Abstract
To investigate the predictability of genetic adaptation, we examined the molecular basis of convergence in hemoglobin function in comparisons involving 56 avian taxa that have contrasting altitudinal range limits. Convergent increases in hemoglobin-oxygen affinity were pervasive among high-altitude taxa, but few such changes were attributable to parallel amino acid substitutions at key residues. Thus, predictable changes in biochemical phenotype do not have a predictable molecular basis. Experiments involving resurrected ancestral proteins revealed that historical substitutions have context-dependent effects, indicating that possible adaptive solutions are contingent on prior history. Mutations that produce an adaptive change in one species may represent precluded possibilities in other species because of differences in genetic background.
A fundamental question in evolutionary genetics concerns the extent to which adaptive convergence in phenotype is caused by convergent or parallel changes at the molecular sequence level. This question has important implications for understanding the inherent repeatability (and, hence, predictability) of molecular adaptation. One especially powerful approach for addressing this question involves the examination of phylogenetically replicated changes in protein function that can be traced to specific amino acid replacements. If adaptive changes in protein function can only be produced by a small number of possible mutations at a small number of key sites—representing “forced moves” in genotype space—then evolutionary outcomes may be highly predictable. Alternatively, if adaptive changes can be produced by numerous possible mutations—involving different structural or functional mechanisms, but achieving equally serviceable results—then evolutionary outcomes may be more idiosyncratic and unpredictable (1–4). The probability of replicated substitution at the same site in different species may be further reduced by context-dependent mutational effects (epistasis), because a given mutation will only contribute to adaptive convergence if it retains a beneficial effect across divergent genetic backgrounds (4).
To assess the pervasiveness of parallel molecular evolution and to investigate its causes, we examined replicated changes in the oxygenation properties of hemoglobin (Hb) in multiple bird species that have independently colonized high-altitude environments. Specifically, we tested whether high-altitude taxa have convergently evolved derived increases in Hb-O2 affinity, and we assessed the extent to which such changes are attributable to parallel amino acid substitutions. We performed comparisons of Hb function in 56 avian taxa making up 28 pairs of high- and low-altitude lineages (table S1). The comparisons involved pairs of species or conspecific populations that are native to different altitudes.
Under severe hypoxia, an increased Hb-O2 affinity can help sustain tissue O2 delivery by safeguarding arterial O2 saturation while simultaneously maintaining the pressure gradient for O2 diffusion from capillary blood to the tissue mitochondria, so altitude-related changes in Hb function likely have adaptive relevance (5, 6). Evolutionary increases in Hb-O2 affinity can be caused by amino acid mutations that increase intrinsic O2 affinity and/or mutations that suppress the sensitivity of Hb to the inhibitory effects of allosteric effectors such as Cl− ions and organic phosphates (5, 7).
In a highly influential paper on biophysical mechanisms of protein evolution, Perutz (7) predicted that adaptive changes in functional properties of vertebrate Hb are typically attributable to “a few replacements at key positions.” According to Perutz, amino acid substitutions that can be expected to make especially important contributions to evolutionary changes in Hb-O2 affinity involve heme-protein contacts (affecting intrinsic heme reactivity), intersubunit contacts (affecting the oxygenation-linked, allosteric transition in quaternary structure), and binding sites for allosteric effectors (7). If Perutz is correct that adaptive modifications of Hb function are typically attributable to a small number of substitutions at key positions, then it follows that the same mutations will be preferentially fixed in different species that have independently evolved Hbs with similar functional properties. For example, in high-altitude vertebrates that have convergently evolved elevated Hb-O2 affinities, Perutz’s hypothesis predicts that parallel amino acid substitutions should be pervasive.
Most bird species express two tetrameric (α2β2) Hb isoforms in adult red blood cells: (i) the major hemoglobin A (HbA) isoform, which incorporates α-chain products of the αA-globin gene, and (ii) the minor HbD isoform, which incorporates products of the closely linked αD-globin gene. Both iso-forms share the same β-chain subunits. By cloning and sequencing the adult-expressed globin genes, we identified all amino acid differences that distinguish the Hbs of each pair of high-and low-altitude taxa. The comparative sequence data revealed phylogenetically replicated replacements at numerous sites in the αA-, αD-, and βA-globins (Fig. 1 and figs. S1 and S2).
Fig. 1. Amino acid differences that distinguish the Hbs of each pair of high- and low-altitude taxa.
Derived (nonancestral) amino acids are shown in red lettering, and rows corresponding to high-altitude taxa are shaded in blue. Subunits of the major HbA isoform are encoded by the αA- and βA-globin genes, whereas those of the minor HbD isoform are encoded by the αD- and βA-globin genes. Phylogenetically replicated β-chain replacements that contribute to convergent increases in Hb-O2 affinity (N/G83S, A86S, D94E, and A116S) are outlined. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; and Y, Tyr.
