Abstract
Modified vaccinia virus Ankara (MVA) is a highly attenuated virus strain being developed as a vaccine for delivery of viral and recombinant antigens. The MVA genome lacks functional copies of numerous genes interfering with host response to infection. The interferon resistance gene E3L encodes one important viral immune defense factor still made by MVA. Here we demonstrate an essential role of E3L to allow for completion of the MVA molecular life cycle upon infection of human HeLa cells. A deletion mutant virus, MVA-ΔE3L, was found defective in late protein synthesis, viral late transcription, and viral DNA replication in infected HeLa cells. Moreover, we detected viral early and continuing intermediate transcription associated with degradation of rRNA, indicating rapid activation of 2′-5′-oligoadenylate synthetase/RNase L in the absence of E3L. Further molecular monitoring of E3L function by microarray analysis of host cell transcription in MVA- or MVA-ΔE3L-infected HeLa cells revealed an overall significant down regulation of more than 50% of cellular transcripts expressed under mock conditions already at 5 h after infection, with a more prominent shutoff following MVA-ΔE3L infection. Interestingly, a cluster of genes up regulated exclusively in MVA-ΔE3L-infected cells could be identified, including transcripts for interleukin 6, growth arrest and DNA damage-inducible protein β, and dual-specificity protein phosphatases. Our data indicate that lack of E3L inhibits MVA antigen production in human HeLa cells at the level of viral late gene expression and suggest that E3L can prevent activation of additional host factors possibly affecting the MVA molecular life cycle.
Modified vaccinia virus Ankara (MVA) was originally obtained from vaccinia virus strain Ankara by over 570 serial passages in chicken embryo fibroblast cells (CEF) (57, 75). The resulting MVA is highly attenuated and cannot replicate in human and most other mammalian cells (18, 33). During this attenuation process, the virus loses 31 kbp of DNA of the parental genome, mainly in six major deletions (58). Consequently, MVA lacks several of the immunomodulatory gene products exploited by other orthopoxviruses such as soluble receptors for gamma interferon (IFN-γ), IFN-α/β, tumor necrosis factor, and CC chemokines (2, 10). However, several further regulatory gene sequences of vaccinia virus are still conserved within the MVA genome, including two genes that encode proteins (K3L and E3L) for intracellular inhibition of IFN-inducible activities of the host cell (2, 60, 66).
Vaccinia virus gene K3L encodes a protein that has homology to the α subunit of the eukaryotic translation initiation factor eIF-2 (eIF-2α) (7, 38). K3L is a pseudosubstrate for double-stranded RNA (dsRNA)-activated protein kinase (PKR), which thus prevents the virus-induced phosphorylation of eIF-2α by PKR (17, 27, 45).
The vaccinia virus E3L IFN resistance gene encodes a 25-kDa polypeptide which is synthesized early during the viral infection cycle (20). A vaccinia virus E3L deletion mutant was shown to be highly sensitive to the antiviral activity of IFN-α/β and replication deficient in Vero and HeLa cell cultures but still of full replicative capacity in CEF, hamster BHK, and rabbit RK13 cells (5, 6, 7, 21, 52). The E3L protein harbors an amino-terminal Z-DNA-binding domain (46, 49, 50) and a carboxyl-terminal domain with a typical dsRNA-binding motif (19, 21, 43, 68). The amino-terminal domain of E3L is dispensable for infection of cells in culture, but both the amino- and carboxy-terminal domains of E3L are required for full pathogenesis in mice (12, 67). By binding and sequestering dsRNA, E3L can inhibit stimulation of PKR and activation of 2′-5′-oligoadenylate synthetase (2′-5′OA synthetase), two enzymes which are activated by dsRNA (19, 20, 65). Upon stimulation with dsRNA, 2′-5′OA synthetase polymerizes oligoadenylates with 2′-5′ linkages (48). These synthesized 2′-5′-oligoadenylates in turn activate an endoribonuclease (RNase L), which is suggested to affect viral replication by cleaving cellular and viral RNAs and general inhibition of protein synthesis (29, 35, 51, 68, 69, 71).
In addition to inhibition of the IFN-inducible PKR and 2′-5′OA synthetase by sequestering dsRNA, it was shown recently that the E3L protein can block phosphorylation and thus activation of IFN regulatory factor 3 (IRF3) and IRF7, required for viral induction of transcripts specific for IFN-α/β (70, 81). The mechanism of inhibition of the IFN system by E3L at this level remains to be elucidated.
An MVA E3L deletion mutant (MVA-ΔE3L) was found to be unable to replicate in CEF, the cell culture used for serial passage in MVA attenuation, but to possess full replicative capacity in mammalian BHK-21 cells (44). This vaccinia virus host range phenotype in CEF was associated with deficient viral DNA and protein synthesis, induction of apoptosis, and enhanced production of chicken IFN-α/β (44).
As MVA vectors serve as candidate vaccines to immunize against infectious diseases and cancer in humans (reviewed in reference 75), it appears particularly relevant to characterize the function of MVA immunomodulatory genes in the context of infection of human cells. Here, we analyzed the role of IFN resistance gene E3L in the MVA molecular life cycle. Our data demonstrate that viral late transcription and late protein biosynthesis is blocked in MVA-ΔE3L-infected HeLa cells. Viral DNA replication is also impaired in the absence of E3L. We detected activation of the 2′-5′OA synthetase/RNase L pathway exclusively in MVA-ΔE3L-infected cells in contrast to the results seen with mock- or MVA-infected cells. Moreover, upon analysis of the host response in a global approach, we found distinct sets of host cell genes with significantly increased signal intensities in MVA-ΔE3L-infected cells including interleukin 6 (IL-6), growth arrest and DNA damage-inducible protein β (Gadd45β), activating transcription factor 3 (ATF3), basic-leucine zipper (bZIP) transcription factor AP-1 (JUN), dual-specificity protein phosphatases (DUSPs), and zinc finger protein SNAIL2, suggesting the possibility for additional cellular targets of E3L function.
MATERIALS AND METHODS
Viruses and cells.
Baby hamster kidney BHK-21 (ATCC CCL-10) and HeLa cells were grown in RPMI 1640 medium or Dulbecco's modified Eagle's medium supplemented with 10 or 8% heat-inactivated fetal calf serum, respectively. Cells were maintained at 37°C in a 5% CO2 atmosphere. MVA (cloned isolate F6 at 582nd CEF passage) (58) and MVA-ΔE3L (44) were routinely propagated and titered by vaccinia virus-specific immunostaining on BHK-21 cells to determine the numbers of infectious units per milliliter.
Analysis of [35S]methionine-labeled polypeptides.
HeLa and BHK-21 cell monolayers in 6-well plates were either mock infected or infected with MVA or MVA-ΔE3L at a multiplicity of infection (MOI) of 20. Following 30 min of adsorption at 4°C, virus inocula were replaced by prewarmed tissue culture medium (RPMI 1640) and incubated at 37°C. At indicated time points postinfection, cells were washed with methionine-free medium and incubated with methionine-free medium at 37°C for 15 min, 50 μCi of [35S]methionine was added to each well, and the mixture was incubated for 30 min at 37°C. Cytoplasmic extracts of infected cell monolayers were prepared by adding 0.2 ml of 1% Nonidet P-40 lysis buffer (50 mM Tris-HCl, 150 mM NaCl [pH 8.0]) for 10 min at 37°C. Polypeptides from cell extracts were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and analyzed by autoradiography.
RNase protection assay (RPA).
DNA fragments encoding the MVA genes 005R, 078R, and 047R and the cellular human GAPDH were amplified via PCR with primer pair HLG15 (5′-GTT TAC ACG GTG ACT GTA TC-3′) and HLG16 (5′-AAG CAT AAT ACC GGG AGA TG-3′), primer pair HLG3 (5′-CGA TAA ACT GCG CCA AAT TG-3′) and HLG10 (5′-TTG GAC ACA GGA AGA TTA AAC-3′), primer pair HLG1 (5′-ATT CTC ATT TTG CAT CTG CTC-3′) and HLG11 (5′-TAT TTT GTA GCA TGT CCG TCC-3′), and primer pair HLG7 (5′-CAT CGC TCA GAA CAC CTA TG-3′) and HLG12 (5′-TCA TTG ATG GCA ACA ATA TCC-3′), respectively. Genomic MVA DNA or plasmid pT-AdvGAPDH served as template DNA for PCR amplification of viral DNAs or GAPDH, respectively. Plasmid pT-AdvGAPDH was created by cloning of a 474-bp PCR fragment representing nucleotides 12 to 485 of human GAPDH mRNA (53) into pT-Adv (Clontech) (M. H. Lehmann, unpublished data). PCR products specific for MVA genes 005R, 078R, and 047R were cloned into plasmid pCR2.1 (Invitrogen) via a TA cloning kit (Invitrogen), yielding plasmids p005R, p078R, p047R, and pGAPDH.
From these plasmids, the gene fragments were reamplified by PCR using the respective gene-specific forward primers (see above) and a modified M13 forward (−20) primer (5′-CGG CCA GTG AAT TGT AAT AC-3′). These PCR products including the T7 promoter sequence at the 3′ end were used as templates for in vitro transcription (IVT) with T7 RNA polymerase (Roche Diagnostics) for synthesis of biotinylated antisense riboprobes by incorporation of biotin-16-UTP as described previously (54). HeLa cells cultured in 35-mm-diameter dishes were mock infected or infected with MVA or MVA-ΔE3L at an MOI of 20. Total RNA was isolated at 0, 1, 2, 3, 4, 7.5, and 10 h postinfection (h p.i.) with TRIzol reagent (Invitrogen) following the manufacturer's instructions, and 5 μg of RNA per reaction was applied for nonradioactive RPA, performed as described previously (54). In all experimental data, time point 0 h refers to the end of the adsorption period, routinely 1 h at 4°C. Electrophoretic separation of RPA samples occurred in precast 6% polyacrylamide urea gels (Anamed), and RNA was transferred onto positively charged nylon membranes via semidry blotting. For detection, a North2South chemiluminescent hybridization and detection kit (PIERCE) was used.
