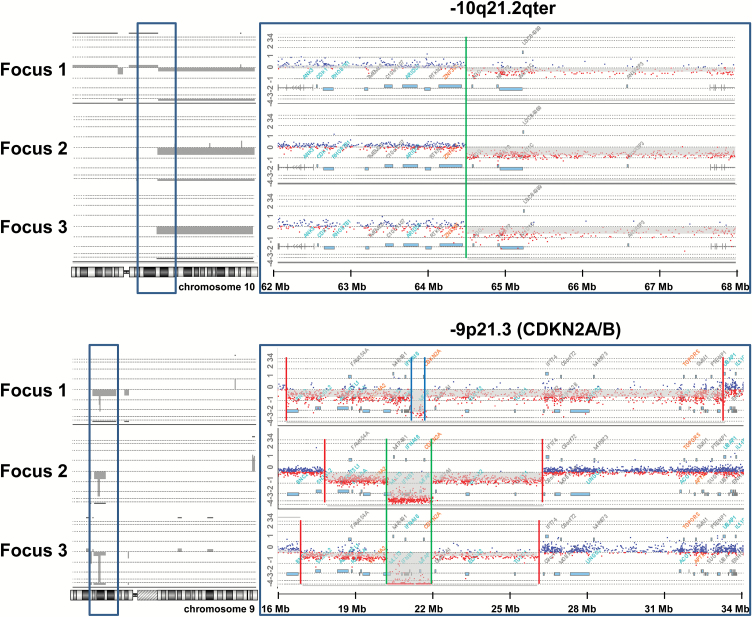
Tumor foci from the same patient always shared distinct aberrations (unique start and stop positions of CNVs, unique translocation breakpoints, or unique single nucleotide variants [SNVs]), clearly proving their monoclonal origin. For example, we found partial loss of the long arm of chromosome 10 with identical breakpoints in all 3 foci from patient 5 (Fig. 3). In the same patient, foci 1 and 2 shared identical breakpoints for the CDKN2A/B deletion, whereas focus 3 differed in its breakpoints (Fig. 3). The identical PTEN point mutation c.518G>A was found in both foci from patient 3 (Table 1, Supplementary Table S4). Using SKY and array-CGH, we could characterize a complex and unbalanced rearrangement between chromosomes 1 and 15 that was identical in all 3 foci from patient 5, previously described as reciprocal translocation t(1;15)(p3?6;q2?5)14 (Supplementary Figure S1 in S1 File).
Fig. 3.
Shared and different copy number variations (CNVs) between 3 tumor foci from patient 5 as detected by array-comparative genome hybridization (array-CGH). Above: Copy number loss of the long arm of chromosome 10 with identical breakpoints in all 3 foci from patient 5. The breakpoint area bordered by the blue frame is enlarged on the right and shows the identical breakpoint in all 3 foci at position [hg19]10q21.2(64,485,714) (indicated by green line). Below: Copy number loss in 9p (log-2-ratio approximately −1) and homozygous deletion of the area containing CDKN2A/B (log-2 ratio approximately −2) (indicated in grey on the left, enlargement of the region on the right). The breakpoints for the homozygous deletion of CDKN2A/B are identical between focus 1 and focus 2 (arr[hg19]9p21.3(21,531,275-22,086,857)x0 indicated by inner green lines) but dissimilar for focus 3 (arr[hg19]9p21.3(21,983,069-22,125,464)x0; blue lines). The region for loss on 9p (indicated by red lines) was different in all 3 foci, indicating 3 different independent and late events in tumor evolution.
The different tumor foci from each patient also shared gross chromosomal aberrations such as loss of whole chromosomes, which in principle could also arise independently. Therefore, we compared available single nucleotide polymorphisms (SNP) on chromosome 10 (from panel-sequencing and Agilent 400K CGH+SNP arrays) between tumor foci from the same patient;they were hemizygous and identical between the foci in all cases, showing that the identical allele of chromosome 10 was lost and indicating the loss of chromosome 10 to be an early event.
Despite their monoclonal origin, the multifocal GBMs showed marked genetic heterogeneity. We always found additional aberrations that were different between foci from the same patient. The tumor foci from one patient shared only between 20% and 51% of aberrations (SNVs and CNVs that were identical between foci based on the criteria explained above); however, this was significantly more when compared with tumor foci from all other patients. Interestingly, Kim et al. found that distant GBM recurrences had an elevated divergence from the initial GBM (<50% of mutations retained) compared with local recurrences.27 The considerable divergence between different tumor foci underlines the genetic instability of GBM leading to marked heterogeneity.