After identifying the complete set of Hb substitutions that distinguish each pair of high- and low-altitude taxa, we experimentally assessed how many of the replicated amino acid replacements actually contributed to convergent changes in Hb function. To characterize the functional mechanisms that are responsible for evolved changes in Hb-O2 affinity, we measured P50 (the O2 partial pressure at which Hb is 50% saturated) of purified Hbs in the presence and absence of Cl− ions and the organic phosphate inositol hexaphosphate (IHP) (8). We focus on measures of P50 in the presence of Cl− and IHP, because this experimental treatment is most relevant to in vivo conditions in avian red blood cells.
HbD exhibited uniformly higher O2 affinities than HbA in all examined taxa (table S2), consistent with results of previous studies (9–13). This consistent pattern of isoform differentiation suggests that up-regulating the expression of HbD could provide a ready means of increasing blood O2 affinity. However, our results demonstrate that this regulatory mechanism does not play a general role in hypoxia adaptation, because there was no consistent trend of increased HbD expression among high-altitude taxa (Wilcoxon signed-rank test, Z = −0.775, P = 0.441, n = 26; table S3 and fig. S3).
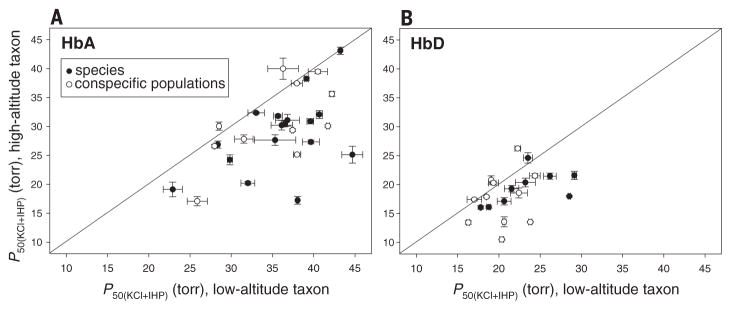
Phylogenetically independent comparisons involving all 28 taxon pairs revealed that highland natives have generally evolved an increased Hb-O2 affinity relative to that of their lowland counterparts, a pattern that is consistent for both HbA (Wilcoxon’s signed-rank test, Z = −4.236, P < 0.0001, n = 28; Fig. 2A and table S2) and HbD (Z = −2.875, P = 0.0041, n = 20; Fig. 2B and table S2). In all pairwise comparisons in which the high-altitude taxa exhibited significantly higher Hb-O2 affinities (n = 22 taxon pairs for HbA and 15 taxon pairs for HbD), the measured differences were almost entirely attributable to differences in intrinsic O2 affinity, rather than differences in sensitivity to Cl− or IHP (table S4). Thus, genetically based increases in Hb-O2 affinity were not generally associated with a diminution of allosteric regulatory capacity (i.e., O2 affinity could still be modulated by erythrocytic changes in anion concentrations), in contrast to the case with some high-altitude mammals (5, 14, 15).
Fig. 2. Convergent increases in Hb-O2 affinity in high-altitude Andean birds.
(A) Plot of P50(KCl+IHP) (± 1 SE) for HbA in 28 matched pairs of high- and low-altitude taxa. Data points that fall below the diagonal line (x = y) denote cases in which the high-altitude member of a given taxon pair possesses a higher Hb-O2 affinity (lower P50). Comparisons involve replicated pairs of taxa, so all data points are phylogenetically independent. (B) Plot of P50(KCl+IHP) (± 1 SE) for the minor HbD isoform in a subset of the same taxon pairs in which both members of the pair express HbD. P50(KCl+IHP), O2 partial pressure at which Hb is 50% saturated in the presence of chloride and inositol hexaphosphate.
Results of experiments based on both native Hb variants and engineered, recombinant Hb mutants revealed that only a subset of replicated replacements actually contributed to convergent increases in Hb-O2 affinity in high-altitude taxa (table S5). These include replicated replacements at just four β-chain sites: N/G83S, A86S, D94E, and A116S. β116 is an α1β1 intersubunit contact, and β94 plays a key role in allosteric proton binding; neither of the other replicated replacements—and few of the affinity-enhancing non-replicated replacements—involved heme contacts, intersubunit contacts, or canonical binding sites for allosteric effectors.