Analysis of viral DNA.
Genomic viral DNA was isolated from infected cells as described previously (32, 34). To assess viral DNA replication, total DNA was transferred by a dot blot procedure to a positively charged nylon membrane (Roche Diagnostics) and hybridized to a digoxigenin (DIG)-labeled MVA 078R-specific riboprobe, generated via PCR with primer pair HLG3 (see above) and HLG4 (5′-CTA ATA CGA CTC ACT ATA GGG AGA CAT AAT AGC CAA ATG CTG ATG-3′) by use of MVA genomic DNA as the template and subsequent IVT for incorporation of DIG-UTP as described for Northern blot analysis (see below). The reverse primer contained a T7 RNA polymerase promoter recognition sequence (underlined). Hybridization and detection were carried out as described for Northern blot analysis (see below), applying 50°C as the temperature for prehybridization, hybridization, and high-stringency washing. Chemiluminescence was quantified with a LumiImager (Roche Diagnostics).
RNase L activity assay.
HeLa cells cultured in 35-mm-diameter dishes were mock infected or infected with MVA or MVA-ΔE3L at an MOI of 20. Total RNA was isolated at 0, 1, 2, 3, 4, 7.5, and 10 h p.i. with TRIzol reagent (Invitrogen) following the manufacturer's instructions. For electrophoresis in a 1% agarose formaldehyde gel, 5 μg of total RNA per lane was used. For visualization of rRNA species, the gel was stained with ethidium bromide (2 μg/ml).
Microarray analysis.
HeLa cells cultured in 35-mm-diameter dishes were mock infected or infected with MVA or MVA-ΔE3L at an MOI of 20 in three independent replicates each. Following 1 h of adsorption at 4°C, virus inocula were replaced by prewarmed tissue culture medium (Dulbecco's modified Eagle's medium with 2% fetal calf serum). After 2.5 and 5 h, total RNA of mock-infected and MVA- and MVA-ΔE3L-infected cells was isolated with TRIzol reagent (Invitrogen) following the manufacturer's instructions. The quality of the RNA was examined by electrophoresis in 1% agarose formaldehyde gels and ethidium bromide staining. By use of one-cycle T7-driven IVT according to Affymetrix protocols, 20 μg of RNA was biotin labeled. Amplified cRNA (15 μg) was hybridized at 45°C overnight on Affymetrix HG-U133A human oligonucleotide expression microarrays, washed, and stained as recommended by Affymetrix. Subsequently the gene chips were scanned and analyzed using Affymetrix Microarray Suite 5.0 software. Arrays were normalized to probe sets AFFX-BioB-3_at, AFFX-BioC-3_at, AFFX-BioDn-3_at, and AFFX-CreX-3_at, with a target value of 1,500. These probe sets represent the signals of four prelabeled controls, which were spiked routinely into the hybridization cocktail.
A standard t test was carried out with Spotfire DecisionSite V7.2 software, and k-means clustering was performed with Genesis software (release 1.1.3), with the Euclidean distance used as the similarity distance measurement (72).
For detection of genes with significant differences in signal intensity between MVA- and MVA-ΔE3L-infected cells at 2.5 or 5 h p.i., a statistic analysis applying paired data sets was performed using SAM algorithm V1.15 (http://www-stat.stanford.edu/∼tibs/SAM/) (77). Mean false discovery rates were below 1%. The stringency of the standard t test and SAM statistical analyses was augmented by applying the following threshold criteria: (i) a severalfold change criterion of more than twofold was used, (ii) a maximum (all mean expression values) minus minimum (all mean expression values) filter of more than 50 was used, and (iii) only probe sets detected as present at least three times were included.
Northern blot analysis.
Total RNA used for microarray analysis (see above) was separated by electrophoresis in 1% agarose formaldehyde gels. Subsequently, RNA was transferred onto a positively charged nylon membrane (Roche Diagnostics) via vacuum blotting. For synthesis of reverse transcription-PCR (RT-PCR) products (Cloned AMV First-Strand cDNA synthesis kit [Invitrogen] and PCR Master [Roche Diagnostics]) specific for IL-6, GTFII, ATF3, CDA, and HIST1H3A, primer pair HLG30 (5′-CAA GCG CCT TCG GTC CAG-3′) and HLG31 (5′-CTA ATA CGA CTC ACT ATA GGG AGA AGG AAC TCC TTA AAG CTG CG-3′), primer pair HLG42 (5′-TAC AAT GCA GGA ATC TGC AG-3′) and HLG43 (5′-CTA ATA CGA CTC ACT ATA GGG AGA ATA CTG CAA CAT AAA GTA CCA G-3′), primer pair HLG34 (5′-ATG ATG CTT CAA CAC CCA GG-3′) and HLG35 (5′-CTA ATA CGA CTC ACT ATA GGG AGA TTA GCT CTG CAA TGT TCC TTC-3′), primer pair HLG38 (5′-TGG CCC AGA AGC GTC CTG-3′) and HLG39 (5′-CTA ATA CGA CTC ACT ATA GGG AGA CTG AGT CTT CTG CAG GTC C-3′), and primer pair HLG26 (5′-CTA AGC AAA CTG CTC GGA AG-3′) and HLG27 (5′-CTA ATA CGA CTC ACT ATA GGG AGA CAT GAT AGT GAC GCG CTT G-3′) were used, respectively.
For synthesis of riboprobes for detection of MVA-encoded mRNAs 005R, 078R, and 047R, PCR products processed with viral DNA as the template and primer pair HLG5 (5′-TTA TCT GAT GTT GTT GTT GTT CGC-3′) and HLG6 (5′-CTA ATA CGA CTC ACT ATA GGG AGA GTT TAG TTC GTC GAG TGA AC-3′), primer pair HLG3 and HLG4, and primer pair HLG1 and HLG2 (5′-CTA ATA CGA CTC ACT ATA GGG AGA CTA GAA GCT ACA TTA TCG CG-3′) were used. Reverse primers contained a T7 RNA polymerase promoter recognition sequence (underlined). DIG-labeled riboprobes were obtained by IVT with T7 RNA polymerase (Roche Diagnostics) by use of the respective RT-PCR- and PCR-generated DNA fragments as templates. In vitro RNA labeling, hybridization, and signal detection were carried out according to the manufacturer's instructions (DIG RNA labeling kit and detection chemicals; Roche Diagnostics), applying 68°C for prehybridization, hybridization, and high-stringency washing.
Quantitative real-time RT-PCR.
Total RNA was isolated as described for microarray analysis in an independent experiment (see above). For RT, 3 μg of RNA was used according to the instructions for a SuperScript III First-Strand synthesis system (Invitrogen) for RT-PCR. Primers, TaqMan 6-carboxyfluorescein-6-carboxytetramethylrhodamine-labeled probes, and two-step amplification protocols were supplied by TIB MOLBIOL. TaqMan assays for IL-6, ATF-3, GADD45β, JUN, and HIST1H3A were performed with FastStart DNA MasterPlus hybridization probes and a LightCycler system (Roche Diagnostics). Corresponding specific RT-PCR products were used as external standards. The thermal cycler conditions were an initial 10 min at 95°C followed by 45 cycles of 5 s at 95°C and 30 s at 58°C and finishing with 30 s at 40°C.
RESULTS
Lack of viral late protein synthesis in MVA-ΔE3L-infected human HeLa cells.
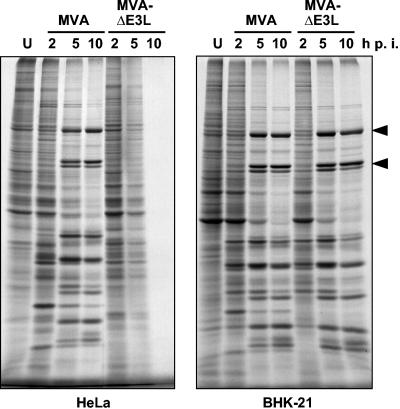
In previous work we were able to show that the E3L early gene product is required for unimpaired MVA replication in CEF (44). Interestingly, when monitoring for vaccinia virus antigen in infected human HeLa cells we observed immunostaining after infection with MVA-ΔE3L weaker than that seen with MVA infection (data not shown). Suspecting a deficiency in viral antigen production, we investigated the pattern of viral protein synthesis during MVA-ΔE3L infection. Human HeLa or hamster BHK-21 cells were metabolically labeled with [35S]methionine at 2, 5, and 10 h after infection with MVA or MVA-ΔE3L. After each labeling period, lysates were prepared and analyzed by SDS-PAGE and autoradiography. Starting at 5 h after infection, we detected typical viral late polypeptides in lysates from MVA-infected HeLa cells, and a general shutoff of cellular protein synthesis became visible (Fig. 1). These data were very comparable to those of previous work (74). In contrast, no late viral proteins were detectable among polypeptides from MVA-ΔE3L-infected HeLa cells, and we noticed a similarly prominent shutoff of cellular protein synthesis. Furthermore, our analysis of lysates from BHK-21 cells confirmed equal capacities of MVA and MVA-ΔE3L for unimpaired protein biosynthesis under E3L-independent conditions (Fig. 1), a result in full agreement with previous data (44). We concluded from this experiment that the E3L gene product is essential for MVA late viral protein synthesis in human HeLa cells.
FIG. 1.
Characterization of viral polypeptide synthesis. HeLa and BHK-21 cells (as indicated) were either mock infected (U) or infected with MVA or MVA-ΔE3L at an MOI of 20 and labeled with [35S]methionine for 30 min at the indicated h p.i. Cytoplasmatic extracts were separated by SDS-10% PAGE and analyzed by autoradiography. Typical viral late polypeptides are indicated by arrowheads.
Arrest of viral intermediate transcription and lack of viral late transcripts in MVA-ΔE3L-infected human HeLa cells.
