Abstract
Background.
Invasion and angiogenesis are major hallmarks of glioblastoma (GBM) growth. While invasive tumor cells grow adjacent to blood vessels in normal brain tissue, tumor cells within neovascularized regions exhibit hypoxic stress and promote angiogenesis. The distinct microenvironments likely differentially affect metabolic processes within the tumor cells.
Methods.
In the present study, we analyzed gene expression and metabolic changes in a human GBM xenograft model that displayed invasive and angiogenic phenotypes. In addition, we used glioma patient biopsies to confirm the results from the xenograft model.
Results.
We demonstrate that the angiogenic switch in our xenograft model is linked to a proneural-to-mesenchymal transition that is associated with upregulation of the transcription factors BHLHE40, CEBPB, and STAT3. Metabolic analyses revealed that angiogenic xenografts employed higher rates of glycolysis compared with invasive xenografts. Likewise, patient biopsies exhibited higher expression of the glycolytic enzyme lactate dehydrogenase A and glucose transporter 1 in hypoxic areas compared with the invasive edge and lower-grade tumors. Analysis of the mitochondrial respiratory chain showed reduction of complex I in angiogenic xenografts and hypoxic regions of GBM samples compared with invasive xenografts, nonhypoxic GBM regions, and lower-grade tumors. In vitro hypoxia experiments additionally revealed metabolic adaptation of invasive tumor cells, which increased lactate production under long-term hypoxia.
Conclusions.
The use of glycolysis versus mitochondrial respiration for energy production within human GBM tumors is highly dependent on the specific microenvironment. The metabolic adaptability of GBM cells highlights the difficulty of targeting one specific metabolic pathway for effective therapeutic intervention.
Keywords: angiogenesis, glioblastoma, glycolysis, hypoxia, invasion.
Aggressive tumor growth of glioblastoma (GBM), the most frequent and most malignant brain tumor, is strongly associated with hypoxia and angiogenesis. In contrast, invasion into the normal brain parenchyma is a key feature of both high-grade as well as low-grade gliomas such as astrocytomas and oligodendrogliomas.1 Invasive and angiogenic tumor cells reside in different niches. While angiogenic tumor cells are found in hypoxic areas, which lack oxygen and nutrients, invasive tumor cells migrate to well-vascularized brain areas with intact vessels and higher levels of oxygen. Tumor cells rely on glycolysis for energy production in the hypoxic niche,2 but Otto Warburg was the first to observe that even under aerobic conditions, malignant tumor cells may use glycolysis instead of mitochondrial respiration.3 The glycolytic switch of tumors has been attributed to the major mutations that occur in cancer, including mutations in oncogenes as well as tumor suppressor genes. These mutations cause proliferation of cancer cells, and the rapid growth in biomass can more efficiently be achieved using anabolic metabolism.4
Warburg′s initial hypothesis that the glycolytic shift is caused by mitochondrial defects in tumor cells has been disproven. Mitochondria in tumor cells are intact and can use oxidative phosphorylation, but they do so to a lesser extent compared with normal cells.5 However, a recent study pointed out that human GBM may rely on mitochondrial glucose oxidation, as shown in an orthotopic GBM xenograft model which did not express significant amounts of hypoxia-inducible factor (HIF)1A and HIF2A.6 Both proteins are key mediators of angiogenesis and are upregulated in hypoxic areas of cancer. Increased mitochondrial activity was also found in an isocitrate dehydrogenase 1 mutant human oligodendroglioma xenograft model.7 The studies together suggest that GBM cells localized in regions with physiological oxygen levels, often corresponding with tumor invasion, can exploit mitochondrial respiration as a major energy source. Whether or not invasive GBM cells utilize a different metabolism compared with angiogenic cells, however, remains elusive. We addressed this important question using a xenograft model derived from a patient biopsy with epidermal growth factor receptor (EGFR) amplification, which grows highly invasive and is independent of angiogenesis. By introducing a dominant-negative (DN) EGFR mutant into the tumor cells, we previously showed that the tumors progress to an angiogenic phenotype.8 In the present study, we demonstrate that the angiogenic progression coincides with a transition from a proneural to a mesenchymal GBM molecular subtype.9 Furthermore, we assessed the metabolic profiles of both invasive and angiogenic tumor cells in this model as well as in patient biopsies.
Materials and Methods
Cell Culture
Biopsy spheroids were cultured as described previously.11 For functional experiments with EGFR-CD533, spheroids with a standardized cell number were generated as described in the section “Lentiviral EGFR-CD533 Production and Infection of GBM Cells.”
The human embryonic kidney cell line 293T (#CRL-11268) and the U87 cell line were obtained from the American Type Culture Collection and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and 1% glutamine. All cell lines were grown at 37°C in a humidified atmosphere comprising 5% CO2.
Cloning of EGFR-CD533
The EGFR-CD533 construct was a gift from Joseph Contessa, Yale University School of Medicine, and cloned into the lentiviral pRRL.sinCMVeGFPpre 12 as described previously.8
Lentiviral EGFR-CD533 Production and Infection of GBM Cells
Lentiviral vectors carrying EGFR-CD533 were produced in 293T cells using FuGene HD transfection reagent (Life Technologies) according to the manufacturer’s instructions. Details of production and infection have been described previously and are provided in the Supplementary materials.8
In vivo Experiments
Nude immunodeficient rats (rnu/rnu Rowett) were fed a standard pellet diet and were provided with water ad libitum. All procedures were approved by the Norwegian National Animal Research Authority. Biopsy spheroids were stereotactically implanted into the right brain hemisphere using 10 spheroids per rat as described previously.13 Rats were euthanized with CO2, perfused intracardially with 0.9% NaCl, and sacrificed when symptoms developed. Brains were removed and fixed in 4% formalin for 1–7 days, or tumors were excised and snap frozen in liquid nitrogen for metabolic mapping.