Our experiments revealed a striking pattern of convergence in the oxygenation properties of Hb in high-altitude natives (Fig. 2, A and B), and, in several cases, convergent increases in Hb-O2 affinity were caused by parallel substitutions at key residues that mediate protein allostery (e.g., D94E in the β-chains of high-altitude ground doves and waterfowl; Fig. 1 and table S5). However, in the majority of cases, convergent increases in Hb-O2 affinity were attributable to nonreplicated substitutions and/or parallel substitutions at sites that are not considered “key residues” (e.g., N/G83S in the β-chains of high-altitude hummingbirds and flowerpiercers; Fig. 1). Clearly, evolutionary increases in Hb-O2 affinity can be produced by amino acid substitutions at numerous sites. These findings expose a clear demarcation between the realms of chance and necessity at different hierarchical levels. At the level of biochemical phenotype, and even at the level of functional mechanism, evolutionary changes are highly predictable. At the amino acid level, in contrast, predictability breaks down.
In addition to the many-to-one mapping of genotype to phenotype, the phylogenetic distribution of affinity-enhancing parallel substitutions suggests another possible explanation for the limited contribution of such substitutions to convergent functional changes in the Hbs of distantly related species. The most striking functional parallelism at the amino acid level was concentrated in the hummingbird clade. Replicated G83S substitutions contributed to convergent increases in Hb-O2 affinity in multiple high-altitude hummingbird species (table S5 and fig. S4) (16), and a convergent substitution at the same site (N83S) occurred in one other (nonhummingbird) high-altitude species: the black-throated flowerpiercer, Diglossa brunneiventris. One possible explanation for this phylogenetically concentrated pattern of parallelism is that the mutation’s phenotypic effect is conditional on genetic background, so the same mutation produces different effects in different species.
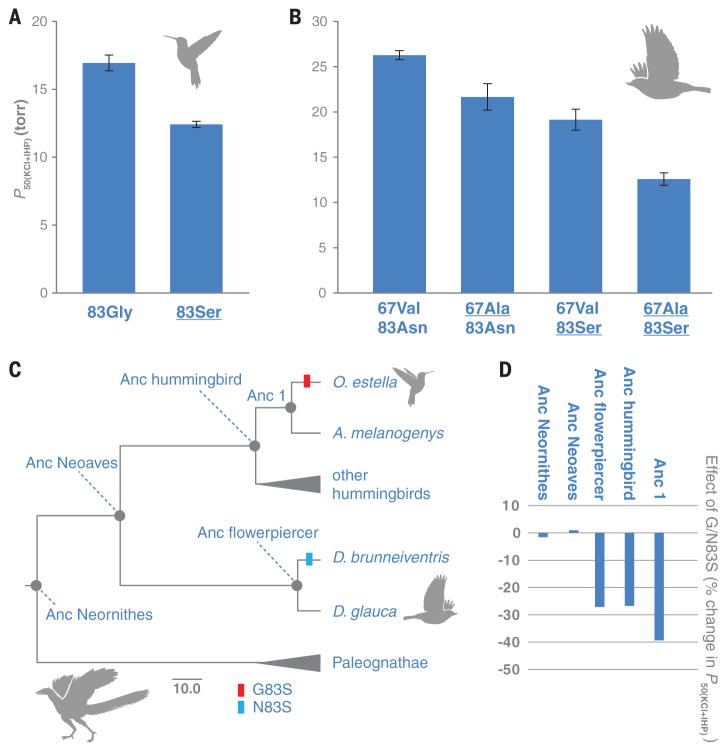
To test this hypothesis, we used ancestral sequence reconstruction in combination with site-directed mutagenesis to test the effect of β83 substitutions in a set of distinct genetic backgrounds. We first resurrected HbA of the common ancestor of hummingbirds (“Anc hummingbird”) (figs. S5 to S7), and we confirmed that G83S has a significant affinity-enhancing effect on this ancestral genetic background (Fig. 3A). This result is consistent with the affinity-enhancing effect of G83S in numerous descendant lineages of high-altitude hummingbirds (table S5 and fig. S4). In similar fashion, we resurrected HbA of the common ancestor of the high- and low-altitude flowerpiercers (“Anc flowerpiercer”) to test the effect of N83S (fig. S7). Hbs of the two flowerpiercers differed at two sites because of substitutions in the D. brunneiventris lineage (V67A in αA-globin, in addition to N83S in βA-globin; Fig. 1). We therefore synthesized a total of four recombinant Hb mutants, representing each possible genotypic combination of the two substituted sites, to measure the relative contributions of V67A and N83S to the evolved increase in Hb-O2 affinity in D. brunneiventris (table S2 and fig. S4). The tests showed that both mutations increased Hb-O2 affinity in an additive fashion (Fig. 3B). We then engineered the same N83S mutation into resurrected ancestral Hbs representing two far more ancient nodes in the avian phylogeny: the reconstructed common ancestor of Neoaves (“Anc Neoaves”) and the common ancestor of all extant birds (“Anc Neornithes”) (Fig. 3C and figs. S5, S7, S8, and S9). In contrast to the highly significant effects of N/G83S in hummingbird and flowerpiercer Hbs, N83S produced no detectable effect in Anc Neoaves or Anc Neornithes (Fig. 3D and table S6). The ancestral hummingbird and flowerpiercer Hbs contained 18 and 32 amino acid states, respectively, that were not present in Anc Neornithes (fig. S7), representing net sequence differences that accumulated over a ~100-million-year time period. The context-dependent effects of N/G83S indicate that lineage-specific substitutions in the ancestry of hummingbirds and flowerpiercers produced a genetic background in which mutations at β83 could contribute to an adaptive increase in Hb-O2 affinity. This adaptive solution was apparently not an option in the deeper ancestry of birds and may also represent a precluded possibility in contemporary, high-altitude members of other avian lineages.