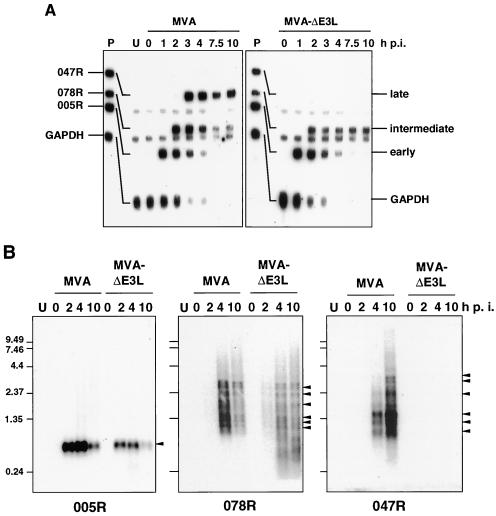
To investigate a possible deficiency on the transcriptional level we wished to characterize levels and kinetics of viral early, intermediate, and late transcription in MVA- or MVA-ΔE3L-infected HeLa cells. For this purpose we designed a probe set to simultaneously monitor transcripts of all three vaccinia virus gene classes in an RPA. Probes specific for MVA genes 005R (vaccinia virus growth factor), 078R (vaccinia virus late transcription factor VLTF-1), and 047R (vaccinia virus 11-kDa DNA-binding protein) were selected, representing well-characterized vaccinia virus early, intermediate, and late genes, respectively, also present in the MVA genome (2, 4). A probe specific for human GAPDH was integrated to monitor transcription of a cellular mRNA.
To determine the relevance of E3L protein for MVA transcription, we infected HeLa cells with MVA or MVA-ΔE3L at 20 IU per cell, and total RNA was isolated at 0, 1, 2, 3, 4, 7.5, and 10 h after infection and analyzed via RPA. In cells infected with MVA we observed the expected cascade-like pattern of vaccinia virus transcription (Fig. 2). Expression of the early MVA transcript 005R started at 1 h after infection and declined to the threshold level at 4 h after infection. A signal corresponding to the intermediate transcript 078R peaked from 2 to 4 h after infection, steadily decreasing in intensity at later times. Transcription of the late gene 047R started at 3 h p.i. and remained at constant levels throughout the time course of the assay. Expression of the GAPDH mRNA was down regulated starting at 3 h p.i., indicating shutoff of cellular transcription after MVA infection of human HeLa cells (Fig. 2), a result in agreement with the observed decrease of cellular protein synthesis (Fig. 1). In HeLa cells infected with MVA-ΔE3L, similar kinetics of early gene transcription and shutoff of GAPDH mRNA synthesis were detectable; however, the decline in the levels of the GAPDH mRNA was slightly faster in the absence of E3L during infection. Expression of the intermediate gene also started at 2 h p.i. but, in contrast to MVA infection results, remained constant until at least 10 h p.i. Interestingly, there was no late gene expression detectable in HeLa cells infected with MVA-ΔE3L (Fig. 2A). The phenotype of the MVA-ΔE3L virus characterized by prolonged intermediate transcription and lack of late mRNA synthesis was confirmed in a complementary Northern blot experiment again using riboprobes specific for 005R, 078R, and 047R (Fig. 2B). Thus, the failure of viral late protein biosynthesis appeared to be clearly a consequence of a preceding inhibition of viral late transcription, and the data suggested an arrest of vaccinia virus gene expression at the stage of intermediate gene transcription in MVA-ΔE3L-infected HeLa cells.
FIG. 2.
Analysis of viral mRNA synthesis. (A) For RPA, HeLa cells were mock infected (U) or infected with MVA or MVA-ΔE3L at an MOI of 20. Total RNA was isolated at 0, 1, 2, 3, 4, 7.5, and 10 h p.i. Probes specific for viral genes 005R, 078R, and 047R were used, representing typical early, intermediate, and late vaccinia virus transcripts, respectively. To monitor cellular transcription activity, a probe specific for GAPDH was used. After RNase digestion, the protected biotin-labeled probe fragments were analyzed by polyacrylamide urea gel electrophoresis followed by detection via a chemiluminescent hybridization and detection kit (PIERCE). Lane P represents probes without RNase digestion. (B) Northern blot analysis of 005R, 078R, and 047R transcripts. Total RNA was isolated at the indicated time points and electrophoretically separated in 1% agarose formaldehyde gels by applying 1 μg of total RNA per lane. Subsequently, RNA was transferred onto a positively charged nylon membrane (Roche Diagnostics) via vacuum blotting and hybridized to riboprobes specific for 005R, 078R, and 047R, respectively. Sizes (in kilobases) of the RNA standards are shown at the left. Detected RNA species are indicated by arrowheads.
Impairment of viral DNA replication in MVA-ΔE3L-infected HeLa cells.
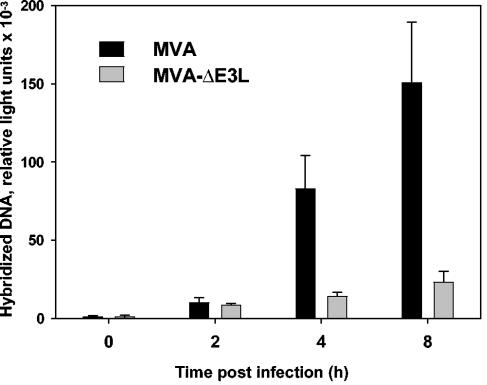
The observation of functional intermediate transcription in MVA-ΔE3L-infected cells suggested that the replication of the viral genome should not be affected, because viral DNA replication is considered a prerequisite for vaccinia virus intermediate and late transcription (13, 47, 59, 79). Even so, we wished to comparatively analyze the capacities for genome replication of MVA and MVA-ΔE3L. Viral DNA synthesis was monitored by isolating total DNA from HeLa cells infected with MVA or MVA-ΔE3L at 0, 2, 4, and 8 h p.i. For detection of viral DNA, a DIG-labeled riboprobe specific for MVA was used for hybridization followed by quantification of chemiluminescence. Viral DNA replication of MVA proceeded as previously described for infection of nonpermissive human cells (Fig. 3) (74). Somewhat to our surprise, however, replication of MVA-ΔE3L genomes was drastically impaired after infection of HeLa cells. These data indicated that in the absence of E3L protein synthesis little or no viral DNA replication can occur during MVA infection of HeLa cells.
FIG. 3.
Comparison of viral DNA synthesis following MVA and MVA-ΔE3L infection. DNA isolated from HeLa cells at 0, 2, 4, and 8 h after infection with MVA or MVA-ΔE3L at an MOI of 10 was immobilized on a positively charged nylon membrane. A DIG-labeled MVA-specific riboprobe was used for hybridization, and chemiluminescence was detected and quantified with a LumiImager (Roche Diagnostics).
Activation of RNase L in MVA-ΔE3L-infected cells.
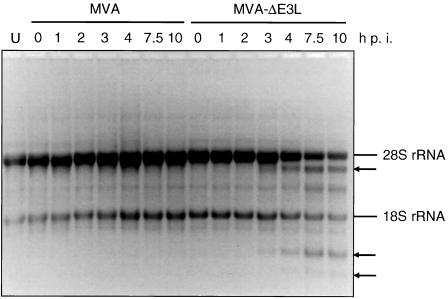
One important characteristic of vaccinia virus E3L gene function is to prevent activation of the antiviral 2′-5′OA synthetase/RNase L pathway resulting in the degradation viral and host cellular RNA including rRNA (19, 20, 52, 65, 71). To determine whether the E3L gene product also had the capacity to block the RNase L activation during MVA infection we infected HeLa cells with MVA or MVA-ΔE3L, isolated total RNA at 0, 1, 2, 3, 4, 7.5, and 10 h p.i., and analyzed the integrity of cellular rRNA. No degradation of rRNA was detected in uninfected cells or in MVA-infected cells (Fig. 4). In contrast, we could clearly notice typical distinct degradation products of 28S and 18S rRNA after infection with MVA-ΔE3L, starting at 3 h p.i. (80). From this result we concluded that inhibition of the dsRNA-activated 2′-5′OA synthetase/RNase L pathway is also one of the relevant E3L functions in MVA infection of human HeLa cells. The intracellular formation of dsRNA during poxvirus infection is attributed to incomplete termination of intermediate and late transcripts leading to mRNAs with heterogeneous 3′ ends, as shown for, e.g., the intermediate G8R vaccinia virus mRNA (4, 11, 25, 78). Thus, because of the absence of viral late transcripts during the MVA-ΔE3L molecular life cycle, intermediate gene transcripts, as shown here for the MVA G8R homologue 078R synthesized starting at 2 h p.i. (Fig. 2), are apparently sufficient for stimulation of the dsRNA-dependent 2′-5′OA synthetase/RNase L pathway accompanied by rRNA cleavage initiated at 3 h after infection (Fig. 4).
FIG. 4.
Activation of RNase L in MVA-ΔE3L-infected cells. HeLa cells were mock infected (U) or infected with MVA or MVA-ΔE3L at an MOI of 20. Total RNA was isolated at 0, 1, 2, 3, 4, 7.5, and 10 h p.i., and 5 μg of total RNA per lane was applied for electrophoresis in a 1% agarose formaldehyde gel. For visualization of rRNA species the gel was stained with ethidium bromide (2 μg/ml). The arrows indicate bands corresponding to characteristic degradation products of rRNA.
General effects of MVA and MVA-ΔE3L infection on the host cell transcriptome.
To take a new approach in looking for host defense pathways possibly modulated by E3L function, we decided to compare the effects of MVA and MVA-ΔE3L infection on steady-state mRNA levels of HeLa cells. We performed microarray experiments by using GeneChip human genome U133A (Affymetrix) to monitor expression levels of about 22,000 probe sets representing 18,000 human cellular transcripts, of which 14,500 are known genes. For transcription analysis, HeLa cells were mock infected or infected with MVA or MVA-ΔE3L in three independent experiments each. Considering our finding of first evidence for RNase L activity at 3 h after MVA-ΔE3L infection, cells were harvested after 2.5 and 5 h of incubation, thus potentially including time points before and after induction of the antiviral 2′-5′OA synthetase/RNase L pathway, respectively (Fig. 4).