Gene Expression Analysis
RNA was purified from tissue samples using Ambion TRI Reagent (Life Technologies) following the manufacturer’s instructions. RNA samples were then DNase treated using Ambion’s Turbo DNA-free Kit to remove any contaminating genomic DNA. Transcriptome profiling assays were performed on xenograft tissue from 3 animals per group using Affymetrix Human GeneChip 1.0 ST arrays. Briefly, 250ng of total RNAs were reverse transcribed into cDNA, then transcribed into cRNAs and labeled into biotinylated cRNA using the GeneChip WT Plus Reagent Kit (Affymetrix) according to the manufacturer’s standard protocols (P/N 4425209 Rev. B 05/2009 and P/N 702808 Rev. 6). Labeled cRNA products were randomly fragmented and hybridized onto Affymetrix GeneChips. Upon hybridization, arrays were washed and stained using the Affymetrix GeneChip WT Terminal Labeling and Hybridization Kit, before being scanned using a GeneChip Scanner 3000.
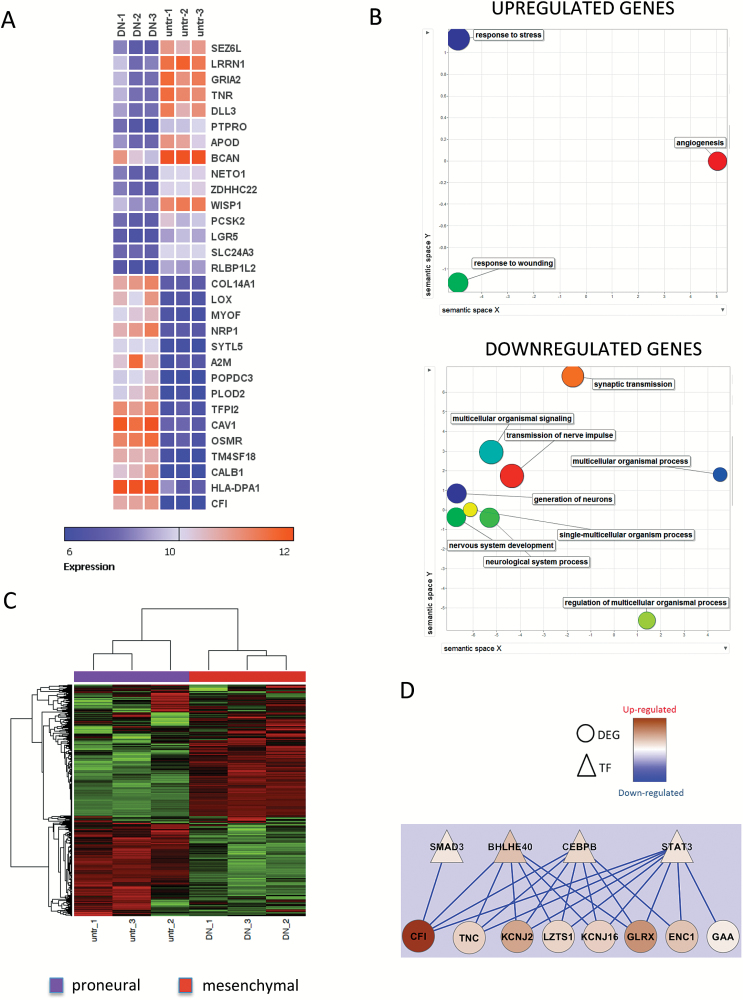
CEL files generated after array scanning were imported into the Partek Genomics Suite for estimating transcript cluster expression levels from raw probe signal intensities using default Partek settings. Resulting expression data were then further processed using R scripts (http://www.R-project.org/). First nonspecific filtering was applied to remove transcript clusters with no specified chromosome location. Then, box plots, density plots, relative log expressions, and sample pairwise correlations were generated to assess the quality of the data. Principal component analysis was also used to identify inter-individual sample variability and outliers within the datasets. Finally, the limma package (Linear Model for Microarray Data; R/Bioconductor) was used to detect differentially expressed genes (DEGs) between control and EGFR-DN groups. The linear model and eBayes methods were used to calculate the statistical significance of the differential mRNA expression values obtained for each contrast as P-values adjusted for multiple testing errors using the Benjamini and Hochberg false discovery rate (FDR). Genes with an FDR <0.05 were considered differentially expressed, irrespective of the fold change. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4898. Expression of the top 30 most differentially expressed genes was depicted as a heatmap generated using gitools.15 Differentially expressed genes (FDR <0.05 and fold change >2) were analyzed for gene ontology enrichment using webgestalt.16 Enriched biological processes were summarized using Revigo.17
Transcriptional Association Models
To acquire deeper insight into potential transcriptional regulatory control distinguishing the angiogenic and invasive phenotypes, we implemented a gene coexpression analysis based on our microarray data and prior knowledge of regulatory associations in GBM. First, we selected genes exhibiting strong differential expression (DEGs) in our microarray data (FDR < 0.01). Independently, we selected transcription factors (TFs) that were previously reported to be specifically altered in GBM tumors.18 We then computed all the pairwise (Pearson) correlations, r, between DEGs and TFs. To focus our attention on the strongest associations while reducing the risk of selecting potential spurious correlations, we filtered out DEG-TF correlations with r ≤ 0.99. The capability of the remaining DEG-TF associations to discriminate between angiogenic and invasive samples was investigated through network visualization and functional enrichment analysis. Network generation and visualization tasks were completed with Cytoscape.19 HEFalMp20 was applied to estimate functional enrichments of the resulting networks.