Fig. 3. Phenotypic effects of substitutions at β83 are conditional on genetic background.
(A) The engineered G83S mutation produced a significant reduction in P50(KCl+IHP) (increase in Hb-O2 affinity) in the reconstructed Hb of the hummingbird ancestor. (B) The engineered A67V and N83S mutations produced additive reductions in P50(KCl+IHP) in the reconstructed Hb of the flowerpiercer ancestor. Underlining indicates derived (nonancestral) amino acids. (C) Diagrammatic tree with time-scaled branch lengths showing internal nodes that we targeted for ancestral protein resurrection. Scale bar, 10 million years. (D) N/G83S mutations produced significant increases in Hb-O2 affinity (expressed as reductions in P50(KCl+IHP)) in the ancestors of hummingbirds and flowerpiercers. Substitutions at the same site produced no detectable effects in Anc Neoaves or Anc Neornithes.
These findings reveal a potentially important role of contingency in adaptive protein evolution. In different species that are adapting to the same selection pressure, the set of possible amino acids at a given site that have unconditionally beneficial effects may be contingent on the set of antecedent substitutions that have independently accumulated in the history of each lineage. Consequently, possible options for adaptive change in one species may be foreclosed options in other species.
Acknowledgments
This work was funded by grants from the U.S. NIH (HL087216), the U.S. NSF (IOS-0949931, MCB-1517636, and MCB-1516660), and the Danish Council for Independent Research (10-084-565 and 4181-00094). We thank E. Petersen, H. Moriyama, and A. Kumar for assistance in the laboratory and C. Meiklejohn and K. Montooth for helpful suggestions. All experimental data are tabulated in the supplementary materials, and sequence data are archived in GenBank under accession numbers KX240692 to KX241466.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/354/6310/336/suppl/DC1
Materials and Methods
Figs. S1 to S9
Tables S1 to S6
References (17–33)
REFERENCES AND NOTES
- 1.Stern DL, Orgogozo V. Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losos JB. Evolution. 2011;65:1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- 3.Stern DL. Nat Rev Genet. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 4.Storz JF. Nat Rev Genet. 2016;17:239–250. doi: 10.1038/nrg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber RE. Respir Physiol Neurobiol. 2007;158:132–142. doi: 10.1016/j.resp.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Storz JF, Scott GR, Cheviron ZA. J Exp Biol. 2010;213:4125–4136. doi: 10.1242/jeb.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perutz MF. Mol Biol Evol. 1983;1:1–28. doi: 10.1093/oxfordjournals.molbev.a040299. [DOI] [PubMed] [Google Scholar]
- 8.Materials and methods are available as supplementary materials on Science Online.
- 9.Grispo MT, et al. J Biol Chem. 2012;287:37647–37658. doi: 10.1074/jbc.M112.375600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheviron ZA, et al. Mol Biol Evol. 2014;31:2948–2962. doi: 10.1093/molbev/msu234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galen SC, et al. Proc Natl Acad Sci USA. 2015;112:13958–13963. doi: 10.1073/pnas.1507300112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Natarajan C, et al. PLOS Genet. 2015;11:e1005681. doi: 10.1371/journal.pgen.1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Opazo JC, et al. Mol Biol Evol. 2015;32:871–887. doi: 10.1093/molbev/msu341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storz JF, Runck AM, Moriyama H, Weber RE, Fago A. J Exp Biol. 2010;213:2565–2574. doi: 10.1242/jeb.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natarajan C, et al. Mol Biol Evol. 2015;32:978–997. doi: 10.1093/molbev/msu403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Projecto-Garcia J, et al. Proc Natl Acad Sci USA. 2013;110:20669–20674. doi: 10.1073/pnas.1315456110. [DOI] [PMC free article] [PubMed] [Google Scholar]