For normalization between arrays, we first applied the standard global scaling procedure using Microarray Suite 5.0 software and a target intensity of 150. We observed a large variation of scaling factors between mock-infected and virus-infected samples, especially at 5 h p.i. (a mean scaling factor of 1.4 for mock-infected cells versus 5.3 for MVA-infected cells). Additionally, the mean percentage of genes expressed above the background level (“present calls”) decreased drastically from 43.8% in mock-infected cells to 32.1% of MVA-infected cell levels at 5 h p.i. These observations suggested a severe reduction in cellular mRNA levels that precluded the use of the standard global scaling method for normalization. To circumvent that problem, we normalized our data by using spike-in control RNAs, which are routinely added to the cRNAs after the labeling process described in Materials and Methods. Scaling factors after normalization to the spike-in controls ranged between 0.42 and 1.44, with a median of 0.70 and a standard deviation of 0.27. A similar normalization strategy was used for microarray analysis of rabbitpox virus infection of human cells (14).
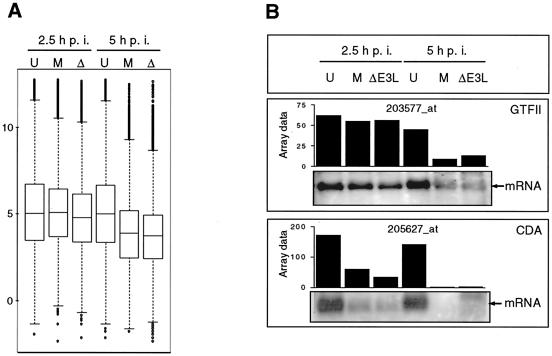
The decrease in the percentage of expressed genes suggested a general down regulation of cellular transcription in MVA- and MVA-ΔE3L-infected cells. By including average signal intensities (log 2) of all probe sets into a box plot analysis, a decrease in the mean signal intensity at 5 h p.i. with MVA was visualized, corroborating the impression of a global down regulation of cellular mRNA levels (Fig. 5A). This effect of overall decrease in signal intensity of cellular transcription was likewise detectable for MVA-ΔE3L-infected cells at 5 h p.i. (Fig. 5A).
FIG. 5.
General effect of MVA and MVA-ΔE3L infection on host cell transcription activity. (A) For box plot analysis, all (22,000) probe sets present on Affymetrix GeneChip human genome U133A were included. The average signal intensities determined as log 2 values for uninfected cells (U) and MVA (M)- and MVA-ΔE3L (Δ)-infected cells at 2.5 and 5 h p.i. (as indicated) are presented with the median and the 50th percentiles in the form of boxes, signifying that 50% of all probe sets are represented within the box. The whiskers extend to the most extreme data points which are no more than 1.5 times the interquartile range from the box. (B) Northern blot analysis for validation of general transcriptional down regulation detected by microarray analysis. Total RNA was isolated from mock-infected (U)- and MVA (M)- and MVA-ΔE3L (ΔE3L)-infected cells at 2.5 and 5 h p.i. and electrophoretically separated in 1% agarose formaldehyde gels by applying 1 μg of total RNA per lane. Subsequently, RNA was transferred onto a positively charged nylon membrane (Roche Diagnostics). Transcripts encoding general transcription factor IIH (GTFIIH4) and cytidine deaminase (CDA) were detected by using DIG-labeled riboprobes specific for respective genes. Results of hybridization and detection by chemiluminescence are shown at the bottom of each panel. Average microarray signal intensities (Array data) of probe sets with the corresponding Affymetrix probe set designations are shown in bar charts.
When the threshold criteria described in Materials and Methods were applied, 6,007 probe sets were regulated following MVA infection at 2.5 or 5 h p.i. compared to the results seen with uninfected cells, whereas when a standard t test algorithm was used in addition, 83% of the probe sets were significantly repressed or induced (P < 0.05). Following MVA-ΔE3L infection, 93% of 6,402 probe sets were significantly regulated. Under our experimental conditions, the signal intensities of 2,904 and 2,231 probe sets did not change at either time point after MVA and MVA-ΔE3L infection, respectively. We detected 469 or 4,897 probe sets as significantly down regulated in comparison to the results seen with untreated cells at 2.5 or 5 h post-MVA infection, representing 5 or 57% of all probe sets detected under mock-infection conditions, respectively, and confirming the above-described effect of general down regulation of cellular transcripts after infection. After 5 h p.i., signal intensities of probe sets corresponding to, e.g., GAPDH, GTFII, and CDA (encoding glyceraldehyde-3-phosphate dehydrogenase, general transcription factor IIH polypeptide 4, and human cytidine deaminase, respectively) were significantly down regulated compared to the results seen with uninfected cells. Interestingly, 42 or 35 probe sets were identified as induced at 2.5 or 5 h post-MVA infection, respectively. These probe sets encode proteins such as connective tissue growth factor (CTGF), early growth response 1 (EGR1), and zinc finger protein 36 (ZFP36) and include further transcripts encoding cellular histones or also ribosomal RNAs (unpublished data). At 2.5 or 5 h p.i. with MVA-ΔE3L, we detected 1,382 or 5,776 probe sets, respectively, as significantly down regulated in comparison to the results seen with mock-infected cells. Compared to the results seen with appropriate mock-infected samples, 54 or 87 probe sets were identified as induced at 2.5 or 5 h after MVA-ΔE3L infection, respectively (unpublished data). In similarity to the results seen with MVA-infected cells, these probe sets also encode CTGF, EGR1, ZFP36, and include transcripts of cellular histones or ribosomal RNAs. Remarkably, some further transcripts exclusively induced following MVA-ΔE3L infection were identified which were characterized in more detail by further bioinformatics below (unpublished data and see below).
First, the effect of general silencing of cellular transcription in response to MVA infection (Fig. 5A) should be confirmed by Northern blot analyses. Using gene-specific riboprobes for GTFIIH4 and CDA, we verified down regulation following infection with MVA or MVA-ΔE3L by Northern blot experiments (Fig. 5B). A further confirmation of the effect of transcriptional down regulation after infection was given by detection of decreasing signal intensities of three Affymetrix GAPDH control probe sets present on the array (data not shown) and the observed shutoff process of respective mRNA after infection detected in the RPA shown above (Fig. 2).
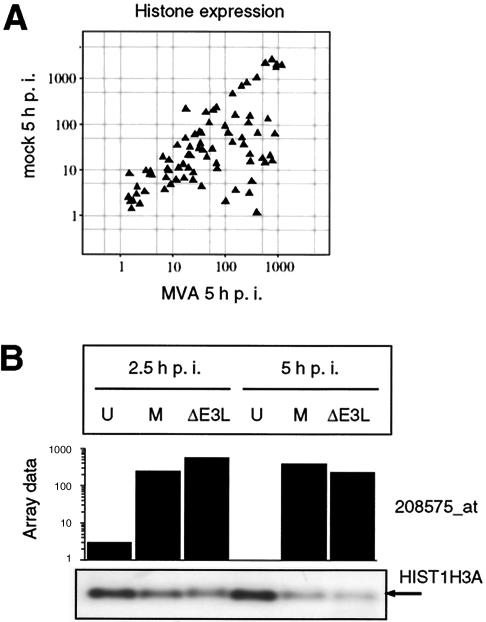
In recent studies, induction of cellular histone genes was detected using microarray technology for analysis of vaccinia virus or rabbitpox virus infection of human cells (14, 40, 41). We also detected increasing signal intensities for numerous probe sets representing histone genes in MVA- and MVA-ΔE3L-infected cells, as visualized in a scatter plot including all histone genes present on Affymetrix microarray HG-U133A (Fig. 6A and unpublished data). In contrast to microarray data, when a HIST1H3A-specific riboprobe was used exemplarily for Northern blot analysis the detectable amount of mRNA encoding an H3 histone was reduced after infection with MVA or MVA-ΔE3L (Fig. 6B). Furthermore, no induction of histone H3 protein following MVA infection compared to that seen with mock-infected cells was detectable by using a histone H3-specific antibody in a Western blot analysis (data not shown). In a quantitative RT-PCR approach using a HIST1H3A gene-specific primer for RT, the down regulation after MVA infection was confirmed. In contrast, after applying oligo(dT) primers for cDNA synthesis as used for labeling procedures during microarray analysis, induction of histone transcription following infection was observed (Table 1). A similar measurement artifact of microarray data was shown for histone regulation in rabbitpox virus-infected human A549 alveolar epithelial cells (see Discussion and reference 14).
FIG. 6.
Effect of MVA infection on histone transcription. (A) Scatter plot of all histone genes monitored by Affymetrix microarray. Normalized signal intensities of histone probe sets from mock- and MVA-infected HeLa cells at 5 h p.i. are shown. (B) Regulation of histone mRNA level. HeLa cells were mock infected (U) or infected with MVA (M) or MVA-ΔE3L (ΔE3L), and total RNA was isolated at 2.5 and 5 h p.i. RNA was electrophoretically separated in 1% agarose formaldehyde gels by applying 1 μg of total RNA per lane. Transcripts encoding H3 histone family member A (HIST1H3A) were detected by using a DIG-labeled riboprobe specific for HIST1H3A gene, as indicated by the arrow. Average microarray signal intensities (Array data) of corresponding probe set 208575_at are shown as a bar chart with a logarithmic scale.
TABLE 1.
Confirmation of microarray data by quantitative real-time RT-PCR
| Gene | Severalfold induction
|
|||
|---|---|---|---|---|
| 2.5 h p.i.a
|
5 h p.i.
|
|||
| MVA | ΔE3Lb | MVA | ΔE3L | |
| IL-6 | 2.70 | 6.14 | 5.71 | 9.49 |
| ATF3 | 3.27 | 3.29 | 1.40 | 8.83 |
| GADD45β | 0.87 | 1.10 | 0.81 | 2.03 |
| JUN | 0.53 | 0.55 | 0.60 | 2.34 |
| HIST1H3A | 4.58 | 6.03 | 6.95 | 2.00 |
| HIST1H3Ac | 0.63 | NAd | 0.73 | NA |
hpi, hours postinfection.