Isolation and Analysis of Mitochondrial DNA
Genomic DNA was extracted from homogenate of frozen tumor tissue using the DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s instructions. Further details are described in the Supplementary materials.
Immunohistochemistry
Tissue labeling for lactate dehydrogenase A (LDH-A) and glucose transporter 1 (GLUT1) was performed using the DiscoveryXT immunohistochemistry system (Ventana/Roche) as previously described.21 Immunohistochemistry for respiratory complex-I 20kDa subunit (NDUFB8) and porin subunit VDAC1 was performed using the MACH4 Universal AP Polymer Kit (Biocare Medical) according to the manufacturer’s protocol. Details are described in the Supplementary materials.
Ethics Statement
The Norwegian Data Inspectorate and the Regional Committee for Ethics in Research have approved this project (REK 013.09). The study was performed in accordance with the Helsinki Declaration.
The use of patient material was endorsed by the local ethical committee of the Goethe University Frankfurt, Germany (GS 04/09).
Metabolic Mapping (Quantitative Enzyme Histochemistry)
The tetrazolium salt method was used to localize activity of LDH in sections of the tumors. This method generates formazan as a precipitate at the site of LDH activity.22 Details are described in the Supplementary materials.
Lactate Measurement
For lactate measurements, spheroids expressing EGFR-CD533 and control spheroids, generated as described in the section “Lentiviral EGFR-CD533 Production and Infection of Glioblastoma Cells” were cultured at 37°C under 5% CO2 (normoxia) or in a hypoxic chamber under 0.5% O2 (hypoxia) for the indicated duration. Lactate measurements were performed as previously described.23 Samples were normalized to a standard curve of 0–4mM L-lactate. All chemicals were acquired from Sigma-Aldrich, and calculations were performed using Microsoft Excel v12.
Magnetic Resonance Imaging and Magnetic Resonance Spectroscopy
MR imaging (MRI) and MR spectroscopy (MRS) were performed using a 7T small animal MR scanner (Bruker Pharmascan). Axial T1 weighted (T1w) rapid acquisition with relaxation enhancement (RARE) sequences (repetition time [TR], 1000ms; echo time [TE], 8.7ms; slice thickness, 1mm; field of view [FOV], 3.2cm; matrix size, 256×256; 20 slices) were acquired before and after of subcutaneous injection of contrast agent (1.0mL of 0.5 mmol/mL; Omniscan, Amersham Health). Axial T2-weighted (T2w) RARE sequences were also acquired before contrast injections (TR, 4200ms; TE, 36ms; slice thickness, 1mm; FOV, 3.2cm; matrix size, 256×256; 20 slices), using the same slice positioning as for the T1w images. During scanning, the animals were kept in anesthesia with 1.5% isoflurane (Schering-Plough) mixed with 50% air and 50% O2.
Single-voxel proton spectroscopy was performed in a voxel size of 3mm within the tumor area as defined on the T2w MR images, using the PRESS (point-resolved spectroscopy) sequence (TR, 2500ms; TE, 6.017ms; number of acquisitions, 512). A quantitative analysis of the different metabolites was performed using LCModel.24
Statistical Analysis
Differences between pairs of groups were determined by Student’s t-test. P < .05 was considered to indicate significant differences.
Results
Increased Expression of BHLHE40, CEBP, and STAT3 Is Associated with the Angiogenic and Mesenchymal Switch in Human Glioblastoma Xenografts
Previously, we have shown that an angiogenic switch can be induced in highly invasive, EGFR amplified GBM xenografts by overexpression of EGFR-DN. This xenograft model is based on short-term 3D spheroid cultures from GBM biopsies. Details about the model and transduction of cells with EGFR-DN are described in the Supplementary materials. Microarray analysis of xenograft tumors revealed that HIF1A is a central regulator of known angiogenic genes significantly upregulated in the angiogenic xenografts.8 Thus, this model represents a unique in vivo setting to study the shift from invasive to angiogenic tumor growth induced by hypoxia/HIF1A. Here, we verified the induction of an angiogenic switch by EGFR-DN in another invasive, EGFR amplified xenograft model (P228) which is of classical subtype (data not shown). EGFR-DN tumors showed greater contrast enhancement on MRI, microvascular proliferation and upregulation of HIF1A and vascular endothelial growth factor A (VEGFA) (Supplementary Fig. 1). Furthermore, we expanded our microarray analysis from the previously published model.8 The Human Gene v1.0 ST microarray was performed on xenograft tissue from 3 animals per group. The tumors were harvested when they reached similar sizes as observed by MRI (data not shown). Tissue harvested for RNA isolation was verified by histology to contain >90% tumor tissue. Microarray analysis revealed 645 genes differentially expressed between the phenotypes (FDR < 0.05; Supplementary Table 1). We extracted the top 30 genes mostly deregulated between the 2 groups (Fig. 1A). By analyzing associations of gene