Although we have demonstrated the monoclonal origin of multifocal GBM in all analyzed patients, of course we cannot rule out the existence of truly independent occurrences of multiple GBM lesions in rare cases (eg, in familial tumor syndromes such as Li-Fraumeni syndrome).28
Comparison of Tumor Foci From the Same Patient Identifies Early and Late Events in Multifocal GBM Development
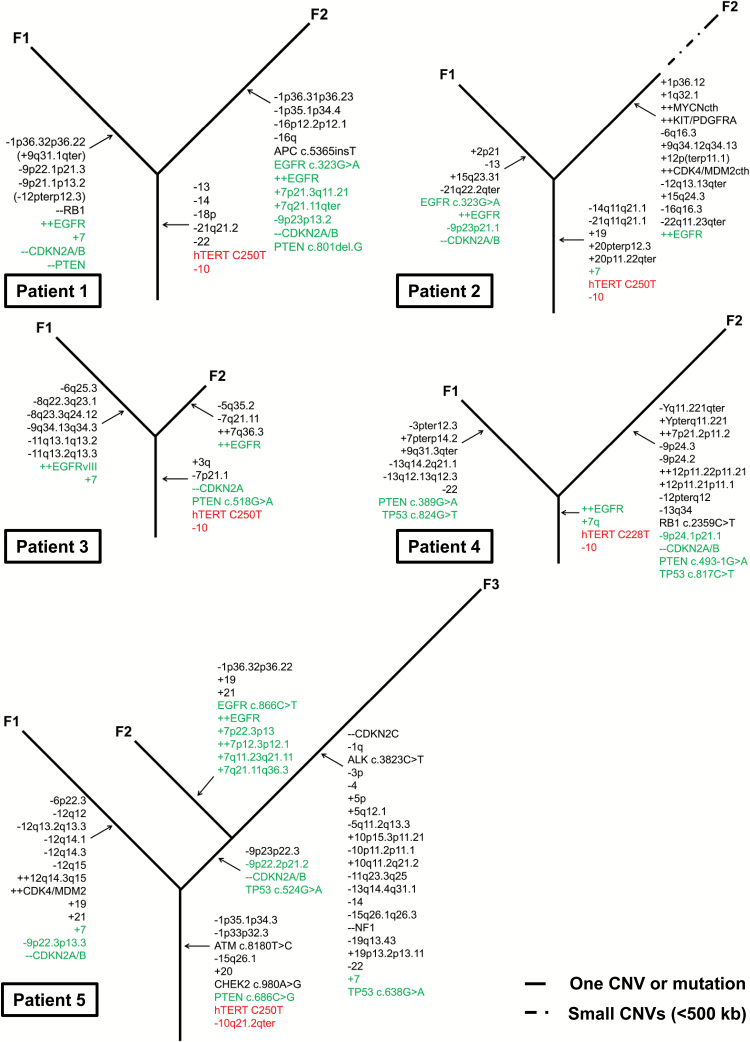
In an effort to better understand the evolution of multifocal GBM, we developed a dendrogram mapping the shared (early) and different (late) aberrations detected in tumor foci from the same patients (Fig. 4). Surprisingly, we found that mutations in frequently affected genes were different between foci from the same patient and therefore must have occurred late and independently during GBM development. For example, EGFR amplifications showed different breakpoints of the amplified regions between the tumor foci in 3 patients (Supplementary Figure S3 in S1 File). We even observed one case with different amplifications of EGFR in the 2 foci, where only one focus showed amplification of EGFRvIII (Supplementary Figures S2 and S3 in S1 File). Although EGFR amplifications were present in different tumor foci of the same patient, the different breakpoints clearly identified them as independent and therefore late events in GBM. This contradicts a recent study, where the authors concluded that EGFR amplification and EGFRvIII are early events in GBM tumorigenesis based on the observation that these alterations were present in different tumor areas.29 However, since the methods used in this study did not define the exact breakpoints of EGFR amplification, it is also possible that the authors observed independent events throughout the tumor. We also detected 2 of the most common oncogenic point mutations in EGFR (ie, c.323G>A and c.866C>T) in only one of the foci in 3 patients and in patient 6 (in whome we had only one focus) (Supplementary Table S4).30 Based on allele frequency (approximately 78%–92%), the mutations must have preceded the amplification of the EGFR locus. The co-occurrence of EGFR amplifications and mutations in a single GBM has been reported previously.2,7 In our cohort, 5/6 patients presented 2–3 independent EGFRalterations, underlying the marked heterogeneity of these tumors and the complexity of EGFR activation in GBM.