MVA-ΔE3L (ΔE3L).
HIST1H3A-specific primer was used for cDNA synthesis by RT.
NA, not analyzed.
Identification of host genes differentially expressed as a consequence of E3L deletion during MVA infection.
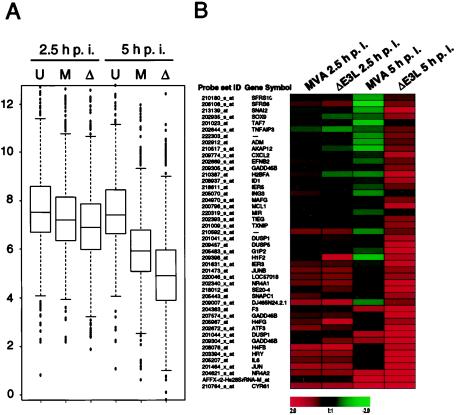
In a second attempt we wished to specifically analyze differences in cellular gene expression between MVA- and MVA-ΔE3L-infected HeLa cells. For that purpose we performed a direct statistical comparison of probe set signal intensities from microarrays of MVA- and MVA-ΔE3L-infected cells. Probe sets were defined as significantly regulated between both viruses when identified by SAM algorithm V1.15 while fulfilling additional criteria for threshold levels as described in Materials and Methods. By applying this stringent approach, 1,102 probe sets were identified as significantly differentially expressed between MVA- and MVA-ΔE3L-infected HeLa cells at both time points of infection. When these E3L-dependent differentially expressed genes were utilized for box plot analysis, a stronger down regulation of cellular transcription level was detectable for MVA-ΔE3L-infected cells than for MVA-infected cells (Fig. 7A).
FIG. 7.
Identification of E3L-dependent differentially expressed host genes. (A) For box plot analysis 1,102 probe sets differentially expressed between MVA- and MVA-ΔE3L-infected cells were included. The average signal intensities determined as log 2 values for uninfected cells (U) and MVA (M)- and MVA-ΔE3L (Δ)-infected cells are presented with the median and the 50th percentiles as the box, signifying that 50% of differentially expressed probe sets are represented within the box. The whiskers extend to the most extreme data point which is no more than 1.5 times the interquartile range from the box. (B) Cluster 1, including genes induced in the absence of E3L during MVA infection of HeLa cells. For K-means cluster analysis, 1,102 probe sets were included, which were identified as significantly regulated in comparisons of MVA and MVA-ΔE3L infection. For clustering log 2 data, severalfold change values compared to mock-infection results were used. Cluster 1 of 10 is shown, representing genes with signal intensities following MVA-ΔE3L infection higher than those seen with MVA infection. The color scale symbolizes up (red) or down (green) regulation of the indicated genes. (C) Northern blot analysis for validation of microarray data. HeLa cells were mock infected (U) or infected with MVA (M) or MVA-ΔE3L (ΔE3L), and total RNA was isolated at 2.5 and 5 h p.i. RNA was electrophoretically separated in 1% agarose formaldehyde gels by applying 1 μg of total RNA per lane. Subsequently, RNA was transferred onto a positively charged nylon membrane (Roche Diagnostics). Transcripts encoding ATF3 and IL-6 were detected by using DIG-labeled riboprobes specific for the respective genes. Results of hybridization and detection by chemiluminescence are shown at the bottom of each panel. Average microarray signal intensities (Array data) of respective probe sets with corresponding Affymetrix probe set ID are shown in bar charts.
To determine characteristic patterns of regulation, we used a K-means clustering providing 10 clusters that included the log 2 severalfold change values versus the respective mock-infected conditions for 1,102 identified regulated probe sets. Candidates of most clusters were characterized by stronger down regulation in MVA-ΔE3L-infected cells than in MVA-infected cells (data not shown). Interestingly, 46 probe sets of cluster 1 were induced exclusively in MVA-ΔE3L-infected cells, especially at 5 h p.i. (Fig. 7B and Table 2). The corresponding genes with higher expression levels in the absence of E3L during infection are involved in cellular signal transduction, transcription processes, and immune response. This cluster includes the proinflammatory cytokine interleukin-6 (IL-6), ATF3, Gadd45β, transcription factor AP-1 (JUN), DUSP1 (MKP-1), DUSP5 (HVH3), and Zinc finger protein SNAIL2.
TABLE 2.
Genes of cluster 1, representing probe sets with increased signal intensities in MVA-ΔE3L infected HeLa cells compared to MVA-infected cell results at 2.5 or 5 h postinfection calculated as severalfold change factors derived from a comparison to corresponding mock-expression values
| Gene group and designation | Probe seta | Accession no. | Description | Severalfold change at:
|
|||
|---|---|---|---|---|---|---|---|
| 2.5 h p.i.b
|
5 h p.i.
|
||||||
| MVA | ΔE3Lc | MVA | ΔE3L | ||||
| Signaling | |||||||
| AKAP12 | 210517_s_at | NM_005100 | A kinase (PRKA) anchor protein (gravin) 12 | 1.08 | 0.72 | 0.47 | 1.82 |
| DUSP1 | 201041_s_at | NM_004417 | DUSP1 (MKP-1) | 1.04 | 1.45 | 1.10 | 2.52 |
| DUSP1 | 201044_x_at | NM_004417 | DUSP1 (MKP-1) | 0.83 | 1.19 | 2.52 | 12.13 |
| DUSP5 | 209457_at | NM_004419 | DUSP5 (HVH3) | 1.31 | 1.10 | 1.15 | 3.10 |
| GADD45β | 209305_s_at | NM_015675 | Gadd4β | 1.21 | 1.25 | 0.81 | 1.87 |
| GADD45β | 207574_s_at | NM_015675 | Gadd45β | 1.48 | 1.55 | 1.38 | 4.49 |
| GADD45β | 209304_x_at | NM_015675 | Gadd45β | 1.30 | 1.73 | 2.14 | 7.13 |
| Transcription | |||||||
| ATF3 | 202672_s_at | NM_001674 | ATF3 | 1.83 | 1.54 | 1.00 | 7.29 |
| ID1 | 208937_s_at | NM_002165 | Inhibitor of DNA binding 1, helix-loop-helix protein | 1.11 | 1.14 | 0.78 | 2.46 |
| SOX9 | 202935_s_at | NM_000346 | SRY (sex determining region Y)-box 9 | 0.80 | 0.69 | 0.69 | 2.14 |
| TAF7 | 201023_at | NM_005642 | TATA box binding protein-associated factor | 1.22 | 1.13 | 0.46 | 1.26 |
| JUNB | 201473_at | NM_002229 | jun B proto-oncogene | 1.74 | 1.90 | 0.85 | 2.05 |
| NR4A1 | 202340_x_at | NM_002135 | Nuclear receptor subfamily 4, group A, member 1 | 1.71 | 1.90 | 0.85 | 3.10 |
| NR4A2 | 204621_s_at | NM_006186 | Nuclear receptor subfamily 4, group A, member 2 | 1.77 | 1.95 | 2.83 | 8.77 |
| SNA12 | 213139_at | NM_003068 | Snail homolog 2 (Drosophila) | 0.79 | 0.83 | 0.56 | 3.65 |
| TIEG | 202393_s_at | NM_005655 | TGFB-inducible early-growth response | 1.14 | 1.08 | 0.91 | 1.91 |
| JUN | 201464_x_at | NM_002228 | v-jun sarcoma virus 17 oncogene homolog | 2.39 | 2.30 | 0.83 | 5.79 |
| MAFG | 204970_s_at | NM_002359 | v-maf homolog G (avian) | 1.21 | 1.01 | 0.81 | 1.78 |
| SNAPC1 | 205443_at | NM_003082 | Small nuclear RNA activating complex | 1.60 | 1.18 | 1.18 | 2.30 |
| Immune response | |||||||
| CXCL2 | 209774_x_at | NM_002089 | Chemokine (C-X-C motif) ligand 2 | 1.02 | 0.91 | 0.76 | 2.70 |
| F3 | 204363_at | NM_001993 | Coagulation factor III | 0.98 | 1.33 | 2.35 | 4.49 |
| IL6 | 205207_at | NM_000600 | IL-6 (IFN, beta 2) | 1.64 | 1.66 | 1.07 | 9.19 |
| mRNA splicing | |||||||
| SFRS10 | 210180_s_at | NM_004593 | Splicing factor, arginine/serine-rich 10 | 1.04 | 0.94 | 0.54 | 1.48 |
| SFRS6 | 206108_s_at | NM_006275 | Splicing factor, arginine/serine-rich 6 | 1.47 | 1.70 | 0.35 | 1.74 |
| Histones | |||||||
| H1F2 | 209398_at | NM_005319 | H1 histone family, member 2 | 0.98 | 4.12 | 0.45 | 2.64 |
| H2BFA | 210387_at | NM_003518 | H2B histone family, member A | 1.46 | 0.66 | 0.62 | 3.40 |
| H4FB | 208076_at | NM_003539 | H4 histone family, member B | 2.49 | 3.36 | 1.18 | 2.83 |
| H4FG | 205967_at | NM_003542 | H4 histone family, member G | 1.57 | 3.24 | 0.85 | 2.30 |
| Miscellaneous | |||||||
| MCL1 | 200796_s_at | NM_021960 | Myeloid cell leukemia sequence 1 (BCL2-related) | 1.39 | 1.24 | 0.79 | 2.89 |
| IER3 | 201631_s_at | NM_003897 | Immediate-early response 3 | 1.55 | 1.59 | 0.78 | 3.25 |
| IER5 | 218611_at | NM_016545 | Immediate-early response 5 | 1.32 | 1.23 | 0.71 | 1.70 |
| ADM | 202912_at | NM_001124 | Adrenomedullin | 0.84 | 0.91 | 0.56 | 1.59 |
| SE20-4 | 218012_at | NM_022117 | T-cell lymphoma-associated tumor antigen | 1.28 | 1.61 | 0.83 | 3.10 |
| CYR61 | 210764_s_at | NM_001554 | Cysteine-rich, angiogenic inducer, 61 | 2.12 | 2.29 | 3.25 | 7.64 |
| EFNB2 | 202669_s_at | NM_004093 | Ephrin-B2 | 1.44 | 0.98 | 0.68 | 1.70 |
| ING3 | 205070_at | NM_019071 | Inhibitor of growth family, member 3 | 1.51 | 1.38 | 0.60 | 1.15 |
| GIP2 | 205483_s_at | NM_005101 | IFN, alpha-inducible protein (clone IF1-15K) | 1.00 | 1.42 | 0.71 | 4.59 |
| MIR | 220319_s_at | NM_013262 | Myosin regulatory light chain interacting protein | 1.34 | 1.26 | 0.69 | 1.23 |
| TXNIP | 201009_s_at | NM_006472 | Thioredoxin interacting protein | 1.23 | 1.29 | 1.02 | 2.05 |
| TNFAIP3 | 202644_s_at | NM_006290 | Tumor necrosis factor, alpha-induced protein 3 | 0.72 | 0.60 | 0.69 | 1.66 |
| LOC57018 | 220046_s_at | NM_020307 | Cyclin L ania-6a | 1.64 | 1.83 | 1.12 | 2.24 |
| DJ465N24.2.1 | 209007_s_at | NM_020317 | Hypothetical protein dj465N24.2.1 | 2.27 | 2.81 | 0.62 | 1.82 |
| HRY | 203394_s_at | NM_005524 | Hairy homolog (Drosophila) | 1.90 | 1.99 | 1.20 | 5.66 |
| 210592_s_at | Spermidine-spermine N1-acetyltransferase | 1.79 | 1.77 | 0.62 | 1.70 | ||
| 222303_at | ESTs | 1.26 | 1.07 | 0.60 | 1.78 | ||
| AFFX-r2-Hs28SrRNA-M_at | 1.20 | 1.37 | 4.09 | 8.19 | |||
Probe set designations correspond to GeneChip Human Genome U133A (Affymetrix).