expression with biological functions, we observed that the genes upregulated in the angiogenic xenografts indeed are involved in angiogenesis, response to wounding, and response to cellular stress. In contrast, the genes downregulated in the angiogenic xenografts are involved in neuronal processes, including nervous system development, generation of neurons, and synaptic transmission (Fig. 1B). According to the Verhaak classification system for GBM molecular subgroups, we demonstrated that the invasive tumors belong to the proneural subgroup, whereas the angiogenic tumors show an expression pattern characteristic of the mesenchymal subgroup (Fig. 1C). We confirmed upregulation of mesenchymal proteins CCAAT-enhancer binding homologous protein beta (CEBPB), basic helix-loop-helix family member e40 (BHLHE40), phosphorylated signal transducer and activator of transcription 3 (phospho-STAT3), and CD44 in angiogenic tumors by immunostaining, while expression of 2 typical proneural genes ASCL1 and OLIG2 were downregulated in angiogenic tumors compared with invasive tumors (Supplementary Fig. 2). Thus, the angiogenic switch and mesenchymal transition seem to be highly linked in our model. To identify transcription factors important for the angiogenic/mesenchymal switch, we used a transcriptional association model and thereby uncovered 2 modules of highly coexpressed transcription factors and differentially expressed genes (27 associations with r > 0.99 involving 5 TFs in total). This analysis showed that the genes BHLHE40, CEBPB, STAT3, and SMAD3 are upregulated in the angiogenic xenografts and are also strongly coexpressed with other genes significantly upregulated in the angiogenic xenografts (Fig. 1D). By analyzing associations of these genes with biological functions, we found that these genes are involved in vasculature development (HEFalMp20, P = 0), glycoprotein metabolism (P = 0), regulation of cell proliferation (P = 0), and migration (P = 0). Three of the TFs (STAT3, CEBPB, and BHLHE40) have previously been identified as master regulators of mesenchymal transition in human GBM,25 which is also associated with a higher degree of hypoxia and a poor prognosis.26 We demonstrated significant upregulation of these factors in angiogenic xenografts by immunohistochemistry (Supplementary Fig. 3). Thus, our analysis indicates that increased expression of the mesenchymal genes CEBPB, STAT3, and BHLHE40 is associated with the phenotypic transition in our model.
Fig. 1.
The angiogenic switch is linked to proneural-to-mesenchymal transition in GBM. (A) Highest differentially expressed genes between invasive (untr) and angiogenic (DN) tumors. (B) Biological processes linked to up- and downregulated genes. (C) Heatmap of Verhaak classification shows that invasive tumors (untr) have a proneural phenotype, while angiogenic (DN) tumors have a mesenchymal phenotype. (D) Transcription factor association to upregulated genes (DN). untr = untransduced EGFR amplified cells.
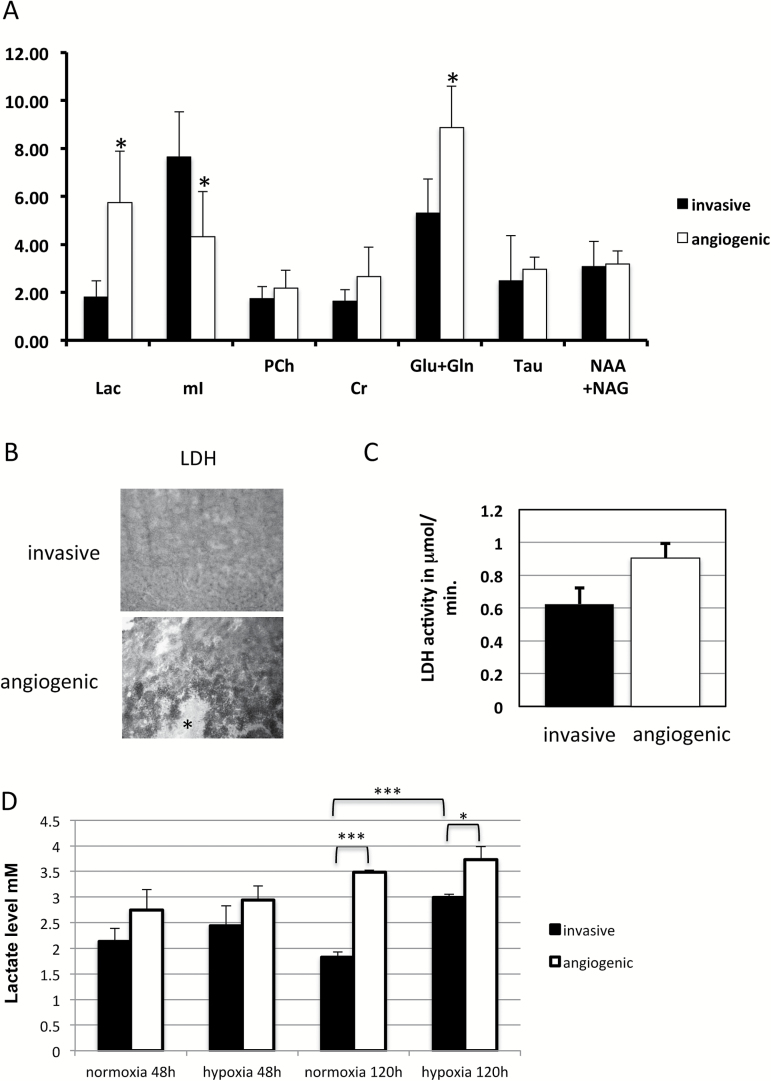
High Levels of Lactate and Glutamine in Angiogenic Xenografts
To assess metabolic changes in our animal model, we first analyzed the microarray data. Ingenuity pathway analysis revealed upregulation of glycolysis in the angiogenic phenotype compared with the invasive phenotype (Supplementary Fig. 4). To confirm this result, we performed in vivo MRS of both invasive and angiogenic xenograft tumors. Angiogenic xenografts clearly showed upregulation of lactate levels indicative of increased glycolysis as well as a significant upregulation of glutamine and glutamate levels (Fig. 2A). To confirm the results from MRS, we performed metabolic mapping on frozen sections from both phenotypes to measure activity of LDH, an enzyme involved in glycolysis. We showed significantly higher activity of LDH in angiogenic (0.9±0.1 µmol lactate converted per mL tissue per min) as compared with invasive xenografts (0.6±0.1 µmol lactate converted per mL tissue per min; P < .05; Fig. 2B, 2C). As hypoxia is an important mediator of angiogenesis, we decided to investigate whether or not hypoxia changes the metabolic fate of GBM cells. We performed in vitro hypoxia experiments with GBM spheroids derived from invasive and angiogenic xenografts and measured lactate levels. No significant difference in lactate levels was observed under normoxia or short-term hypoxia (48h); however, there was a trend of higher levels in angiogenic spheroids under both conditions (Fig. 2D). The difference became more evident and highly significant upon extended culturing (120h) under normoxia. Interestingly, the increase in lactate levels in tumor cells of the angiogenic phenotype cultured under long-term hypoxia (120h) was not significant when compared with normoxia, indicating a Warburg effect. In contrast, the lactate levels of the invasive phenotype significantly increased under long-term hypoxia, indicating that invasive cells can also upregulate glycolysis in a hypoxic microenvironment.