Fig. 4.
Evolution in 5 patients with multifocal glioblastoma (GBM): A phylogenetic dendrogram showing the genetic evolution based on the shared and different alterations in the tumor foci from each patient. Loss of chromosome 10 and TERT promoter mutation were the only events that were shared between tumor foci in all patients (red). Frequent affected genes/regions considered important for mGBM development are marked in green. The dashed line in patient 2 focus 2 indicates the presence of numerous (14) small duplications and deletions (<500 kb). Abbreviations: -, deletion; -- , homozygous deletion; +, gain; ++, amplification; F, focus.
Similarly, homozygous CDKN2A/B deletions were found in all of our patients but were different between the tumor foci in most cases (Supplementary Figure S3 in S1 File). We also observed independent occurrence of TP53 and PTEN mutations and gain of chromosome 7 (see Fig. 4, Table 1). Furthermore, we found aberrations that always occurred in only one of the foci and therefore always later during tumor development, which are likely progression markers (eg, RB1 mutation or deletion and amplification of MDM2/CDK4 and PDGFR).
Large-scale studies of glioblastomas found 8 genes to be significantly mutated across hundreds of samples, including TP53, PTEN, RB1, NF1, and EGFR, and hence identified them as necessary drivers.2,7,31,32 Ozawa et al. used mathematical modeling of the TCGA dataset and predicted gain of chromosome 7 and loss of chromosome 10 to be early events, followed by CDKN2A loss and/or TP53 mutation, followed by subtype specific late events (EGFR or PDGFR amplification, loss of NF1).33 Kim et al. analyzed the TCGA dataset to identify clonal (early) and subclonal (late) events by integrating variant allele fractions and concluded that alterations in TP53 and PIK3CA/PIK3R1 were likely founder events, while EGFR, PDGFRA, and PTEN alterations could occur at different time points.34 However, our results show that common events are not necessarily early events in glioblastomagenesis but rather could have occurred later and independently in different subclones. In fact, the only alterations that were always shared between foci in all patients—and therefore might be early founder events—were loss of one copy of chromosome 10 including PTEN and TERT promoter mutations. This is in line with the recent work by Patel et al., who found chromosome 10 LOH in all 672 individual tumor cells from 5 primary GBMs, leading them to propose it as an early event in glioblastomagenesis.5TERT promoter mutations have been theorized to be an early event in the progression of hepatocellular carcinoma and follicular thyroid adenoma.35,36 A recent study has even found them to precede chromosome 7 and 10 alterations.37 We suggest that TERT promoter mutations constitute an early event in the development of (multifocal) GBMs based on their high frequency and their shared occurrence in different tumor foci from the same patient.38 That EGFR mutations and amplifications follow the loss of chromosome 10 in glioblastomagenesis has been previously hypothesized based on the frequency of both events and their overlap,33,39 and our data now confirm this hypothesis.
The early loss of one copy of PTEN and the occurrence of the second hit in PTEN after divergence of the tumor foci suggests that loss of function of one allele of PTEN might be sufficient for tumor initiation. This is supported by recent studies showing that loss of one copy of PTEN can cause tumorigenicity by haploinsufficiency.40 Ozawa et al. computationally predicted that loss of PTEN is the driving initial non-disjunction event in GBM on chromosome 10.33 Another possibility is that a gross chromosomal lesion can be the initial event in tumorigenesis due to deregulation of expression of a very large number of genes, which is a matter of debate in the scientific community.41,42 One could argue that we may have missed other early (initiating) events since we did not perform genome-wide sequencing. However, since primary GBMs have been largely analyzed by whole-genome and exome sequencing,2,3,7 it is somewhat unlikely that there are are still unidentified common early founder events in primary GBM.
Parallel Genetic Evolution of Multifocal GBM
There are 2 possible scenarios for the development of multifocal GBMs: (i) separation of a subpopulation of tumor cells within a primary GBM tumor mass at a later stage of tumor development or (ii) parallel genetic evolution of GBM tumors from a common tumor precursor cell clone through accumulation of further aberrations (early separation of cell clones). Our results indicate that the latter scenario is more likely. First of all, there were only 2 common early events found in all patients: loss of one copy of PTEN/chromosome 10 and point mutations in the TERT promoter. Moreover, the alterations in the 3 main GBM pathways (RTK/MAPK, p53, and RB1) that were present in all tumors—and therefore presumably necessary drivers—occurred independently (in parallel) in the different tumor foci from the same patient (Fig 1 and Fig 4).