hpi, hours postinfection.
MVA-ΔE3L (ΔE3L).
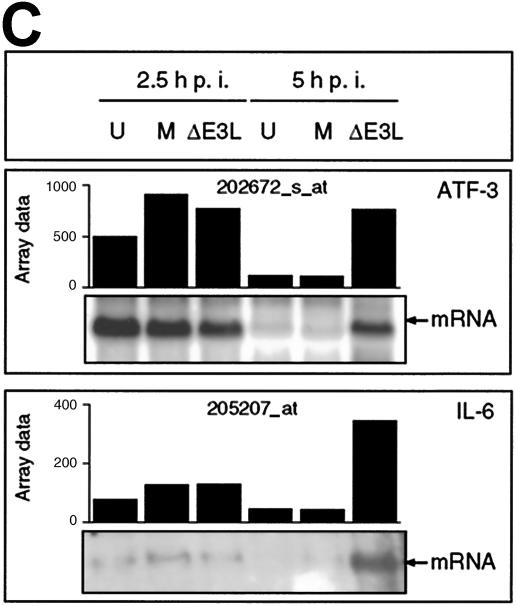
The increased signal intensity level of the IL-6 probe set at 5 h p.i. with MVA-ΔE3L was confirmed by the presence of elevated amounts of IL-6-specific mRNA, as detected by a IL-6 riboprobe in Northern blot analysis (Fig. 7C). In addition, in the absence of E3L during infection elevated amounts of IL-6 mRNA were also determined by quantitative RT-PCR; however, significant induction was also detected for MVA infection, as described previously (41) (Table 1). Thus, the absence of E3L during MVA infection may result in an altered activation status of the JAK/STAT pathway. The expression pattern of ATF3 was also included for cluster 1; however, there was a particular down regulation result detectable in mock-infected cells at 5 h p.i. This observation may have been due to the incubation protocol (designed to allow for virus adsorption at 4°C) which was also applied to mock-infected cells, particularly as involvement of ATF3 in cellular stress response has been described previously (22, 42). Importantly, the effect of higher ATF3 expression levels in MVA-ΔE3L-infected cells than in MVA-infected cells at 5 h could be verified upon Northern blot and RT-PCR analysis (Fig. 7C and Table 1).
We also detected an MVA-ΔE3L-specific induction of transcripts specific for Gadd45β and JUN encoding basic-leucine zipper (bZIP) transcription factor AP-1; both results were confirmed by RT-PCR (Table 1). Furthermore, we detected in MVA-ΔE3L-infected cells increased levels of transcripts encoding two cellular DUSPs, enzymes described to dephosphorylate phosphothreonine and phosphotyrosine residues within mitogen-activated protein (MAP) kinases (16). The induction of SNAI2 transcription in MVA-ΔE3L-infected cells suggested the synthesis of increased amounts of the encoded zinc finger transcription factor SNAIL2 involved in melanocyte development and hematopoiesis (62). Thus, the up-regulated transcripts being included in cluster 1 encode potential candidate host cell genes, which may be part of an antiviral response normally repressed by intact E3L function during MVA infection of human cells.
DISCUSSION
A key feature of CEF-adapted, replication-deficient MVA is the generally unimpaired synthesis of early, intermediate, and late viral gene products upon infection of human and other nonpermissive mammalian cells (18, 33, 74). As abundant production of viral and recombinant antigens requires late viral gene expression, the maintenance of an undisturbed MVA molecular life cycle under nonpermissive conditions is considered one advantage of MVA-based candidate vaccines (75). Despite numerous deletions and mutations being present in the MVA genome after more than 570 passages on CEF, the IFN resistance gene E3L had been fully conserved during this attenuation process (2). Recent characterization of an MVA deletion mutant lacking the E3L gene (MVA-ΔE3L) revealed a host range phenotype upon CEF infection, with defects in viral DNA and protein synthesis being associated with induction of apoptosis and elevated chicken type I IFN activity (44).
The severe deficiency of MVA-ΔE3L to produce late viral polypeptides in human HeLa cells was an important initial finding in this study. The possibility that removal of the immune evasion factor E3L might be beneficial for improvement of MVA vaccines appears hampered by the obvious paucity of viral antigen being made in the absence of E3L. Yet this phenotype pointed towards a possibly distinct regulatory role of this viral protein in the MVA life cycle. Indeed, when assessing the molecular function of E3L in MVA-infected human HeLa cells by RPA and Northern blot analysis we detected late viral RNA synthesis only in the presence of E3L. Monitoring for expression of well-characterized early, intermediate, and late viral mRNAs represented by vaccinia virus growth factor (MVA ORF 005R), vaccinia virus late transcription factor VLTF-1 (MVA ORF 078R), and vaccinia virus 11k DNA-binding protein (MVA ORF 047R), respectively, we found the typical cascade-like pattern of vaccinia virus RNA synthesis after infection with wild-type MVA (13, 26, 59). Obviously, the absence of late viral RNA in MVA-ΔE3L-infected HeLa cells explains our failure to detect late viral protein synthesis. Remarkably, despite the absence of late viral RNA in MVA-ΔE3L-infected HeLa cells, we detected normally initiated intermediate transcription which was active until the end of our assay. This could be explained by the failure of onset of late transcription which somehow limits intermediate transcription (13). In the absence of the E3L gene product we detected very little if any synthesis of viral DNA, in contrast to the results of infection with wild-type MVA. This was surprising, because intermediate transcription is normally only initiated after viral genome replication has started. Inhibition of DNA synthesis results in the persistence of early transcription but the inhibition of intermediate (79) and late transcription. We assume that minimal residual genome replication or a somehow aborted initiation of viral DNA replication in MVA-ΔE3L-infected HeLa cells is still sufficient to allow for the onset of intermediate transcription but not for initiation of late transcription. Alternatively, reduced translation of intermediate transcripts encoding late transcription factors may limit late transcription.
Concomitantly with synthesis of MVA-specific mRNAs during infection of HeLa cells, we showed inhibition of cellular transcription activity (i) by an RPA visualizing decreased amounts of GAPDH transcript and (ii) by microarray detection at 2.5 or 5 h p.i. of probe sets significantly down regulated by 5% or more than 50%, respectively, compared to untreated cell results. These results are in agreement with the induced degradation and, most notably, inhibition of cellular RNA synthesis following vaccinia virus infection (9, 61, 64). Furthermore, overall silencing of cellular transcription was shown when a cDNA microarray approach was used for analysis of vaccinia virus-infected HeLa cells (40). Comparably strong down regulation of host cell transcription was observed for rabbitpox virus infection of human A549 alveolar epithelial cells (14). Decreased expression of GTFIIH4 and CDA transcripts encoding human polypeptide 4 of general transcription factor IIH and cytidine deaminase, respectively, after MVA infection was confirmed by Northern blot analysis. Due to the essential role of GTFIIH in cellular RNA polymerase II transcription initiation (31), this down regulation might be one of several causes for the general shutoff of cellular transcription in consequence of vaccinia virus infection. Direct interaction of vaccinia virus proteins with GTFIIH, as shown for Epstein-Barr virus nuclear antigen 2 (EBNA 2), has not been described (76).
In this study we have shown degradation of cellular rRNA, one hallmark of activation of 2′-5′OA synthetase/RNase L pathway, early during infection of HeLa cells with MVA lacking the E3L gene and without additional stimulation by type I IFNs. Degradation of cellular rRNA was also observed after infection of human fibroblasts, human glioblastoma, monkey BSC-40, or HeLa cells with vaccinia virus strain Copenhagen lacking the E3L gene (vP1080) (23, 52, 65). No RNase L induction was observed during infection of BHK-21 or mouse L929 cells with this E3L deletion mutant vP1080 (8, 52). Interestingly, another E3L deletion mutant of vaccinia virus strain Western Reserve did not induce rRNA degradation during infection of human fibrosarcoma HT-1080 cells without the addition of type I IFNs (81). After infection of HeLa cells with MVA-ΔE3L we observed activation of 2′-5′OA synthetase/RNase L, obviously on the enzymatic level only, as there was rRNA degradation but no detectable induction of transcripts encoding 2′-5′OA synthetase or RNase L in our microarray experiments (unpublished data). Posttranslational activation of these IFN response enzymes requires the presence of dsRNA. During vaccinia virus infection, viral late mRNAs are believed to produce high levels of dsRNA molecules due to read-through transcription by the viral RNA polymerase in the late phase of infection, generating heterogeneous 3′ termini (11, 25, 78). Instead of late viral RNA, we detected expression of early and intermediate transcripts (MVA 078R, encoding the vaccinia virus G8R homologue) in MVA-ΔE3L-infected cells. Interestingly, formation of RNA 3′-end heterogeneity was also demonstrated for vaccinia virus intermediate transcript G8R (4). Thus, the synthesis of intermediate transcripts during MVA-ΔE3L infection seemed to be sufficient for dsRNA formation and for activation of RNase L.