Fig. 2.
Glycolysis is upregulated in angiogenic compared with invasive tumors. (A) MRS of invasive and angiogenic tumors. * P < .005; Lac=Lactate; mI=myo Inositol; Cr=creatine; Ch=choline; NAA+NAAG=; Glu+Gln=glutamine and glutamate; Tau=taurine; NAA+NAAG=N-acetylaspartate + N-acetyl-aspartylglutamate (B) Metabolic mapping of LDH activity in angiogenic compared with invasive tumors. Asterisk indicates necrotic area. (C) Quantification of metabolic mapping. (D) Lactate levels of invasive (P8) and angiogenic (P8-EGFRDN) human GBM cells in 48h and 120h of normoxia and hypoxia. *P < .05; ***P < .001.
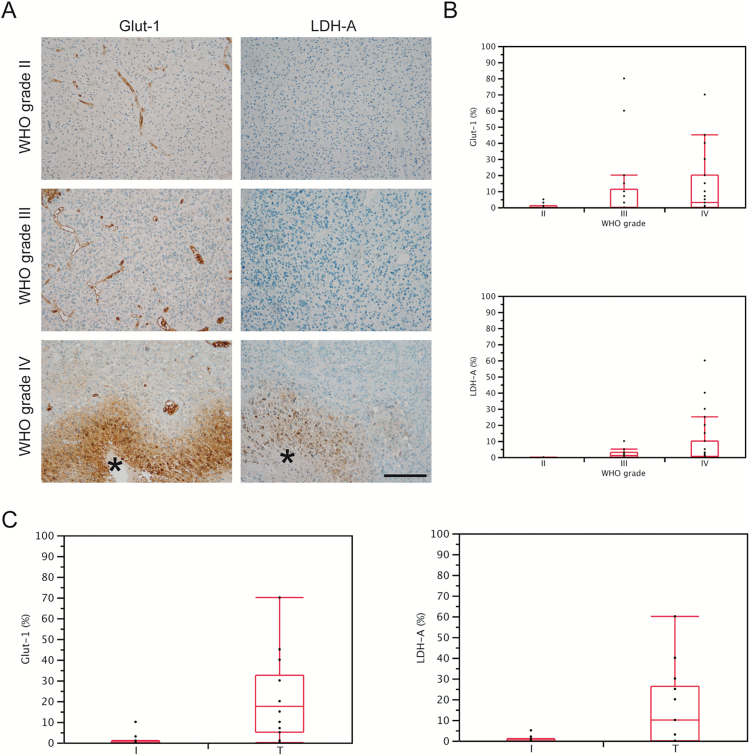
In line with our xenograft results, we showed that the glycolytic proteins LDH-A and GLUT1 were upregulated in particular around necrotic areas in GBM tumors representing hypoxic areas. Meanwhile, low-grade gliomas and the invasive edges of GBM tumors generally exhibited a low or undetectable expression of LDH-A and GLUT1 (Fig. 3A–C).
Fig. 3.
LDH-A and GLUT1 are overexpressed in hypoxic areas of human GBM compared with nonhypoxic areas and low-grade glioma. (A) Immunohistochemistry for LDH-A and GLUT1 in glioma with different World Health Organization grades. Asterisk indicate necrotic areas. (B) Quantification of immunohistochemistry from (A). GLUT1: grade IV versus grade II, P=.0243; other comparisons n.s. LDH-A: grade IV versus II, P=.0087; grade IV versus grade III P=.0088. (C) Quantification of LDH-A and GLUT1 immunohistochemistry in invasive (I) and central (T) tumor areas of GBM. GLUT1, P=.0021; LDH-A, P=.0083.