Accordingly, we found an enrichment in the same signaling pathways in the 6 tumors that were available for expression analysis (including EGF, mechanistic target of rapamycin [mTOR], and ErbB signaling pathways as well as cell cycle and Rb-mediated pathways) (Supplementary Figure S4 in S1 File). This again suggests that dysregulation of a defined set of core pathways might be important for the evolution of multifocal GBMs, which can be achieved via parallel genetic evolution.
Using a mathematical model based on mutation frequencies in a GBM cohort, Gerstung et al. proposed that there is no clear linear temporal order of mutation accumulation in GBM.43 The authors concluded that the first mutations occur in TP53, parallel to which EGFR, NF1, and PTEN mutate, sequentially followed by RB1 and other mutations.43 Our results for the first provide experimental data for a parallel genetic evolution model in GBM. In line with Gerstung et al., we identified RB1 alterations as a late event in GBM development that sequentially follows aberrations in the 3 main pathways. However, our results suggest that TP53, PTEN, EGFR, and NF1 alterations are not founder events but rather later events in GBM development. While multiple events in a single pathway could be seen (eg, c-KIT, PDGFRA, and EGFR amplification; CDKN2A/B deletion and TP53 mutation; CDKN2A/B and RB1 mutation/deletion), we confirmed that some events were mutually exclusive (eg, MDM2/CDK4 amplification and TP53 mutation or RB1 deletion/mutation; NF1 deletion and EGFR amplification) that have also been described in primary GBM.2,7
Since multifocal GBMs genetically resemble primary GBMs, as we show here and as recently shown,20 we assume that our findings can be transferred to primary GBMs and that parallel genetic evolution might be accountable for the marked heterogeneity in GBMs. Recent findings in recurrent GBM, showing different alterations in key GBM genes and pathways compared with the initial tumor in some cases, would support this hypothesis.27,34 This might have important implications for targeted therapies, especially if directed against specific mutations.30,44
Chromothripsis Is a Late Event That Occurred Without TP53 Mutations
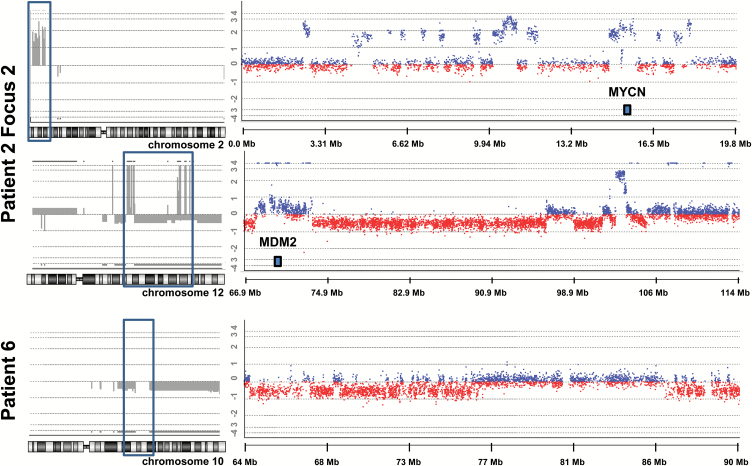
Two tumors showed regions with an exceptionally large number of CNVs indicating chromothripsis (Fig. 5).45 Focus 2 from patient 2 had CNVs alternating between 3 copy number states on the short arm of chromosome 2 (34 breaks) and the long arm of chromosome 12 (64 breaks) leading to amplification of the oncogenes MYCN as well as CDK4 and MDM2, respectively. Interestingly, previous studies on chromothripsis in neuroblastoma and medulloblastoma identified their association with high-level amplification of MYCN.45 The other focus from patient 2 did not show these CNVs, indicating that chromothripsis happened late in the development of this tumor. Additionally, chromothripsis was identified in the one focus available from patient 6 in chromosome 10 (Fig. 5). None of the tumors had mutations in TP53 (Supplementary Table S4), nor were there mutations in the 25 DNA damage sensing and repair genes included in the Illumina TruSight Cancer Panel. This is in line with recent results of large scale genomic studies.46
Fig. 5.