While not designed for this purpose our microarray experiments also indicated altered host cell transcription activity following high-dose infection with MVA or MVA-ΔE3L in comparison to mock-infection results. We found a more drastic overall shutdown of cellular transcripts compared to the results of another recent analysis of MVA-infected HeLa cells (41), and we assume that our choice of a high MOI (an MOI of 20 compared to an MOI of 5) is responsible for this difference. Using our infection protocol we had expected particularly stringent selection of possibly directly or indirectly increased cellular gene expression signals, especially when comparing MVA and MVA-ΔE3L infections. Under these conditions we were pleased to corroborate the MVA-specific induction of the transcripts for CTGF and ETR101, as already indicated by Guerra and coworkers. Recently, activation of the mitogenic signal transmission pathway MEK/ERK1/2/RSK2 was specifically described for vaccinia virus Western Reserve infection of mouse A31 3T3 cells and suggested to be relevant for formation of virus progeny (1, 28). Hereby, the transcriptional induction of EGR1 was detected as an important consequence of MEK/ERK1/2/RSK2 activation after infection (1).
Interestingly, we also identified a clearly increased signal of EGR1 expression after MVA infection of HeLa cells, as demonstrated by our microarray data (unpublished data). Another remarkable observation was the increased level of signal obtained with probe sets corresponding to cellular histone mRNAs as well as 28S rRNA, 18S rRNA, and a small nuclear RNA (RNU2) (unpublished data). An important collective feature of all these RNA species is the absence of polyadenylation at their 3′ ends; this includes histone mRNAs, known as the only cellular mRNAs terminating at the 3′ end with a stem-loop structure instead of a typical poly(A) tail (30). Interestingly, there was no induction but there was repression of histone mRNA detectable after MVA infection in Northern blot analysis, exemplarily shown for the transcript encoding HIST1H3A. A similar discrepancy between microarray data and Northern blot analysis concerning histone transcripts was shown for rabbitpox virus-infected human A549 alveolar epithelial cells (14). We suppose that the amount of histone mRNAs was not induced after MVA infection but turned out to become accessible for the oligo(dT)-based labeling procedure during microarray analysis because of nonspecific polyadenylation performed by vaccinia virus-included poly(A) polymerase (37). This vaccinia virus-induced phenomenon was originally described for small nontranslated polyadenylated RNAs (3, 15, 73). Recently, nonspecific polyadenylation by vaccinia virus was also shown for cellular tRNAs, for small nuclear RNAs, and also for mRNAs (56). The latter effect is the likely reason for the apparent high-level induction of histone mRNAs detected by our and other recent microarray analyses following MVA or vaccinia virus Western Reserve infection (40, 41). Corresponding findings after rabbitpox virus infection were recently interpreted in a similar manner (14).
Finally, we succeeded in identifying induction patterns of cellular transcripts specifically restricted to MVA-ΔE3L-infected cells. Interestingly, we detected in our microarray experiment an increase in the levels of two transcripts encoding DUSP1 (MKP-1/HVH1) and DUSP5 (HVH3). Cellular DUSPs, representing enzymes originally identified as a vaccinia virus-encoded protein (39), can dephosphorylate phosphothreonine and phosphotyrosine residues within MAP kinases (reviewed in reference 16). As MKP-1 preferentially inhibits JNK/SAPK and p38 MAP kinases (24, 36) and the transcript encoding MKP-1 was induced in MVA-ΔE3L-infected cells compared to MVA-infected cell results, one can speculate about a specific role of E3L in the activation of these signal transduction pathways. Moreover, we found significantly enhanced expression of transcripts encoding the cytokine IL-6. These data point towards an increased inflammatory host response after MVA-ΔE3L infection, even though induction of IL-6 could be also observed after in vitro (41) (Table 1) and in vivo (63) infection with nonmutated MVA. Note that we also detected an MVA-ΔE3L-specific induction of Gadd45β, a cellular factor involved in cell cycle arrest, apoptosis, and signal transduction and recently described as being directly involved in the regulation of innate and acquired immune response (55). In addition, in the absence of E3L during infection we detected increased transcription of SNAI2, also referred to as the SLUG gene. This suggests enhanced production of the SNAI2-encoded zinc finger transcription factor SNAIL2, which is proposed as an important factor of the stem cell factor c-kit signaling pathway engaged in melanocyte development and hematopoiesis (62). Thus, in future experiments it should be interesting to take further advantage of E3L function to elucidate the cellular pathways regulating host response to poxvirus infection.
Acknowledgments
We thank Angela Servatius for expert technical assistance. We also thank Michael Chudy, Johannes Blümel, and Claudia Schopper for helpful advice and support in LightCycler technology.
This work was supported by grants from the European Commission (QLK2-CT-2002-01867 and QLK2-CT-2002-01034) and the Deutsche Forschungsgemeinschaft (SFB 455-A10).
REFERENCES
- 1.Andrade, A. A., P. N. G. Silva, A. C. T. C. Pereira, L. P. de Sousa, P. C. P. Ferreira, R. T. Gazzinelli, E. G. Kroon, C. Ropert, and C. A. Bonjardim. 2004. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. Biochem. J. 381(Pt. 2):437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 3.Bablanian, R., S. Scribani, and M. Esteban. 1993. Amplification of polyadenylated nontranslated small RNA sequences (POLADS) during superinfection correlates with the inhibition of viral and cellular protein synthesis. Cell. Mol. Biol. Res. 39:243-255. [PubMed] [Google Scholar]
- 4.Baldick, C. J., Jr., and B. Moss. 1993. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J. Virol. 67:3515-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie, E., E. B. Kauffman, H. Martinez, M. E. Perkus, B. L. Jacobs, E. Paoletti, and J. Tartaglia. 1996. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes 12:89-94. [DOI] [PubMed] [Google Scholar]
- 6.Beattie, E., E. Paoletti, and J. Tartaglia. 1995. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L− and E3L− mutant viruses. Virology 210:254-263. [DOI] [PubMed] [Google Scholar]
- 7.Beattie, E., J. Tartaglia, and E. Paoletti. 1991. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology 183:419-422. [DOI] [PubMed] [Google Scholar]
- 8.Beattie, E., K. L. Denzler, J. Tartaglia, M. E. Perkus, E. Paoletti, and B. L. Jacobs. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker, Y., and W. K. Joklik. 1964. Messenger RNA in cells infected with vaccinia virus. Proc. Natl. Acad. Sci. USA 51:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchard, T. J., A. Alcami, P. Andrea, and G. L. Smith. 1998. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: implications for use as a human vaccine. J. Gen. Virol. 79:1159-1167. [DOI] [PubMed] [Google Scholar]
- 11.Boone, R. F., R. P. Parr, and B. Moss. 1979. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. J. Virol. 30:365-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt, T. A., and B. L. Jacobs. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broyles, S. S. 2003. Vaccinia virus transcription. J. Gen. Virol. 84:2293-2303. [DOI] [PubMed] [Google Scholar]
- 14.Brum, L. M., M. C. Lopez, J.-C. Varela, H. V. Baker, R. W. Moyer. 2003. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology 315:322-334. [DOI] [PubMed] [Google Scholar]
- 15.Cacoullos, N., and R. Bablanian. 1993. Role of polyadenylated RNA sequences (POLADS) in vaccinia virus infection: correlation between accumulation of POLADS and extent of shut-off in infected cells. Cell. Mol. Biol. Res. 39:657-664. [PubMed] [Google Scholar]
- 16.Camps, M., A. Nichols, and S. Arkinstall. 2000. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 14:6-16. [PubMed] [Google Scholar]
- 17.Carroll, K., O. Elroy-Stein, B. Moss, and R. Jagus. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J. Biol. Chem. 268:12837-12842. [PubMed] [Google Scholar]
- 18.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238:198-211. [DOI] [PubMed] [Google Scholar]
- 19.Chang, H. W., and B. L. Jacobs. 1993. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 194:537-547. [DOI] [PubMed] [Google Scholar]
- 20.Chang, H. W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, H. W., L. H. Uribe, and B. L. Jacobs. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, B. P., C. D. Wolfgang, and T. Hai. 1996. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell. Biol. 16:1157-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Child, S. J., S. Jarrahian, V. M. Harper, and A. P. Geballe. 2002. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J. Virol. 76:4912-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu, Y., P. A. Solski, R. Khosravi-Far, C. J. Der, and K. Kelly. 1996. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J. Biol. Chem. 271:6497-6501. [DOI] [PubMed] [Google Scholar]
- 25.Colby, C., C. Jurale, and J. R. Kates. 1971. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. J. Virol. 7:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Condit, R. C., and E. G. Niles. 2002. Regulation of viral transcription elongation and termination during vaccinia virus infection. Biochim. Biophys. Acta 1577:325-336. [DOI] [PubMed] [Google Scholar]
- 27.Davies, M. V., O. Elroy-Stein, R. Jagus, B. Moss, and R. J. Kaufman. 1992. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 66:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Magalhães, J. C., A. A. Andrade, P. N. G. Silva, L. P. Sousa, C. Ropert, P. C. P. Ferreira, E. G. Kroon, R. T. Gazzinelli, and C. A. Bonjardim. 2001. A mitogenic signal triggered at an early stage of vaccinia virus infection: implication of MEK/ERK and protein kinase A in virus multiplication. J. Biol. Chem. 276:38353-38360. [DOI] [PubMed] [Google Scholar]
- 29.Dìaz-Guerra, M., C. Rivas, and M. Esteban. 1997. Inducible expression of the 2-5A synthetase/RNase L system results in inhibition of vaccinia virus replication. Virology 227:220-228. [DOI] [PubMed] [Google Scholar]
- 30.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 31.Drapkin, R., and D. Reinberg. 1994. The multifunctional TFIIH complex and transcriptional control. Trends Biochem. Sci. 19:504-508. [DOI] [PubMed] [Google Scholar]
- 32.Drexler, I., E. Antunes, M. Schmitz, T. Wölfel, C. Huber, V. Erfle, P. Rieber, M. Theobald, and G. Sutter. 1999. Modified vaccinia virus Ankara for delivery of human tyrosinase as melanoma-associated antigen: induction of tyrosinase- and melanoma-specific human leukocyte antigen A*0201-restricted cytotoxic T cells in vitro and in vivo. Cancer Res. 59:4955-4963. [PubMed] [Google Scholar]
- 33.Drexler, I., K. Heller, B. Wahren, V. Erfle, and G. Sutter. 1998. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J. Gen. Virol. 79:347-352. [DOI] [PubMed] [Google Scholar]
- 34.Earl, P. L., and B. Moss. 1991. Expression of proteins in mammalian cells using vaccinia viral vectors, p. 16.15.1-16.18.10. In F. M. Ausbel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley and Sons, Inc., New York, N.Y.