Mitochondrial Complex I Is Highly Reduced in Angiogenic Xenografts and Hypoxic Areas of GBM Tumors
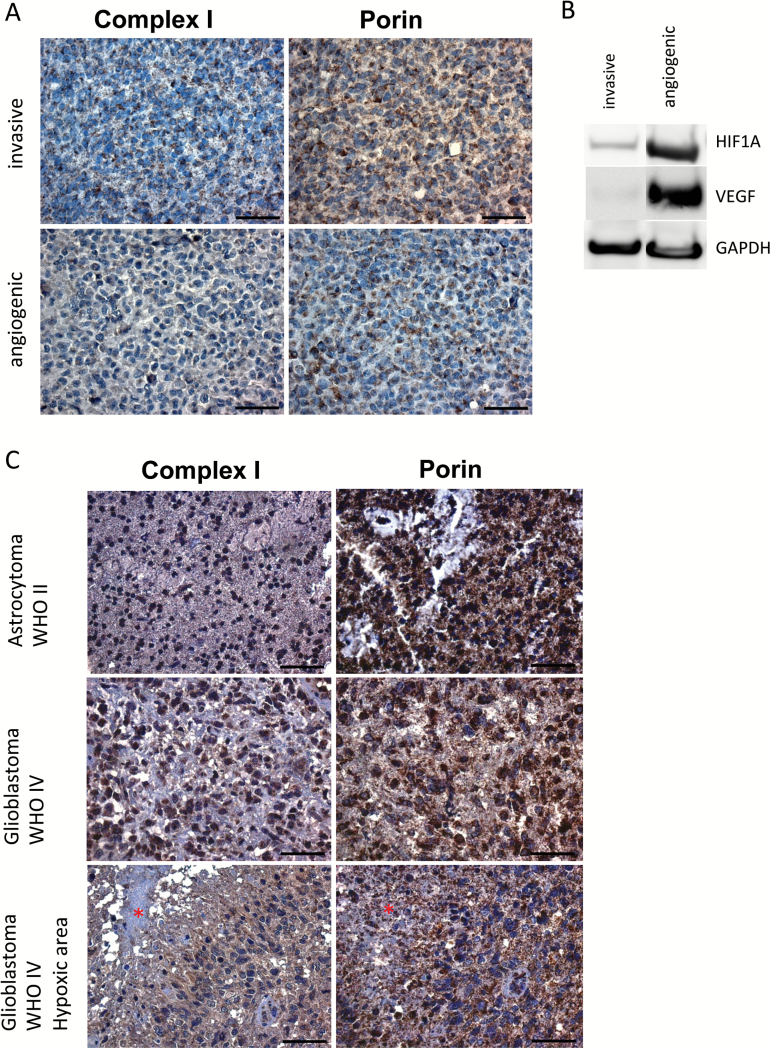
The upregulation of glycolytic activity in angiogenic xenografts and hypoxic areas of GBM tumors may have an impact on mitochondrial respiration. Therefore, paraffin sections of angiogenic and invasive tumor xenografts were immunostained with antibodies against mitochondrial complex I, which is partly encoded by mitochondrial DNA (mtDNA), and porin, which is a general mitochondrial marker reflecting amounts of mitochondria per cell. Expression of complex I was substantially reduced in angiogenic compared with invasive xenografts, while there was no difference in porin expression (Fig. 4A). Upregulation of HIF1A and VEGFA as shown on western blots verified the angiogenic switch (Fig. 4B). To identify the cause for complex I loss, we compared mtDNA quantity and structural integrity in the angiogenic compared with invasive xenografts. However, no mtDNA deletions or quantitative changes were found which could explain the reduction/loss of complex I in the angiogenic tumors (data not shown).
Fig. 4.
Complex I is downregulated in angiogenic compared with invasive tumors. (A) Immunostaining for complex I and porin in invasive and angiogenic xenografts. Complex I is downregulated in angiogenic compared with invasive xenografts while porin is not differentially expressed. (B) Western blot with antibodies against HIF1A and VEGF. (C) Immunostaining for complex I and porin in low-grade glioma and GBM. Asterisks indicate necrotic areas.
Next, we assessed expression of mitochondrial complex I and porin in 5 low-grade and 10 high-grade gliomas. While there was variability in complex I expression between individual patients, we observed that complex I was reduced around necrotic/hypoxic areas in GBM, compared with low-grade tumors and nonnecrotic/nonhypoxic areas of GBM (Fig. 4C, Supplementary Fig. 5). Thus, complex I reduction was associated with hypoxia/angiogenesis in our xenograft model, which was confirmed on patient biopsies.
Discussion
Our study shows that GBM cell metabolism is distinctly affected by the microenvironment. Tumor cells in hypoxic areas are dependent on glycolysis and exhibit strongly reduced expression of mitochondrial complex I. The glycolytic dependency of tumor cells has been shown for many different cancer types.5 However, in GBM it has been unclear whether the predominant use of glycolysis is a general phenomenon of all tumor cells or whether different metabolic programs are dependent on specific tumor areas/microenvironments. Most studies have shown that glycolysis is the primary source of energy production in GBM cells.27–29 Recently, however, Marin-Valencia et al observed, using a xenograft model, that GBM cells can utilize mitochondrial glucose oxidation in addition to glycolysis.6 This finding is quite surprising considering the general view of diminished mitochondrial function in tumor cells. One explanation may be the very low amounts of HIF1A and HIF2A in the invasive xenograft model that was used in this study. Indeed, we also observed low to undetectable levels of HIF1A in our invasive xenograft model.8 Furthermore, high levels of mitochondrial complex I were detected in the invasive phenotype, but low levels in the angiogenic phenotype. Deighton et al recently showed that 23 complex I proteins are reduced in GBM.30 However, we report for the first time that low complex I levels in GBM correlate with hypoxic and angiogenic tumor areas. Complex I deficiency has been previously observed in cancer which is caused by either mutations in the mitochondrial genome31 or impairment through reactive oxygen species.32 However, analysis of mtDNA did not reveal somatic deletions or depletion that could explain the severe reduction of complex I in our model. It is possible that complex I reduction in GBM is caused by oncogenic signaling affecting metabolic regulators such as HIF1A or AMPK33 or by somatic mtDNA point mutations or that it may be the result of hypoxia-induced production of reactive oxygen species.