Chromothripsis in 2 tumors using array-comparative genome hybridization (array-CGH). A total of 34 breaks (17 copy number variations (CNVs) between 2 states (0 and 2) were observed on the short arm of chromosome 2 in focus 2 from patient 2 (topmost on the left framed in blue and enlarged on the right). The long arm of chromosome 12 in the same patient carried a total of 64 breaks (32 CNVs between 3 copy states: −1, 0, and 4; middle figure framed in blue on the left and enlarged on the right). Amplifications of MYCN (chromosome 2) and MDM2 (chromosome 12) are indicated in the figure. The tumor from patient 6 showed a large number of CNVs between 2 copy number states (0 and −1) involving loss on chromosome 10 (bottommost framed in blue on the left and enlarged on the right).
Conclusion
Studies addressing clonal evolution and heterogeneity of cancer usually analyze several fragments or subpopulations obtained from different regions of one tumor or even single cells, assuming that they represent different clonal subpopulations.6,47,48 However, this does not guarantee that these tumor fragments represent individual and independent clonal populations that are biologically relevant, and artifacts may arise from whole genome amplification necessary for single-cell analyses. Another approach is the comparison of primary and recurrent tumors; however, patients with glioblastoma will usually have received chemo- and radiotherapy. The advantage of using multifocal GBMs in our study is that the different tumor foci are independent, spatially separated entities that have gone through a “biological” selection and have given rise to individual tumor masses without the influence of treatment. We therefore propose that multifocal GBMs serve as a unique model for studying the tumor evolution and heterogeneity of GBM. Although we did not analyze subpopulations within one single focus—and therefore might have missed events that occurred only in small subclones—this does not alter our conclusion about late and early events identified by comparison of the different foci within one patient.
We propose that multifocal GBM develops early on from a common precursor with loss of at least one copy of PTEN and a TERT promoter mutation (and possibly further unknown founder alterations) through parallel alteration of the 3 main pathways—RTK/PI3K, p53, and RB1—in different cell clones as necessary drivers (but not founders) of GBM evolution. Therefore, the order of occurrence of these events might not be important. The parallel genetic evolution ultimately leads to different tumor cell clones with the necessary aggregation of alterations in critical pathways for the clinical presentation of these highly malignant tumors. This could then be followed by further alterations in RB1, MDM2/CDK4, and PDGFRA during GBM progression. The parallel genetic evolution might also be responsible for the marked intratumoral heterogeneity of primary GBMs. Since multifocal GBMs are rare tumors, our study is limited by the small number of cases, and our results need to be confirmed in future studies.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by a grant (MeDDrive program) of the Medizinische Fakultät Carl Gustav Carus, Technische Universität Dresden.
Conflict of interest statement. The authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgements
We thank Katja Robel (Klinik und Poliklinik für Neurochirurgie, Universitätsklinikum Carl Gustav Carus Dresden) and Eva-Maria Gerlach, Arleta Käßner-Frensel, and Petra Freitag (Institut für Klinische Genetik, Medizinische Fakultät Carl Gustav Carus) for their excellent technical assistance. The authors are members of the University Cancer Center Dresden Network Brain Tumors.
References
- 1. Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. [DOI] [PubMed] [Google Scholar]
- 2. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 5. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. The Cancer Genome Atlas Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas RP, Xu LW, Lober RM, Li G, Nagpal S. The incidence and significance of multiple lesions in glioblastoma. J Neurooncol. 2013;112(1):91–97. [DOI] [PubMed] [Google Scholar]
- 9. Patil CG, Yi A, Elramsisy A, et al. Prognosis of patients with multifocal glioblastoma: a case-control study. J Neurosurg. 2012;117(4):705–711. [DOI] [PubMed] [Google Scholar]
- 10. Djalilian HR, Shah MV, Hall WA. Radiographic incidence of multicentric malignant gliomas. Surg Neurol. 1999;51(5):554–7; discussion 557. [DOI] [PubMed] [Google Scholar]
- 11. Barnard RO, Geddes JF. The incidence of multifocal cerebral gliomas. A histologic study of large hemisphere sections. Cancer. 1987;60(7):1519–1531. [DOI] [PubMed] [Google Scholar]
- 12. Giannopoulos S, Kyritsis AP. Diagnosis and management of multifocal gliomas. Oncology. 2010;79(3-4):306–312. [DOI] [PubMed] [Google Scholar]