- 35.Floyd-Smith, G., E. Slattery, and P. Lengyel. 1981. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science 212:1030-1032. [DOI] [PubMed] [Google Scholar]
- 36.Franklin, C. C., and A. S. Kraft. 1997. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-activated protein kinase in U937 cells. J. Biol. Chem. 272:16917-16923. [DOI] [PubMed] [Google Scholar]
- 37.Gershon, P. D., B. Y. Ahn, M. Garfield, and B. Moss. 1991. Poly(A) polymerase and a dissociable polyadenylation stimulatory factor encoded by vaccinia virus. Cell 66:1269-1278. [DOI] [PubMed] [Google Scholar]
- 38.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266. [DOI] [PubMed] [Google Scholar]
- 39.Guan, K. L., S. S. Broyles, and J. E. Dixon. 1991. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature 350:359-362. [DOI] [PubMed] [Google Scholar]
- 40.Guerra, S., L. A. López-Fernández, A. Pascual-Montano, M. Munoz, K. Harshman, M. Esteban. 2003. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 77:6493-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerra, S., L. A. López-Fernández, R. Conde, A. Pascual-Montano, K. Harshman, and M. Esteban. 2004. Microarray analysis reveals characteristic changes of host cell gene expression in response to attenuated modified vaccinia virus Ankara infection of human HeLa cells. J. Virol. 78:5820-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hai, T., C. D. Wolfgang, D. K. Marsee, A. E. Allen, and U. Sivaprasad. 1999. ATF3 and stress responses. Gene Expr. 7:321-335. [PMC free article] [PubMed] [Google Scholar]
- 43.Ho, C. K., and S. Shuman. 1996. Physical and functional characterization of the double-stranded RNA binding protein encoded by the vaccinia virus E3 gene. Virology 217:272-284. [DOI] [PubMed] [Google Scholar]
- 44.Hornemann, S., O. Harlin, C. Staib, S. Kisling, V. Erfle, B. Kaspers, G. Häcker, and G. Sutter. 2003. Replication of modified vaccinia virus Ankara in primary chicken embryo fibroblasts requires expression of the interferon resistance gene E3L. J. Virol. 77:8394-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jagus, R., and M. M. Gray. 1994. Proteins that interact with PKR. Biochimie 76:779-791. [DOI] [PubMed] [Google Scholar]
- 46.Kahmann, J. D., D. A. Wecking, V. Putter, K. Lowenhaupt, Y.-G. Kim, P. Schmieder, H. Oschkinat, A. Rich, and M. Schade. 2004. The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro. Proc. Natl. Acad. Sci. USA 101:2712-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keck, J. G., C. J. Baldick, Jr., and B. Moss. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late trans-activator genes. Cell 61:801-809. [DOI] [PubMed] [Google Scholar]
- 48.Kerr, I. M., and R. E. Brown. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. USA 75:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim, Y.-G., K. Lowenhaupt, D.-B. Oh, K. K. Kim, and A. Rich. 2004. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: Implications for development of a therapy for poxvirus infection. Proc. Natl. Acad. Sci. USA 101:1514-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim, Y.-G., M. Muralinath, T. Brandt, M. Pearcy, K. Hauns, K. Lowenhaupt, B. L. Jacobs, and A. Rich. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. USA 100:6974-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar, R., D. Choubey, P. Lengyel, and G. C. Sen. 1988. Studies on the role of the 2′-5′-oligoadenylate synthetase-RNase L pathway in beta interferon-mediated inhibition of encephalomyocarditis virus replication. J. Virol. 62:3175-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langland, J. O., and B. L. Jacobs. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133-141. [DOI] [PubMed] [Google Scholar]
- 53.Lehmann, M. H., J. Weber, O. Gastmann, and H. H. Sigusch. 2002. Pseudogene-free amplification of human GAPDH cDNA. BioTechniques 33:766. [DOI] [PubMed] [Google Scholar]
- 54.Lehmann, M. H., S. Schreiber, H. Vogelsang, and H. H. Sigusch. 2001. Constitutive expression of MCP-1 and RANTES in the human histiocytic lymphoma cell line U-937. Immunol. Lett. 76:111-113. [DOI] [PubMed] [Google Scholar]
- 55.Lu, B., A. F. Ferrandino, and R. A. Flavell. 2004. Gadd45β is important for perpetuating cognate and inflammatory signals in T cells. Nat. Immunol. 5:38-44. [DOI] [PubMed] [Google Scholar]
- 56.Lu, C., and R. Bablanian. 1996. Characterization of small nontranslated polyadenylylated RNAs in vaccinia virus-infected cells. Proc. Natl. Acad. Sci. USA 93:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayr, A., V. Hochstein-Mintzel, and H. Stickel. 1975. Abstammung, Eigenschaften und Verwendung des attenuierten Vaccinia virus-Stammes MVA. Infection 3:6-14. [Google Scholar]
- 58.Meyer, H., G. Sutter, and A. Mayr. 1991. Mapping of deletions in the genome of the highly attenuated vaccinia MVA and their influence on virulence. J. Gen. Virol. 72:1031-1038. [DOI] [PubMed] [Google Scholar]
- 59.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 60.Moss, B., and J. L. Shisler. 2001. Immunology 101 at poxvirus U: immune evasion genes. Semin. Immunol. 13:59-66. [DOI] [PubMed] [Google Scholar]
- 61.Pedley, S., and R. J. Cooper. 1984. The inhibition of HeLa cell RNA synthesis following infection with vaccinia virus. J. Gen. Virol. 65:1687-1697. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Losada, J., M. Sánchez-Martín, A. Rodríguez-García, M. L. Sánchez, A. Orfao, T. Flores, and I. Sánchez-García. 2002. Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood 100:1274-1286. [PubMed] [Google Scholar]
- 63.Ramírez, J. C., M. M. Gherardi, and M. Esteban. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 74:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rice, A. P., and B. E. Roberts. 1983. Vaccinia virus induces cellular mRNA degradation. J. Virol. 47:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rivas, C., J. Gil, Z. Mĕlková, M. Esteban, and M. Díaz-Guerra. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (IFN)-induced 2-5A synthetase enzyme. Virology 243:406-414. [DOI] [PubMed] [Google Scholar]
- 66.Samuel, C. E. 1991. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology 183:1-11. [DOI] [PubMed] [Google Scholar]
- 67.Shors, S. T., E. Beattie, E. Paoletti, J. Tartaglia, and B. L. Jacobs. 1998. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J. Interferon Cytokine Res. 18:721-729. [DOI] [PubMed] [Google Scholar]
- 68.Shors, T., K. V. Kibler, K. B. Perkins, R. Seidler-Wulff, M. P. Banaszak, and B. L. Jacobs. 1997. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology 239:269-276. [DOI] [PubMed] [Google Scholar]
- 69.Silverman, R. H., P. J. Cayley, M. Knight, C. S. Gilbert, and I. M. Kerr. 1982. Control of the ppp(a2′p)nA system in HeLa cells. Effects of interferon and virus infection. Eur. J. Biochem. 124:131-138. [DOI] [PubMed] [Google Scholar]
- 70.Smith, E. J., I. Marié, A. Prakash, A. García-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 71.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 72.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207-208. [DOI] [PubMed] [Google Scholar]
- 73.Su, M. J., and R. Bablanian. 1990. Polyadenylated RNA sequences from vaccinia virus-infected cells selectively inhibit translation in a cell-free system: structural properties and mechanism of inhibition. Virology 179:679-693. [DOI] [PubMed] [Google Scholar]
- 74.Sutter, G., and B. Moss. 1992. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. USA 89:10847-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sutter, G., and C. Staib. 2003. Vaccinia vectors as candidate vaccines: the development of modified vaccinia virus Ankara for antigen delivery. Curr. Drug Targets Infect. Disord. 3:263-271. [DOI] [PubMed] [Google Scholar]
- 76.Tong, X., R. Drapkin, D. Reinberg, and E. Kieff. 1995. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 92:3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tusher, V. G., R. Tibshirani, and G. Chu. 1999. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varich, N. L., I. V. Sychova, N. V. Kaverin, T. P. Antonova, and V. I. Chernos. 1979. Transcription of both DNA strands of vaccinia virus genome in vivo. Virology 96:412-430. [DOI] [PubMed] [Google Scholar]
- 79.Vos, J. C., and H. G. Stunnenberg. 1988. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 7:3487-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wreschner, D. H., T. C. James, R. H. Silverman, and I. M. Kerr. 1981. Ribosomal RNA cleavage, nuclease activation and 2-5A (ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 9:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]