In addition to increased glycolysis and reduced complex I expression, the angiogenic phenotype showed accumulation of glutamine compared with the invasive phenotype. The elevated levels of glutamine were detected by in vivo MRS and may indicate that tumor cells in angiogenic regions because of the high glycolytic activity have a lower demand to consume glutamine.34
Interestingly, we have previously shown that anti-angiogenic treatment leading to increased GBM invasion was also associated with increased glycolysis.35 In the study by Keunen et al, the invasive and glycolytic phenotype could be explained by upregulation of HIF1A after bevacizumab treatment.35 In contrast to our results, Beckner and colleagues detected abundance of glycolytic enzymes in pseudopodia of U87 glioma cells under normoxic conditions, which may indicate that glycolysis is upregulated during tumor invasion.36 However, the U87 cell line is a more artificial model compared with our tumor model, which more closely resembles the patient situation.13,37 In this regard, we detected increased expression of glycolytic enzymes LDH-A and GLUT1 in hypoxic areas of patient GBM, but not in invasive tumor areas. In other tumor types (ie, melanoma, breast cancer and prostate cancer), glycolysis seems to be the primary energy source for tumor invasion,38,39 which may be explained by the microenvironment as well. Metastatic tumor cells have to explore new microenvironments, while invasive brain tumor cells reside in the same niche, which is highly vascularized.
In our model the invasive phenotype did not exhibit hypoxia. However, when exposed to long-term hypoxia in vitro, a substantial upregulation of glycolysis was observed in the invasive tumor cells, confirming that tumor cell metabolism responds to microenvironmental changes. We did not analyze whether these hypoxia-primed cells induce angiogenic tumor growth in vivo. However, we observed that spheroids of the invasive phenotype may exhibit central necroses in vitro indicating hypoxia (unpublished observations). Upon implantation of these spheres, we did not observe a switch to angiogenesis, which indicates that the hypoxia state is reversible upon change of the microenvironment.
The xenograft model that we used in the present study is derived from a human GBM classified as a proneural subtype. Proneural subtype GBM are generally associated with a better prognosis due to the high frequency of isocitrate dehydrogenase–mutated tumors which also harbor extensive methylation in the cytosine–phosphate–guanine (CpG) islands of a large number of genetic loci (glioma CpG island methylator phenotype positive [GCIMP+])9,40 and in addition show downregulation of EGFR expression and signaling.41 However, the proneural GBM from our xenograft model was a primary GBM and showed high levels of EGFR amplification as well as gain of chromosome 7 and loss of chromosome 10.8,42 Thus, this tumor most likely belongs to a subgroup of proneural tumors associated with a worse prognosis which have been classified as non-GCIMP tumors.40 The proneural xenograft showed a purely invasive phenotype that could be switched to an angiogenic phenotype and a mesenchymal subclass upon overexpression of EGFR-DN. The switch from the proneural to the mesenchymal subtype is a common finding and often induced upon postoperative treatment of GBM.43–45 Interestingly, recent work from Ozawa et al suggests that most non-GCIMP mesenchymal tumors are derived from a proneural-like precursor, again implicating the shift from the proneural to the mesenchymal subtype upon tumor progression.46
The mesenchymal subclass has been associated with increased hypoxia and worse outcome.26 We have previously shown that HIF1A is a central regulator of the genes upregulated in the angiogenic/mesenchymal tumors of our model.8 When comparing our dataset with a previously published hypoxia gene cluster comprising of 45 genes analyzed in glioma patient samples,47 we observed a substantial overlap with 19 genes. In this publication, the authors identified genes that previously have not been related to hypoxia. Most of the regulated genes in our dataset most likely reflect expression changes in the human tumor cells due to high purity of our tumor samples. However, expression changes related to the rat microenvironment could be identified by using species-specific transcriptome-based masks as described previously.48
In conclusion, we have shown in an animal model and in patient biopsies that the use of glycolysis versus mitochondrial respiration is dependent on the specific microenvironment in GBM. The xenograft model that we used is valuable to study proneural-to-mesenchymal subtype transition, progression from invasion to angiogenesis, and the associated metabolic changes in GBM.
Supplementary material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by the Research Council of Norway, the Norwegian Cancer Society, Helse Vest, Haukeland University Hospital, the K.G. Jebsen Research Foundation, and the Bergen Medical Research Foundation.
Supplementary Material
Acknowledgments
We thank B. Nordanger, I. Gavlen, and B. Hansen for expert technical assistance and the Molecular Imaging Center (MIC) in Bergen, Norway for technical support. K.M.T. and G.V.R. were supported by a PhD fellowship from the University of Bergen, Norway. E.E was supported by a PhD fellowship from the Norwegian Cancer Society.
Conflict of interest statement. The authors declare that they have no conflict of interests.