- 13. Batzdorf U, Malamud N. The problem of multicentric gliomas. J Neurosurg. 1963;20:122–136. [DOI] [PubMed] [Google Scholar]
- 14. Krex D, Mohr B, Appelt H, Schackert HK, Schackert G. Genetic analysis of a multifocal glioblastoma multiforme: a suitable tool to gain new aspects in glioma development. Neurosurgery. 2003;53(6):1377–84; discussion 1384. [DOI] [PubMed] [Google Scholar]
- 15. Schröck E, Veldman T, Padilla-Nash H, et al. Spectral karyotyping refines cytogenetic diagnostics of constitutional chromosomal abnormalities. Hum Genet. 1997;101(3):255–262. [DOI] [PubMed] [Google Scholar]
- 16. Seifert M, Garbe M, Friedrich B, Mittelbronn M, Klink B. Comparative transcriptomics reveals similarities and differences between astrocytoma grades. BMC Cancer. 2015;15:952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seifert M, Abou-El-Ardat K, Friedrich B, Klink B, Deutsch A. Autoregressive higher-order hidden Markov models: exploiting local chromosomal dependencies in the analysis of tumor expression profiles. PLoS One. 2014;9(6):e100295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klink B, Schlingelhof B, Klink M, Stout-Weider K, Patt S, Schrock E. Glioblastomas with oligodendroglial component—common origin of the different histological parts and genetic subclassification. Anal Cell Pathol (Amst). 2010;33(1):37–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park CK, Lee SH, Kim JY, et al. Expression level of hTERT is regulated by somatic mutation and common single nucleotide polymorphism at promoter region in glioblastoma. Oncotarget. 2014;5(10):3399–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Q, Liu Y, Li W, et al. Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma. Acta Neuropathol. 2015;130(4):587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126(6):931–937. [DOI] [PubMed] [Google Scholar]
- 22. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Talasila KM, Soentgerath A, Euskirchen P, et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol. 2013;125(5):683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ekstrand AJ, Longo N, Hamid ML, et al. Functional characterization of an EGF receptor with a truncated extracellular domain expressed in glioblastomas with EGFR gene amplification. Oncogene. 1994;9(8):2313–2320. [PubMed] [Google Scholar]
- 27. Kim J, Lee IH, Cho HJ, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28(3):318–328. [DOI] [PubMed] [Google Scholar]
- 28. Paunu N, Syrjäkoski K, Sankila R, et al. Analysis of p53 tumor suppressor gene in families with multiple glioma patients. J Neurooncol. 2001;55(3):159–165. [DOI] [PubMed] [Google Scholar]
- 29. Del Vecchio CA, Giacomini CP, Vogel H, et al. EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene. 2013;32(21):2670–2681. [DOI] [PubMed] [Google Scholar]
- 30. Lee JC, Vivanco I, Beroukhim R, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3(12):e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ozawa T, Riester M, Cheng YK, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim H, Zheng S, Amini SS, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nault JC, Calderaro J, Di Tommaso L, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology. 2014;60(6):1983–1992. [DOI] [PubMed] [Google Scholar]
- 36. Wang N, Liu T, Sofiadis A, et al. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA. Cancer. 2014;120(19):2965–2979. [DOI] [PubMed] [Google Scholar]
- 37. Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. von Deimling A, Louis DN, von Ammon K, et al. Association of epidermal growth factor receptor gene amplification with loss of chromosome 10 in human glioblastoma multiforme. J Neurosurg. 1992;77(2):295–301. [DOI] [PubMed] [Google Scholar]
- 40. Ding L, Chen S, Liu P, et al. CBP loss cooperates with PTEN haploinsufficiency to drive prostate cancer: implications for epigenetic therapy. Cancer Res. 2014;74(7):2050–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13(3):189–203. [DOI] [PubMed] [Google Scholar]
- 42. Duesberg P. Does aneuploidy or mutation start cancer? Science. 2005;307(5706):41. [DOI] [PubMed] [Google Scholar]
- 43. Gerstung M, Eriksson N, Lin J, Vogelstein B, Beerenwinkel N. The temporal order of genetic and pathway alterations in tumorigenesis. PLoS One. 2011;6(11):e27136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27(34):4702–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rausch T, Jones DT, Zapatka M, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148(1-2):59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kloosterman WP, Koster J, Molenaar JJ. Prevalence and clinical implications of chromothripsis in cancer genomes. Curr Opin Oncol. 2014;26(1):64–72. [DOI] [PubMed] [Google Scholar]
- 47. Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105(11):4283–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.