References
- 1. Louis D, Ohgaki H, Wiestler O, et al. WHO Classification of Tumours of the Central Nervous System.Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeng W, Liu P, Pan W, et al. Hypoxia and hypoxia inducible factors in tumor metabolism. Cancer Lett. 2015;356(2 Pt A):263–267. [DOI] [PubMed] [Google Scholar]
- 3. Warburg O. On the origin of cancer cells. Science. 1956; 123(3191):309–314. [DOI] [PubMed] [Google Scholar]
- 4. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. [DOI] [PubMed] [Google Scholar]
- 6. Marin-Valencia I, Yang C, Mashimo T, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15(6):827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Navis AC, Niclou SP, Fack F, et al. Increased mitochondrial activity in a novel IDH1-R132H mutant human oligodendroglioma xenograft model: in situ detection of 2-HG and alpha-KG. Acta Neuropathol Commun. 2013;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Talasila KM, Soentgerath A, Euskirchen P, et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol. 2013;125(5):683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Madhavan S, Zenklusen JC, Kotliarov Y, et al. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res.: MCR. 2009;7(2):157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bjerkvig R, Tonnesen A, Laerum OD, et al. Multicellular tumor spheroids from human gliomas maintained in organ culture. J Neurosurg. 1990;72(3):463–475. [DOI] [PubMed] [Google Scholar]
- 12. Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. [DOI] [PubMed] [Google Scholar]
- 13. Sakariassen PO, Prestegarden L, Wang J, et al. Angiogenesis-independent tumor growth mediated by stem-like cancer cells. Proc Natl Acad Sci U S A. 2006;103(44):16466–16471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Philippidou D, Schmitt M, Moser D, et al. Signatures of micro RNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70(10):4163–4173. [DOI] [PubMed] [Google Scholar]
- 15. Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One. 2011;6(5):e19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Duncan D, Shi Z, et al. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Supek F, Bosnjak M, Skunca N, et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun J, Gong X, Purow B, et al. Uncovering microRNA and transcription factor mediated regulatory networks in glioblastoma. PLoS Comput Biol. 2012;8(7):e1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huttenhower C, Haley EM, Hibbs MA, et al. Exploring the human genome with functional maps. Genome Res. 2009;19(6):1093–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baumgarten P, Harter PN, Tonjes M, et al. Loss of FUBP1 expression in gliomas predicts FUBP1 mutation and is associated with oligodendroglial differentiation, IDH1 mutation and 1p/19q loss of heterozygosity. Neuropathol Appl Neurobiol. 2014;40(2):205–216. [DOI] [PubMed] [Google Scholar]
- 22. Molenaar RJ, Botman D, Smits MA, et al. Radioprotection of IDH1-mutated cancer cells by the IDH1-mutant Inhibitor AGI-5198. Cancer Res. 2015;75(22):4790–4802. [DOI] [PubMed] [Google Scholar]
- 23. Yang Y, Balcarcel RR. 96-well plate assay for sublethal metabolic activity. Assay Drug Dev Technol. 2004;2(4):353–361. [DOI] [PubMed] [Google Scholar]
- 24. Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14(4):260–264. [DOI] [PubMed] [Google Scholar]
- 25. Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cooper LA, Gutman DA, Chisolm C, et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol. 2012;180(5):2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wolf A, Agnihotri S, Micallef J, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y, Zhou Y, Shingu T, et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286(37):32843–32853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jelluma N, Yang X, Stokoe D, et al. Glucose withdrawal induces oxidative stress followed by apoptosis in glioblastoma cells but not in normal human astrocytes. Mol Cancer Res: MCR. 2006;4(5):319–330. [DOI] [PubMed] [Google Scholar]
- 30. Deighton RF, Le Bihan T, Martin SF, et al. Interactions among mitochondrial proteins altered in glioblastoma. J Neurooncol. 2014;118(2):247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iommarini L, Calvaruso MA, Kurelac I, et al. Complex I impairment in mitochondrial diseases and cancer: parallel roads leading to different outcomes. Int J Biochem Cell Biol. 2013;45(1):47–63. [DOI] [PubMed] [Google Scholar]
- 32. Barrientos A, Moraes CT. Titrating the effects of mitochondrial complex I impairment in the cell physiology. J Biol Chem. 1999;274(23):16188–16197. [DOI] [PubMed] [Google Scholar]
- 33. Hagland H, Nikolaisen J, Hodneland LI, et al. Targeting mitochondria in the treatment of human cancer: a coordinated attack against cancer cell energy metabolism and signalling. Expert Opin Ther Targets. 2007;11(8):1055–1069. [DOI] [PubMed] [Google Scholar]
- 34. Yang C, Sudderth J, Dang T, et al. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69(20):7986–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keunen O, Johansson M, Oudin A, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci U S A. 2011;108(9):3749–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beckner ME, Chen X, An J, et al. Proteomic characterization of harvested pseudopodia with differential gel electrophoresis and specific antibodies. Lab Invest. 2005;85(3):316–327. [DOI] [PubMed] [Google Scholar]
- 37. Wang J, Miletic H, Sakariassen PO, et al. A reproducible brain tumour model established from human glioblastoma biopsies. BMC Cancer. 2009;9:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shiraishi T, Verdone JE, Huang J, et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget. 2015;6(1):130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beckner ME, Stracke ML, Liotta LA, et al. Glycolysis as primary energy source in tumor cell chemotaxis. J Natl Cancer Inst. 1990;82(23):1836–1840. [DOI] [PubMed] [Google Scholar]
- 40. Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li J, Taich ZJ, Goyal A, et al. Epigenetic suppression of EGFR signaling in G-CIMP+ glioblastomas. Oncotarget. 2014;5(17):7342–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Talasila KM, Brekka N, Mangseth K, et al. Tumor versus stromal cells in culture—survival of the fittest? PLoS One. 2013;8(12):e81183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Halliday J, Helmy K, Pattwell SS, et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A. 2014;111(14):5248–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bhat KP, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piao Y, Liang J, Holmes L, et al. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res. 2013;19(16):4392–4403. [DOI] [PubMed] [Google Scholar]
- 46. Ozawa T, Riester M, Cheng YK, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murat A, Migliavacca E, Hussain SF, et al. Modulation of angiogenic and inflammatory response in glioblastoma by hypoxia. PLoS One. 2009;4(6):e5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naef F, Huelsken J. Cell-type-specific transcriptomics in chimeric models using transcriptome-based masks. Nucleic Acids Res. 2005;33(13):e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.